Abstract
The diazotrophic communities play an important role in sustaining primary productivity through adding new nitrogen to oligotrophic marine ecosystems. Yet, their composition in the oligotrophic Indian Ocean is poorly understood. Here, we report the first observation of phylogenetic diversity and distribution of diazotrophs in the Eastern Indian Ocean (EIO) surface water (to 200 m) during the pre-southwest monsoon period. Through high throughput sequencing of nifH genes, we identified diverse groups of diazotrophs in the EIO including both non-cyanobacterial and cyanobacterial phylotypes. Proteobacteria (mainly Alpha-, Beta-, and Gamma-proteobacteria) were the most diverse and abundant groups within all the diazotrophs, which accounted for more than 86.9% of the total sequences. Cyanobacteria were also retrieved, and they were dominated by the filamentous non-heterocystous cyanobacteria Trichodesmium spp. Other cyanobacteria such as unicellular diazotrophic cyanobacteria were detected sporadically. Interestingly, our qPCR analysis demonstrated that the depth-integrated gene abundances of the diazotrophic communities exhibited spatial heterogeneity with Trichodesmium spp. appeared to be more abundant in the Bay of Bengal (p < 0.05), while Sagittula castanea (Alphaproteobacteria) was found to be more dominating in the equatorial region and offshores (p < 0.05). Non-metric multidimensional scaling analysis (NMDS) further confirmed distinct vertical and horizontal spatial variations in the EIO. Canonical correspondence analysis (CCA) indicated that temperature, salinity, and phosphate were the major environmental factors driving the distribution of the diazotroph communities. Overall, our study provides the first insight into the diversity and distribution of the diazotrophic communities in EIO. The findings from this study highlight distinct contributions of both non-cyanobacteria and cyanobacteria to N2 fixation. Moreover, our study reveals information that is critical for understanding spatial heterogeneity and distribution of diazotrophs, and their vital roles in nitrogen and carbon cycling.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Biological nitrogen fixation (BNF), the enzyme-catalyzed reduction of dinitrogen (N2) from air to bioavailable ammonium, is identified a major source of nitrogen in N-starving marine ecosystems [1, 2]. Multiple studies have highlighted the significance of N2 fixation in sustaining marine primary production, powering the biological carbon pump, and eventually accelerating carbon sequestration to the ocean [3,4,5]. Based on in situ experiments, the fixed nitrogen by the diazotrophic communities was estimated to fuel up to half or more of primary production in the subtropical North Pacific and tropical North Atlantic regions [6, 7]. BNF is a process catalyzed by a nitrogenase enzyme complex in a selected group of microorganisms termed as diazotrophs. Nitrogenase is a highly conserved enzyme which consists of two multisubunit metallo-proteins, namely, dinitrogenase reductase and dinitrogenase encoded by nifH and nifKD [8]. Due to its highly conserved sequences, nifH gene has been widely used for phylogenetic and ecological studies of the diazotrophic communities in marine ecosystems [9].
The molecular techniques have revolutionized the traditional method of identifying N2 fixers via a microscope, and a wide range of diazotrophs have been separated and identified from marine environments [10]. Ever since, related studies of diazotrophs on biogeochemical, physio-ecological, and molecular aspects have been thriving and prosperous. Among all the diazotrophic communities, Proteobacteria and cyanobacteria are identified as the most abundant groups inhabiting euphotic layer of marine ecosystems [11]. Cyanobacteria have a broad geographic distribution and contain a variety of morphologic and phylogenetic groups including Trichodesmium spp., diatom–diazotroph symbioses, and unicellular N2 fixing cyanobacteria (groups A, B, and C) [12]. Previous studies have been mainly focused on cyanobacterial diazotrophs which are widely distributed in tropical and subtropical oceans. More recently, Proteobacteria, including Alpha-, Beta-, Gamma-, and Delta-proteobacteria, were reported dominant in open oceans such as the northern South China Sea, the Southern Indian Ocean, and the Eastern Tropical South Pacific region [13,14,15]. The dominance of Proteobacteria in open oceans reveals that our understanding of the diazotrophic community is still limited. Further studies focusing on physiological and ecological characterizations of these widely distributed bacteria and their interactions with environmental gradients have important implications for understanding their contribution to nitrogen fixation and global nitrogen cycling [16].
As the third largest ocean in the world, the Indian Ocean plays a significant role in global climatic change, energy flow, and material cycles [17]. The Indian Ocean is divided into two semi-enclosed basins in the north by Indian subcontinent and extends to at least 26°N in the south. North Indian Ocean is heavily influenced by the South Asian monsoon system. The surface circulation changes seasonally in response to the prevalent monsoons, with Somali Current flowing equatorward during the winter monsoon and poleward during summer monsoon [18, 19]. Upwelling and convective mixing induced by seasonally reversing monsoon winds can increase upward supply of nutrients and cause algal blooms seasonally in the northern Indian Ocean [20, 21]. It is generally believed that the Bay of Bengal (BOB) is less productive than the Arabian Sea (AS), partly due to the freshwater input which decrease the salinity on the surface and aggravate stratification [20, 22,23,24]. Winds over the northern Indian Ocean are generally weak during the period of pre-southwest monsoon, and stratified water results in low supplement of the nutrient from deeper water. Strong stratification, calm water, sufficient solar heating, and extreme oligotrophic condition make the northern Indian Ocean an ideal habitat for the diazotroph community during the pre-southwest monsoon period. In fact, surface blooms of Trichodesmium spp. are well documented in both the AS and the BOB [25,26,27], and heterotrophic bacteria have been reported dominant among diazotrophs in the AS during the winter monsoon [14, 28]. However, no detailed studies have been conducted in investigating composition and distribution of the diazotrophic communities at the BOB and the eastern Equator region.
Compared to the North Pacific [29,30,31] and North Atlantic [32,33,34,35], the diazotrophic community in Indian Ocean is understudied, and currently available data are primarily focused on the AS [14, 28]. In the present study, we collected water samples in the BOB and the open region of eastern equatorial Indian Ocean during the pre-southwest monsoon of 2017. In order to investigate composition and spatial distribution of the diazotrophic communities in the EIO, molecular approaches including high throughput sequencing analysis and real-time fluorescent quantitative polymerase chain reaction (qPCR) assay were applied to samples collected from different locations and depths. The high throughput sequencing analysis provided detailed information of the diazotrophic community structures including dominant and rare species, as well as uncultured populations. Based on the results of high throughput sequencing, qPCR assay was used to quantify the dominant diazotrophic groups in the EIO. In addition, multivariate statistics were conducted between communities and environmental parameters to determine the controlling factors in structuring diazotrophs.
Materials and Methods
Station Location, Sampling, and Physicochemical Analysis
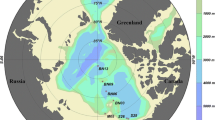
The cruise was carried out in the EIO onboard R/V “Shiyan 3” from 9 March to 7 April 2017. Samples for molecular analyses were collected from 10 sampling sites at seven depths (0 m, 25 m, 50 m, 75 m, 100 m, 150 m, and 200 m) for each site. The sampling sites were located along four main transects targeting the BOB (I1, I2), the Equator region (I4), and the transect (I5) parallel to the coastline of the Sumatra (Fig. 1). Among all the stations, we chose six stations (I105, I504, I203, I210, I405, and I413) with three depths (0 m, 75 m, and 200 m) for high throughput sequencing (Fig. 1). Whereas for qPCR analysis, all samples were included (10 stations and 7 depths as shown in Fig. 1 and Table 1). Surface water was collected by using 10% HCl-rinsed polyethylene (PE) bucket at all the stations. Vertical seawater was collected by using 5 L Teflon-coated Go-Flo bottles (General Oceanics, Miami, Florida, USA) attached to a rosette multisampler, on which conductivity, temperature, and depth (CTD) probes were installed (Seabird SBE 911Plus, Sea-Bird Electronics, Washington, USA). Temperature and salinity were measured and recorded vertically by a CTD profiler.
The map showing the sampling stations in the EIO. Miniature map at lower left described the transects and stations for hydrography measurement (blue dots in red box). In enlarged map of the EIO, 10 sites (red dots) were used for qPCR analysis and 6 sites (circled) were subjected to high throughput sequencing
Sample Collection and Environmental Parameter Measurements
Water samples from different layers were transferred individually into 15 L PE buckets that were previously washed with 10% HCl and rinsed thrice with Milli-Q water. Samples for determination of nutrients were subsampled into 100 mL of 10% HCl-rinsed PE bottles and stored in 4 °C until analysis. In the laboratory, nutrient concentrations including ammonium (NH4+), nitrate (NO3−), nitrite (NO2−), phosphate (PO43−), and silicate (SiO32−) were determined by using a Technicon AA3 Auto-Analyzer (Bran+ Luebbe, Norderstedt, Germany) based on the classical colorimetric methods [36]: NH4+, dissolved inorganic nitrogen (DIN), PO43−, and SiO32− are measured by indophenol blue method, the copper-cadmium column reduction methods, phosphor-molybdate complex methods, and silico-molybdate complex methods, respectively.
For Chl a analysis, 500 mL water from each layer were vacuum-filtered (< 10 mmHg) through a 25-mm GF/F filter (Waterman, Florham Park, NJ, USA). The filters were placed into aluminum foil bags and stored in the dark at − 20 °C until analyzed. In the lab, filters were kept in 20 mL vials and pigments were extracted with 90% acetone (Guanghui, Yixing, Jiangsu, China) for 24 h at 4 °C. Chlorophyll concentrations were then determined using a Turner® Trilogy (CHL NA, model no. 046) fluorometer (Turner designs, San Jose, CA, USA). For molecular analyses, 2–4 L of seawater were filtered through 0.22-μm GTTP filters (Millipore, Eschborn, Germany) under low pressure vacuum. The filters were placed into 2-mL microtubes, and flash frozen immediately in liquid nitrogen on board. The filters were transferred to − 80 °C freezer in the lab until DNA extraction.
DNA Extraction, nifH Gene Amplification, and Sequencing
Genomic DNA were extracted by CTAB method as previously described with minor modifications [37, 38]. Briefly, filters were cut into small pieces and placed into 1.5-mL sterile centrifuge tubes. For each extraction, 600-μL CTAB extraction buffer and 15 μL 2% β-mercaptoethanol (Goaobio, Beijing, China) were added and incubated at 60 °C in a water bath for 60 min. Proteins, polysaccharides, and other impurities were separated by adding equal volume of 24:1 chloroform:isoamyl alcohol (Solarbio, Beijing, China), and then centrifuged at 14000 rpm for 20 min at 4 °C. Supernatant was transferred into a new clean tube, and this step was repeated once to insure complete removal of impurities. The genomic DNA were precipitated overnight at − 20 °C by addition of 2/3 volume of isopropanol (Sangon Biotech, Shanghai, China) followed by washing with 75% ethanol (Hushi, Shanghai, China) twice, then air-dried genomic DNA pellet was eluted with 70 μL sterile ultra-pure water and stored at − 20 °C until further processing. Note that the solutions used in our experiment were all marked with molecular grade. The quantity and quality of genomic DNA were checked by using a ND-2000 Nanodrop spectrometer (Thermo Fisher Scientific, Wilmington, Delaware, USA).
The nifH gene fragments were amplified from the genomic DNA through nested polymerase chain reaction (nested PCR) according to a previous protocol [10]. PCRs were performed using a Veriti 9902 thermocycler (Applied Biosystems, Foster City, CA, USA) with 10 μL reaction volumes containing 1× PCR buffer, 4 mM MgCl2, 400 mM dNTPs, 1 μM forward, and reverse primers (nifH3 and nifH4 for primary, nifH1 and nifH2 for secondary PCR), 0.2-unit KOD FX Neo polymerase (Toyobo, Osaka, Japan), and 1 μL of template DNA (genomic DNA for the first round, and PCR products from the primary PCR for the second round). Negative controls were set up by replacing template DNA with nuclease-free water. The thermal profile used for the nifH gene amplification was initial denaturation (95 °C, 5 min), followed by 38 (primary) or 40 (secondary) cycles of denaturation (94 °C, 1 min), annealing (52 °C for the first round and 59 °C for the second round, 1 min), and extension (72 °C, 1 min) with a final extension (72 °C, 7 min). Remarkably, primers used in secondary PCR were composed of dual-indexed barcodes, Illumina linkers, a sequencing primer binding region, and gene-specific sites. PCR products were checked by 1.8% agarose gel (BioWest, Castropol, Spain) electrophoresis after amplification, and products with approximately 360 bp bands were used for high throughput sequencing. All libraries were constructed and sequenced via paired-end chemistry (PE250) on an Illumina Hiseq2500 platform (Illumina, San Diego, CA, USA) at Biomarker Technologies, Beijing, China.
Quality Control and Sequencing Date Processing
The raw image data file obtained from Illumina Hiseq2500 platform were transformed into sequenced reads via base calling, and the results were stored in the format of FASTQ file which include the information of raw sequence data and corresponding sequencing quality. The raw sequence data were then separated by samples based on their barcodes, permitting up to one mismatch [39]. The raw sequence data were de-multiplexed, quality filtered, and analyzed through the open-source software pipeline QIIME [40], and the paired reads were merged into full-length sequences by FLASH v1.2.7 software [41]. For each sample, the raw tags were quality filtered to get high-quality clean tags via Trimmomatic v0.33 software [42], including removal of sequences less than 300 bases, homopolymers containing sequences (homopolymers ≥ 8 bases), and ambiguous base containing sequences [43, 44]. The barcodes, linker sequences, and primers were also removed in this process. Effective tags were subsequently obtained after removing chimera by UCHIME v4.2 software [45]. The remaining effective tags were clustered using USEARCH v10.0 at 97% similarity to generate operational taxonomic units (OTUs) [46]. Low abundance OTUs which containing less than 20 sequences across all samples were excluded from further processing.
To identify dominant taxa, top OTUs in each sample were selected for subsequent phylogenetic analysis. The most common sequences in each OTU were selected as the representative sequences. These sequences were first translated into amino acid sequences and blasted in the protein database at National Center for Biotechnology Information (NCBI) via BLASTX v2.8.1+ to identify the most closely related sequences [47] The representative sequences and the most closely related sequences were then aligned with ClustalW and used to construct a phylogenetic neighbor-joining tree by MEGA v7.0 [48, 49]. Cluster stability was tested by bootstrap resampling for 1000 times, and the phylogenetic tree was further edited by an online webpage iTOL [50]. The raw sequences obtained from this study were deposited in NCBI Sequence Read Archive with accession no. SUB3781024.
Quantification of Two Cyanobacterial and Two Non-Cyanobacterial nifH Phylotypes
According to the results of high throughput sequencing, abundance of four top diazotrophs, namely, Trichodesmium spp., Crocosphaera watsonii, Sagittula castanea, and γ-24774A11, were quantified by qPCR using an ABI Step One Plus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The specific primers and probes shown in Table 2 were designed by previous studies with minor modifications to correct mismatch [33, 51]. The specificity of the primers and probes were all checked in the abovementioned studies, and no significant cross-reactivity was observed. The TaqMan probes were 5′-labeled with the fluorescent reporter FAM (6-carboxyfluorescein) and 3′-labeled with the quenching dye TAMRA (6-carboxytetramethylrhodamine). All primers and probes used in the present study were synthesized in Sangon Biotech, Shanghai, China. For all TaqMan PCRs, duplicate or triplicate 10 μL reactions were performed with 5 μL of 2 × Premix Ex Taq™ (Takara Bio, Tokyo, Japan), 0.2 μM of the forward and reverse primers, 0.4 μM of TaqMan probe, 0.2 μL of 50 × ROX reference dye, 1 μL of template DNA, and 3 μL of nuclease-free water. The PCR conditions were 95 °C for 30 s, followed by 45 cycles of 95 °C for 5 s and 60 °C for 30 s. Standard curves were determined by analyzing 10-fold serial dilutions of the target nifH gene inserted plasmids with a final gene copy numbers ranged from 101 to 107 for each reaction. The r2 values of each standard curve ranged between 0.98 to 1.00, and PCR amplification efficiency ranged from 90 to 110%. The copy numbers of each diazotroph in the environmental samples were calculated based on mean Ct values. Furthermore, non-target templates were also tested in the same conditions, and gene copies in negative controls less than 10 or undetectable were considered contamination-free. The detection limit of qPCR reactions was approximately 50 nifH genes in 1-μL template DNA, which was equivalent to approximately 3500 copies with a final volume of 70 μL in the present study.
Statistical Analysis
Richness and diversity indices including Chao1 richness estimator, Ace richness estimator, Shannon diversity indices, and Simpson diversity index were calculated with R v 3.3.2 software [52]. To evaluate the coverage of sequencing, the abundance data was standardized and calculated based on entropy (Q statistics) in the online software iNEXT [53, 54]. Rarefaction curves were also done in this online software based on the standard operating procedure shown in the website (https://chao.shinyapps.io/iNEXTOnline/). Non-metric multidimensional scaling (NMDS) analysis and cluster analysis were used in the present study to demonstrate vertical and horizontal distribution patterns of the diazotrophic communities in PRIMER V6.0 software [55]. The diazotroph community data was first square-root transformed in the software, and then, a lower triangular resemblance matrix was created based on Bray–Curtis similarity. Subsequently, a hierarchical cluster tree and a NMDS biplot were constructed in the software based on the matrix. To reveal the correlations between the diazotrophic communities and environmental factors, detrended correspondence analysis (DCA) was first carried out by decorana function in vegan to determine whether redundancy analysis (RDA) or canonical correspondence analysis (CCA) was more suitable [56, 57]. Because the length of gradient value of the first axis was greater than 3.0, CCA was selected to explore correlations between community structure and environmental gradients and run in R v 3.3.2 software. Highly correlated environmental factors were removed by co-linearity test conducted with vif.cca function in vegan and variance inflation factors (VIFs) of remaining environmental factors were all less than 20 [58]. Depth-integrated (0–200 m) gene abundances were calculated by trapezoidal integration over depths of the euphotic zone, and the results were plotted by ODV V5.0.0 [59]. The significant differences of depth-integrated gene abundances (log-transformed) for diazotrophs among different regions were evaluated by t test in IBM SPSS Statistics 25.
Results
Hydrography and Environmental Parameters
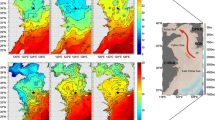
The surface hydrographic characteristics for all stations were shown in Table 1 and Table S1. During the study period, sea surface temperature (SST) varied from 28.5 to 31.1°C, and BOB had higher SST than other regions (p < 0.01). Sea surface salinity (SSS) ranged from 30.4 to 34.3, and BOB had higher SSS than other regions (p < 0.05). The maximum and minimum value of SSS was 34.3 ppt at Sta. I404 and 30.4 ppt at Sta. I201, respectively. The surface regime could be distinguished by T-S properties of the upper 200 m at sampling stations where lower salinity and slightly higher temperature were observed at the BOB stations (Fig. 2). The potential density (σ0) was less than 21.0 kg m−3 at near surface water at Sta. I110, I203, I206, and I210, followed by nearly 21.0 kg m−3 at other stations except for some unstable data. Chl a concentrations ranged from 0.067 to 0.407 μg L−1, and there was no significant difference of surface Chl a concentrations in the area study. Across all the sampling stations, concentrations for dissolved inorganic nutrients were quite low in the study area, suggesting the EIO was a typical oligotrophic ocean. For instance, nitrate, nitrite, ammonium, and phosphate concentration ranged from 0 to 2.043 μM, 0.129 to 0.2 57 μM, 0 to 1.379 μM, and 0 to 0.155 μM, respectively (Table S1).
Vertical profiles of temperature, salinity, dissolved inorganic nutrients, and Chl a are shown in Fig. 3 and Fig. S1. The study region was characterized by stable surface waters with higher temperature and lower salinity than deep waters. Obviously, the thermocline and halocline in the Equator region was shallower than other areas and increased gradually from the open oceans to the offshore region. The halocline was clearly observed at the BOB and the Equator region, but not obvious in the offshore section. Vertical profiles of nitrate and phosphate in survey stations showed consistent patterns with lower concentrations in the top 50 m and increased dramatically in deep waters. Nitrite concentrations peaked at 75 m but remained nearly the same in other layers. The Chl a maximum layer was at 75 m except for Stn. I105 (50 m instead), and integrated Chl a concentrations for top 200 m water column ranged from 27.83 μg L−1 (Stn. I409) to 40.70 μg L−1 (Stn. I105). Compared to other nutrients, no consistent patterns were observed for vertical distribution of ammonium.
Sequencing Statistics and Diversity Estimates
In total, 1,185,461 effective tags were included in our study after quality control, and the detailed information for each sample was listed in Table 3. The sequencing coverage (C) was all greater than 99.9%, suggesting the sequencing effort was deep enough to cover nifH gene diversity. On average, 65,859 sequences per sample were obtained with an average length at 320 bp. Based on 97% similarity, a total of 218 OTUs were obtained after excluding rare OTUs (< 20 sequences across all samples). The rarefaction curve plateau are shown in Fig. S2. The OTU richness ranged from 17 to 72, and the minimum and maximum OTUs were observed at Stn. I405 (200 m) and Stn. I210 (75 m), respectively. Combined all alpha diversity indices, the lowest and highest diversities occurred at the same station Stn. I203 but at different layers, 0 m and 75 m, respectively. Overall, Stn. I210 had the highest diversity indices (H = 2.39, D = 0.86) while the lowest indices occurred in Stn. I405 (H = 1.28, D = 0.63).
Phylogeny and Composition of Diazotrophs
Due to large numbers of OTUs recovered and most of them were unidentifiable, we only focused on the top OTUs in the present study. The OTUs contained ≥ 500 sequences across all the samples were defined as top OTUs in our study. Top 30 OTUs accounted for more than 97% of all the sequences, and they were included in subsequent phylogenetic analysis (Fig. 4). Reference sequences from NCBI were all retrieved from marine environments, and the unknown species were labeled with ocean names where they originated. Top 30 OTUs were grouped into three defined clusters of nifH genes [2], in which 28 OTUs belonged to cluster I, 1 OTU (Phasecolarcto bacterium) belonged to cluster II, and 1 OTU (Verrucomicrobiae bacterium) belonged to cluster III (Fig. 4). Nearly half of the OTUs were closely related to Alphaproteobacteria (14/30 OTUs), followed by Betaproteobacteria (5/30 OTUs), Gammaproteobacteria (5/30 OTUs), and Cyanobacteria (4/30 OTUs).
A neighbor-joining phylogenetic tree (left) and a bubble map to show OTU relative abundance (right) at each site. The tree was constructed by nifH gene amino acid sequences obtained from this study and reference sequences from GenBank. The topology of the tree was inferred from 1000 bootstrap resampling, and bootstrap values greater than 50% were shown with red labels at branches. The bubble sizes corresponded to relative abundance of each OTU
Relative abundance of diazotrophs is shown in Figs. 4 and 5. Overall, Proteobacteria was clearly the most dominant group in the EIO, followed by Cyanobacteria. Within all the diazotrophic community, OTU 550 shared 100% similarity with a recently isolated Sagittula (Rhodobacteraceae, Alphaproteobacteria) [60] and dominated diazotrophs across all samples. OTU 550 was detected in all the samples but was more abundant in the Equator region (Stn. I405 and I413) and deep layers (Fig. 4). Besides OTU 550, other Alphaproteobacteria such as OTU 115 (Novosphingobium malaysiense), OTU 881, and OTU 537 (Bradyrhizobium sp.) were also commonly detected. OTU 702 was the second most dominant diazotroph which shared 100% similarity with an uncultured Betaproteobacterium, and its occurrence was noted in deep waters. OTU 536, closely related to Gammaproteobacterium γ-24774A11, was also recovered in the present study though in low abundance. Apart from γ-24774A11, other Gammaproteobacteria including OTU 1136, OTU 1128 (Vibrio diazotrophicus), OTU 659, and OTU 388 (Pseudomonas stutzeri) were also detected.
In addition, four OTUs were clustered with cyanobacteria, including OTU 229 (Trichodesmium spp.), OTU 325 (Crocosphaera watsonii), OTU 888 (UCYN-A3) and OTU 301 (Cyanothece sp. WH 8904). Among which, Trichodesmium spp. was the most abundant cyanobacteria, while the other cyanobacteria exhibited low abundances (Fig. 4). Trichodesmium sequences were recovered from all surface waters except for Stn. I405. Compared to the Equator area, Trichodesmium was more abundant at the BOB, especially at Stn. I203 where Trichodesmium blooms might have occurred. Meanwhile, Crocosphaera watsonii, UCYN-A3, and Cyanothece sp. WH 8904 were mainly detected in surface waters at Stn. I210 and I413.
Quantification of Dominant nifH Phylotypes
Abundances for four major nifH phylotypes (Sagittula castanea, γ-24774A11, Trichodesmium spp. and UCYN-B) were quantified by qPCR. The Ct values of non-target template were all undiscovered or in high values for Sagittula castanea (not detected), γ-24774A11 (Ct = 38), Trichodesmium (not detected), and UCYN-B (Ct = 35.4). The sensitivity and accuracy of standard curves for different targets were shown in Table 4.
Depth profiles of nifH genes for the four dominant phylotypes are shown in Fig. 5, and the depth-integrated abundances (up to 200 m) were shown in Fig. 6. The highest abundances were usually detected in the upper layer from 0 to 50 m. Sagittula castanea was detected at all stations with all depths, and peaked at 25 m across all sampling sites (Fig. 5). Combined with depth-integrated gene abundances, Sagittula castanea was more abundant in the Equator region and the offshore than the BOB (p < 0.05) (Fig. 6). Trichodesmium, UCYN-B, and γ-24774A11 were generally concentrated in the upper water layers. For instance, nifH genes for Trichodesmium spp. reached up to 3 × 108 copies L−1 in the surface water at Stn. I203 (Fig. 5). Trichodesmium and UCYN-B showed similar patterns regarding depth-integrated gene abundances, with higher gene copy numbers in the BOB and lower in the Equator region (p < 0.05) (Fig. 6). For γ-24774A11, no significant differences were observed at different regions. In summary, our results demonstrated contrasting spatial patterns for Trichodesmium (higher at the BOB) and Sagittula castanea (higher in the Equator region and the offshore) in the EIO (Fig. 6).
Statistical Analysis
Spatial distribution patterns (both horizontal and vertical) of the diazotrophic communities in the EIO are presented in Fig. 7. Horizontally, the surface samples from the same transect were almost grouped together (Fig. 7a), showing similar community structures in the same area. The surface samples in the BOB were mainly composed of Cyanophyceae, while samples in the Equator region and the offshore were mainly composed of Alphaproteobacteria (Fig. 7b). Vertically, we observed that the surface samples and the deeper samples were separated at 30% similarity (Fig. 7a). It indicated that the surface samples were distinct from other deeper samples, and the composition of the diazotrophic communities varied along the vertical gradient. From Fig. 7a, samples at 75 m and 200 m were almost grouped together except for samples from Sta. I405. The two samples at Sta.I405 were mainly composed of Alphaproteobacteria. Whereas for other deeper samples, Betaproteobacteria was also an important component (Fig. 7b).
Correlations of the diazotrophic community and associated environmental factors were analyzed by CCA (Fig. 8). Temperature, salinity, phosphate, and ammonia were included in CCA after excluding environmental factors with VIFs > 20. The environmental factors in the first two axis explained > 83.63% of the total variance in the diazotrophic community distributions. Temperature (p = 0.001), salinity (p = 0.001), and phosphate (p = 0.001) contributed significantly to the total variance and were closely associated with the first and second axes (999 times Monte Carlo permutations). The sample distributions in CCA were similar to NMDS which also presented a vertical separation. Temperature positively correlated with the diazotrophic distributions in surface, while salinity and phosphate were driving distribution of the diazotrophic communities in deep water samples. Cyanobacteria were all positively correlated with temperature, while most of Proteobacteria were positively related to salinity and phosphate. Interestingly, we observed γ-24774A11 was plotted together with cyanobacteria, suggesting γ-24774A11 possibly shared same ecological niches with cyanobacteria.
Canonical correspondence analysis (CCA) based on diazotrophic composition and biotic/abiotic parameters as explanatory variables. The two CCA axes (CCA1 and CCA2) explained 83.63% of total variations in abundance data. Arrows represented environmental parameters. Different colors of circle and triangle symbols represented different samples and taxa, respectively. Only the top 30 OTUs were included. Significance (**) was determined by 999 Monte Carlo permutation tests with R V.3.0 software
The correlations between nifH gene abundances and various environmental parameters are listed in Table 5. Among which, γ-24774A11, Trichodesmium, and UCYN-B related nifH gene abundance were all exhibiting similar trends to the environmental parameters: significant negative correlations with water depth (p < 0.01), salinity (p < 0.01) and nutrients (p < 0.01), and a positive correlation with temperature (p < 0.01). In contrast, Sagittula castanea was homogeneously distributed in the water column and therefore exhibited no correlation with depth and temperature, but showed significant negative correlations with nitrate and phosphate (p < 0.05) (Table 5).
Discussion
Recent studies have shown that putative N2-fixing phylotypes of heterotrophic diazotrophs appeared to be ubiquitous in diverse marine and estuarine environments, even in deep, cold, or high-latitude waters where cyanobacteria were low or absent [61,62,63,64]. Due to their widespread distribution, heterotrophic diazotrophs are potentially important N2 fixers, and global N inputs solely based on cyanobacterial diazotrophs is likely underestimated. Similar to previous studies, our data elaborate that Proteobacteria are the most abundant diazotrophs in the EIO, especially in the equatorial region and deep waters. The prevalence of diverse heterotrophic N2-fixing bacteria in oceans has been attributed to the overproduction of phytoplankton and accumulation of DOC in previous study [65]. During our cruise, we observed an unusually high phytoplankton primary productivity (Liu HJ et al. unpublished data) in the equatorial region which could be influenced by Wyrtki Jets [66]. The periodic Wyrtki Jets bring high-salinity and high-nutrient waters from 60° E to east along the equator [67]. Rahav et al. explained that addition of polysaccharide may increase efficiency of ectoenzymes such as β-glucosidase, and generated bioavailable molecules with enhanced polysaccharide hydrolysis [68]. Further, numerous studies have demonstrated that heterotrophic diazotrophs were stimulated by addition of nutrients or dissolved organic matter (DOM) [69, 70]. However, distribution of heterotrophic diazotrophs and their environmental drivers in marine ecosystems are still poorly understood. Further studies on biogeochemical, physio-ecological, and molecular aspects are needed to evaluate roles that heterotrophic diazotrophs play in nutrient and carbon cycling in global oceans.
Alphaproteobacterial genes were contributing the biggest fraction to the nifH gene diversity in our study, although previous studies have revealed that Gammaproteobacteria dominated global oceans [11, 16]. The dominance of Alphaproteobacteria and Betaproteobacteria in the diazotrophic communities was also reported in the Arabian Sea and the Western Equatorial region during northeast monsoon [14]. In the present study, OTU550 was the most abundant Alphaproteobacteria, as well as the dominant diazotroph in the EIO. The representative sequence of OTU550 shared 100% similarity with a new species named Sagittula castanea which was isolated from the oxygen minimum zone off Peru in the eastern tropical South Pacific (ETSP) [60]. Sagittula castanea has been confirmed capable of fixing N2 in the laboratory experiments, while it is still uncertain if this species is actively fixing N2 in natural environments [60]. In fact, Sagittula-related nifH sequences have been reported dominant in both shelf and open oceans of the ETSP, surface waters in the Indian Ocean, and the deep waters at the South China Sea, where N2 fixation was measurable [14, 51]. Presumably, non-cyanobacterial diazotrophs were responsible for N2 fixation in these regions. Sagittula castanea was recognized as a particle-attached lifestyle based on metabolic pathway analysis [60]. High C:N ratio contained in the particles provides sufficient organic carbon to heterotrophic diazotrophs. Due to Sagittula castanea was universally distributed in the euphotic zone (Figs. 4 and 5), this species exhibited no correlations with water depth, temperature, or salinity (Table 5). However, the negative correlations between Sagittula castanea and nitrate/phosphate revealed that the growth of Sagittula castanea might be limited by other factors rather than nutrients (Table 5).
In this study, a typical Gammaproteobacteria species with a broad geographic distribution, γ-24774A11, was also retrieved. Our qPCR results showed that the depth-integrated gene abundance of γ-24774A11 was two or three orders of magnitude lower than that of Sagittula castanea and Trichodesmium spp. (Fig. 6). Though γ-24774A11 was not dominant diazotroph in our study, its abundance was still comparable to other studies in global oceans [14, 16]. γ-24774A11 always occurred in upper layers of the water column and became undetected below approximately 100 m (Fig. 5). The correlation analysis showed that γ-24774A11 abundances were controlled by depth and dissolved inorganic nitrogen (Table 5), which agreed well with a previous study conducted in the South Pacific Ocean [15]. Interestingly, we observed that the distribution of depth-integrated gene abundances of γ-24774A11 was similar to Trichodesmium and UCYN-B, but in contrast to Sagittula castanea. Our CCA also demonstrated the concurrent ecological niches between γ-24774A11 and cyanobacteria. As stated above, availability of DOC can be a limiting factor for the growth of heterotrophic N2-fixing bacteria, and therefore, a positive correlation was expected between phytoplankton production and the abundance of γ-24774A11. However, in this study, γ-24774A11 had a negative correlation with Chl a. The same observation was also reported by Moisander et al. (2014), who suggested that environmental conditions in favor of Trichodesmium and Crocosphaera watsonii would benefit growth of γ-24774A11 [15].
Higher abundance of Trichodesmium spp. were found at BOB than the Equator region and the transect parallel to the coastline of the Sumatra, although hydrological conditions seemed favorable for Trichodesmium in the whole study area. Especially at Sta. I203, Trichodesmium nifH genes was up to 1.5 × 108nifH gene copies L−1 at the surface water according to the result of qPCR (Fig. 5). It was hard to explain the difference of Trichodesmium distribution in such similar hydrological conditions across the study area. Many studies have revealed that iron (Fe) is an important cofactor for nitrogenase, and it plays a crucial role in synthesis and expression of nitrogenase in diazotrophs [14, 30]. According to literature, dissolved Fe (dFe) in surface waters ranged from 0.1–0.62 nM in the north-eastern Indian Ocean, and the concentration decreased dramatically from the BOB to the Equator region [71, 72]. It was hypothesized that Fe availability might limit growth of Trichodesmium in the Equator regions, while Fe was not a limiting factor in the BOB. The similar result was also reported by Shiozaki et al. (2014), who observed high Fe concentration coupled with high abundance of Trichodesmium in the AS, while severely limited Fe concentration coupled with undetectable Trichodesmium in the Equator region [14]. Another possible explanation was mesoscale eddy circulations frequently occurred in the BOB during pre-southwest monsoon. Anti-cyclonic (warm-core) eddies are a common phenomenon in the Indian Ocean during pre-southwest monsoon, and they could influence the distribution of phytoplankton via regulating vertical environmental parameters [73]. Jyothibabu reported that most Trichodesmium blooms recorded in the Indian Ocean were during the pre-southwest monsoon, and warm-core eddies caused downwelling of the surface waters and provided optimal growing conditions for Trichodesmium [74]. Furthermore, the specific gas vesicles in Trichodesmium could provide buoyancy and help them migrate vertically in water columns to obtain phosphate and other nutrients from deep waters, and help them float on the sea surface forming patches, bands, or mats depending upon the status of the sea [75].
Except for Trichodesmium, other cyanobacteria such as UCYN-A3, Crocosphaera watsonii, and Cyanothece sp. WH 8904 were also retrieved in the present study via high throughput sequencing. UCYN-A were reported widely distributed in tropical and subtropical oceans and made significant contribution to BNF in these regions [76, 77]. According to nifH phylogeny, the UCYN-A lineage was divided into at least four main sublineages, namely, UCYN-A1, UCYN-A2, UCYN-A3, and UCYN-A4 [78]. However, in the present study, only UCYN-A3 was recovered via high throughput sequencing. Turk-Kubo et al. (2016) reported that the UCYN-A3 sublineage is more widely distributed in oligotrophic waters than other three sublineages [79]. It has been reported that optimum temperature for UCYN-A lineage was 24 °C, but water temperature in the present study ranged from 29.3 to 31.1°C, which was not a favorable condition in general [31]. Therefore, water temperature and nutrient levels likely explained the low occurrence of UCYN-A in our study. In addition, Crocosphaera watsonii was also presented low abundance in our study. Fu et al. (2014) reported that thermal limits for Crocosphaera watsonii ranged from 24 to 32 °C, and the optimum growth temperatures was 30 °C [80]. Our correlation analysis also showed gene abundance of Crocosphaera watsonii was positively related to water temperature and negatively related to salinity and phosphate. Though the temperature in the EIO seemed favorable for the growth of Crocosphaera watsonii, it was still presented in low abundance. Shiozaki et al. [14] suggested that the shallower nitracline depths, which could result in higher upward fluxes of nutrients to the surface water, were responsible for the low abundance of UCYN-B in the EIO [52]. Until now, little is known about controlling factors that drive distributions of Cyanothece sp. WH8904.
Our results indicated that temperature, salinity, and phosphate were major environmental factors to explain variation and distribution of the diazotrophic community in the EIO. Similarly, it was reported in the northern South China Sea where salinity and phosphate were responsible for spatio-temporal variations of the diazotrophic communities [11]. Also, the diazotrophic communities may exhibit vertical distribution patterns along water depths; however, Loescher et al. (2014) suggested that water depth integrated temperature, salinity, and other physical variables, as they were all highly collinear with depth [70]. Nevertheless, distribution and composition of the diazotrophic communities were strongly influenced by these environmental variables. Ammonia was also included in our CCA analysis, in spite of its influence on the diazotrophic communities was not significant. In addition, due to highly collinearity with other factors, nitrate and nitrite were not included in our CCA analysis. But, this could not eliminate potential roles of such inorganic nutrients in structuring spatial heterogeneity in the diazotrophic communities. In fact, it is generally believed that reactive inorganic nutrient concentrations regulate the abundance of diazotrophs, as well as N2 fixation [16]. Presumably, these widespread heterotrophic diazotrophs may have many adaptive mechanisms to deal with high concentration of various inorganic nutrients. For instance, a recent comparative genomics study revealed that nitrogenase in an isolated heterotrophic bacterium can be activated with high ammonia concentration in lack of functional CbbP or DraT2 proteins [81]. Nevertheless, how heterotrophic diazotrophs respond to their ambient nutrients warrant future studies.
Conclusion
To the best of our knowledge, this study presented the first evidence of composition and distribution of the diazotrophic communities in the EIO during the pre-southwest monsoon. Due to logistical constraints, our results were all generated from molecular data, and direct cell counts were not included in our study. Also due to possibly low abundance of diazotrophs, we applied high numbers of PCR cycles in gene amplification. Although it has been a well-developed protocol [e.g., 59], we realized that high PCR cycles might generate PCR bias and skew community structure characterization. In addition, primers used in our nested PCR were recently reported containing one mismatch that might influence the results of high throughput sequencing [64]. Despite these challenges, we observed diverse groups of diazotrophs that belong to Alpha-, Beta-, Gamma-proteobacterial groups; cyanobacterial cluster; and Firmicutes. Among which, Proteobacteria was the most dominant diazotroph in the EIO during pre-southwest monsoon, potentially contributing significantly to BNF in this oligotrophic ecosystem. Our results were in good agreement with previous studies conducted in the Western Indian Ocean, where heterotrophic bacteria were also reported dominant within the euphotic zone. Up to date, it is still poorly understood about ecology of marine heterotrophic diazotrophs and their actual contribution to BNF in the EIO. Further investigations on niche specialization and eco-physiological characterization of diazotrophs are greatly needed in order to better understand interactions of heterotrophic and cyanobacterial diazotrophs and how these interactions influence global nitrogen cycling.
References
Dugdale RC, Goering JJ (1967) Uptake of new and regenerated forms of nitrogen in primary productivity. Limnol Oceanogr 12:196–206. https://doi.org/10.4319/lo.1967.12.2.0196
Zehr JP (2011) Nitrogen fixation by marine cyanobacteria. Trends Microbiol 19:162–173. https://doi.org/10.1016/j.tim.2010.12.004
Gradoville MR, Bombar D, Crump BC, Letelier RM, Zehr JP, White AE (2017) Diversity and activity of nitrogen-fixing communities across ocean basins. Limnol Oceanogr 62:1895–1909. https://doi.org/10.1002/lno.10542
Levitan O, Rosenberg G, Setlik I et al (2007) Elevated CO2 enhances nitrogen fixation and growth in the marine cyanobacterium Trichodesmium. Glob Chang Biol 13:531–538. https://doi.org/10.1111/j.1365-2486.2006.01314.x
Benavides M, Voss M (2015) Five decades of N2 fixation research in the North Atlantic Ocean. Front Mar Sci 2:1–40. https://doi.org/10.3389/fmars.2015.00040
Yang L, Wang DX, Huang J, Wang X, Zeng L, Shi R, He Y, Xie Q, Wang S, Chen R, Yuan J, Wang Q, Chen J, Zu T, Li J, Sui D, Peng S (2015) Toward a mesoscale hydrological and marine meteorological observation network in the South China Sea. Bull Am Meteorol Soc 96(7):1117–1135. https://doi.org/10.1175/BAMS-D-14-00159.1
Carpenter EJ, Subramaniam A, Capone DG (2004) Biomass and primary productivity of the cyanobacterium Trichodesmium spp. in the tropical N Atlantic Ocean. Deep-Sea Res I 51:173–203. https://doi.org/10.1016/j.dsr.2003.10.006
Sohm JA, Webb EA, Capone DG (2011) Emerging patterns of marine nitrogen fixation. Nat Rev Microbiol 9(7):499–508. https://doi.org/10.1038/nrmicro2594
Zehr JP, Jenkins BD, Short SM, Steward GF (2003) Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol 5:539–554. https://doi.org/10.1046/j.1462-2920.2003.00451.x
Zehr JP, Mellon MT, Zani S (1998) New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl Environ Microbiol 64:3444–3450
Kong LL, Jing HM, Kataoka T, Sun J, Liu H (2011) Phylogenetic diversity and spatio-temporal distribution of nitrogenase genes (nifH) in the northern South China Sea. Aquat Microb Ecol 65(1):15–27. https://doi.org/10.3354/ame01531
Monteiro FM, Follows MJ, Dutkiewicz S (2010) Distribution of diverse nitrogen fixers in the global ocean. Glob Biogeochem Cycles 24:GB3017. https://doi.org/10.1029/2009GB003731
Cheung SY, Suzuki K, Saito H, Umezawa Y, Xia X, Liu H (2017) Highly heterogeneous diazotroph communities in the Kuroshio current and the Tokara Strait, Japan. PLoS One 12(10):e0186875. https://doi.org/10.1371/journal.pone.0186875
Shiozaki T, Ijichi M, Kodama T, Takeda S, Furuya K (2014) Heterotrophic bacteria as major nitrogen fixers in the euphotic zone of the Indian Ocean. Glob Biogeochem Cycles 28:1096–1110. https://doi.org/10.1002/2014GB004886
Moisander PH, Serros T, Paerl RW, Beinart RA, Zehr JP (2014) Gammaproteobacterial diazotrophs and nifH gene expression in surface waters of the South Pacific Ocean. ISME J 8(10):1962–1973. https://doi.org/10.1038/ismej.2014.49
Bombar D, Paerl RW, Riemann L (2016) Marine non-cyanobacterial diazotrophs: moving beyond molecular detection. Trends Microbiol 24(11):916–927. https://doi.org/10.1016/j.tim.2016.07.002
Schott FA, Xie SP, McCreary JP (2009) Indian ocean circulation and climate variability. Rev Geophys 47(1):RG1002. https://doi.org/10.1029/2007RG000245
Wang J, Kan JJ, Zhang XD et al (2017) Archaea dominate the ammonia-oxidizing community in deep-sea sediments of the Eastern Indian Ocean-from the equator to the Bay of Bengal. Front Microbiol 8:1–16. https://doi.org/10.3389/fmicb.2017.00415
Singh A, Jani RA, Ramesh R (2010) Spatiotemporal variations of the δ18O-salinity relation in the northern Indian Ocean. Deep-Sea Res I 57(11):1422–1431. https://doi.org/10.1016/j.dsr.2010.08.002
Singh A, Ramesh R (2015) Environmental controls on new and primary production in the northern Indian Ocean. Prog Oceanogr 131(3):138–145. https://doi.org/10.1016/j.pocean.2014.12.006
Lévy M, Shankar D, André JM, Shenoi SSC, Durand F, de Boyer Montégut C (2009) Basin-wide seasonal evolution of the Indian Ocean’s phytoplankton blooms. J Geophys Res Oceans 112:C12014. https://doi.org/10.1029/2007JC004090
Gomes HR, Goes JI, Saino T (2000) Influence of physical processes and freshwater discharge on the seasonality of phytoplankton regime in the Bay of Bengal. Cont Shelf Res 20:313–330. https://doi.org/10.1016/S0278-4343(99)00072-2
Kumar SP, Muraleedharan PM, Prasad TG et al (2002) Why is the Bay of Bengal less productive during summer monsoon compared to the Arabian Sea? Geophys Res Lett 29(24):2235–3770. https://doi.org/10.1002/2016JC012639
Madhupratap M, Gauns M, Ramaiah N et al (2003) Biogeochemistry of the Bay of Bengal: physical, chemical and primary productivity characteristics of the central and western Bay of Bengal during summer monsoon. Deep-Sea Res II 50(5):881–896. https://doi.org/10.1016/S0967-0645(02)00611-2
Hegde S, Anil AC, Patil JS et al (2008) Influence of environmental settings on the prevalence of Trichodesmium spp. in the Bay of Bengal. Mar Ecol Prog Ser 356(01):93–101. https://doi.org/10.3354/meps07259
Gandhi N, Singh A, Prakash S, Ramesh R, Raman M, Sheshshayee MS, Shetye S (2011) First direct measurements of N2 fixation during a Trichodesmium bloom in the eastern Arabian Sea. Glob Biogeochem Cycles 25:GB4014. https://doi.org/10.1029/2010GB003970
Ahmed A, Gauns M, Kurian S (2017) Nitrogen fixation rates in the eastern Arabian Sea. Estuar Coast Shelf Sci 191:74–83. https://doi.org/10.1016/j.ecss.2017.04.005
Bird C, Wyman M (2012) Transcriptionally active heterotrophic diazotrophs are widespread in the upper water column of the Arabian Sea. FEMS Microbiol Ecol 84(1):189–200. https://doi.org/10.1111/1574-6941.12049
Kitajima S, Furuya K, Hashihama F, Takeda S, Kanda J (2009) Latitudinal distribution of diazotrophs and their nitrogen fixation in the tropical and subtropical western North Pacific. Limnol Oceanogr 54(2):537–547. https://doi.org/10.4319/lo.2009.54.2.0537
Shiozaki T, Bombar D, Riemann L, Hashihama F, Takeda S, Yamaguchi T, Ehama M, Hamasaki K, Furuya K (2017) Basin scale variability of active diazotrophs and nitrogen fixation in the North Pacific, from the tropics to the subarctic Bering Sea. Glob Biogeochem Cycles 31(6):996–1009. https://doi.org/10.1002/2017GB005681
Church MJ, Mahaffey C, Letelier RM, Lukas R, Zehr JP, Karl DM (2009) Physical forcing of nitrogen fixation and diazotroph community structure in the North Pacific subtropical gyre. Glob Biogeochem Cycles 23:GB2020. https://doi.org/10.1029/2008GB003418
Benavides M, Moisander PH, Daley MC, Bode A, Arístegui J (2016) Longitudinal variability of diazotroph abundances in the subtropical North Atlantic Ocean. J Plankton Res 38(3):662–672. https://doi.org/10.1093/plankt/fbv121
Goebel NL, Turk KA, Achilles KM, Paerl R, Hewson I, Morrison AE, Montoya JP, Edwards CA, Zehr JP (2010) Abundance and distribution of major groups of diazotrophic cyanobacteria and their potential contribution to N2 fixation in the tropical Atlantic Ocean. Environ Microbiol 12(12):3272–3289. https://doi.org/10.1111/j.1462-2920.2010.02303.x
Shiozaki T, Nagata T, Ijichi M (2015) Nitrogen fixation and the diazotroph community in the temperate coastal region of the northwestern North Pacific. Biogeosciences 12:4751–4764. https://doi.org/10.5194/bg-12-4751-2015
Foster RA, Subramaniam A, Mahaffey C (2007) Influence of the Amazon River plume on distributions of free-living and symbiotic cyanobacteria in the western tropical North Atlantic Ocean. Limnol Oceanogr 52(2):517–532. https://doi.org/10.4319/lo.2007.52.2.0517
. Grasshoff K, Ehrhardt M, Kremling K (1982) Methods of seawater analysis. ISBN (Verlag Chemie), 3– 527, 25,998–26,008
Schmidt TM, Delong EF, Pace NR (1991) Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol 173:4371–4378. https://doi.org/10.1128/jb.173.14.4371-4378.1991
Kan JJ, Clingenpeel S, Macur RE et al (2011) Archaea in Yellowstone Lake. ISME J 5(11):1784–1795. https://doi.org/10.1038/ismej.2011.56
Zhang Y, Yang Q, Ling J et al (2017) Diversity and structure of diazotrophic communities in mangrove rhizosphere, revealed by high-throughput sequencing. Front Microbiol 8(2032). https://doi.org/10.3389/fmicb.2017.02032
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Magoč T, Salzberg SL (2010) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123. https://doi.org/10.1111/j.1462-2920.2009.02051.x
Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8:R143. https://doi.org/10.1186/gb-2007-8-7-r143
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17(3):377–386. https://doi.org/10.1101/gr.5969107
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Letunic I, Bork P (2011) Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:475–478. https://doi.org/10.1093/nar/gkr201
Zhang Y, Zhao ZH, Sun J (2011) Diversity and distribution of diazotrophic communities in the South China Sea deep basin with mesoscale cyclonic eddy perturbations. FEMS Microbiol Ecol 78(3):417–427. https://doi.org/10.1111/j.1574-6941.2011.01174.x
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3–900051–07-0, URL http://www.R-project.org. Accessed 31 Oct 2016
Chao A, Jost L (2012) Coverage-based rarefaction and extrapolation: standardizing samples by completeness rather than size. Ecology 93:2533–2547. https://doi.org/10.1890/11-1952.1
Hsieh TC, Ma KH, Chao A (2016) iNEXT: an R package for rarefaction and extrapolation of species diversity (hill numbers). Methods Ecol Evol 7:1451–1456. https://doi.org/10.1111/2041-210X.12613
. Clarke KR, Gorley RN (2006) PRIMER v6: user manual/tutorial. Marine Laboratory, Plymouth, UK, pp 190
ter Braak CJF, Šmilauer P (2002) Canoco for Windows, version 4.5. Biometris-Plant Research International, Wageningen
Lepš J, Šmilauer P (2003) Multivariate analysis of ecological data using CANOCO. Cambridge University Press, Cambridge
Dixon P (2003) VEGAN, a package of R functions for community ecology. J Veg Sci 14(6):927–930. https://doi.org/10.1111/j.1654-1103.2003.tb02228.x
Schlitzer R (2018) Ocean data view. https://odv.awi.de. Accessed 19 May 2018
Martínez-Pérez C, Mohr W, Schwedt A, Dürschlag J, Callbeck CM, Schunck H, Dekaezemacker J, Buckner CRT, Lavik G, Fuchs BM, Kuypers MMM (2018) Metabolic versatility of a novel N2-fixing Alphaproteobacterium isolated from a marine oxygen minimum zone. Environ Microbiol 20(2):755–768. https://doi.org/10.1111/1462-2920.14008
Riemann L, Farnelid H, Steward GF (2010) Nitrogenase genes in non-cyanobacterial plankton: prevalence, diversity and regulation in marine waters. Aquat Microb Ecol 61:235–247. https://doi.org/10.3354/ame01431
Farnelid H, Andersson AF, Bertilsson S, al-Soud WA, Hansen LH, Sørensen S, Steward GF, Hagström Å, Riemann L (2011) Nitrogenase gene amplicons from global marine surface waters are dominated by genes of non-cyanobacteria. PLoS One 6(4):e19223. https://doi.org/10.1371/journal.pone.0019223
Bentzon-Tilia M, Traving SJ, Mantikci M, Knudsen-Leerbeck H, Hansen JLS, Markager S, Riemann L (2015) Significant N2 fixation by heterotrophs, photoheterotrophs and heterocystous cyanobacteria in two temperate estuaries. ISME J 9(2):273–285. https://doi.org/10.1038/ismej.2014.119
Delmont TO, Quince C, Shaiber A et al (2018) Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat Microbiol 3:804–813. https://doi.org/10.1038/s41564-018-0176-9
Halm H, Lam P, Ferdelman TG, Lavik G, Dittmar T, LaRoche J, D'Hondt S, Kuypers MMM (2012) Heterotrophic organisms dominate nitrogen fixation in the South Pacific Gyre. ISME J 6(6):1238–1249. https://doi.org/10.1038/ismej.2011.182
Duan YL, Liu L, Han G, Liu H, Yu W, Yang G, Wang H, Wang H, Liu Y, Zahid, Waheed H (2016) Anomalous behaviors of Wyrtki Jets in the equatorial Indian Ocean during 2013. Sci Rep 6:29688. https://doi.org/10.1038/srep29688
Qian G, Wang J, Kan JJ, Zhang X, Xia Z, Zhang X, Miao Y, Sun J (2018) Diversity and distribution of anammox bacteria in water column and sediments of the Eastern Indian Ocean. Int Biodeterior Biodegrad 133:52–62. https://doi.org/10.1016/j.ibiod.2018.05.015
Rahav E, Giannetto MJ, Bar-Zeev (2016) Contribution of mono and polysaccharides to heterotrophic N2 fixation at the eastern Mediterranean coastline. Sci Rep 6:27858. https://doi.org/10.1038/srep27858
Moisander PH, Zhang RF, Boyle EA, Hewson I, Montoya JP, Zehr JP (2012) Analogous nutrient limitations in unicellular diazotrophs and Prochlorococcus in the South Pacific Ocean. ISME J 6:733–744. https://doi.org/10.1038/ismej.2011.152
Loescher CR, GroβKopf T, Desai FD et al (2014) Facets of diazotroph in the oxygen minimum zone waters off Peru. ISME J 8(11):2180–2192. https://doi.org/10.1038/ismej.2014.71
Grand MM, Measures CI, Hatta M, Hiscock WT, Buck CS, Landing WM (2015) Dust deposition in the eastern Indian Ocean: the ocean perspective from Antarctica to the Bay of Bengal. Glob Biogeochem Cycles 29(3):357–374. https://doi.org/10.1002/2014GB004898
Grand MM, Measures CI, Hatta M, Hiscock WT, Landing WM, Morton PL, Buck CS, Barrett PM, Resing JA (2015) Dissolved Fe and Al in the upper 1000 m of the eastern Indian Ocean: a high-resolution transect from the Antarctic margin to the bay of Bengal. Glob Biogeochem Cycles 29(3):375–396. https://doi.org/10.1002/2014GB004920
He Q, Zhan H, Shuai Y (2017) Phytoplankton bloom triggered by an anticyclonic eddy: the combined effect of eddy-Ekman pumping and winter mixing. J Geophys Res-Oceans 122:4886–4901. https://doi.org/10.1002/2017JC012763
Jyothibabu R, Karnan C, Jagadeesan L, Arunpandi N, Pandiarajan RS, Muraleedharan KR, Balachandran KK (2017) Trichodesmium blooms and warm-core ocean surface features in the Arabian Sea and the Bay of Bengal. Mar Pollut Bull 121(1–2):201–215. https://doi.org/10.1016/j.marpolbul.2017.06.002
Wu C, Fu FX, Sun J et al (2018) Nitrogen fixation by Trichodesmium and unicellular diazotrophs in the northern South China Sea and the Kuroshio in summer. Sci Rep 8(2415). https://doi.org/10.1038/s41598-018-20743-0
Cabello AM, Cornejo-Castillo FM, Raho N, Blasco D, Vidal M, Audic S, de Vargas C, Latasa M, Acinas SG, Massana R (2016) Global distribution and vertical patterns of a prymnesiophyte–cyanobacteria obligate symbiosis. ISME J 10(3):693–706. https://doi.org/10.1038/ismej.2015.147
Stenegren M, Caputo A, Berg C, Bonnet S, Foster RA (2018) Distribution and drivers of symbiotic and free-living diazotrophic cyanobacteria in the Western Tropical South Pacific. Biogeosciences 15(5):1559–1578. https://doi.org/10.5194/bg-15-1559-2018
Cornejo-Castillo FM, Cabello AM, Salazar G, Sánchez-Baracaldo P, Lima-Mendez G, Hingamp P, Alberti A, Sunagawa S, Bork P, de Vargas C, Raes J, Bowler C, Wincker P, Zehr JP, Gasol JM, Massana R, Acinas SG (2016) Cyanobacterial symbionts diverged in the late cretaceous towards lineage-specific nitrogen fixation factories in single-celled phytoplankton. Nat Commun 7:11071. https://doi.org/10.1038/ncomms11071
Turk-Kubo KA, Farnelid HM, Shilova IN, Henke B, Zehr JP (2016) Distinct ecological niches of marine symbiotic N2-fixing cyanobacterium Candidatus atelocyanobacterium thalassa sublineages. J Phycol 53(2):451–461. https://doi.org/10.1111/jpy.12505
Fu FX, Yu E, Garcia NS, Gale J, Luo Y, Webb EA, Hutchins DA (2014) Differing responses of marine N2-fixers to warming and consequences for future diazotroph community structure. Aquat Microb Ecol 72:33–46. https://doi.org/10.3354/ame01683
Heiniger EK, Oda Y, Samanta SK, Harwood CS (2012) How posttranslational modification of nitrogenase is circumvented in Rhodopseudomonas palustris strains that produce hydrogen gas constitutively. Appl Environ Microbiol 78:1023–1032. https://doi.org/10.1128/AEM.07254-11
Acknowledgements
We thank Prof. Dongxiao Wang from South China Sea Institute of Oceanology, Chinese Academy of Sciences for providing hydrographic (CTD) data. Dr. Liangliang Kong at McGill University and Dr. Xiaomin Xia at the Hong Kong University of Science and Technology are also acknowledged for their help and technical support during experiments. We also gratefully acknowledge the crew of R/V “Shiyan 3” and all participants for their assistance during the cruise.
Funding
This study was supported by the National Natural Science Foundation of China (41876134, 41276124, and 41676112) and NSFC open research cruises (NORC2017-10), the Science Fund for University Creative Research Groups in Tianjin (TD12-5003), the Changjiang Scholar Program of Chinese Ministry of Education to JS, and endowment support from Stroud Water Research Center to JK.
Author information
Authors and Affiliations
Contributions
This work was designed by JS. Samples were collected by CW, HL, and XW. CW performed experiments and analyzed data. CG performed the nutrients analysis. CW drafted the paper and revised by JK and PL. All authors contributed to the writing of the manuscript.
Corresponding author
Electronic Supplementary Material
Fig. S1
Depth profiles of dissolved inorganic nutrients (nitrate, nitrite, ammonium, phosphate, silicate) and Chl a in the EIO (PNG 5281 kb)
High resolution image
(TIF 1530 kb)
Fig. S2
Rarefaction curves comparing the number of reads with the number of phylotypes (OTUs) found in the DNA libraries in the EIO. The last digit represented water depth: 1, 2 and 3 represent 0m, 75m and 200m, respectively. (PNG 1240 kb)
High resolution image
(TIF 2708 kb)
Supplementary Table 1
(DOCX 22 kb)
Rights and permissions
About this article
Cite this article
Wu, C., Kan, J., Liu, H. et al. Heterotrophic Bacteria Dominate the Diazotrophic Community in the Eastern Indian Ocean (EIO) during Pre-Southwest Monsoon. Microb Ecol 78, 804–819 (2019). https://doi.org/10.1007/s00248-019-01355-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01355-1