Abstract
The current information on the polymorphism variation and haplotype structure of the domestic dog leukocyte antigen (DLA) genes is limited in comparison to other experimental animals. In this paper, to better elucidate the degree and types of polymorphisms and genetic differences for DLA-88, DLA-12 and DLA-64, we genotyped four families of 38 beagles and another 404 unrelated dogs representing 49 breeds by RT-PCR based Sanger sequencing. We also sequenced and analyzed the genomic organization of the DLA-88 and DLA-12 gene segments to better define these two-gene DLA haplotypes more precisely. We identified 45 alleles for DLA-88, 15 for DLA-12 and six for DLA-64, of which 20, 14 and six, respectively, were newly described alleles. Therefore, this study shows that the DLA-12 and DLA-64 loci are far more polymorphic than previously reported. Phylogenetic analysis strongly supported that the DLA-88, DLA-12 and DLA-64 alleles were independently generated after the original divergence of the DLA-79 alleles. Two distinct DLA-88 and DLA-12 haplotype structures, tentatively named DLA-88–DLA-12 and DLA-88–DLA-88L, were identified, and the novel haplotype DLA-88–DLA-88L contributed to 32.7% of the unrelated dogs. Quantitative real-time PCR analysis showed that the gene expression levels of DLA-88L and DLA-88 were similar, and that the gene expression level of DLA-12 was significantly lower. In addition, haplotype frequency estimations using frequently occurring alleles revealed 45 different DLA-class I haplotypes (88-88L/12-64) overall, and 22 different DLA-class I haplotypes in homozygous dogs for 18 breeds and mongrels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Experimental animal models have been developed to advance biomedical research on various aspects of the Major Histocompatibility Complex (MHC) such as gene expression and the mechanisms of peptide presentation in the mouse and rat, and diversity in the dog, pig and primate species (Shiba et al. 2016). As one of the experimental animal models, domestic dogs (Canis lupus familiaris) are often used for biomedical research for various diseases such as systemic lupus erythematosus-related disease complex (Wilbe et al. 2009), immune-mediate hematopoietic anemia (Friedenberg et al. 2016; Kennedy et al. 2006), diabetes (Kennedy et al. 2007), chronic hepatitis (Bexfield et al. 2012), hypoadrenocorticism (Massey et al. 2013a), polymyositis (Massey et al. 2013b), exocrine pancreatic insufficiency (Tsai et al. 2013) and for transplantation studies such as using peripheral stem cells (Sandmaier et al. 1996), bone marrow cells (Shi et al. 1998; Storb et al. 1997) and mesenchymal stem cells (Arinzeh et al. 2003; Jung et al. 2009; Kim et al. 2013) in transplantation. Most of these diseases and treatment outcomes are influenced by polymorphisms within the MHC genomic region that encodes the MHC transplantation and immune regulatory molecules (Shiina et al. 2009; Shiina et al. 2004). Matching the MHC polymorphisms between donor and recipient is believed to be an essential factor to avoid acute graft rejection. However, transplantation studies so far have not been performed on canine donors and recipients that were matched for dog MHC (known as Dog Leukocyte Antigen: DLA) polymorphisms, although in some cases microsatellite markers located in the DLA genomic region were used to select donors and recipients for transplantation (Wagner et al. 1996). Hence, detailed information on the polymorphisms and haplotypes of the DLA genes are necessary to better understand major genetic factors of MHC-related diseases and to advance MHC matched canines in transplantation studies.
The human MHC (known as the Human Leucocyte Antigen; HLA) genomic region is located on the short arm of chromosome 6 with the class I region located at the telomeric end and the class II region located at the centrometric end. The HLA genomic regions encode the highly polymorphic gene complex of the HLA-class I and HLA-class II genes. The class I region is additionally divided into three genomic blocks, alpha, beta and kappa, that include duplicated HLA genes (Kulski et al. 2002) and two framework gene blocks that include well-conserved non-MHC genes in mammalian species (Kumanovics et al. 2003). HLA-A, HLA-G and HLA-F are in the alpha block, HLA-B and HLA-C are in the beta block, and HLA-E is in the kappa block (Fig. 1). The classical HLA-class I genes, HLA-A, HLA-B and HLA-C, and the classical HLA-class II genes, HLA-DR, HLA-DQ and HLA-DP, are distinguished by their extraordinary polymorphisms, whereas the non-classical HLA-class I genes, HLA-E, HLA-F and HLA-G, and the non-classical HLA-class II genes, HLA-DM and HLA-DO, are distinguished by their tissue-specific expression, their specific function and/or limited polymorphism (Shiina et al. 2009; Shiina et al. 2004).
Comparative map of the HLA and DLA genomic regions. The genetic maps are based on the genomic information of the NCBI map viewer. The regions are divided into three sub-regions, class I, class III, and class II. The class I region is separated into three blocks, the alpha, beta, and kappa blocks (Kulski et al. 2002; Kumanovics et al. 2003; Shiina et al. 2017), as indicated by blue letters and horizontal arrows. Dark and light blue boxes indicate classical and non-classical MHC class I genes, respectively, dark and light pink boxes indicate classical and non-classical MHC class II genes, respectively, and white boxes indicate the non-MHC genes that are the landmarks for defining the comparative MHC sub-regions and blocks between the HLA and DLA regions
In comparison to the HLA, the DLA genomic region is located on chromosome 12, except for one divergent DLA-class I gene DLA-79 that is on chromosome 18. The major DLA genomic region on chromosome 12 contains three transcribed DLA-class I genes (DLA-88, DLA-12 and DLA-64) (Burnett et al. 1997; Burnett and Geraghty 1995; Sarmiento and Storb 1990a; Wagner et al. 2005) and four DLA-class II genes (DLA-DR, DLA-DQ, DLA-DM and DLA-DO) (Debenham et al. 2005; Sarmiento et al. 1992; Sarmiento et al. 1993; Sarmiento and Storb 1990b). A small fragment of the DLA genomic region is also located on chromosome 35. All of the DLA-class I genes are located in the beta block that is orthologous to the HLA-B and HLA-C segment, whereas the HLA-DP orthologous gene is absent from the whole genome sequence of a female boxer breed (Lindblad-Toh et al. 2005) (Fig. 1). Therefore, the basic organizational structure of the MHC sub-region is largely conserved between the dog and the human with the exception that the dog class I region is distributed on three different chromosomes (Fig. 1) and that the class I and class II genes probably were remodeled between the species by “birth and death” evolution in response to environmental pathogens (Nei et al. 1997).
From the previous DLA polymorphism research, 73 DLA-88, one DLA-12, one DLA-64 and six DLA-79 were released in the IPD and NCBI databases. Of the class I genes, DLA-88 is considered to have classical functions, and DLA-12 and DLA-64 are defined as non-classical genes in the DLA nomenclature (IPD-MHC Canines). However, comparative polymorphism and gene expression analyses of DLA-88, DLA-12 and DLA-64 have not been performed in much detail so far, although DLA-79, located on chromosome 18, is highly expressed in muscle and in various other tissues (Burnett and Geraghty 1995; Graumann et al. 1998; Ross et al. 2012; Venkataraman et al. 2013; Wagner et al. 2000). Furthermore, Ross et al. (2012) reported the possibility of gene duplication of two DLA-88 genes, DLA-88*028:03 and DLA-88*029:01, but they did not provide any detailed genomic and genetic analyses. A total of 16 DLA haplotypes that are composed of DLA-class I and -class II genes (DLA-88–DLA-DRB1 or DLA-88–DLA-DRB1–DLA-DQA1–DLA-DQB1) were estimated from only two breeds, the Beagle and the German Shepherd (Hardt et al. 2006; Tsai et al. 2013). In addition, most of the DLA-class I polymorphisms and haplotypes have been limited to large-sized breeds and/or European breeds such as the Boxer, Doberman and Labrador Retriever, although genotyping of DLA-88 using 428 dogs from 92 different breeds was reported (Kennedy et al. 2012). Hence, detailed comparison of polymorphisms and relative gene expression levels among the DLA-class I genes and estimation of the DLA-class I haplotypes (DLA-88–DLA-12–DLA-64) in small-sized breeds and/or Asian breeds such as Chihuahua, Dachshund and Pomeranian are still necessary for various MHC-controlled biomedical research in domestic dogs.
In this study, to better elucidate the degree and types of allele and haplotype diversity of the DLA-class I genes, we identified 45 DLA-88, 15 DLA-12 and six DLA-64 alleles of which 20 DLA-88, 14 DLA-12 and six DLA-64 alleles were novel alleles in 38 related Beagles (four families) and 404 unrelated animals by reverse transcriptase-polymerase chain reaction (RT-PCR) based Sanger sequencing. During the course of this study, we identified a newly duplicated DLA-88 gene either as a gene replacement or gene conversion within the DLA-12 gene locus and named it unofficially as “DLA-88 like” (DLA-88L) to distinguish it from DLA-88 and DLA-12. To elucidate the genomic organization of the DLA-88, DLA-88L and DLA-12 genes, we sequenced a complete genomic region of 95-kb including the DLA-88, DLA-88L and DLA-12 genes, and identified two distinct DLA-88 and DLA-12 haplotype structures, tentatively named as DLA-88–DLA-12 and DLA-88–DLA-88L. The former haplotype is composed of the originally known DLA-88 and DLA-12 genes and the latter novel haplotype is composed of duplicated DLA-88 genes as described in this paper. We analyzed the diversity and variability of the allele and haplotype DLA-class I sequences by performing gene expression, amino acid variability, phylogenetic and population genetic analyses.
Material and methods
Terminology and gene nomenclature
We have used the terms ‘major’ and ‘minor’ alleles and haplotypes in this paper as synonyms with ‘common’ or ‘frequent’ and ‘rare’ alleles, respectively. In all cases, ‘major’ refers to the number of alleles and haplotypes that are present in more than three dogs and ‘minor’ refers to the number of alleles and haplotypes that are present in three dogs or less. The description ‘official’ allele refers to the DLA allele that has been released by IPD-MHC and/or by the GenBank databases.
The use of the gene names DLA-88 and DLA-12 refers to any gene sequence or gene allele previously designated as such. We have found in this study that some of the gene alleles that were previously designated as DLA-88 are more likely to belong to the newly identified DLA-locus, DLA-88L (“DLA-88 like”). We have used the official allele designations for all previously discovered alleles (e.g., DLA-88*016:03, DLA-12*1) and our own allele designations for the new alleles that we have identified and reported in this paper (e.g., DLA-88*nov12, DLA-12*nov1-2). In this paper, we refer to the previously reported DLA-88L allele in GenBank (HQ340122) as DLA-88*L in order to specify that it is part of the DLA-88L gene. The DLA-12 and DLA-88L genes are hypothesized to be located at the same genomic position (locus), but they are different genes because of their differences in gene expression levels, evolutionary history and phylogenetic lineages that are described in the Results and Discussion sections.
Animals
Peripheral white blood cells (PWBCs) were obtained from 38 related dogs from four Beagle families (Oriental Yeast Co. Ltd. Tokyo, Japan) and 404 unrelated dogs from 49 breeds and mongrels (Animal Medical Center (ANMEC) at Nihon University and the Marble Veterinary Medical Center) (Table 1). The blood collection and dog studies were conducted in accordance with the guidelines for animal experiments specific to each location.
Total RNA and genomic DNA isolation
Total RNA and genomic DNA samples were isolated from the PWBCs of each of the 442 dogs in this study by using the TRIzol LS Reagent (Invitrogen/Life Technologies/Thermo Fisher Scientific, Carlsbad, CA) according to the manufacturer’s instructions.
Primers for RT-PCR and DLA allele sequence determination
Three pairs of primers were used to amplify the DLA-88, DLA-12 and DLA-64 genes as previously reported (Ross et al. 2012) and new primers were designed incorporating sequences from exons 1 and 4 of DLA-88 (PCR product size: 654–657 bp), DLA-12 (660 bp) and DLA-64 (653 bp). The primer names, the 5′ to 3′ nucleotide sequence, and gene locations, and the PCR annealing temperatures that were used for RT-PCR amplification are presented in Supplementary table 1A. The exact primer locations and comparison of the primer sequences among the DLA-class I loci are shown in Supplementary figure 1. Of these three primer pairs, 88/88L/12-F and 88/88L/12-R were previously reported (Ross et al. 2012), and 12-F and 64-R were newly designed for this study. The three DLA genes, DLA-88, DLA-12 and DLA-64, share high similarity, therefore it was not possible to simply design the specific forward and reverse primers individually for each gene. To amplify the DLA-88 gene, we designed the forward primer (88/88L/64-F) to anneal with the DLA-88 and DLA-64 loci and the reverse primer (88/88L/12-R) to anneal with the DLA-88 and DLA-12 loci (Ross et al. 2012). We also designed a DLA-12 gene specific forward primer (12-F) and a DLA-64 gene specific reverse primer (64-R). The two primer pairs, 12-F and 88/88L/12-R, and 88/88L/64-F and 64-R, were used separately for the amplification of DLA-12 and DLA-64, respectively. DLA-88L was amplified by the same primer pair that we used for DLA-88 (88/88L/64-F and 88/88L/12-R), but its gene location was the same as the DLA-12 locus. The DLA-12 locus (RefSeq NM_001014378.1) was genotyped with the 12-F and 88/88L/12-R primer pair. All the alleles were identified and confirmed by Sanger sequencing the PCR products as described in the following sections. All the primer sequences have significant nucleotide differences with the DLA-79 RefSeq (NM_001020810.1).
Reverse-transcriptase (RT) reaction and RT-PCR amplification
cDNA was synthesized with the oligo-dT primer using the RevaTra Ace reverse transcriptase reaction (TOYOBO, Osaka, Japan) after treatment of the isolated RNA with DNase I (Invitrogen/Life Technologies/Thermo Fisher Scientific, Carlsbad, CA). In brief, the 20 μl amplification reaction contained 10 ng of cDNA, 0.4 units of KOD FX polymerase (TOYOBO, Osaka, Japan), 1 × PCR buffer, 2 mM of each dNTP and 0.4 μM of each primer. The cycling parameters were as follows: an initial denaturation of 94 °C/2 min followed by 33 cycles of 98 °C/10 s, 63 °C/30 s and 68 °C/45 s for DLA-88, DLA-12 and DLA-64. PCR reactions were performed with the thermal cycler GeneAmp PCR system 9700 (Applied Biosystems/Life Technologies/Thermo Fisher Scientific, Foster City, CA). The specificity of the primers was confirmed by Sanger sequencing of the PCR products.
Sub-cloning and Sanger sequencing
RT-PCR products were cloned into the pTA2 cloning vector with the TA cloning kit according to the protocol provided by the manufacturer (TOYOBO, Osaka, Japan) and sequenced by using the ABI3130 genetic analyzer (Applied Biosystems/Life Technologies/Thermo Fisher Scientific, Foster City, CA) in accordance with the protocol of the Big Dye terminator method. To avoid PCR and sequencing artifacts generated by polymerase errors, eight to 32 clones per dog were sequenced. The nucleotide sequences of all the dogs’ cDNA also were sequenced by direct-sequencing of the RT-PCR products using the primers that we used for PCR amplification.
Determination of the DLA-class I allele sequences
Allele sequences were determined using Sequencher Ver. 5.0.1 DNA sequence assembly software (Gene Code Co., Ann Arbor, MI) and by comparing them with known DLA-class I allele sequences released in the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and the IPD-MHC Canines databases. Allele sequences were also assigned using the MHC allele assignment software, Assign ATF ver. 1.0.2.45 (Conexio, Australia) from direct sequencing data. Published and newly determined DLA-class I sequences were used as references. When allele sequences were found to be unique by both methods, we confirmed the sequences of the new alleles by sub-cloning and Sanger sequencing them again.
Genomic sequencing of the DLA-88–DLA-88–DLA-64 haplotype (LC271133)
The 95 kb genomic segment including the DLA-88 duplicated haplotype, DLA-88–DLA-88–DLA-64, was independently amplified by long-range PCR method using ten kinds of long-range primer pairs (Fig. 2a and Supplementary table 1)B1 and one DLA-class I homozygous dog that has the DLA-88*028:03–DLA-88*029:01–DLA-64*nov2 haplotype (LC271133). For PCR amplification, the 20 μL PCR amplification-reaction-volume contained 25 ng of genomic DNA, 1 unit of PrimeSTAR GXL DNA polymerase (TaKaRa Bio, Shiga, Japan), 4.0 μL of 5 × PrimeSTAR GXL Buffer (5 mM Mg2+), 1.6 μL of 2 mM of each dNTP and 0.4 μM of each primer. The cycling parameters were as follows: primary denaturation 94 °C/2 min, followed by 30 cycles for 98 °C/10 s and 68 °C/10 min using the GeneAmp PCR system 9700 (Applied Biosystems/Life Technologies/Thermo Fisher Scientific). The PCR products were purified by the Agencourt AMPure XP (Beckman Coulter, Fullerton, CA) and quantified by the PicoGreen assay (Invitrogen/Life Technologies/Thermo Fisher Scientific) with a Fluoroskan Ascent micro-plate fluorometer (Thermo Fisher Scientific, Waltham, MA). One hundred nanograms of the pooled PCR products was used for the preparation of the DNA library that was prepared with an Ion Xpress Plus Fragment Library Kit according to the manufacturer’s protocol for 400 base-read sequencing (Life Technologies/Thermo Fisher Scientific). Emulsion PCR (emPCR) was performed using the library with the Ion 520 & 530 Kit - OT2 on an Ion OneTouch 2 automated system (Life Technologies/Thermo Fisher Scientific). After the emulsion was automatically broken with the OneTouch 2 instrument, the beads carrying the single-stranded DNA templates were enriched according to the manufacturer’s recommendation. Sequencing was performed using the Ion S5 and an Ion 530 Chip (Life Technologies/Thermo Fisher Scientific). The raw data processing and base-calling, trimming and output of quality-filter sequence reads were all performed with the Torrent Suite 4.2.1 software (Life Technologies/Thermo Fisher Scientific) and with full processing for shotgun analysis. This file was further quality trimmed to remove poor sequence at the end of the reads with QVs of less than 20. The trimmed sequence reads were used for mapping of the sequence reads and the reference genome sequence (Accession number: NC_006594) using the CLC Genomics Workbench 8.5.1 software (QIAGEN, Hilden, Germany) with default settings for alignments of 3 mismatch cost, 3 insertion cost, 3 deletion cost, 0.9 length fraction and 0.9 similarity fraction parameters. Remaining gaps or ambiguous nucleotides were determined by the direct sequencing of PCR products obtained with appropriate primers. The completed and annotated genomic sequence was submitted to DDBJ (DNA databank) with the accession number LC271133.
Genomic comparison of DLA-class I segments by dot-matrix analysis. a Sequence-ready map of the 95-kb genomic segment, ranging from DLA-88 to DLA-64, for the genomic comparison between the DLA-88–DLA-12–DLA-64 and the DLA-88–DLA-88L–DLA-64 segments. The exact primer names and primer sequences are shown in Supplementary table 1B1. b Dot-matrix of the 95-kb gene segments between DLA-88–DLA-12–DLA-64 and the DLA-88–DLA-88L–DLA-64 segments. Horizontal and vertical axes show the reference DLA-88–DLA-12 genome sequence (accession number: NC_006594) and the novel DLA-88–DLA-88L genome sequence (LC271133). Green dots represent alignments of forward reads and red dots correspond to alignments between the reverse complement of one sequence and the forward read of the other. Dark blue, light blue, and orange triangles indicate positions of DLA-88, DLA-88L/12, and DLA-64 loci, respectively, and the black vertical bars between these triangles (DLA-class I genes) indicate the position of LINE interspersed sequences. Parenthesis indicates DLA-class I allele name. The three large circles in the dot-matrix indicate the location of the orthologous DLA-class I genes in the two genomic sequences
Dot-matrix analysis
Dot-matrix analysis of the reference DLA genomic sequence NC_006594 and the novel DLA genomic sequence LC271133 was performed by using a genomic similarity search tool (YASS) (Noe and Kucherov 2005). NC_006594 is a part of the dog genome reference sequence derived from the dog genome project Canis lupus familiaris breed boxer chromosome 12, CanFam3.1, whole genome shotgun sequence (Lindblad-Toh et al. 2005). LC271133 is the novel DLA genomic sequence that we sequenced as described above.
Reclassification of DLA-88 alleles as DLA-12 and DLA-88L alleles
The genomic DNA samples that were used as templates to characterize the DLA-88 alleles for reclassification as DLA-12 and DLA-88L were extracted from three dogs: (1) a dog with the homozygous haplotype DLA-88–DLA-12 carrying one DLA-88 allele and one DLA-12 allele (DLA-88*006:01 and tentatively named DLA-12*1 that was reported by Burnett et al. (1997) as “DLA-12”), (2) a dog with a homozygous haplotype DLA-88–DLA-88 carrying two DLA-88 alleles (DLA-88*003:02 and DLA-88*017:01) and no DLA-12 allele, and (3) a dog with the heterozygous haplotypes DLA-88–DLA-88 and DLA-88–DLA-12 carrying three DLA-88 alleles (DLA-88*016:03, DLA-88*025:01, DLA-88*501:01) and one DLA-12 allele (DLA-12*1). The 5.6 kb PCR product included the entire DLA-12 (DLA-12 and/or DLA-88L) locus, ranging from the promoter-enhancer region to the 3′-untranslated region, and it was amplified using the long-range primer pair 88L/12-seg-F and 88L/12-seg-R (Fig. 3a and Supplementary table 1B2). The cycling parameter was as follows: an initial denaturation of 94 °C/2 min followed by 33 cycles of 98 °C/10 s, 58 °C/30 s and 68 °C/5 min. The PCR product was used for the nested PCR with the genotyping primer pairs, DLA-12 (12-F and 88/88L/12-R) and DLA-88L (88/88L/12-F and 88/88L/12-R) (Fig. 3a and Supplementary table 1A). The 5.6 kb genomic sequence of the PCR product from the DLA-88*003:02–DLA-88*017:01 homozygous dog was determined using the DLA-88L primers, the 88L/12-seg-F and 88L/12-seg-R primer pair and six direct-sequencing primers (Supplementary table 1A, 1B2 and 1B3).
Development of a PCR nested method for detection of polymorphism at DLA-12 and DLA-88L using genomic DNA samples. a Schematic diagram shows the location of the DLA-88L/12 gene, the exon/intron structure of DLA-88L/12, and the primer sites and operational map for the genomic analysis of the haplotype structural segments. Numbers around the gene structure indicate exon numbers. White and black boxes show the coding exons and the 5′ and 3′ untranslated regions, respectively. Primer sequences (sequencing primers 2F, 3F, 4F, 2R, 3R and 4R, 88L/12-seg-F and 88L/12-seg-R, 12-F, 88/88L/12-R, and 88/88L/64-F) are shown in Supplementary table 1A, 1B2 and 1B3. Of them, the primer specificity of 12-F, 88/88L/12-R and 88/88L/64-F are shown in Supplementary figure 1. b, c Electrophoresis images of the PCR products that were amplified using the 88L/12-seg-F and 88L/12-seg-R primer pair (5.6 kb) and the DLA-12 or DLA-88L specific primer pairs (1.6 kb), respectively. Two types of DNA ladder markers, Quick-Load 1 Kb DNA Ladder (New England BioLabs) and Quick-Load 2-Log DNA Ladder (New England BioLabs), were used as DNA size markers for the image B and image C, respectively
In total, 105 genomic DNAs, 102 unrelated dogs with three DLA-88 alleles, two unrelated dogs with four DLA-88 alleles and one Beagle in family 2 with three DLA-88 alleles, were used for the genotyping of DLA-12 and DLA-88L at the DLA-12 locus.
Phylogenetic analysis among the DLA-class I allele sequences
Multiple DLA-class I nucleotide sequences were aligned using the ClustalW Sequence Alignment program of the Molecular Evolution Genetics Analysis software 6 (MEGA6) (Tamura et al. 2011). The phylogenetic tree, consisting of 52 DLA-88 (45 major and seven minor official alleles), 15 major DLA-12, six major DLA-64 sequences determined in this study, and six published DLA-79 sequences (Venkataraman et al. 2013), was constructed with only synonymous substitutions used to identify differences by the Neighbor-joining (NJ) method in MEGA6 (Saitou and Nei 1987) using exons 1 to 3 (alignment length: 610 bp excluding gap sites) with the modified Nei-Gojobori model. The tree was reconstructed using only synonymous substitution sites to identify differences in order to remove the influence of positive selection. We used mouse H2-D1 (DNA accession numbers: NM_010380) and H2-K1 (NM_001001892) as the outgroup sequences.
Measurement of gene expression level in the DLA-class I genes by quantitative real-time PCR
Nine allele-specific primer pairs (Supplementary figure 1 and Supplementary table 1C) were used by real-time PCR for gene expression analysis of DLA-88, DLA-12, DLA-88L and DLA-64 in 22 (ten 88-12-64/88-12-64 and 12 88-88L-64/88-88L-64) homozygous dogs. The allele-specific primer pairs were designed in exons 3 and 4 (amplified length: 124 to 131 bp) (Supplementary table 1C). A pair of primers designed from the dog glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene sequence was used for the analysis as an internal control (Livak and Schmittgen 2001). Expression levels were measured by real-time PCR using the Thermal Cycler Dice Real Time System II (TaKaRa Bio) with SYBR Premix Ex Taq II (TaKaRa Bio). The 25 μl amplification reaction volume contained 50 ng of cDNA, 12.5 ul SYBR Premix Ex Taq II and 0.2 μM of each primer. The cycling parameter was as follows: 40 cycles of 95 °C/10 s, 60 °C/20 s. Melting curve analysis showed that there was no primer dimer formation. The relative quantitative values in each sample were normalized and calibrated by the 2 (-DeltaDeltaC(T)) method (Livak and Schmittgen 2001; Schmittgen and Livak 2008). The paired t test program with two-samples assuming an unequal variances model in Excel determined the statistical differences of expression levels between DLA-88 and DLA-12 and between DLA-88 and DLA-88L.
Estimation of the DLA-class I allelic haplotypes
Of 404 unrelated dogs, 355 dogs with more than three heterozygous dogs, or at least two homozygous dogs per dog breed were used for allelic haplotype estimation. The DLA-class I haplotype (88-88L/12-64) was initially characterized by manually sorting the major 38 DLA-88, seven DLA-88L, 15 DLA-12 and six DLA-64 alleles based on the 88-88L/12-64 homozygous dogs. Estimations of the 88-88L/12-64 allelic haplotype frequencies were performed by using the PHASE 2.1.1 program (Stephens et al. 2001) and the 88-88L/12-64 homozygous haplotype data.
Results
Evaluation of the DLA-class I polymorphism detection method using four Beagle families
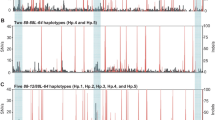
To evaluate our newly designed DLA-class I gene-specific primers and PCR conditions for amplifying and sequencing of DLA-88, DLA-12 and DLA-64, we initially performed genotyping and haplotyping using 38 related Beagles from four families that had well defined familial relationships (Supplementary figure 2). Although no polymorphism was detected in DLA-64, seven DLA-88 and three DLA-12 alleles were identified in these families. Of the 11 alleles, six of them, DLA-64*nov2, DLA-12*nov1-2, DLA-12*nov1-3, DLA-88*nov2, DLA-88*nov9 and DLA-88*nov19, were novel. A total of six DLA-class I allelic haplotypes was estimated without any pedigree discrepancies. One DLA-class I haplotype that was composed of two DLA-88 alleles (DLA-88*nov2 and DLA-88*nov19) was observed in one Beagle in family 2 (Supplementary figure 2).
Polymorphism analysis of the DLA-class I genes using cDNA samples of 404 unrelated dogs
In our initial characterization of the DLA-88, DLA-12 and DLA-64 allele sequences (exons 1 to 4 in DLA-88, DLA-12 and DLA-64) in 404 unrelated dogs including 49 breeds and mongrels by RT-PCR based sub-cloning and Sanger sequencing methods, we identified 76 alleles for DLA-88, 21 for DLA-12 and seven for DLA-64. Of them, 45 DLA-88, 15 DLA-12 and six DLA-64 were “major” alleles identified in more than three heterozygous dogs, or at least two homozygous dogs. On the other hand, 31 DLA-88, six DLA-12 and one DLA-64 were “minor” alleles identified in three or less heterozygous dogs, or a single homozygous dog. All the major and minor alleles were identified by matching our sequences with all of the known DLA-class I allele sequences released in the GenBank and the IPD-MHC Canines databases using Sequencher and Assign ATF software (Table 2 and Supplementary table 2A and 2B). Of all the DLA class I alleles detected in this study, 25 DLA-88 alleles and one DLA-12 allele were previously reported in the IPD-MHC Canines database and NCBI database (Burnett et al. 1997; Graumann et al. 1998; Ross et al. 2012; Venkataraman et al. 2007) and the remaining 20 DLA-88, 14 DLA-12 and six DLA-64 were newly identified in this study (DDBJ/EMBL/GenBank accession numbers LOC130502-LC130511, LC130513-LC130547, LC171419-LC171437 and LC171439-LC171445, Table 2 and Supplementary table 2A). The most frequent alleles for each gene were DLA-88*006:01 (85 of the 404 dogs), DLA-12*1 (275 dogs) and DLA-64*nov2 (380 dogs) that were observed in 15, 40, 48 and 20 breeds and/or mongrels, respectively (Supplementary table 2A).
Genomic organization of the DLA-88 and DLA-12 gene segment
Two previous reports described three DLA-88 alleles in one dog that inferred that there were duplicated DLA-88 genes, DLA-88*028:03 and DLA-88*029:01, located at a single chromosome as a DLA-88 haplotype (DLA-88*028:03–DLA-88*029:01) (Kennedy et al. 2012; Ross et al. 2012). In our RT-PCR analysis, we identified three types of DLA-88 and one type of DLA-12 allele per dog in 102 unrelated dogs and one Beagle derived from the familial analysis, and four types of DLA-88 per dog were detected in two unrelated dogs. In total, we found that 105 dogs had unusual DLA-88 allele numbers. On the basis of our DLA-88–DLA-88 haplotype estimations using DLA-88 genotypes in the 105 dogs, we identified the following eight DLA-88 allelic haplotypes: DLA-88*003:02–DLA-88*017:01, DLA-88*025:01–DLA-88*016:03, DLA-88*028:01–DLA-88*029:01, DLA-88*028:03–DLA-88*029:01, DLA-88*nov2–DLA-88*nov19, DLA-88*nov10–DLA-88*L, DLA-88*nov15–DLA-88*029:01 and DLA-88*nov23–DLA-88*nov12. There were 25 dogs in the 404 unrelated dogs that were homozygous for any one of those DLA-88–DLA-88 haplotypes. Furthermore, we could not detect the DLA-12 gene by PCR in 27 dogs including two dogs with four types of DLA-88 alleles and 25 homozygous dogs with the DLA-88–DLA-88L haplotype. Hence, we hypothesized that a genomic arrangement event had occurred that involved the DLA-88 and DLA-12 genes and that had generated two distinct DLA-class I haplotype structures, such as DLA-88–DLA-12 and DLA-88–DLA-88, in some of the DLA-class I haplotypes.
To elucidate the DLA-88 and DLA-12 genomic structures in more detail, we sequenced the complete 95-kb-nucleotide sequence of a DLA-88–DLA-88 haplotype in a homozygous dog with the allelic haplotype DLA-88*028:03–DLA-88*029:01, ranging from DLA-88 to DLA-64, by long-range PCR and next generation sequencing (NGS) methods using ten primer pairs (Fig. 2a and Supplementary table 1B1). Sequence read information was obtained after sequencing of the pooled PCR products using the Ion S5 system in a single sequencing run. Draft read numbers in total were 1,794,204 sequence reads that were high quality sequence reads with more than 20 quality values (QV). The average read length of 304 bases and an overall mode read length of 416 bases. Therefore, the sequence reads had high quality and sufficient sequence volume for further genomic analysis. After the NGS run, gap filling and validation of the assembled sequences, we determined a 94,790 bp complete genomic sequence and deposited it in a DNA databank with the accession ID of LC271133. We then compared our LC271133 sequence with the reference genome sequence (NC_006594, 96,115 bp) that has the DLA-88–DLA-12–DLA-64 genomic structure.
In the genomic sequence the two DLA-88 genes (DLA-88*028:03 and DLA-88*029:01) were identified to be located in the DLA-88 and DLA-12 orthologous regions, respectively, when compared to the dog genome RefSeq (NC_006594) (Fig. 2b). However, we tentatively named the DLA-88 gene locating in DLA-12 orthologous region as DLA-88L because of its stronger sequence similarity to DLA-88 than to DLA-12. Nevertheless, the DLA-12 genotyping primer pair (12-F and 88/88L/12-R) and the DLA-88 genotyping primer pair (88/88L/64-F and 88/88L/12-R) amplify DLA-12 and DLA-88L, respectively.
Figure 2b shows the dot-matrix analysis of the genomic comparison between the DLA-88–DLA-12–DLA-64 segment and the DLA-88–DLA-88L–DLA-64 segment. The dot-matrix plot reveals that the two genomic sequences have similar structural features, proportions and lengths such as the dense insertions of long interspersed elements (LINE) in approximately 30 kb around the DLA-88L/DLA-12 gene region, suggesting that the genomic position of DLA-88L is at the same location as DLA-12. Dot-matrix analysis using the previously released 5726 bp of DLA-88 (DLA-88*034:01, NC_006594, position: 891280-897005), 5649 bp of DLA-12 (identified and tentatively named in this study as DLA-12*nov6 corresponds to RefSeq NC_006594, position: 932034-937682) and 5585 bp of the newly determined DLA-88L (LC189199) sequences showed that the DLA-12 vs DLA-88L comparison with 93.5% similarity was more conserved than either the DLA-88 vs DLA-88L comparison with 71.0% similarity or the DLA-88 vs DLA-12 comparison with 69.3% similarity (Supplementary Table 3). These comparisons incorporated both the gene regions (approximately 3.4 kb) and the upstream and downstream regions of the gene. In contrast, comparisons of the nucleotide similarities of DLA-class I gene regions using 3347 bp of DLA-88, 3451 bp of DLA-12 and 3401 bp of DLA-88L nucleotide sequences showed 93.8% similarity between DLA-88 and DLA-88L, 92.2% similarity between DLA-12 and DLA-88L, and 88.7% similarity between DLA-88 and DLA-12. The greatest gene divergence and sequence dissimilarity occurred in the 1.1 kb of the gene regions between exon 1 and exon 3 including introns with 83.4% similarity between DLA-12 and DLA-88L and 82.7% similarity between DLA-88 and DLA-12, compared to the 95.1% similarity between DLA-88 and DLA-88 L for the same region. Therefore, it is evident that the DLA-88L gene has higher sequence similarity with DLA-88 in the region of exons 1 to 3, whereas DLA-88L has higher sequence similarity with DLA-12 in the genomic region outside of DLA class I gene.
Reclassification of DLA-88 alleles as DLA-88L and estimated frequency of the DLA-88-DLA-88L haplotypes
The dot-matrix analysis suggests that the some of the alleles initially classified as DLA-88 alleles are part of the DLA-12 locus that is composed of one of two distinct gene types, DLA-12 or DLA-88L (Fig. 2b). In order to differentiate between the true DLA-88 alleles that originated from DLA-88 locus and the DLA-88L alleles, initially classified as DLA-88, but that were generated by the DLA-12 locus, we investigated the 5.6 kb genomic segment that included the DLA-12 and/or DLA-88L genes using one DLA-88–DLA-12 haplotype in a homozygous dog, one DLA-88–DLA-88 haplotype in a homozygous dog and a DLA-88–DLA-12 and DLA-88–DLA-88 haplotype in one heterozygous dog as structural templates. Although the 5.6 kb bands obtained using the long-range primer pair for DLA-12 and DLA-88L genes (88L/12-seg-F and 88L/12-seg-R) (Fig. 3a and Supplementary table 1B2) were observed in all samples without non-specific products (Fig. 3b), the 1.6 kb PCR product obtained by nested PCR using the DLA-12 primer pair (12-F and 88/88L/12-R) was amplified from the DLA-88–DLA-12 haplotype of a homozygous dog, but not from the DLA-88–DLA-88 haplotype of a homozygous dog that has the allelic haplotype DLA-88*003:02 and DLA-88*017:01. On the other hand, the 1.6 kb PCR product obtained by nested PCR using the DLA-88L primer pair (88/88L/64-F and 88/88L/12-R) was amplified from the DLA-88–DLA-88 haplotype of a homozygous dog, but not from the DLA-88–DLA-12 haplotype of a homozygous dog that has DLA-88*006:01 and DLA-12*1 (Fig. 3c). Therefore, this experiment showed that the DLA-12 and DLA-88L primer pairs were specific for the detection of the DLA-12 and DLA-88L genotypes, respectively, in a nested PCR reaction. These results strongly point to two distinct DLA-class I haplotype structures in domestic dogs, the DLA-88–DLA-12 haplotype and the DLA-88–DLA-88L haplotype.
To distinguish between the true DLA-88 and DLA-88L alleles from the 45 major DLA-88 alleles that had been previously classified in this study, we performed sequence-based genotyping of 105 DNA samples that have three or four DLA-88 alleles by using the long-range primer pair for DLA-12 and DLA-88L genes (88L/12-seg-F and 88L/12-seg-R), and then the nested PCR was performed using the primer pairs for the exons 1–4 segments of DLA-88L genes (88/88L/64-F and 88/88L/12-R) (Fig. 3a and Supplementary table 1A and 1B2). Of the 45 major DLA-88 alleles, seven (15.6%) were identified to be from the DLA-88L locus (Table 3).
To summarize the reclassification of the DLA-class I alleles, 38 DLA-88, seven DLA-88L, 15 DLA-12 and six DLA-64 were identified as the major alleles. Consequently, eight DLA-88–DLA-88L major allelic haplotypes (16 haplotypes in total) were inferred in 105 dogs, and the most frequent allelic haplotype observed in nine breeds was DLA-88*003:02–DLA-88*017:01 with an estimated 12.4% frequency (44 of the 355 dogs) (Table 4 and Supplementary table 4). Only one of the previously reported DLA-88–DLA-88L haplotypes (DLA-88*028:03–DLA-88*029:01) (Ross et al. 2012) was detected in our study.
Phylogenetic relationship among the DLA-class I alleles
We constructed a phylogenetic tree using the neighbor-joining method to examine the inter- and intra-relationships of the DLA-88, DLA-12 and DLA-64 allele sequences along with six of the previously detected DLA-79 allele sequences (Venkataraman et al. 2013). The phylogenetic tree of the 79 aligned DLA-class I allele sequences supported a gene-specific evolution of the DLA-class I genes. Namely, the DLA-class I allele sequences could be separated into four lineages (DLA-88, DLA-12, DLA-64 and DLA-79) after the divergence of the mouse MHC class I (H2-D1 and H2-K1) sequences (Fig. 4). Both the DLA-88L and the DLA-88 alleles were widely dispersed in the DLA-88 lineage.
Nucleotide sequence-based phylogenetic tree of DLA-class I alleles constructed by the Neighbor joining method. 66 major and seven minor official DLA-class I cDNA sequences that were identified in this study and eight released sequences (six DLA-79 and two mouse H-2D and H-2K) were used for constructing the tree. Light blue background and white letters indicate the newly identified DLA-class I alleles. Of the DLA-class I alleles that are composed of DLA-88–DLA-88L haplotypes detected in this study, the DLA-88 and DLA-88L allele names are framed by dark blue and red rectangular lines, respectively. DLA-64*nov2 is identical with DLA-64 sequence of the dog genome reference sequence (NC_006594). DLA-88L alleles are shown in Table 3
Relative gene expression level of the DLA class I genes in PWBCs
To compare the gene expression levels among the DLA-class I genes in PWBCs, we performed a relative quantification assay of the DLA-class I genes by the real-time PCR method using newly designed gene-specific primer pairs (Supplementary figure 1 and Supplementary table 1C). Figure 5 and Supplementary figure 3 show the mean differences in the relative gene expression levels of 22 RNA samples isolated from ten DLA-88–DLA-12–DLA-64 and 12 DLA-88–DLA-88L–DLA-64 haplotype homozygous dogs. In the case of both haplotype structures, the relative expression levels were observed as follows: DLA-88 > DLA-88L> DLA-12 > DLA-64, with a significant difference (P < 0.01) in the mean expression level between DLA-88 and DLA-12, and no significant difference between DLA-88 and DLA-88L (Fig. 5). Taken together, these results show that the gene expression levels of DLA-88 and DLA-88L were significantly higher than those of DLA-12 and DLA-64 in the PWBCs.
Relative gene expression levels of DLA-class I genes. a, b Summary of the mean differences between the relative gene expression levels of DLA-88, DLA-88L/12 and DLA-64 genes of DLA-88–DLA-12–DLA-64 and DLA-88–DLA-88L–DLA-64 haplotypes using ten and 12 dogs for each haplotype, respectively. Vertical axis shows the relative quantitative values by the real-time PCR method. Primer sequences and specificity are shown in Supplementary table 1C and Supplementary fig 1, respectively. Thin bars show standard errors, and P values indicate statistical significant difference between DLA-88 and DLA-12 and DLA-88 and DLA-88L genes. The detail relative gene expression levels of each DLA haplotype are shown in Supplementary figure 3
Comparison of amino acid sequences among the DLA-class I genes
A total of 45 DLA-88, seven DLA-88L, 11 DLA-12 and two DLA-64 nucleotide sequences that were classified as major and minor official alleles in this study were translated as amino acid sequences that yielded 45 DLA-88, seven DLA-88L, 15 DLA-12 and six DLA-64 different allele sequences (Supplementary table 2A and 2B). Of the DLA-88 alleles, three signature nucleotides coding for the amino acid residue leucine, L, at position 155 were inserted in eight of the known DLA-88 alleles (DLA-88*501:01, DLA-88*502:01, DLA-88*503:01, DLA-88*504:01, DLA-88*505:01, DLA-88*506:01, DLA-88*507:01 and DLA-88*508:01) in the exon 3 region (Kennedy et al. 1999). The same location for the three signature nucleotide insertions was observed in five of the newly identified DLA-88 alleles, DLA-88*nov25, DLA-88*nov26, DLA-88*nov27, DLA-88*nov37 and DLA-88*nov41. Therefore, we defined these 13 DLA-88 alleles as a DLA-88*50X group of alleles.
Supplementary figure 4 shows that amino acid sequence logos of the T cell recognition sites (TRSs) and the peptide binding regions (PBRs) that were deduced from the structural analyses of the HLA-class I and DLA-88 molecules using the newly identified major alleles in this study and six previously published DLA-79 alleles (Bjorkman et al. 1987; Parham et al. 1988; Xiao et al. 2016; Venkataraman et al. 2013). Amino acid sequences of the TRSs translated from each DLA-class I gene were well-conserved with only one (DLA-79) to six (DLA-88) amino acid differences, and of them, three amino acid differences (positions 62, 72 and 169 or 170) were commonly observed in the DLA-88, DLA-12 and DLA-88L sequences. In contrast, amino acid sequences of the PBRs on DLA-88 and DLA-88L were more variable with 19 (DLA-88) and nine (DLA-88 L) amino acid differences, although the amino acid sequences of DLA-12 were relatively more conserved than those of DLA-88 and DLA-88 L (Supplementary figure 4). On the other hand, amino acid sequences were well conserved for DLA-64 and DLA-79 with one and four amino acid differences for the DLA-64 and DLA-79 antigens, respectively. The results of this amino acid sequence analysis show that DLA-88, DLA-12 and DLA-88L have relatively high amino acid variability at TRSs and PBSs when compared to DLA-64 and DLA-79 (Supplementary figure 4). Moreover, one TRS residue at position 154 and four PBR residues at positions 97, 99, 114 and 147 were different amino acids between the DLA-12 and DLA-88L protein sequences.
Estimation of DLA-class I allelic haplotypes and their detection in homozygous dogs
A total of 45 different DLA-class I major allelic haplotypes (Hp. 01 to Hp. 45) were composed of DLA-88-DLA-88L/12-DLA-64 (88-88L/12-64); 37 were 88-12-64 and eight were 88-88L-64 (Supplementary table 4). Some DLA-88 alleles such as DLA-88*501:01 were observed in several different haplotypes and DLA-88L alleles were observed in mostly different haplotypes. The combinations of the two major structural haplotypes, 88-12-64/88-12-64, 88-12-64/88-88L-64 and 88-88L-64/88-88L-64, were observed in 239, 90 and 26 unrelated dogs, respectively. The haplotype frequencies of the 88-12-64 and 88-88L-64 were 80.0 and 20.0%, respectively. Of the major 45 DLA-class I allelic haplotypes, Hp.04, Hp.07, Hp.18, Hp.21 and Hp.38 collectively showed high frequencies with 36 to 76 dogs in the 355 unrelated dogs, and the DLA-class I haplotypes were also widely observed in seven to ten breeds (Supplementary table 4).
Table 5 shows the DLA-class I allelic haplotype information in five representative breeds, 48 dogs of Miniature Dachshund, 42 of Toy Poodle, 41 of Yorkshire Terrier, 37 of Shiba and 36 of Chihuahua. Six to 14 DLA-class I haplotypes were identified in each breed. A comparison of the inferred DLA haplotypes among the five breeds revealed the presence of breed-specificity with high frequency haplotypes such as Hp.15 (haplotype frequency of 0.107) and Hp.32 (0.131) in the Toy Poodle, Hp.04 (0.268) and Hp.05 (0.183) in the Yorkshire Terrier, and Hp.33 (0.162) and Hp.37 (0.189) in the Shiba (Table 5 and Supplementary table 5).
An examination of the DLA-class I allelic haplotypes in homozygous dogs of the 355 unrelated dogs and 38 related dogs in four Beagle families revealed that 86 homozygous dogs in 18 breeds had 22 different DLA-class I allelic haplotypes by DLA-class I gene genotyping data and DLA-88–DLA-88L allelic haplotype information. In comparison, these 22 DLA-class I allelic haplotypes were also detected in 275 heterozygous dogs (70% of the 393 dogs) from 20 breeds and mongrels (Table 6).
Inferred evolutionary model for the genomic structures of the DLA-88–DLA-12 and DLA-88–DLA-88L haplotypes
Figure 6 shows the inferred model for the generation of the genomic structures of the DLA-88–DLA-12 and DLA-88–DLA-88L haplotypes. Essentially, a gene transfer (unequal crossing over) or gene conversion has replaced the DLA-12 gene locus with a DLA-88 gene that we have unofficially designated here to be DLA-88L thereby resulting in a DLA-88–DLA-88L structural haplotype. The original DLA-88–DLA-12 haplotype is retained in various dog populations at a frequency of 80%, whereas the newer haplotype has emerged at a lower frequency of 20%. This haplotype frequency was similar to the previously reported frequency (18%) that has duplicated DLA-88 alleles in at least one haplotype (Kennedy et al. 2012). Also, while the DLA-12 allelic sequences have diverged over time to form their own single lineage separate from the DLA-88 lineage, the more recent DLA-88L allelic sequences have not diverged sufficiently from the DLA-88 allelic sequences to establish their own unique lineage. Consequently, the DLA-88 and DLA-88L alleles are intermixed in the same DLA-88 lineage as seen in the phylogenetic tree in Fig. 4. A hypothetical DLA-12–DLA-12 structural haplotype that might have arisen from the same gene crossover event that generated the DLA-88–DLA-88L haplotype (Fig. 6) has not been identified in any dog populations and it is therefore extremely rare, extinct or it was never generated.
Inferred evolutionary model for the genomic structures of DLA-88–DLA-12 and DLA-88–DLA-88L haplotypes. Dark and light blue boxes show the DLA-88 and DLA-12 genes, respectively, and the light blue box with a dark blue rectangular line shows DLA-88L. “Hp. Freq.” indicates the haplotype frequency. Because the DLA-88 gene in a gene conversion or crossing over event replaced the DLA-12 locus, we have unofficially named the replacement gene DLA-88L despite its location
Discussion
Recent genomic studies suggest that the dog species was first domesticated in Asia (Pang et al. 2009; Vonholdt et al. 2010), but with a possible dual origin of domestic dogs in Europe and Asia (Frantz et al. 2016). Domestic dogs that originated from Asia have a much greater genetic diversity of the DLA-DRB1 gene than those that originated from Europe (Niskanen et al. 2013). From this viewpoint, we identified 20 DLA-88, 14 DLA-12 and six DLA-64 novel and major alleles in 38 related and 404 unrelated dogs (Table 2) with a greater focus on small-sized dogs such as the Chihuahua, Miniature Dachshund and Toy Poodle, and Asian dogs such as the Japanese Spitz, Shiba and Shih Tzu than had been previously studied. Namely, 61% of them were novel alleles (Table 2 and Supplementary table 2A). Whereas previous studies reported numerous polymorphisms within the DLA-88 locus and few or none at DLA-12 and DLA-64, we found that there are also many polymorphisms within the DLA-12 and DLA-64 gene loci. This suggests that future polymorphism analysis of the DLA-class I genes using a greater number of dogs and different breeds is likely to result in the discovery of many more DLA-class I alleles at the four DLA loci, DLA-88, DLA-88L, DLA-12 and DLA-64.
A serious problem for performing a polymorphism analysis by Sanger sequencing of the DLA-class I genes is the difficulty of obtaining clear genotyping and haplotyping results because 1–4 types of DLA-88 alleles per dog can be identified within two types of DLA-class I structural haplotypes, DLA-88–DLA-12 and DLA-88–DLA-88L (Fig. 2). The existence of two types of DLA-class I haplotype structures was first indicated by the observation of three DLA-88 alleles in one dog in a previous report (Ross et al. 2012). Although the DLA-12 locus was identified to express a protein that classified as the probable class Ib gene (Wagner et al. 2005), our study is the first to highlight that the DLA-12 locus is moderately to highly polymorphic with the identification of at least 22 major DLA-12 alleles (seven DLA-88L and 15 DLA-12 alleles) whereby some of them had been previously misclassified as DLA-88 alleles at the DLA-88 locus. As our genomic and phylogenetic analyses suggests, the possible reason for this misclassification is that the alleles at the DLA-88L locus have very high sequence similarities with the DLA-88 alleles at the DLA-88 locus (Figs. 2 and 4 and Supplementary table 3), and that it is difficult to separate the DLA-88 and DLA-88L alleles by simple PCR amplification of sequences shared between the two loci. For example, the intron 2 sequence of DLA-88*016:03 (Venkataraman et al. 2017) showed 100% match with DLA-88*034:01 and 99.5% match with the novel DLA-88 L (DLA-88*029:01), although the allele showed 62.6% with DLA-12*nov6. Consequently, we developed a nested method to specifically detect the DLA-88 L alleles by performing the PCR amplification using one primer pair for the exons 1–4 segment of the DLA-88L gene (88/88L/64-F and 88/88L/12-R) after PCR amplification of the 5.6 kb segment using the long-range primer for the DLA-12 and DLA-88L genes (88L/12-seg-F and 88 L/12-seg-R) (Fig. 3). This new PCR nested genotyping method will be most helpful for differentiating between the DLA-12 and DLA-88L alleles. This method can be modified and applied to genotyping DLA-88 alleles by simply changing the primer sequences of the 88L/12-seg-F and 88L/12-seg-R primer sites and instead use DLA-88 specific primers such as the DLA88-12-64_F1 and DLA88-12-64_R1 pair (Fig. 2a and Supplementary table 1B). Although, at least seven DLA-88 alleles were classified as DLA-88L major alleles in our study (Table 3), a large-scale haplotype analysis will be required to elucidate the diversity and frequency of the DLA-88–DLA-88L haplotype structures among various dog breeds and mongrels, which was beyond the scope of our current study.
The DLA-88–DLA-88L haplotype appears to have been generated by (1) duplication of the DLA-88 gene with the production of DLA-12 after divergence from the Feliformia species (52.9 Mya) (Hedges and Kumar 2009), and (2) a gene conversion or an unequal crossing over between DLA-88 and DLA-12, resulting in the production of the DLA-88–DLA-88L and the DLA-88–DLA-12 haplotypes (Fig. 6). Although the DLA-12 allele lineage has a close evolutionary relationship with the DLA-88 allele lineage, all of the ten DLA-88L allele sequences (exons 1 to 3) were included in the DLA-88 lineage (Fig. 4). If mutations were evolving at the same rate between DLA-12 and DLA-88L, then the allele numbers for DLA-12 (15 alleles) would suggest that they have evolved and diverged over a longer time period than those for DLA-88L (seven alleles) (Table 3 and Supplementary table 2A). Although the overall allele number for DLA-88L was less than that for DLA-12, the amino acids on PBRs and TRSs in DLA-88L were more polymorphic than those in DLA-12 (Supplementary figure 4). This suggests that these two genes were derived from different DLA-class I ancestral genes and/or that they were subjected to different natural selection pressures during their evolution (Fig. 6).
The DLA-88 and DLA-88L showed slightly higher nucleotide sequence similarity with 93.6% than those of DLA-12 and DLA-88L (92.2%) and DLA-88 and DLA-12 (88.7%) at the 3.3–3.4 kb of the DLA-class I gene region. However, the sequence similarity comparisons show that the 5.6 kb of genomic structures between the DLA-12 and DLA-88L segments are more conserved than those between the DLA-88 and DLA-12 segments and between the DLA-88 and DLA-88L segments (Supplementary table 3). This suggests that the DLA-12 and DLA-88L genes were exchanged with each other from the upstream region to at least the 3’untranslated region. However, it is noteworthy from the DLA-12 and the DLA-88L primers and the PCR experiment shown in Fig. 3 that there are nucleotide sequence differences between the DLA-12 and DLA-88L in the regions of exon 1 and exon 4. Furthermore, the amino acid variability analysis supports the view that DLA-12 and DLA-88L have a classical MHC class I function because the amino acid variations at PBRs and at the TRSs of both genes are similar to those of DLA-88 that is known to have a classical MHC class I function (Supplementary figure 4). Although new MHC genes have been inferred to have been generated by duplication, crossing over and gene conversion events in the MHC class I genomic structure of some vertebrate species (Holmes et al. 2003; Hosomichi et al. 2008), this is the first report attributing unequal crossing over or gene conversion at a single working DLA locus contributing to the MHC class I diversity of the domestic dog species.
The DLA-88–DLA-88L haplotype was observed in 17 Asian and European breeds and mongrels (Table 4). On the other hand, although the DLA-12–DLA-12 haplotype may have been generated immediately after the first unequal crossing over event in domestic dogs, the detection of the DLA-88 alleles in all of the dogs supports the premise that the DLA-12–DLA-12 haplotype without DLA-88 and DLA-88L alleles was negatively selected against by birth and death evolution (Fig. 6) (Nei et al. 1997).
The level of HLA gene expression activity such as the association of the highly expressed HLA-C*14:02 allele and progression after infection of HIV (Apps et al. 2013), and the association of HLA-C*14:02 and HLA-DP5 alleles and acute graft versus host disease (GVHD) (Petersdorf et al. 2014; Petersdorf et al. 2015) has become an important topic in human MHC genetics in recent years. In regard to the dog MHC, one of the main differences between DLA-12 and DLA-88L is that the gene expression level of DLA-88L is significantly higher than that of DLA-12, and almost the same as DLA-88 (Fig. 5 and Supplementary figure 3). The DLA-88L positive dogs are thought to have a higher peptide presentation ability and an higher gene expression level than the DLA-88L negative dogs. Therefore, the functional difference in gene expression levels may have originated from the unequal crossing over event and this difference may influence the resistance and susceptibility of dogs to various viral infections such as canine distemper virus (CDV), canine adenovirus (CAV) and canine parvovirus (CPV) (Laurenson et al. 1998), and various inherited diseases and those that may be associated with transplantation outcome in dogs.
Of the DLA-class I genes, DLA-64 was confirmed to have a non-classical function with low polymorphisms and gene expression levels in the PWBCs, and limited amino acid variations in the PBRs and TRSs domains compared to DLA-88 and DLA-88L that have a classical class I function as described in Supplementary figure 4. In contrast, although DLA-79 was reportedly expressed at high levels in muscle (Burnett and Geraghty 1995), the detailed expression patterns are unknown. Therefore, gene expression analysis of the DLA-class I genes including DLA-88, DLA-12, DLA-88L, DLA-64 and DLA-79 are necessary for future biomedical studies.
In humans, HLA-G is a non-classical class I gene that is highly expressed in the trophoblasts of the placenta and it has acquired a maternal immunity function by inhibiting cytotoxic activity of maternal natural killer (NK) cells (Kovats et al. 1990; Rouas-Freiss et al. 1997). Moreover, the human and mouse non-classical MHC class I genes HLA-E and Qa-1 b respectively, are recognized by NK cells and result in the inhibition of the cytotoxic activity (Braud et al. 1998; Vance et al. 1998). As inhibitory receptors for NK cells, the killer immunoglobulin receptors (KIRs) in human and the Ly49 receptor in mouse play an especially important role for the recognition of the MHC molecules and their genomic regions are extremely diverged and polymorphic (Anfossi et al. 2006; Rahim and Makrigiannis 2015). However, the dog genome surprisingly does not have any KIR genes and it has only one Ly49 gene (Gagnier et al. 2003; Hammond et al. 2009). Therefore, domestic dogs may have a different mechanism to inhibit NK activity that is not seen in the human and mouse. In this regard, the elucidation of the essential functions of DLA-64 and DLA-79 and their functional relationships with NK cells is necessary for a better understanding of the mechanisms of acquired immunity and transplantation rejection in domestic dogs.
Our study of DLA-class I allelic haplotypes among five representative breeds (Table 5 and Supplementary table 5) supports previous observations that there are breed-specific DLA haplotypes, although genetic bias of DLA allelic haplotype numbers was observed in some breeds such that only three haplotypes (Hp. 07, Hp. 33 and Hp. 37) occupied 87.9% of all haplotypes (six haplotypes) in the Shiba breed used in this study (Supplementary table 5). The English Bulldog breed was reported recently to have low genetic diversity because of inbreeding and a small founder population (Pedersen et al. 2016). This low genetic diversity may endanger the health and well-being of the breed by increasing the number and prevalence of inherited diseases. On the other hand, a small number of polymorphisms and low diversity are thought to be one of the advantages for the allo-transplantation model by allowing the easier detection and graft acceptance of DLA matched donor and recipient dogs. Namely, grafts from DLA haplotype homozygous dogs as donors are theoretically more likely to be accepted in homozygous dogs with the same haplotype and in heterozygous dogs with the recipient having one of the donor’s haplotypes. In this study, we identified 86 dogs with DLA homozygous allelic haplotypes and 275 dogs with DLA heterozygous allelic haplotypes (Table 6). If we can stock stem cells such as dedifferentiated fat (DFAT) cells and induced pluripotent stem (iPS) cells representing the 22 major DLA-class I allelic haplotypes from homozygous dogs, then 91.9% of all dogs might be useful as animal models for studying transplantation mechanisms and benefit regenerative medicine for dogs and humans (Matsumoto et al. 2008; Yamanaka 2009).
In conclusion, we have identified and presented a large number of novel alleles for the DLA-class I genes and provided some further insights into the demographic and selection factors on the genomic structure, gene expression level, nucleotide diversity and phylogenetic relationships of DLA-class I alleles and haplotypes in 49 domestic dog breeds. This DLA polymorphism information and genetic differences among the dog breeds could be used as a standard internal control of the MHC genetic background for the benefit of biomedical research into regeneration medicine using the most common DLA allelic haplotypes as models for human MHC-related diseases.
References
Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, Vivier E (2006) Human NK cell education by inhibitory receptors for MHC class I. Immunity 25:331–342
Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, Yuki Y, Del Prete GQ, Goulder P, Brumme ZL, Brumme CJ, John M, Mallal S, Nelson G, Bosch R, Heckerman D, Stein JL, Soderberg KA, Moody MA, Denny TN, Zeng X, Fang J, Moffett A, Lifson JD, Goedert JJ, Buchbinder S, Kirk GD, Fellay J, McLaren P, Deeks SG, Pereyra F, Walker B, Michael NL, Weintrob A, Wolinsky S, Liao W, Carrington M (2013) Influence of HLA-C expression level on HIV control. Science 340:87–91
Arinzeh TL, Peter SJ, Archambault MP, van den Bos C, Gordon S, Kraus K, Smith A, Kadiyala S (2003) Allogeneic mesenchymal stem cells regenerate bone in a critical-sized canine segmental defect. J Bone Joint Surg Am 85-A:1927–1935
Bexfield NH, Watson PJ, Aguirre-Hernandez J, Sargan DR, Tiley L, Heeney JL, Kennedy LJ (2012) DLA class II alleles and haplotypes are associated with risk for and protection from chronic hepatitis in the English Springer spaniel. PLoS One 7:e42584
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC (1987) The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329:512–518
Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ (1998) HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391:795–799
Burnett RC, DeRose SA, Wagner JL, Storb R (1997) Molecular analysis of six dog leukocyte antigen class I sequences including three complete genes, two truncated genes and one full-length processed gene. Tissue Antigens 49:484–495
Burnett RC, Geraghty DE (1995) Structure and expression of a divergent canine class I gene. J Immunol 155:4278–4285
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Debenham SL, Hart EA, Ashurst JL, Howe KL, Quail MA, Ollier WE, Binns MM (2005) Genomic sequence of the class II region of the canine MHC: comparison with the MHC of other mammalian species. Genomics 85:48–59
Frantz LA, Mullin VE, Pionnier-Capitan M, Lebrasseur O, Ollivier M, Perri A, Linderholm A, Mattiangeli V, Teasdale MD, Dimopoulos EA, Tresset A, Duffraisse M, McCormick F, Bartosiewicz L, Gal E, Nyerges EA, Sablin MV, Brehard S, Mashkour M, Balasescu A, Gillet B, Hughes S, Chassaing O, Hitte C, Vigne JD, Dobney K, Hanni C, Bradley DG, Larson G (2016) Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science 352:1228–1231
Friedenberg SG, Buhrman G, Chdid L, Olby NJ, Olivry T, Guillaumin J, O'Toole T, Goggs R, Kennedy LJ, Rose RB, Meurs KM (2016) Evaluation of a DLA-79 allele associated with multiple immune-mediated diseases in dogs. Immunogenetics 68:205–217
Gagnier L, Wilhelm BT, Mager DL (2003) Ly49 genes in non-rodent mammals. Immunogenetics 55:109–115
Graumann MB, DeRose SA, Ostrander EA, Storb R (1998) Polymorphism analysis of four canine MHC class I genes. Tissue Antigens 51:374–381
Hammond JA, Guethlein LA, Abi-Rached L, Moesta AK, Parham P (2009) Evolution and survival of marine carnivores did not require a diversity of killer cell Ig-like receptors or Ly49 NK cell receptors. J Immunol 182:3618–3627
Hardt C, Ferencik S, Tak R, Hoogerbrugge PM, Wagner V, Grosse-Wilde H (2006) Sequence-based typing reveals a novel DLA-88 allele, DLA-88*04501, in a beagle family. Tissue Antigens 67:163–165
Hedges SB, Kumar S (2009) The timetree of life. Oxford University Press, New York
Holmes EC, Roberts AF, Staines KA, Ellis SA (2003) Evolution of major histocompatibility complex class I genes in Cetartiodactyls. Immunogenetics 55:193–202
Hosomichi K, Miller MM, Goto RM, Wang Y, Suzuki S, Kulski JK, Nishibori M, Inoko H, Hanzawa K, Shiina T (2008) Contribution of mutation, recombination, and gene conversion to chicken MHC-B haplotype diversity. J Immunol 181:3393–3399
Jung DI, Ha J, Kang BT, Kim JW, Quan FS, Lee JH, Woo EJ, Park HM (2009) A comparison of autologous and allogenic bone marrow-derived mesenchymal stem cell transplantation in canine spinal cord injury. J Neurol Sci 285:67–77
Kennedy LJ, Altet L, Angles JM, Barnes A, Carter SD, Francino O, Gerlach JA, Happ GM, Ollier WE, Polvi A, Thomson W, Wagner JL (1999) Nomenclature for factors of the dog major histocompatibility system (DLA), 1998. First report of the ISAG DLA Nomenclature Committee. International Society for Animals Genetics. Tissue Antigens 54:312–321
Kennedy LJ, Barnes A, Ollier WE, Day MJ (2006) Association of a common dog leucocyte antigen class II haplotype with canine primary immune-mediated haemolytic anaemia. Tissue Antigens 68:502–508
Kennedy LJ, Barnes A, Short A, Brown JJ, Seddon J, Fleeman L, Brkljacic M, Happ GM, Catchpole B, Ollier WE (2007) Canine DLA diversity: 3. Disease studies. Tissue Antigens 69(Suppl 1):292–296
Kennedy LJ, Ollier WER, Marti R, Wagner JL, Storb R (2012) Immunogenetics. In: Ruvinsky A, Ostrander E (eds) The Genetics of the Dog, 2nd, Edition edn. CAB International, Wallingford
Kim JW, Lee JH, Lyoo YS, Jung DI, Park HM (2013) The effects of topical mesenchymal stem cell transplantation in canine experimental cutaneous wounds. Vet Dermatol 24:242–e53
Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R (1990) A class I antigen, HLA-G, expressed in human trophoblasts. Science 248:220–223
Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H (2002) Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev 190:95–122
Kumanovics A, Takada T, Lindahl KF (2003) Genomic organization of the mammalian MHC. Annu Rev Immunol 21:629–657
Laurenson K, Sillero-Zubiri C, Thompson H, Shiferaw F, Thirgood S, Malcolm J (1998) Disease as a threat to endangered species: Ethiopian wolves, domestic dogs and canine pathogens. Anim Conserv 1:273–280
Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ 3rd, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, PJ DJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli KP, Parker HG, Pollinger JP, Searle SM, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M et al (2005) Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438:803–819
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Massey J, Boag A, Short AD, Scholey RA, Henthorn PS, Littman MP, Husebye E, Catchpole B, Pedersen N, Mellersh CS, Ollier WE, Kennedy LJ (2013a) MHC class II association study in eight breeds of dog with hypoadrenocorticism. Immunogenetics 65:291–297
Massey J, Rothwell S, Rusbridge C, Tauro A, Addicott D, Chinoy H, Cooper RG, Ollier WE, Kennedy LJ (2013b) Association of an MHC class II haplotype with increased risk of polymyositis in Hungarian Vizsla dogs. PLoS One 8:e56490
Matsumoto T, Kano K, Kondo D, Fukuda N, Iribe Y, Tanaka N, Matsubara Y, Sakuma T, Satomi A, Otaki M, Ryu J, Mugishima H (2008) Mature adipocyte-derived dedifferentiated fat cells exhibit multilineage potential. J Cell Physiol 215:210–222
Nei M, Gu X, Sitnikova T (1997) Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci U S A 94:7799–7806
Niskanen AK, Hagstrom E, Lohi H, Ruokonen M, Esparza-Salas R, Aspi J, Savolainen P (2013) MHC variability supports dog domestication from a large number of wolves: high diversity in Asia. Heredity (Edinb) 110:80–85
Noe L, Kucherov G (2005) YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res 33:W540–W543
Pang JF, Kluetsch C, Zou XJ, Zhang AB, Luo LY, Angleby H, Ardalan A, Ekstrom C, Skollermo A, Lundeberg J, Matsumura S, Leitner T, Zhang YP, Savolainen P (2009) mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol 26:2849–2864
Parham P, Lomen CE, Lawlor DA, Ways JP, Holmes N, Coppin HL, Salter RD, Wan AM, Ennis PD (1988) Nature of polymorphism in HLA-A, −B, and -C molecules. Proc Natl Acad Sci U S A 85:4005–4009
Pedersen NC, Pooch AS, Liu H (2016) A genetic assessment of the English bulldog. Canine Genet Epidemiol 3:6
Petersdorf EW, Gooley TA, Malkki M, Bacigalupo AP, Cesbron A, Du Toit E, Ehninger G, Egeland T, Fischer GF, Gervais T, Haagenson MD, Horowitz MM, Hsu K, Jindra P, Madrigal A, Oudshoorn M, Ringden O, Schroeder ML, Spellman SR, Tiercy JM, Velardi A, Witt CS, O'Huigin C, Apps R, Carrington M, International Histocompatibility Working Group in Hematopoietic Cell T (2014) HLA-C expression levels define permissible mismatches in hematopoietic cell transplantation. Blood 124:3996–4003
Petersdorf EW, Malkki M, O'HUigin C, Carrington M, Gooley T, Haagenson MD, Horowitz MM, Spellman SR, Wang T, Stevenson P (2015) High HLA-DP expression and graft-versus-host disease. N Engl J Med 373:599–609
Rahim MM, Makrigiannis AP (2015) Ly49 receptors: evolution, genetic diversity, and impact on immunity. Immunol Rev 267:137–147
Ross P, Buntzman AS, Vincent BG, Grover EN, Gojanovich GS, Collins EJ, Frelinger JA, Hess PR (2012) Allelic diversity at the DLA-88 locus in Golden Retriever and Boxer breeds is limited. Tissue Antigens 80:175–183
Rouas-Freiss N, Goncalves RM, Menier C, Dausset J, Carosella ED (1997) Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc Natl Acad Sci U S A 94:11520–11525
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sandmaier BM, Storb R, Santos EB, Krizanac-Bengez L, Lian T, McSweeney PA, Yu C, Schuening FG, Deeg HJ, Graham T (1996) Allogeneic transplant of canine peripheral blood stem cells mobilized by recombinant canine hematopoietic growth factors. Blood 87:3508–3513
Sarmiento UM, DeRose S, Sarmiento JI, Storb R (1992) Allelic variation in the DQ subregion of the canine major histocompatibility complex: I DQA. Immunogenetics 35:416–420
Sarmiento UM, DeRose S, Sarmiento JI, Storb R (1993) Allelic variation in the DQ subregion of the canine major histocompatibility complex: II DQB. Immunogenetics 37:148–152
Sarmiento UM, Storb R (1990a) Nucleotide sequence of a dog class I cDNA clone. Immunogenetics 31:400–404
Sarmiento UM, Storb R (1990b) Nucleotide sequence of a dog DRB cDNA clone. Immunogenetics 31:396–399
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108
Shi Q, Rafii S, Wu MH, Wijelath ES, Yu C, Ishida A, Fujita Y, Kothari S, Mohle R, Sauvage LR, Moore MA, Storb RF, Hammond WP (1998) Evidence for circulating bone marrow-derived endothelial cells. Blood 92:362–367
Shiba Y, Gomibuchi T, Seto T, Wada Y, Ichimura H, Tanaka Y, Ogasawara T, Okada K, Shiba N, Sakamoto K, Ido D, Shiina T, Ohkura M, Nakai J, Uno N, Kazuki Y, Oshimura M, Minami I, Ikeda U (2016) Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 538:388–391
Shiina T, Blancher A, Inoko H, Kulski JK (2017) Comparative genomics of the human, macaque and mouse major histocompatibility complex. Immunology 150:127–138
Shiina T, Hosomichi K, Inoko H, Kulski JK (2009) The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet 54:15–39
Shiina T, Inoko H, Kulski JK (2004) An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens 64:631–649
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Storb R, Yu C, Wagner JL, Deeg HJ, Nash RA, Kiem HP, Leisenring W, Shulman H (1997) Stable mixed hematopoietic chimerism in DLA-identical littermate dogs given sublethal total body irradiation before and pharmacological immunosuppression after marrow transplantation. Blood 89:3048–3054
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tsai KL, Starr-Moss AN, Venkataraman GM, Robinson C, Kennedy LJ, Steiner JM, Clark LA (2013) Alleles of the major histocompatibility complex play a role in the pathogenesis of pancreatic acinar atrophy in dogs. Immunogenetics 65:501–509
Vance RE, Kraft JR, Altman JD, Jensen PE, Raulet DH (1998) Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1(b). J Exp Med 188:1841–1848
Venkataraman G, Kennedy L, Little MT, Graves S, Harkey M, Torok-Storb B, Storb R (2017) Thirteen novel canine dog leukocyte antigen-88 alleles identified by sequence-based typing. HLA 90:165–170
Venkataraman GM, Geraghty D, Fox J, Graves SS, Zellmer E, Storer BE, Torok-Storb BJ, Storb R (2013) Canine DLA-79 gene: an improved typing method, identification of new alleles and its role in graft rejection and graft-versus-host disease. Tissue Antigens 81:204–211
Venkataraman GM, Stroup P, Graves SS, Storb R (2007) An improved method for dog leukocyte antigen 88 typing and two new major histocompatibility complex class I alleles, DLA-88*01101 and DLA-88*01201. Tissue Antigens 70:53–57
Vonholdt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A, Reynolds A, Bryc K, Brisbin A, Knowles JC, Mosher DS, Spady TC, Elkahloun A, Geffen E, Pilot M, Jedrzejewski W, Greco C, Randi E, Bannasch D, Wilton A, Shearman J, Musiani M, Cargill M, Jones PG, Qian Z, Huang W, Ding ZL, Zhang YP, Bustamante CD, Ostrander EA, Novembre J, Wayne RK (2010) Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464:898–902
Wagner JL, Burnett RC, DeRose SA, Francisco LV, Storb R, Ostrander EA (1996) Histocompatibility testing of dog families with highly polymorphic microsatellite markers. Transplantation 62:876–877
Wagner JL, Creer SA, Storb R (2000) Dog class I gene DLA-88 histocompatibility typing by PCR-SSCP and sequencing. Tissue Antigens 55:564–567
Wagner JL, Palti Y, DiDario D, Faraco J (2005) Sequence of the canine major histocompatibility complex region containing non-classical class I genes. Tissue Antigens 65:549–555
Wilbe M, Jokinen P, Hermanrud C, Kennedy LJ, Strandberg E, Hansson-Hamlin H, Lohi H, Andersson G (2009) MHC class II polymorphism is associated with a canine SLE-related disease complex. Immunogenetics 61:557–564
Xiao J, Xiang W, Chai Y, Haywood J, Qi J, Ba L, Qi P, Wang M, Liu J, Gao GF (2016) Diversified anchoring features the peptide presentation of DLA-88*50801: first structural insight into domestic dog MHC class I. J Immunol 197:2306–2315
Yamanaka S (2009) A fresh look at iPS cells. Cell 137:13–17
Acknowledgments
We thank Toshihiro Watari in Animal Medical Center (ANMEC) at Nihon University, Shinichi Namba in Marble Veterinary Medical center (Fujisawa, Japan) and Oriental Yeast Co. Ltd. (Tokyo, Japan) for providing the dog blood samples. We also thank Atsuko Shigenari and Sayaka Ito in Department of Molecular Life Science in Tokai University School of Medicine for technical assistance. This work was supported by Nihon University President’s Grant for Multidisciplinary Studies.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary figure 1
Nucleotide alignment for DLA-class I cDNA sequences and primer locations. The nucleotide alignment of DLA-class I genes was constructed using DLA-88 (NM_001014767.1), DLA-12 (NM_001014379.1) and DLA-64 (NM_001014378.1) cDNA and DLA-88 L (LC189199) genomic sequences. The DLA-79 cDNA sequence was not included in the alignment due to too many nucleotide differences. 5’UTR, SP, TM, CY and 3’UTR indicate 5′ untranslated region, signal peptide region, transmembrane region, cytoplasmic region and 3′ untranslated region, respectively. Hyphen indicates insertion or deletion site, and asterisk and period indicate matched and mismatched site among three sequences, respectively. Yellow background (around positions 100 bp) indicates sense primers for RT-PCR and genomic analyses, blue background (around positions 630 bp) indicates sense primers for quantitative PCR analysis, and green background (around positions 730 bp) indicates anti-sense primers for RT-PCR, genomic and quantitative PCR analyses. (DOCX 37 kb).
Supplementary figure 2
Inheritance of DLA-class I haplotypes based on DLA-class I cDNA analysis in four beagle families. A total of 38 beagles from four families were used for this study. Colored bars show the DLA haplotypes that are indicated in the table below the four pedigree charts. (PPTX 81 kb).
Supplementary figure 3
Relative gene expression levels of DLA-class I genes in each haplotype. (A) Relative gene expression levels of the DLA-88, DLA-12 and DLA-64 genes of the DLA-88-DLA-12-DLA-64 haplotypes using ten dogs. The DLA-88*501:01-DLA-12*1-DLA-64*nov2 (N = 4), DLA-88*006:01-DLA-12*1-DLA-64*nov2 (N = 3) and DLA-88*012:01-DLA-12*1-DLA-64*nov2 (N = 3) were analyzed. (B) Relative gene expression levels of the DLA-88, DLA-88 L and DLA-64 genes of DLA-88-DLA-88L-DLA-64 haplotypes using 12 dogs. The DLA-88*028:01-DLA-88*029:01-DLA-64*nov2 (N = 3), DLA-88*028:03-DLA-88*029:01-DLA-64*nov2 (N = 3), DLA-88*003:02-DLA-88*017:01-DLA-64*nov2 (N = 3) and DLA-88*nov10-DLA-88*L-DLA-64*nov2 (N = 3) were analyzed. Vertical axis shows the relative quantitative values by the real-time PCR method. Thin bars show standard errors. The vertical bars represent the mean of the relative quantitative values. (PPTX 58 kb).
Supplementary figure 4
Logos of amino acid consensus sequences of T cell recognition sites (TRSs) and peptide binding regions (PBRs) of translated DLA-class I genes. The logos plots were analyzed using 65 allele sequences (58 major and seven minor official alleles) with 45 DLA-88 (41 DLA-88 + four DLA-88*50X), seven DLA-88 L, 11 DLA-12 and two DLA-64 and six released DLA-79 sequences. (A) and (B) show logos for TRSs and PBRs, respectively. The overall height of each stack indicates the amino acid conservation at that position and the height of the letters within each stack indicated the relative frequency of the conforming amino acid at that position (Crooks et al. 2004). The overall height of DLA-64 is underestimated due to the low number of alleles. Yellow box shows an extra residue (leucine, L) in the DLA-88*50X allele group at position 155, which was attributed as part of the TRSs in a recent study (Xiao et al. 2016). Blue, black and green letters indicate hydrophilic, hydrophobic and green neutral amino acid residues, respectively. (PPTX 635 kb).
Supplementary Table 1
(XLSX 16 kb).
Supplementary Table 2A
(XLSX 20 kb).
Supplementary Table 2B
(XLSX 16 kb).
Supplementary Table 3
(XLSX 9 kb).
Supplementary Table 4
(XLSX 21 kb).
Supplementary Table 5
(XLSX 16 kb).
Rights and permissions
About this article
Cite this article
Miyamae, J., Suzuki, S., Katakura, F. et al. Identification of novel polymorphisms and two distinct haplotype structures in dog leukocyte antigen class I genes: DLA-88, DLA-12 and DLA-64 . Immunogenetics 70, 237–255 (2018). https://doi.org/10.1007/s00251-017-1031-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-017-1031-5