Abstract
CD8+ T lymphocytes can reduce the production of human immunodeficiency virus 1 (HIV-1) by CD4+ T cells by cytotoxic and non-cytotoxic mechanisms. To investigate the involvement of human leukocyte antigen (HLA) class I compatibility in anti-HIV responses, we co-cultured primary CD8+ T cells, isolated from the peripheral blood of HIV-1-infected individuals, with panels of autologous and heterologous acutely HIV-1-infected primary CD4+ T cells. Altogether, CD8+ T cell anti-HIV activity was evaluated in more than 200 co-cultures. Marked heterogeneity in HIV-1 replication levels was observed among the co-cultures sharing a common CD8+ T cell source. The co-cultures that exhibited greater than 50% reduction in HIV production were found to have significantly increased numbers of matching HLA class I alleles (Yates chi-square = 54.21; p < 0.001). With CD8+ T cells from HIV controllers and asymptomatic viremic individuals, matching HLA-B and/or HLA-C alleles were more predictive of strong anti-HIV activity than matching HLA-A alleles. Overall, HLA class I genotype matches were more closely associated with CD8+ T cell anti-HIV activity than supertype pairings. Antibodies against HLA class I and CD3 reduced the CD8+ T cell anti-HIV activity. Stimulated CD8+ T cells exhibited increased anti-HIV activity and reduced dependency on HLA compatibility. These findings provide evidence that the maximal suppression of HIV replication by CD8+ T cells requires the recognition of multiple epitopes. These studies provide insight for HIV vaccine development, and the analytic approach can be useful for the functional characterization of HLA class I alleles and tentative HLA class I supertypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thirty-five years ago, the human immunodeficiency virus 1 (HIV-1) was identified as the causative agent of acquired immune deficiency syndrome (AIDS) (Barre-Sinoussi et al. 1983). Since then, many studies have found that protection from HIV-1 infection and disease progression is associated with strong CD8+ T cell antiviral responses (Freel et al. 2011). These responses include the CD8+ T cell non-cytotoxic anti-HIV response (CNAR) (Walker et al. 1986) and the killing of virus-infected cells by antigen-specific cytotoxic T lymphocytes (CTL) (Walker et al. 1987). The relative contributions of these two mechanisms to CD8+ T cell-mediated anti-HIV immunity have not been fully determined (Davenport and Petravic 2010).
Besides CTL activity (Addo et al. 2003; Ho et al. 1993; Makadzange et al. 2010; Migueles et al. 2008; Walker et al. 1987), primary CD8+ T cells from HIV-1-infected individuals can potently suppress the replication of divergent HIV strains in CD4+ T cells without eliminating the infected cells (Mackewicz and Levy 1992; Mackewicz et al. 1998; Walker et al. 1991a; Walker et al. 1986; Walker et al. 1991b; Wiviott et al. 1990). Although maximal suppression levels are generally observed in cell-to-cell contact assays, CD8+ T cells can secrete factors that reduce HIV replication levels. These soluble factors include the CD8+ cell antiviral factor (CAF) that inhibits HIV transcription (Mackewicz et al. 1995; Walker and Levy 1989), a variety of β-chemokines that block virus attachment (Cocchi et al. 1995), and possibly other cytokines and cellular products (Cocchi et al. 2012; DeVico and Gallo 2004; Levy 2015). In comparison to the cell culture conditions generally required for measuring cytotoxic activity (Ho et al. 1993; Migueles et al. 2008), the non-cytotoxic suppression of HIV-1 replication can be observed with relatively low input ratios of ex vivo CD8+ T cells (Levy 2003). Nonetheless, both HIV-1-specific CTL (Makadzange et al. 2010) and anti-HIV-1-non-cytotoxic CD8+ T cells (Killian et al. 2011) exhibit a “memory cell” phenotype (e.g., CD45RA−, CD27+, CCR7−, PD-1+ cells). Moreover, IL-2, IL-15, and CD3/CD28 co-stimulation enhance the anti-HIV activity (Barker et al. 1997; Castelli et al. 2004), while IL-4 and IL-10 reduce this CD8+ T cell response (Barker et al. 1995). These attributes suggest that inhibition of HIV-1 production is carried out by memory CD8+ T cells having a Th1 profile and perhaps that the same cells can mediate both cytotoxic and non-cytotoxic activity.
The CD8+ T cell non-cytotoxic anti-HIV activity arises during the acute stage of HIV-1 infection (Mackewicz et al. 1994; Streeck and Nixon 2010) and correlates with a slower course of disease progression (Castelli et al. 2002; Ferbas et al. 1995; Gomez et al. 1994). In comparison to CD8+ T cells from most uninfected individuals, those from HIV-1-infected subjects exhibit greatly increased non-cytotoxic anti-HIV activity (Killian et al. 2011; Killian et al. 2005a; Rosok et al. 1997). This anti-HIV activity rapidly declines following antiretroviral therapy (Killian et al. 2009; Stranford et al. 2001; Wilkinson et al. 1999), likely due to decreases in the levels of cognate antigens, CD8+ T cell activation, and the frequency of circulating memory CD8+ T cells that accompany drug-suppressed virus replication (Giorgi et al. 1998). Notably, studies of SIV-infected rhesus macaques suggest that CD8+ T cell-mediated non-cytotoxic activity, rather than CTL activity, is largely responsible for controlling virus replication in the animals (Andrieu et al. 2014; Klatt et al. 2010; Wong et al. 2010).
Human leukocyte antigen (HLA) class I is expressed on all nucleated cells and functions to display intracellular protein fragments on the cell surface, particularly for recognition by CD8+ T cells (Abbas and Lichtman 2009; Marsh et al. 2000). The HLA class I-A, I-B, and I-C alleles are highly polymorphic, and individual HLA molecules present distinct peptides that are 8–12 amino acids long (Falk et al. 1991; Natarajan et al. 1999). Memory CD8+ T cells are antigen-experienced clonal populations of cells that rapidly respond to a restricted array of HLA class I-bound peptides (Killian et al. 2002). However, divergent HLA class I molecules can be clustered into groups called “supertypes” that present peptides having similar motifs and potential cross-reactivity with clonal CD8+ T cell populations (Sidney et al. 2008). Here, we report studies showing that inhibition of HIV-1 production by bulk CD8+ T cells that have not been artificially stimulated in vitro (hereafter referred to as unstimulated) is directly associated with the number of matching HLA class I alleles on the CD4+ target cells, suggesting that the maximal suppression of HIV replication requires the recognition of multiple HIV epitopes. We also show that the anti-HIV activity of CD8+ T cells that have been stimulated in vitro is less dependent on HLA compatibility.
Materials and methods
Study subjects
The HIV-1-infected subjects (n = 26; Table 1 ) who participated in this study were men reporting sex with men as their primary risk for having acquired HIV-1 infection. Most HIV-1-infected participants were asymptomatic, not receiving antiretroviral therapy, and had normal CD4+ T cell counts (>400 CD4+ T cells/μl). Included were 11 HIV controllers who exhibited low viral loads in the absence of antiretroviral therapy (6 elite controllers who had <75 HIV-1 RNA copies/ml of plasma and 5 low viremia controllers who had <650 HIV-1 RNA copies/ml). Blood from healthy HIV-negative donors was obtained from a local blood bank (Blood Centers of the Pacific, San Francisco, CA). Each participant signed informed consent forms, and the study received approval from the Committee for Human Research at the University of California San Francisco (UCSF).
Measurement of the CD8+ T cell anti-HIV response
To measure CD8+ T cell anti-HIV activity, incremental numbers of CD8+ T cells were co-cultured with autologous (i.e., from the same donor) or heterologous (i.e., from a different donor) acutely HIV-1-infected CD4+ T cells and the ensuing level of HIV replication was measured (Fig. 1) (Killian et al. 2009). Briefly, whole blood was collected from each subject by venipuncture into sodium heparin vacutainer tubes (BD). The blood was separated over Ficoll (Sigma) to obtain peripheral blood mononuclear cells (PBMCs), and purified into populations of primary CD8+ T and CD4+ T cells using immunomagnetic beads (Miltenyi). The purities of the isolated CD8+ T and CD4+ T cell populations were consistently >95% in routine flow cytometric assessments (data not shown). The freshly isolated CD8+ T cells were resuspended in freezing medium (90% fetal calf serum (FCS), 10% dimethyl sulfoxide, v/v) and cryopreserved in liquid nitrogen. The CD4+ T cells were resuspended (3 × 106 cells/ml) in growth medium [RPMI 1640 medium supplemented with FCS (heat-inactivated at 56 °C for 30 min, 10% v/v), penicillin (100 U/ml), streptomycin (100 μg/ml), l-glutamine (2 mM), and recombinant human IL-2 (100 U/ml; Invitrogen)] and stimulated for 3 days in the presence of phytohemagglutinin-leucoagglutinin (PHA-L, 3 μg/ml, Sigma) in a 37 °C humidified incubator. Subsequently, 107 stimulated CD4+ T cells were washed with RPMI medium and resuspended in 1 ml of HIV-1SF33 (10,000 TCID50/ml in PBMC) for 1 h at 37 °C with periodic mixing. HIV-1SF33 is a syncytium-inducing (SI), CXCR4-tropic (X4) isolate that has been maintained in primary cells since its isolation, exhibits rapid replication kinetics with a high degree of cytopathicity, and is not sensitive to β-chemokine-mediated antiviral effects (Mackewicz et al. 1997). The acutely HIV-1-infected CD4+ T cells were then washed and cryopreserved as described above. Thawed CD4+ T cells were resuspended at 106 cells/ml of growth medium, and 100-μl aliquots were placed into wells of a flat-bottom 96-well tissue culture plate (Falcon 3072, BD) in triplicate. Co-cultures were established by adding thawed CD8+ T cells (100 μl) to wells containing acutely infected CD4+ T cells at 1:1 and 0.5:1 CD8+ T cell to CD4+ T cell input ratios. In all experiments, unstimulated CD8+ T cells were co-cultured with the acutely HIV-infected CD4+ T cells. In some experiments, parallel co-cultures were established with CD8+ T cells that had been stimulated in vitro for 3 days with either phytohemagglutinin (PHA; Sigma) or anti-CD3 antibody (BD) conjugated to magnetic beads (Dynal).
Overview of the approach. Whole blood was collected from HIV-1-infected study participants, and peripheral blood mononuclear cells (PBMCs) were then isolated using a Ficoll gradient and manipulated as follows: i the resulting PBMCs were labeled with immunomagnetic beads and processed to obtain purified populations of CD8+ and CD4+ T cells; ii the purified CD4+ T cells were stimulated with PHA-L for 3 days and then were iii acutely infected for 1 h with HIV-1SF33; iv co-cultures were established by plating CD8+ T cells and acutely HIV-1-infected CD4+ T cells together in a combinatorial manner. Performing panels of co-cultures was facilitated by cryopreserving freshly isolated CD8+ T cells and acutely HIV-infected CD4+ T cells
To measure HIV replication levels in the cultures, 100-μl aliquots of the cell culture supernatant from each well were collected on days 3 and 6 of culture, centrifuged at 16,000×g for 1 h at 4 °C, and the resulting virus pellets were assayed for reverse transcriptase (RT) activity (Hoffman et al. 1985). In ongoing cultures, the supernatant removed for measurement of HIV levels was replaced with an equal volume of fresh growth medium. To enumerate the CD8+ T cell anti-HIV activity in each culture, the percent decrease in virus replication was calculated as the ratio of RT activity in the co-culture containing CD8+ T cells to the RT activity in the corresponding culture containing only CD4+ T cells.
HLA-class I and CD3 blocking experiments
To determine the need for HLA class I binding, a purified monoclonal antibody specific for human HLA-A,B,C (clone W6/32, azide-free, AbD Serotec, Raleigh, NC) (Slingluff et al. 1994; Ware et al. 1995) or matched isotype control antibody (IgG2a, AbD Serotec) was evaluated for the potential ability to block the CD8+ T cell anti-HIV activity. The antibodies (final concentration of 50 μg/ml) were added daily during the first 3 days of cell culture. To determine the need for T cell receptor (TCR) complex binding, anti-CD3 antibody (final concentration of 1 μg/ml; clone SK7; Becton Dickinson, BD) was added immediately after the CD8+ T cells and CD4+ T cells were plated.
HLA typing
High-resolution (allele level) HLA class I and II genotyping was carried out by using the sequence-based typing method as recommended by the 13th International Histocompatibility Workshop (Thio et al. 2003; Tilanus and Group 2000). The HLA class I and II genotypes of the study subjects are provided in Table 1. HLA class I and class II supertypes were assigned to the four-digit HLA class I alleles based on previously defined clusters (Doytchinova and Flower 2005; Sidney et al. 2008). The 12 possible HLA class I supertypes were A01, A02, A03, A24, B07, B08, B27, B44, B58, B62, C01, and C02. The 9 possible HLA class II supertypes were DQ1, DQ2, DQ3 and DR1, DR2, DR3, DR4, DR5, and DR9.
Statistical analyses
All data were compiled in an Access database (Microsoft). The associations between HLA class I and II genotypes and CD8+ T cell anti-HIV activity were assessed using χ 2 tests corrected for continuity and Mann–Whitney rank-sum tests. The Cochran–Armitage test for trend was used to assess for the presence of an association between categories of antiviral activity and categories of matching numbers of HLA genotypes. Results from experiments with anti-HLA class I antibodies were compared using Student’s t tests. HLA linkage disequilibrium was evaluated using an online calculator made available by the Los Alamos National Laboratory (http://www.hiv.lanl.gov). Excel (Microsoft), Total Access Statistics X.8 (FMS), and R (R-Core-Team 2013) were used to perform the statistical analyses, and SigmaPlot 11.2 (Systat) was used create the graphs.
Results
CD8+ T cells exhibit marked heterogeneity in their ability to decrease HIV production by differing infected CD4+ T cells
This study began with the observation that unstimulated (i.e., without in vitro stimulation) CD8+ T cells from HIV-1-infected individuals can exhibit heterogeneity in their ability to reduce HIV-1 replication in parallel co-cultures containing CD4+ T cells from different healthy HIV-1 seronegative blood bank donors (range = 0 to 99% decrease in HIV production at 1:1 CD8+ T cell to CD4+ cell input ratios; Fig. 2a). To further investigate the generalizability of this CD8+ T cell anti-HIV activity, HIV replication levels were measured in panels of autologous and heterologous co-cultures that contained unstimulated CD8+ T cells and acutely HIV-infected CD4+ T cells from various HIV-1-infected donors. The CD4+ T cells from HIV-1-infected donors were used for two reasons: they allowed for assessments of autologous co-cultures, and HLA-typing data were available for these cells. Unstimulated CD8+ T cells from each of the 23 asymptomatic HIV-1-infected subjects (including 3 subjects receiving HAART) were co-cultured with acutely HIV-infected CD4+ T cells from 2 to 20 different HIV-1-infected donors. Altogether, the CD4+ T cells isolated from 25 HIV-1-infected donors were used among 231 co-culture experiments. The unstimulated CD8+ T cells from the asymptomatic HIV-1-infected subjects potently reduced HIV replication in autologous CD4+ T cells (e.g., generally greater than 95% suppression at 1:1 CD8+ T cell to CD4+cell input ratios). In comparison, at 1:1 CD8+ T cell to CD4+ T cell input ratios, unstimulated CD8+ T cells from HIV-negative subjects were not observed to exhibit greater than 50% reduction in HIV replication in any co-cultures, including those containing autologous CD4+ T cells infected in vitro (data not shown).
CD8+ T cells from HIV-1-infected individuals exhibit marked heterogeneity in their ability to reduce HIV production by CD4+ T cells from different individuals. a Shown are the antiviral activities of CD8+ T cells from five HIV-1-infected study participants when co-cultured at 1:1 cell input ratios with acutely HIV-1-infected CD4+ T cells from different HIV-negative blood donors. b Shown is an ordered (by subject number) heat map representation of anti-HIV activity in more than 200 co-cultures containing variable CD8+ T cells and acutely HIV-infected CD4+ T cells from HIV-1-infected study participants (1:1 cell input ratio). Individual rows share CD8+ T cells from a common study subject, while individual columns have CD4+ T cells from a common study subject. Bright green and bright red squares indicate low (<30%) and high (>70%) level of suppression, respectively. Lightly shaded areas indicate data not available (N/A; co-cultures not performed) (color figure online)
When the CD8+ T cells from the HIV-1-infected subjects were co-cultured with acutely HIV-infected heterologous CD4+ T cells derived from HIV-1-infected individuals, marked variation was again observed in the CD8+ T cell antiviral activity (Fig. 2b). Maximal CD8+ T cell anti-HIV activity was consistently observed in co-cultures containing autologous CD4+ T cells. The CD8+ T cells in all of the autologous co-cultures exhibited greater than 50% reduction in HIV production at 1:1 and 0.5:1 CD8+ T cell to CD4+ T cell input ratios. In many cases, CD8+ T cells that were able to reduce HIV production in autologous CD4+ T cells by greater than 98% inhibited HIV replication by less than 25% in heterologous CD4+ T cells at 1:1 cell input ratios. Overall, 91 of the 209 (43.5%) heterologous co-cultures exhibited greater than 50% reduction in HIV production at 1:1 CD8+ T cell to CD4+ T cell input ratios. Furthermore, the variable ability of the CD8+ T cells to decrease HIV production by greater than 50% was consistently observed within each panel of co-cultures containing a shared source of CD8+ T cells (e.g., across rows in Fig. 2b). These results demonstrate that the observed level of CD8+ T cell anti-HIV activity is highly dependent on a feature(s) of the HIV-infected CD4+ T cells.
CD8+ T cell-mediated reduction in HIV replication is strongly associated with parity of HLA class I genotype
Among the 26 HIV-1-infected subjects included in this study, 62 unique HLA class I alleles (15 HLA-A, 25 HLA-B, and 22 HLA-C) were present. The most prevalent HLA class I alleles were A0201 (allele frequency = 0.19), A0301 (0.17), C0802 (0.14), A3201 (0.12), B0801 (0.12), B1402 (0.12), and C0602 (0.12). The most prevalent HLA class I haplotypes were A0201/B4402/C0501, A0101/B5701/C0602, and A0101/B0801/C0701, each present in 4 of 26 study subjects. These allele and haplotype frequencies are consistent with those reported for US populations (Cao et al. 2001), although B0702 appears to be underrepresented in our study, while the C0802 frequency is elevated. Genotype homozygosity at the HLA-A, HLA-B, or HLA-C locus was rare in this study group.
To evaluate the relationships between the CD8+ T cell-mediated reduction in HIV replication and the HLA class I and II relatedness of the co-cultured CD4+ T cells, these factors were co-analyzed. Anti-HIV activity was categorized as either (i) <50% decrease or (ii) >50% decrease and then evaluated (Table 2). Only CD8+ T cells that demonstrated the ability to reduce HIV replication by >50% in at least one co-culture at a 1:1 CD8+ T cell to CD4+ T cell input ratio were included in the analyses. This criterion for inclusion was used to avoid the analysis of CD8+ T cells that lacked any appreciable anti-HIV activity, such as those from HIV-1-infected individuals who are receiving antiretroviral therapy and/or have advanced to disease (Killian et al. 2005a; Killian et al. 2009; Stranford et al. 2001).
Among the 209 heterologous co-cultures, the proportion of co-cultures having at least one HLA class I matching allele was significantly elevated among those exhibiting greater than 50% reduction in HIV production (84 vs. 31%; Yates χ 2 = 54.21, p < 0.001). The presence of some co-cultures having a single HLA class I match in the absence of a strong reduction of virus replication is consistent with previous reports showing that CD8+ T cells from HIV-1-infected individuals only respond to a fraction of the theoretically recognizable epitopes (Addo et al. 2003; Killian et al. 2005b). In this regard, a direct monotonic trend (Fig. 3a) was evident between the number of HLA class I matching alleles and the magnitude of the CD8+ T cell-mediated reduction in HIV production (R S = 0.663, p < 0.001). Thus, strong CD8+ T cell anti-HIV activity was associated with an increased probability of presentation of an immunogenic epitope. These results demonstrate that maximal CD8+ T cell anti-HIV activity is strongly associated with HLA class I compatibility between CD8+ T cells and HIV-infected CD4+ T cells.
Direct association between the CD8+ T cell-mediated reduction of HIV production and the HLA class I and class II relatedness of the HIV-infected CD4+ T cells. Box plots show the distributions of the CD8+ T cell anti-HIV activity data categorized by a the number of HLA class I four-digit allelotype matches (0, 1, 2, 3–5, and autologous) and b the number of HLA class II four-digit allelotype matches (0, 1, 2+, and autologous) between the co-cultured CD8+ T cells and the acutely HIV-infected CD4+ T cells (1:1 cell input ratio). The lower, middle, and upper boundaries of each box reveal the 25th, 50th, and 75th percentiles, respectively. Whiskers indicate the 10th and 90th percentiles. A Kruskal-Wallis one-way analysis of variance (ANOVA) on ranks was performed (excluding the autologous data), and Dunn’s method was used to make pairwise multiple comparisons
CD8+ T cell anti-HIV activity is more closely associated with HLA class I than HLA class II
Additional analyses were performed to investigate the potential involvement of HLA class II-restricted CD8+ T cell responses (Ranasinghe et al. 2016). Among the cells included in this study, there were 12 unique HLA-DQ alleles and 21 unique HLA-DR alleles. A direct monotonic relationship was observed (Fig. 3b) between CD8+ T cell antiviral activity and the frequency of matching HLA class II alleles (R S = 0.402, p < 0.001). However, in comparison to the relationship with HLA class I matches, the Spearman correlation coefficient for the relationship with HLA class II matches was lower. Sensitivity analyses, based on logistic regression models for predicting percent suppression, did not reveal any improvement over HLA class I genotype matches when HLA class II genotype matches were added to the model. Moreover, several (29/87 = 33%; HLA class II data not available for 4/91 co-cultures) co-cultures exhibited >50% reduction in HIV production without having any HLA class II matches. The occurrence of HLA-B*1402 and HLA-DR*0102 was linked in our study population (p = 4.34e−06), with 6 of the 26 HIV-infected individuals expressing both alleles and none expressing only one of these alleles. Thus, at least part of the observed association between CD8+ T cell antiviral activity and the frequency of HLA class II matches is likely due to strong linkage disequilibrium with HLA class I (Ness et al. 1981).
HLA-B and HLA-C matches are associated with a strong CD8+ T cell anti-HIV response
In addition to evaluating the relationship between the frequency of HLA class I matches and CD8+ T cell anti-HIV activity, we also assessed potential associations with individual HLA class I loci. Present among the 209 heterologous co-culture experiments were 79 co-cultures that featured a single matching HLA class I allele (Fig. 4a). Among these 79 co-cultures were 35 and 44 co-cultures that exhibited <50 and >50% reduction in HIV production, respectively. The large majority (64/79) of matches were present at the HLA-A locus. Remarkably, none of the 35 co-cultures in this subset that exhibited a <50% decrease had matching HLA-B alleles; 4 of the co-cultures with >50% reduction had matching HLA-B alleles. At the HLA-A locus, the CD8+ T cells from 11 study subjects exhibited differential anti-HIV activity while sharing the same matching allele with variable heterologous CD4+ T cells. The matching alleles were HLA-A*0101, 0201, 0301, 1101, and 3201. At the HLA-B and HLA-C loci, none of the CD8+ T cells exhibited differential anti-HIV activity while sharing the same matching alleles. Notably, the CD8+ T cells from four separate individuals shared a single HLA-C0802 matching allele with variable CD4+ T cells. Despite the identified presence of optimal epitopes in the Nef and Gag sequences of HIV-1SF33, such as AAVDLSHFL and TPQDLNTML, respectively (Llano et al. 2009), none of the co-cultures having only a matching C0802 allele exhibited >50% reduction in HIV production.
Relationship between CD8+ T cell anti-HIV activity and HLA class I loci. a Shown are the frequencies of heterologous co-cultures (y-axis) having no matching HLA class I alleles or matching alleles at specific loci. b Shown are the frequencies of heterologous co-cultures having any matching alleles at the HLA-A locus and at the HLA-B and/or HLA-C loci. For both panels, black bars and gray bars denote co-cultures exhibiting ≤50 and >50% suppression of HIV replication, respectively
While sizable percentages of both anti-HIV activity categories contained only matching HLA-A alleles, relatively few matches were present at only the HLA-B or HLA-C loci, consistent with linkage disequilibrium findings (Hiller et al. 1978). Therefore, we compared the frequencies of matches at the HLA-A locus with those having matches at the HLA-B and/or HLA-C loci among co-cultures exhibiting discordant CD8+ T cell anti-HIV activity (Fig. 4b). Among the co-cultures exhibiting a <50% reduction in HIV production, 25 and 6% had matching HLA-A and HLA-B/C alleles, respectively. Among the co-cultures exhibiting a >50% reduction in HIV production, 59 and 46% had matching HLA-A and HLA-B/C alleles, respectively. Consequently, the relative ratio (RR) of co-cultures having strong CD8+ T cell anti-HIV activity and matching HLA-B/C (RR = 7.7) alleles was more than three times higher than the corresponding ratio for matching HLA-A alleles (RR = 2.4). Thus, HLA-B/C matching alleles are more predictive of strong anti-HIV activity than matching HLA-A alleles (Yates χ 2 = 5.07, p = 0.024).
Similar relationships between CD8+ T cell anti-HIV activity and HLA class I compatibility among HIV controllers and asymptomatic viremic individuals
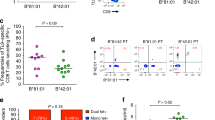
To investigate potential differences in the CD8+ T cells from HIV-1-infected individuals having different viral loads (VL), we compared the patterns of variability in anti-HIV activity in heterologous co-cultures containing CD8+ T cells from HIV controllers (six elite controllers and five viremic controllers; see “Materials and methods” section) with those containing CD8+ T cells from asymptomatic viremic individuals (n = 9, VL ≥2000). The anti-HIV activity exhibited by CD8+ T cells from both HIV controllers and viremic individuals was directly proportional to the number of matching HLA class I alleles, with two or more matching alleles generally being required for maximal control of virus replication (Fig. 5a). Furthermore, for both groups, the presence of matching HLA-B/C alleles was more predictive of strong CD8+ T cell anti-HIV activity than was the presence of matching HLA-A alleles (Fig. 5b). These results demonstrate that the described relationships between CD8+ T cell anti-HIV activity and HLA class I are similar among asymptomatic HIV-1-infected individuals having disparate viral loads.
Similar relationships between CD8+ T cell anti-HIV activity and HLA class I matching among CD8+ T cells from HIV controllers and viremic individuals. a Box plots show the distributions of the CD8+ T cell anti-HIV activity data categorized by the number of HLA class I four-digit allelotype matches (0, 1, 2–5, and autologous) between the co-cultured CD8+ T cells and the acutely HIV-infected CD4+ T cells (1:1 cell input ratio). Box plots for HIV controllers and viremic individuals are shaded gray and white, respectively. The lower, middle, and upper boundaries of each box reveal the 25th, 50th, and 75th percentiles, respectively. Whiskers indicate the 10th and 90th percentiles. b Shown are the frequencies of heterologous co-cultures having any matching alleles at the HLA-A locus and at the HLA-B and/or HLA-C loci for HIV controllers and viremic individuals. Black bars and gray bars denote co-cultures exhibiting ≤50 and >50% suppression of HIV replication, respectively
CD8+ T cells can reduce HIV production in heterologous CD4+ T cells having multiple compatible HLA class I supertypes
To evaluate the import of HLA class I families that are able to present similarly recognizable peptides, supertype designations were assigned to the HLA class I genotypes and then analyzed with respect to the observed CD8+ T antiviral activity in the co-cultures (Fig. 6a). A direct monotonic relationship was present between the number of HLA class I supertype matches and the magnitude of the CD8+ T cell anti-HIV activity (R S = 0.516, p < 0.001). Overall, 206 of the 209 (99%) heterologous co-cultures had at least one HLA class I supertype match at the -A, -B, or -C loci. However, notable supertype differences were present among the co-cultures having discordant antiviral activity. Each of the three co-cultures that lacked any HLA class I supertype matches exhibited less than 50% reduction in HIV production, suggesting that supertype matches are a necessary, but not a sufficient, condition for the CD8+ T cell antiviral activity. All (15 of 15) of the heterologous co-cultures that exhibited greater than 50% anti-HIV activity without having any strict HLA class I four-digit genotype matches at the -A, -B, and -C loci featured HLA class I supertype matches; most had HLA-B matches (13 of 15 = 87%) and two or more distinct HLA class I supertype matches (11 of 15 = 73%). In comparison, fewer than half (40 of 81 = 49%) of the co-cultures that exhibited less than 50% anti-HIV activity without having any strict HLA class I four-digit genotype matches exhibited HLA-B supertype matches, but many (62 of 81 = 77%) exhibited two or more supertype matches at the HLA-A and HLA-C loci. The relationship between HLA class II supertype matches and CD8+ T cell anti-HIV activity (Fig. 6b; R S = 0.317, p < 0.001) was not as strong as the relationship with HLA class I supertype matches. These results indicate that CD8+ T cells can reduce HIV replication in CD4+ T cells having composite genotypes of closely related HLA class I alleles and provide further support for our observed associations with the number of HLA-matching alleles and matches at the HLA-B/C loci.
HLA class I and class II supertype matches in relation to CD8+ T cell anti-HIV activity. Box plots show the distributions of the CD8+ T cell anti-HIV activity data categorized by a the number of HLA class I supertype matches and b the number of HLA class II supertype matches between the co-cultured CD8+ T cells and the acutely HIV-infected CD4+ T cells (1:1 cell input ratio). The lower, middle, and upper boundaries of each box reveal the 25th, 50th, and 75th percentiles, respectively. Whiskers indicate the 10th and 90th percentiles
Anti-HLA-class I and anti-CD3 antibodies can block the CD8+ T cell-mediated anti-HIV activity
To further investigate the observed association between CD8+ T cell anti-HIV activity and HLA class I, anti-HLA class I antibodies were added to the co-cultures (Fig. 7). Notably, the very short half-life (≤1 h) of HLA class I molecules at the cell surface (Su and Miller 2001) necessitates the frequent addition of relatively large quantities of anti-HLA class I antibody. When anti-HLA class I antibody (50 μg/ml) was added daily, the anti-HIV activity of CD8+ T cells otherwise excellent was reduced (Fig. 7a). The anti-IgG antibody of the appropriate isotype had no appreciable effect on HIV replication. Substantial reductions in CD8+ T cell anti-HIV activity were noted in four of four experiments at both the 0.5:1 (p < 0.001) and 1:1 (p = 0.013) CD8+ T cell to CD4+ T cell input ratios (Fig. 7b). Additional experiments were performed to evaluate the potential requirement for T cell receptor (TCR) binding (Fig. 7c, d). In two of two experiments, the addition of anti-CD3 antibody to the co-cultures completely abrogated the ability of CD8+ T cells to reduce HIV production. These results demonstrate that maximal CD8+ T cell anti-HIV activity requires HLA class I and TCR interactions.
Anti-HLA-class I antibody and anti-CD3 antibody block CD8+ T cell anti-HIV activity. a Shown is a representative example of the levels of HIV replication in co-cultures containing medium alone, mouse anti-human IgG (50 μg/ml), or anti-HLA class I antibody (50 μg/ml). For each culture condition, 100,000 HIV-infected CD4+ T cells were cultured alone (0:1) or with 50,000 (0.5:1) or 100,000 (1:1) CD8+ T cells. b The effects of anti-HLA class I antibody on CD8+ T cell antiviral activity are shown for the four experiments that were performed with the daily addition of antibody (50 μg/ml). c Shown is a representative example of the levels of HIV replication in co-cultures containing medium alone or anti-CD3 antibody. For each culture condition, 100,000 HIV-infected CD4+ T cells were cultured alone (0:1) or with 50,000 (0.5:1) or 100,000 (1:1) CD8+ T cells. d The effects of anti-CD3 antibody on CD8+ T cell antiviral activity are shown for the two experiments that were performed. a–d Results from autologous co-cultures
CD8+ T cells that are stimulated in vitro exhibit enhanced and unrestricted anti-HIV activity in co-culture assays
To determine their relative anti-HIV activities, unstimulated and PHA-stimulated CD8+ T cells were evaluated for their abilities to reduce HIV replication levels in acutely HIV-1-infected autologous and heterologous CD4+ T cells (Fig. 8). In parallel co-cultures with unstimulated CD8+ T cells, the CD8+ T cells that were stimulated in vitro with PHA exhibited significantly increased anti-HIV activity (Fig. 8a; Wilcoxon signed-rank test, p < 0.001). The anti-HIV activity increased with stimulation of the CD8+ T cells in nearly all (24/26) experiments. In 10 of 14 heterologous co-cultures where the unstimulated CD8+ T cells exhibited very low anti-HIV activity, the stimulated CD8+ cells reduced HIV production by greater than 50% (Fig. 8b). Moreover, in most (3/5) heterologous co-cultures lacking any HLA class I genotype matches, those containing in vitro stimulated CD8+ T cells exhibited greater than 50% reduction in HIV production, while the co-cultures containing unstimulated CD8+ T cells exhibited very little or no anti-HIV activity. The already high level anti-HIV activity observed in all (7/7) of the autologous co-cultures was further increased with stimulation of the CD8+ T cells (Wilcoxon signed-rank test, p = 0.016). We also observed that stimulated, but not unstimulated, CD8+ T cells from an HIV-2-infected individual exhibited appreciable anti-HIV activity (64% reduction; see s99 in Fig. 8b) in co-cultures with acutely HIV-1-infected heterologous CD4+ cells, indicating that the anti-HIV activity of stimulated CD8+ T cells is not virus specific. These results demonstrate that stimulated CD8+ cells, unlike unstimulated CD8+ T cells, are able to reduce virus production levels in acutely HIV-1-infected CD4+ T cells without a requirement for HLA class I compatibility.
CD8+ T cells stimulated in vitro reduce HIV-1 production in HLA class I discordant CD4+ T cells. a Parallel co-cultures were established with unstimulated and stimulated CD8+ T cells. Shown are the anti-HIV activities of co-cultures containing 1:1 input ratios of CD8+ T cells and acutely HIV-infected CD4+ T cells. Horizontal bars indicate median values. b Representative examples are shown of the anti-HIV activities of unstimulated and stimulated CD8+ T cells co-cultured with acutely HIV-infected autologous (squares) or heterologous (circles) CD4+ T cells lacking any HLA class I genotype matches. Symbols are shaded gray to denote results with stimulated CD8+ T cells
Discussion
CD8+ T lymphocytes from HIV-1-infected individuals can potently reduce HIV-1 replication in CD4+ T cells in vitro, and this antiviral activity is directly associated with a positive clinical prognosis (Castelli et al. 2002; Ferbas et al. 1995; Gomez et al. 1994; Streeck et al. 2014; Streeck and Nixon 2010). A previous study reported that the measurement of the CD8+ T cell non-cytotoxic antiviral activity in vitro can vary with the source of the CD4+ T cells (Mackewicz et al. 1998), but a conclusive association with HLA class I was not demonstrated. Several studies have indicated that the CD8+ cell cytotoxic responses require HLA compatibility (Yang and Walker 1997). In this present large study, our data do not directly address whether the requirement for HLA class I compatibility is associated with a cytotoxic or non-cytotoxic mechanism. However, we have demonstrated that bulk CD8+ cells taken from HIV-infected subjects and not stimulated in vitro can reduce HIV production by CD4+ cells in a manner that is directly proportional to the number of HLA class I matching alleles.
Analysis of the HIV replication levels in 231 (22 autologous and 209 heterologous) co-cultures with respect to the degree of HLA class I relatedness between the co-cultured CD8+ T and CD4+ T cells revealed that the anti-HIV activity of unstimulated CD8+ T cells is strongly associated with concordance in the HLA class I genotype (Fig. 3 and Table 2). The diminished CD8+ T cell anti-HIV activity in the presence of anti-CD3 and anti-HLA class I antibodies (Fig. 7) supports the conclusion that an interaction between the T cell receptor (TCR) complex and HLA class I is associated with the mechanism by which unstimulated CD8+ T cells reduce HIV production by acutely HIV-infected CD4+ T cells. We also observed that the HLA class I-restricted anti-HIV activity observed with unstimulated CD8+ T cells can be overcome following the in vitro stimulation of the CD8+ T cells (Fig. 8), reasonably due a mechanism that mimics the natural TCR-peptide-HLA class I interaction (Valentine et al. 1985).
Associations between HLA class I and HIV-1 disease progression have been observed in numerous epidemiologic studies (Carrington et al. 1999; Pereyra et al. 2010; Saah et al. 1998; Trachtenberg et al. 2003). Given the well-characterized binding of CD8 to HLA class I (Cole and Gao 2004), these findings implicate a role for CD8+ T cells. However, the biologic mechanism(s) underlying these associations remain unclear. One theory is that variable fitness costs are associated with mutations across the HIV genome and that protective HLA types are those that present relatively immutable viral epitopes (Carlson et al. 2015; Kloverpris et al. 2015). Consistent with this theory is the longstanding observation that heterozygosity of HLA class I alleles is associated with a protective advantage in terms of HIV-1 survival (Carrington et al. 1999). Based on instances of reduced HIV production in the presence of a single HLA class I match, as well as the direct association between the numbers of HLA class I matches and the CD8+ T cell anti-HIV activity, our findings provide further evidence for a HLA class I heterozygosity advantage.
A fundamental question that has relevance to HIV vaccine design, and perhaps to the development of cell-based therapies, pertains to whether particular HLA genes are more important for the control of HIV. HLA-B types (e.g., B57, B58, B27, B51, and B81) are most frequently associated with improved prognosis (Carrington and O’Brien 2003; Pereyra et al. 2010). Possible mechanisms for the protective role of HLA-B include its relative resistance to downregulation by the HIV-1-negative regulatory factor (Nef) (Rajapaksa et al. 2012) and its rapid display of peptides on the cell surface (Neefjes et al. 2011). Our findings with unstimulated CD8+ cells provide further support for a dominant role of HLA-B in HIV control by showing that matching HLA-B and/or C alleles are more predictive of strong CD8+ T cell anti-HIV activity than matching HLA-A alleles (Fig. 4). This relationship was similarly observed among CD8+ T cells from HIV controllers and viremic individuals and therefore appears to be independent of viral load (Fig. 5). Importantly, we show that bulk CD8+ T cells from both HIV controllers and non-controllers require multiple matching HLA class I alleles, and presumably the recognition of multiple HIV epitopes, for the maximal suppression of HIV replication. Therefore, optimal vaccines may require an ability to elicit CD8+ T cell responses to multiple HIV epitopes in each person vaccinated.
To our knowledge, this is the first study to evaluate the anti-HIV activity of CD8+ T cells with respect to composite HLA class I supertypes. Nearly all of the heterologous co-cultures in this study had matching HLA class I supertype alleles, whereas fewer than half of the co-cultures exhibited greater than 50% reduction in HIV replication (Fig. 6 and Table 2). In comparison, the frequencies of co-cultures having matching HLA class I genotypes and exhibiting greater than 50% anti-HIV activity were more closely aligned (36 and 44%, respectively). Together, these observations strongly suggest that CD8+ T cells are unable to functionally respond to many theoretically compatible HLA class I supertypes. Thus, the classification of HLA class I supertypes, as defined by others (Lund et al. 2004; Sidney et al. 2008), seems to have limited utility for predicting functional CD8+ T cell responses.
A large number of studies of the CD8+ T cell response to HIV-1 infection have been based on the use of synthetic peptides alone and/or in the context of HLA class I tetrameric complexes or antigens expressed by immortalized cell lines. A vaccine conceptualized from those study results was ineffective (Sekaly 2008). The approach used in the present study can provide a more natural system for the examination of antigen-specific responses in terms of antigen processing, the amount of antigen present, and the interactions with co-stimulatory and inhibitory molecules. In this regard, our approach can potentially be used to reverse-engineer HIV peptides having the greatest capacity to induce CD8+ T cell anti-HIV activity across discordant HLA class I types. This strategy would entail performing panels of heterologous co-cultures and determining the over-represented HLA types among co-cultures exhibiting strong CD8+ T cell anti-HIV activity in the absence of matching alleles. The peptides presented by those HLA types could then be identified using bioinformatics (Larsen et al. 2007) and advanced mass spectrometry procedures (Mommen et al. 2014). The same approach can be used for validation and refinement of proposed HLA class I supertypes. Accordingly, co-cultured cells that exhibit anti-HIV activity in the absence of matching allotypes are hypothesized to share at least one HLA class I supertype. Alternatively, co-cultured cells that do not exhibit anti-HIV activity are likely void of any common biologically relevant HLA class I supertype. Reasonably, the approach can be adapted to investigations of other viral infections and cancer by performing panels of co-cultures with CD8+ T cells from patients and variable heterologous target cells.
In summary, our studies show a direct relationship between CD8+ T cell anti-HIV activity and the number of compatible (recognized) HLA class I alleles, as evidenced by the variable HIV replication levels in co-cultures containing autologous, partially matched, or completely mismatched CD8+ T and CD4+ T cells. Our experimental approach using unstimulated CD8+ T cells and heterologous acutely HIV-infected CD4+ T cells can be useful for vaccine development and the refinement of HLA class I supertype designations.
References
Abbas AK, Lichtman AH (2009) Basic immunology : functions and disorders of the immune system, 3rd edn. Saunders/Elsevier, Philadelphia
Addo MM et al (2003) Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol 77:2081–2092
Andrieu JM, Chen S, Lai C, Guo W, Lu W (2014) Mucosal SIV vaccines comprising inactivated virus particles and bacterial adjuvants induce CD8(+) T-regulatory cells that suppress SIV-positive CD4(+) T-cell activation and prevent SIV infection in the macaque model front. Immunol 5:297. doi:10.3389/fimmu.2014.00297
Barker E, Mackewicz CE, Levy JA (1995) Effects of TH1 and TH2 cytokines on CD8+ cell response against human immunodeficiency virus: implications for long-term survival. Proc Natl Acad Sci USA 92:11135–11139
Barker E, Bossart KN, Fujimura SH, Levy JA (1997) CD28 costimulation increases CD8+ cell suppression of HIV replication. J Immunol 159:5123–5131
Barre-Sinoussi F et al (1983) Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868–871
Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernandez-Vina MA (2001) Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol 62:1009–1030
Carlson JM, Le AQ, Shahid A, Brumme ZL (2015) HIV-1 adaptation to HLA: a window into virus-host immune interactions. Trends Microbiol 23:212–224. doi:10.1016/j.tim.2014.12.008
Carrington M, O’Brien SJ (2003) The influence of HLA genotype on AIDS. Annu Rev Med 54:535–551. doi:10.1146/annurev.med.54.101601.152346
Carrington M et al (1999) HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748–1752
Castelli JC, Deeks SG, Shiboski S, Levy JA (2002) Relationship of CD8(+) T cell noncytotoxic anti-HIV response to CD4(+) T cell number in untreated asymptomatic HIV-infected individuals. Blood 99:4225–4227
Castelli J, Thomas EK, Gilliet M, Liu YJ, Levy JA (2004) Mature dendritic cells can enhance CD8+ cell noncytotoxic anti-HIV responses: the role of IL-15. Blood 103:2699–2704
Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P (1995) Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811–1815
Cocchi F et al (2012) Soluble factors from T cells inhibiting X4 strains of HIV are a mixture of beta chemokines and RNases. Proc Natl Acad Sci USA 109:5411–5416. doi:10.1073/pnas.1202240109
Cole DK, Gao GF (2004) CD8: adhesion molecule, co-receptor and immuno-modulator. Cell Mol Immunol 1:81–88
Davenport MP, Petravic J (2010) CD8+ T cell control of HIV—a known unknown. PLoS Pathog 6:e1000728. doi:10.1371/journal.ppat.1000728
DeVico AL, Gallo RC (2004) Control of HIV-1 infection by soluble factors of the immune response. Nat Rev Microbiol 2:401–413
Doytchinova IA, Flower DR (2005) In silico identification of supertypes for class II MHCs. J Immunol 174:7085–7095
Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG (1991) Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351:290–296. doi:10.1038/351290a0
Ferbas J et al (1995) Virus burden in long-term survivors of human immunodeficiency virus (HIV) infection is a determinant of anti-HIV CD8 + lymphocyte activity. J Infect Dis 172:329–339
Freel SA, Saunders KO, Tomaras GD (2011) CD8(+)T-cell-mediated control of HIV-1 and SIV infection. Immunol Res 49:135–146. doi:10.1007/s12026-010-8177-7
Giorgi JV, Majchrowicz MA, Johnson TD, Hultin P, Matud J, Detels R (1998) Immunologic effects of combined protease inhibitor and reverse transcriptase inhibitor therapy in previously treated chronic HIV-1 infection. AIDS 12:1833–1844
Gomez AM, Smaill FM, Rosenthal KL (1994) Inhibition of HIV replication by CD8+ T cells correlates with CD4 counts and clinical stage of disease. Clin Exp Immunol 97:68–75
Hiller C, Bischoff M, Schmidt A, Bender K (1978) Analysis of the HLA-ABC linkage disequilibrium: decreasing strength of gametic association with increasing map distance. Hum Genet 41:301–312
Ho HN et al (1993) Circulating HIV-specific CD8+ cytotoxic T cells express CD38 and HLA-DR antigens. J Immunol 150:3070–3079
Hoffman AD, Banapour B, Levy JA (1985) Characterization of the AIDS-associated retrovirus reverse transcriptase and optimal conditions for its detection in virions. Virology 147:326–335
Killian MS, Matud J, Detels R, Giorgi JV, Jamieson BD (2002) MaGiK method of T-Cell receptor repertoire analysis. Clin Diagn Lab Immunol 9:858–863
Killian MS, Ng S, Mackewicz CE, Levy JA (2005a) A screening assay for detecting CD8(+) cell non-cytotoxic anti-HIV responses. J Immunol Methods 304:137–150
Killian MS, Sabado RL, Kilpatrick S, Hausner MA, Jamieson BD, Yang OO (2005b) Clonal breadth of the HIV-1-specific T-cell receptor repertoire in vivo as determined by subtractive analysis. AIDS 19:887–896
Killian MS, Roop J, Ng S, Hecht FM, Levy JA (2009) CD8+ cell anti-HIV activity rapidly increases upon discontinuation of early antiretroviral therapy. J Clin Immunol 29:311–318
Killian MS, Johnson C, Teque F, Fujimura S, Levy JA (2011) Natural suppression of human immunodeficiency virus type 1 replication is mediated by transitional memory CD8+ T cells. J Virol 85:1696–1705. doi:10.1128/JVI.01120-10
Klatt NR et al (2010) CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog 6:–e1000747. doi:10.1371/journal.ppat.1000747
Kloverpris HN, Leslie A, Goulder P (2015) Role of HLA adaptation in HIV evolution. Front Immunol 6:665. doi:10.3389/fimmu.2015.00665
Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M (2007) Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinf 8:424. doi:10.1186/1471-2105-8-424
Levy JA (2003) The search for the CD8+ cell anti-HIV factor (CAF). Trends Immunol 24:628–632
Levy JA (2015) Discovery of another anti-HIV protein in the search for the CD8+ cell anti-HIV factor. Proc Natl Acad Sci USA 112:7888–7889. doi:10.1073/pnas.1509324112
Llano A, Frahm N, Brander C (2009) How to optimally define optimal cytotoxic T lymphocyte epitopes in HIV infection? Los Alamos National Laboratory, Los Alamos
Lund O et al (2004) Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics 55:797–810. doi:10.1007/s00251-004-0647-4
Mackewicz C, Levy JA (1992) CD8+ cell anti-HIV activity: nonlytic suppression of virus replication. AIDS Res Hum Retrovir 8:1039–1050
Mackewicz CE, Yang LC, Lifson JD, Levy JA (1994) Non-cytolytic CD8 T-cell anti-HIV responses in primary HIV-1 infection. Lancet 344:1671–1673
Mackewicz CE, Blackbourn DJ, Levy JA (1995) CD8+ T cells suppress human immunodeficiency virus replication by inhibiting viral transcription. Proc Natl Acad Sci USA 92:2308–2312
Mackewicz CE, Barker E, Greco G, Reyes-Teran G, Levy JA (1997) Do beta-chemokines have clinical relevance in HIV infection? J Clin Invest 100:921–930
Mackewicz CE, Garovoy MR, Levy JA (1998) HLA compatibility requirements for CD8(+)-T-cell-mediated suppression of human immunodeficiency virus replication. J Virol 72:10165–10170
Makadzange AT et al (2010) Characterization of an HLA-C-restricted CTL response in chronic HIV infection. Eur J Immunol 40:1036–1041. doi:10.1002/eji.200939634
Marsh SGE, Parham P, Barber LD (2000) The HLA factsbook. Factsbook series. Academic Press, San Diego
Migueles SA et al (2008) Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009–1021
Mommen GP, Frese CK, Meiring HD, van Gaans-van den Brink J, de Jong AP, van Els CA, Heck AJ (2014) Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD). Proc Natl Acad Sci USA 111:4507–4512. doi:10.1073/pnas.1321458111
Natarajan K, Li H, Mariuzza RA, Margulies DH (1999) MHC class I molecules, structure and function. Rev Immunogenet 1:32–46
Neefjes J, Jongsma ML, Paul P, Bakke O (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 11:823–836. doi:10.1038/nri3084
Ness DB, Cann HM, Grumet FC (1981) Strong linkage disequilibria among HLA-Bw50, BfS1, and HLA-DR3/DR7, and mapping of the Bf locus. Hum Immunol 2:225–234
Pereyra F et al (2010) The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi:10.1126/science.1195271
Rajapaksa US, Li D, Peng YC, McMichael AJ, Dong T, Xu XN (2012) HLA-B may be more protective against HIV-1 than HLA-A because it resists negative regulatory factor (Nef) mediated down-regulation. Proc Natl Acad Sci USA 109:13353–13358. doi:10.1073/pnas.1204199109
Ranasinghe S et al (2016) Antiviral CD8+ T cells restricted by human leukocyte antigen class II exist during natural HIV infection and exhibit clonal expansion. Immunity 45:917–930. doi:10.1016/j.immuni.2016.09.015
R-Core-Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna URL http://www.R-project.org/
Rosok B, Voltersvik P, Larsson BM, Albert J, Brinchmann JE, Asjo B (1997) CD8+ T cells from HIV type 1-seronegative individuals suppress virus replication in acutely infected cells. AIDS Res Hum Retrovir 13:79–85
Saah AJ et al (1998) Association of HLA profiles with early plasma viral load, CD4+ cell count and rate of progression to AIDS following acute HIV-1 infection. Multicenter AIDS Cohort Study. AIDS 12:2107–2113
Sekaly RP (2008) The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J Exp Med 205:7–12. doi:10.1084/jem.20072681
Sidney J, Peters B, Frahm N, Brander C, Sette A (2008) HLA class I supertypes: a revised and updated classification. BMC Immunol 9:1. doi:10.1186/1471-2172-9-1
Slingluff CL Jr, Cox AL, Stover JM Jr, Moore MM, Hunt DF, Engelhard VH (1994) Cytotoxic T-lymphocyte response to autologous human squamous cell cancer of the lung: epitope reconstitution with peptides extracted from HLA-Aw68. Cancer Res 54:2731–2737
Stranford SA, Ong JC, Martinez-Marino B, Busch M, Hecht FM, Kahn J, Levy JA (2001) Reduction in CD8+ cell noncytotoxic anti-HIV activity in individuals receiving highly active antiretroviral therapy during primary infection. Proc Natl Acad Sci USA 98:597–602
Streeck H, Nixon DF (2010) T cell immunity in acute HIV-1 infection. J Infect Dis 202(Suppl 2):S302–S308. doi:10.1086/655652
Streeck H et al (2014) Emergence of individual HIV-specific CD8 T cell responses during primary HIV-1 infection can determine long-term disease outcome. J Virol 88:12793–12801. doi:10.1128/JVI.02016-14
Su RC, Miller RG (2001) Stability of surface H-2K(b), H-2D(b), and peptide-receptive H-2K(b) on splenocytes. J Immunol 167:4869–4877
Thio CL et al (2003) Comprehensive analysis of class I and class II HLA antigens and chronic hepatitis B virus infection. J Virol 77:12083–12087
Tilanus MGJ, Group SIHW (2000) IHWG technical manual : genomic analysis of the human MHC; DNA-based typing for HLA alleles & linked polymorphisms; HLA 2000. Fred Hutchinson Cancer Research Center
Trachtenberg E et al (2003) Advantage of rare HLA supertype in HIV disease progression. Nat Med 9:928–935. doi:10.1038/nm893
Valentine MA, Tsoukas CD, Rhodes G, Vaughan JH, Carson DA (1985) Phytohemagglutinin binds to the 20-kDa molecule of the T3 complex. Eur J Immunol 15:851–854. doi:10.1002/eji.1830150821
Walker CM, Levy JA (1989) A diffusible lymphokine produced by CD8+ T lymphocytes suppresses HIV replication. Immunology 66:628–630
Walker CM, Moody DJ, Stites DP, Levy JA (1986) CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science 234:1563–1566
Walker BD et al (1987) HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature 328:345–348
Walker CM, Erickson AL, Hsueh FC, Levy JA (1991a) Inhibition of human immunodeficiency virus replication in acutely infected CD4+ cells by CD8+ cells involves a noncytotoxic mechanism. J Virol 65:5921–5927
Walker CM, Thomson-Honnebier GA, Hsueh FC, Erickson AL, Pan LZ, Levy JA (1991b) CD8+ T cells from HIV-1-infected individuals inhibit acute infection by human and primate immunodeficiency viruses. Cell Immunol 137:420–428
Ware R, Jiang H, Braunstein N, Kent J, Wiener E, Pernis B, Chess L (1995) Human CD8+ T lymphocyte clones specific for T cell receptor V beta families expressed on autologous CD4+ T cells. Immunity 2:177–184
Wilkinson J, Zaunders JJ, Carr A, Cooper DA (1999) CD8+ anti-human immunodeficiency virus suppressor activity (CASA) in response to antiretroviral therapy: loss of CASA is associated with loss of viremia. J Infect Dis 180:68–75
Wiviott LD, Walker CM, Levy JA (1990) CD8+ lymphocytes suppress HIV production by autologous CD4+ cells without eliminating the infected cells from culture. Cell Immunol 128:628–634
Wong JK et al (2010) In vivo CD8+ T-cell suppression of siv viremia is not mediated by CTL clearance of productively infected cells. PLoS Pathog 6:e1000748. doi:10.1371/journal.ppat.1000748
Yang OO, Walker BD (1997) CD8+ cells in human immunodeficiency virus type I pathogenesis: cytolytic and noncytolytic inhibition of viral replication. Adv Immunol 66:273–311
Acknowledgements
The research was supported by NIH Grant 3P30AI027763-19S2 and California HIV/AIDS Research Program Grant ID09-SF-058. We thank Sharon Ng, Sue Fujimura, Jeremy Roop, Carl Johnson, and Kaitlin Killian for their technical assistance and Dr. Jay A. Levy for his contributions and helpful review of the manuscript. We also thank the study participants for their dedication to these studies.
Author information
Authors and Affiliations
Contributions
M.S.K. designed the studies, performed experiments and data analyses, and drafted the manuscript. F.T. performed experiments and data analyses. R.S. performed statistical analyses. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Each participant signed informed consent forms, and the study received approval from the Committee for Human Research at the University of California San Francisco (UCSF).
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Killian, M.S., Teque, F. & Sudhagoni, R. Analysis of the CD8+ T cell anti-HIV activity in heterologous cell co-cultures reveals the benefit of multiple HLA class I matches. Immunogenetics 70, 99–113 (2018). https://doi.org/10.1007/s00251-017-1021-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-017-1021-7