Abstract
In soilless culture, slow filtration is used to eliminate plant pathogenic microorganisms from nutrient solutions. The present study focused on the characterization and the potential functions of microbial communities colonizing the nutrient solutions recycled on slow filters during a whole cultivation season of 7 months in a tomato growing system. Bacterial microflora colonizing the solutions before and after they flew through the columns were studied. Two filters were amended with Pseudomonas putida (P-filter) or Bacillus cereus strains (B-filter), and a third filter was a control (C-filter). Biological activation of filter unit through bacterial amendment enhanced very significantly filter efficacy against plant potential pathogens Pythium spp. and Fusarium oxysporum. However, numerous bacteria (103–104 CFU/mL) were detected in the effluent solutions. The community-level physiological profiling indicated a temporal shift of bacterial microflora, and the metabolism of nutrient solutions originally oriented towards carbohydrates progressively shifted towards degradation of amino acids and carboxylic acids over the 7-month period of experiment. Single-strand conformation polymorphism fingerprinting profiles showed that a shift between bacterial communities colonizing influent and effluent solutions of slow filters occurred. In comparison with influent, 16S rDNA sequencing revealed that phylotype diversity was low in the effluent of P- and C-filters, but no reduction was observed in the effluent of the B-filter. Suppressive potential of solutions filtered on a natural filter (C-filter), where the proportion of Proteobacteria (α- and β-) increased, whereas the proportion of uncultured candidate phyla rose in P- and B-filters, is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In hydroponic greenhouses, the soil is replaced by organic or inorganic substrates in order to avoid root diseases caused by soilborne pathogens. However, numerous bacteria, fungi, and oomycetes generally colonize the water used for supplying plants, and among them some are pathogenic to plants. Oomycetes, e.g., Pythium and Phytophthora, and fungi, e.g., Olpidium, producing zoospores are particularly adapted to this system of cultivation [1, 2]. Once introduced in the greenhouse, these microorganisms spread easily over the whole culture via the recirculation of solutions and cause root diseases that are the major pathogenic problem in greenhouses. Pathogenic bacteria, e.g., Clavibacter michiganense, Pseudomonas solanacearum, are also responsible for the bacterial canker and wilt of tomato in soilless cultures [3,4,5,6]. Another factor that concomitantly increases the risks of pathogen attacks is recycling. This technique is done to minimize pollution by re-using the runoff solution, a development driven by legislation and public concerns. However, recycling of nutrient solutions contaminated by pathogenic microorganisms induced risks for plants [2, 7, 8]. The use of methods dedicated to avoid the dispersal of root-infecting pathogens has therefore become a major challenge.
Several active control methods have been developed to disinfect the circulating water before re-use [5, 9]: heat treatment, UV radiation, and chemical treatments (chlorination, hydrogen peroxide, ozonization, etc.). They have been efficiently used to control target pathogens, such as Pythium spp., from nutrient solutions of tomato and cucumber soilless cultures [10, 11]. However, they have a side effect by eliminating the non-pathogenic microorganisms. Tu et al. [12] reported that resident bacteria of nutrient solutions were shown to reduce Pythium root rot in a closed soilless system. In the same line, Zhang and Tu [11] impute the lack of control of P. aphanidermatum on tomatoes roots to the reduction of bacterial communities caused by UV radiation. Chave et al. [13] reported that chlorine disinfection has an effect on well-established bacterial communities of the rhizosphere by reducing the microbial diversity. Thus, destruction or alteration of non-target microflora is at concern for preservation of non-pathogenic microorganisms occurring naturally in the rhizosphere. Postma et al. [10, 14] have demonstrated that, under certain cultural conditions, indigenous microflora have the capacity to suppress P. aphanidermatum attacks. Disease suppressiveness showed correlation with some specific communities such as the ones formed by filamentous actinomycetes and sometimes by Trichoderma spp. The overall agreement today is that active disinfecting methods negatively affect the suppressive potential of natural microflora and that passive methods which have a relative low impact on bacterial microflora, such as slow filtration, have a better potential to achieve this goal.
In the early 1990s, the slow filtration system originally used for the purification of drinking water was successfully adapted to greenhouses [15, 16]. This technique consists in filtering the nutrient solutions through a substrate-filled filter unit before their delivery to the root system. Slow filters were demonstrated to be effective for the control of various plant pathogens such as bacteria, fungi, nematodes, and viruses [9, 17, 18]. Pathogen elimination relies on a physical filtration but also on a biological process. The colonization of filtering media by bacterial populations was shown to contribute to the removal of plant pathogens [19, 20]. In order to promote the installation of bacterial populations in filtering columns, some filters have been amended with massive amount of bacteria with suppressive traits (i.e., antagonistic and plant growth-promoting bacteria) before the start of nutrient solution recycling. As reported by Renault et al. [21], this type of bacteria frequently colonizes the substrates of filters. Once these bacteria, e.g., Pseudomonas putida and Bacillus cereus group strains, were inoculated in a pozzolana biofilter, the fungal elimination of Fusarium oxysporum from nutrient solutions of a tomato soilless culture was successfully enhanced over the whole cultural season of plants [22].
In that context, one may consider that biofilters can be included in the global strategy of suppressive microflora management for controlling plant pathogens in soilless culture: first, in inoculating bacteria with suppressive traits inside the filter units; second, biofiltration is often considered as a reliable method in the maintenance of the disease suppression because it is harmless to several bacteria. Several reports [22, 23] demonstrated that after the biofiltration process, significant natural bacterial microflora (103–104 CFU/mL) can be detected in biofilter effluents. However, the main gap with this microflora is that available information on the bacterial communities colonizing effluent nutrient solutions is limited. The main issue is that the pathogenic or suppressive potential of this bacterial microflora is relatively unknown. Most of the studies reported in the literature mainly dealt with the cultured aerobic bacteria [22, 24, 25]. Van Os and Postma [8, 26] studied during 12 weeks the impact of a natural slow filter on the metabolic behavior and the 16s rDNA structure of the natural microflora of nutrient solutions. They showed that certain bacterial groups were preserved. Additional experiments are thus needed to understand more about the bacterial communities, their composition and shift, and the changes in trophic behavior. Such baseline information could help in the management of suppressive microflora in soilless cultures.
Therefore, the present study focused on the characterization of microbial communities colonizing the nutrient solutions recycled on slow filters during a whole cultivation season of 7 months in a tomato growing system. To this effect, the polyphasic approaches used by Renault et al. [27] to study the diversity of bacterial communities that colonize the filter units was applied to characterize the bacterial communities of influent and effluent solutions of biofilters. To get a global insight into the structure and the diversity of bacterial communities, microbial investigations, community-level carbon source utilization profile assessment by Biolog®, and single-strand conformation polymorphism analysis coupled to the sequencing of clone libraries were used.
Materials and Methods
Filter Units and Bacterial Amendments
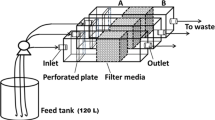
Three filter units were set at room temperature within the same compartment of an experimental greenhouse (CATE, Saint-Pol-de-Léon, France) used to grow tomato plants (cv. Tradiro, De Ruiter Graines, France). Temperature in the greenhouse was regularly measured and ranged from 18 °C (±2 °C) during the night to 21 °C (±2 °C) during the day. Each filter consisted of a plastic pipe (220 cm in length and 40 cm in inner diameter) filled with a 100-cm-thick layer of pozzolana particles (1–4 mm diameter) deposited above a 40-cm-thick layer of different graded gravel stones (2–8, 8–16, 16–32 mm). The upper water layer was regulated by a float switch placed at a distance of 40–50 cm above the pozzolana surface. The filtration rate ranged from 100 to 150 L/h/m2. The three filter units were connected to three independent irrigation circuits. The nutrient solution (Kemira, France) was delivered to each plant (136 tomato plants per condition) through a capillary system. The pH was regularly monitored and found to be between 5.5 and 6.2.
The three filters were flooded in March. The C-filter was used as control (no bacteria added), whereas the two other filters were inoculated with bacterial strains previously selected for their antagonistic properties [22] and supplied by the Brittany Microbial Collection (ESIAB, Plouzané, France). The P-filter was inoculated with three strains of Pseudomonas putida (LMSA3.06.045, LMSA3.06.046, and LMSA3.06.047) and the B-filter with two strains of the Bacillus cereus group (LMSA3.06.043, LMSA3.06.044). Each bacteria-amended filter was inoculated twice, when the filter was flooded and 2 weeks after. The total bacterial densities poured out over the top of filtering columns were, each time, between 2 and 4.1012 colony-forming units (CFU).
Quantification of Bacterial Populations, Pythium spp., and Fusarium Oxysporum in Nutrient Solutions
Samples of nutrient solution were collected in sterile containers from influent and effluent solutions in each filtration unit, from April to September. To count bacterial colonies, 50 μL of nutrient solution was plated with a spiral plater on PCA (plate count agar, AES, France). The number of bacterial colonies was determined after a 48-h incubation in the dark at 30 °C (four replicates per modality). The concentration of Pythium spp. and Fusarium oxysporum was determined on 150 and 100 mL, respectively, of nutrient solution samples filtered through 0.45-μm cellulose acetate filters (Sartorius, Germany). The filters were plated on selective media coded CMA-PARP (Corn Meal Agar implemented with four antibiotics) [28] for Pythium spp. and Komada [29] for F. oxysporum (four replicates per modality). Pythium propagules were counted after a 48-h incubation at 25 °C in the dark, whereas F. oxysporum propagules were counted 7 days after incubation under the same conditions. Final results are expressed as the percentage of eliminated microorganisms.
Assessment of Community-Level Physiological Profiles
Nutrient solutions were monthly collected from May to November upstream and downstream from each filter. Each nutrient solution sample was distributed (150 μL/well) in two 96-well Biolog® GN2 microplates (AES, France). Microplates contain one reference well (negative control) and 95 different carbon sources. They were incubated at 30 °C for 48 h. The rate of conversion of tetrazolium violet associated with the 95 Biolog®GN2 carbon sources was determined by measurement of the optical density (OD) at 620 nm with a microplate reader (Titertek Multiskan® MCC1340). To minimize the effect of difference in bacterial mixture densities between plates, data were standardized as follows: the average well color development (AWCD) corresponding to the mean of the blanked absorbance values of the 95 wells was calculated for each plate; then, the blanked absorbance value of each well was divided by the AWCD of the corresponding plate to get a corrected OD value [30, 31]. In accordance with Grove et al. [32], all corrected OD values were set to fall within 0 and 2 (boundary limits). The averages of corrected OD values were subsequently calculated for each community-level physiological profile.
Microbial DNA Extraction
Two hundred milliliters of each nutrient solution sample monthly collected from May to November was filtered through 0.45-μm cellulose acetate filters (Sartorius, Germany), which were stored at − 80 °C. Total DNA was extracted as reported by Godon et al. [33] with slight changes. Briefly, the frozen filter was ground with a sterile plastic stick and transferred to a 2-mL polypropylene microcentrifuge tube kept frozen in a − 20 °C freezer rack. Then, 400 μL of 4 M guanidine thiocyanate-0.1 M Tris (pH 7.5), 120 μL of 10% N-lauroyl sarcosine, and 500 μL of 5% N-lauryl sarcosine-0.1 M phosphate buffer (pH 8.0) were added and the sample heated to 95 °C for 10 min. An aliquot of 500 μL of zirconium beads (0.1 mm diameter), sterilized by autoclaving, was added, and the mixture was shaken for 10 min at maximal speed in a Vibro shaker (Retsch). Nucleic acids were precipitated with 1 volume of isopropanol added concomitantly with 5 μL of Carrier RNA (Qiagen, Hilden, Germany) to enhance DNA extraction yield. Finally, a QiaAmp DNA Micro Kit (Qiagen) was used for purification of total DNA.
PCR-SSCP Analyses
The V3 region of the 16S rDNA gene was amplified from total DNA with primers W49 (5′-ACGGTCCAGACTCCTACGGG-3′) and W104 (5′-6-FAM-TTACCGCGGCTGCTGGCAC-3′). PCR amplifications were performed with a thermal cycler. The reaction mixtures contained 1× polymerase buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 200 ng of each primer, 1.25 U of Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA, USA), 1 μL of DNA extract, and water added to obtain a final volume of 50 μL. The PCR conditions were an initial denaturation step of 2 min at 94 °C; 25 cycles of a three-stage program of 30 s at 94 °C, 30 s at 61 °C, and 30 s at 72 °C; and a final elongation for 10 min at 72 °C. Amplification product sizes (about 200 pb) were confirmed by electrophoresis on a 2% (w/v) agarose gel. PCR products were analyzed using capillary electrophoresis single-strand conformation polymorphism (CE-SSCP) performed on an ABI 310 Genetic Analyzer (Applied Biosystems, France). The sample mixtures contained 1 μL of PCR product, 18 μL of formamide, and 1 μL of internal size standard 400 HD ROX (Applied Biosystems, Foster City, CA, USA) and were denatured for 5 min at 95 °C and immediately cooled on ice prior to CE-SSCP analysis. The migration was set at 15 kV, 32 °C, and 30 min per sample. CE-SSCP is based on the electrophoresis mobility of single-stranded DNA fragments, which reflects their three-dimensional conformation. Profiles were aligned to the ROX standard internal size marker with SAFUM [34] and normalized with MATLAB 6.5 software (MathWorks).
Global Bacterial Communities Description by Multiple Factor Analysis
The data of (i) CLPP assessment and (ii) SSCP structure description were analyzed by a multiple factor analysis (MFA) with the Rcmdr R package [35]. The two quantitative variable groups named “metabolism” (95 carbon sources) and “structure” (650 SSCP scans) were defined as active, and their variables were scaled. Six descriptive and qualitative variables were added in the analysis to discriminate samples according to the month of sampling (“time”), the filter type (“filter”), and the position of the nutrient solution upstream or downstream filters (“effluent”) as well as all the combinations of these factors (“time.filter,” “time.effluent,” “filter.effluent”).
Microbial Clone Library Construction, Screening, and Sequencing
Six clone libraries were constructed from the DNA extracts of the influent and effluent nutrient solutions sampled in July from C-filter (iClib and eClib), B-filter (iBlib and eBlib), and P-filter (iPlib and ePlib). Bacterial 16S rRNA genes were amplified by PCR with the universal reverse primer W31 (5′-TTACCGCGGCTGCTGGCAC-3′) and the forward primer W18 (5′-GAGTTTGATCMTGGCTCAG-3′). The final PCR mixture (50 μL) contained 1 μL of DNA template, 2 μL of each primer (100 ng/μL), 4 μL of dNTP (2.5 mM), 5 μL of 1× Taq reaction buffer, 1 μL of red Taq DNA polymerase (Sigma), and 35 μL of water. An initial DNA denaturation step at 94 °C for 2 min was followed by 25 cycles at 94 °C for 1 min, 55 °C for 1 min, and 72 °C at 1 min before a final 10-min elongation step at 72 °C. PCR products were purified with a QIAquick® PCR Purification Kit (Qiagen) in accordance with the manufacturer’s instructions. The purified PCR products were cloned and transformed using the pCR4-TOPO plasmid and TOP10 Escherichia coli competent cells from the TOPO TA Cloning Kit as indicated by the manufacturer (Invitrogen, Groningen, the Netherlands). Recombinant cells were selected by kanamycin resistance and ccd gene killer inactivation before cultivation at 37 °C for 24 h in LB2X medium (tryptone 20 g/L, yeast extract 10 g/L, NaCl 10 g/L). Finally, the plasmid DNA was purified with a Montage SEQ96 Sequencing Reaction Cleanup Kit (Millipore). The sequence reaction was performed in a GeneAmp® PCR System 9700 thermocycler (Applied Biosystems) under the following conditions: a DNA denaturation step at 96 °C for 5 min followed by 50 cycles at 96 °C for 30 s, 57 °C for 30 s, and 60 °C for 4 min. Each 5-μL reaction mixture was composed of 1 μL purified DNA template, 1 μL sequencing primer T7 (5′-TAATACGACTCACTATAGGG-3′), 0.5 μL BigDye Terminator V3.1, 0.75 μL 5× buffer, and 1.75 μL sterile water. DNA sequences were obtained with an Applied Biosystems 3130xl Genetic Analyzer (GIS Ouest Génopole—GENOMER, Roscoff, France). They were trimmed and cleaned with Phred [36] and SeqClean (TGIR, the Institute for Genomic Research, Rockville, MP, USA [http://www.tigr.org/tdb/tgi/software]) softwares and subsequently compared to GenBank databases (www.ncbi.nlm.nih.gov/blast) using the Blast program [37]. Thresholds used to assign sequences to the different taxonomic levels, i.e., phylum, family (order), and genus, were, respectively, 78, 87, and 95% of similarity [38]. For each phylotype affiliation, the closest described relative from the GenBank database with its accession number is provided.
Various diversity indexes were estimated. Coverage percentage was determined with Good’s coverage estimator (Cgood). This non-parametric estimator of the proportion of phylotypes within a library of infinite size that would be represented in a smaller library is calculated from the relation CGood = 1 − (n 1 / N) where n 1 is the number of phylotypes appearing only once in the clone library, and N is the total number of analyzed clones [39]. Other diversity and similarity estimators were produced by the EstimateS software package [40]. The total number of phylotypes was estimated by calculating the Chao1 richness estimator (Chao1) and its 95% confidence interval. The phylogenetic compositions of libraries were compared using Chao’s Sorensen abundance-based similarity index being corrected for unseen species [41].
Results
Efficacy of Biofiltration to Eliminate Pythium spp. and Fusarium oxysporum
Pythium spp. and Fusarium oxysporum were regularly detected in nutritive solutions before biofiltration. Density averages were respectively 1.102 and 8.103 propagules/L, and F. oxysporum was particularly more abundant in August and September.
The three filters always reduced the fungal densities, whatever the sampling month (Table 1). Bacteria-amended filters had elimination rates always equal to or greater than the C-filter. According to the Kruskal-Wallis test (α = 0.05), the P-filter and, even more, the B-filter significantly enhanced the removal of Pythium spp. the first month of filtration and of F. oxysporum in July. The P-filter was statistically more efficient than the C-filter in F. oxysporum removal during all the cultivation seasons except in August.
The removal of fungal risks by the C-filter was minimal at the beginning of filtration: 57% for Pythium spp. and 76.5% for F. oxysporum. It reached its maximum efficacy only after 3−5 months of filtration whereas bacteria-amended filters were optimized from the start of utilization. For instance, the P-filter removed from 99.2 to 99.9% of F. oxysporum populations from April and during all the cultivation season.
Impact of Biofiltration on the Bacterial Density of Nutrient Solutions
The sampling of April was not included in the data comparison because the effluent solutions of the two amended filters were more bacteria-colonized than the influent ones. This phenomenon was attributed to the leaching of inoculated bacteria that did not colonize the filtering substrates of columns. This bacterial leaching was never detected after the 2nd month of stabilization in May until September. The biofiltration process always reduced the bacterial density of the nutrient solution (Fig. 1). The average density of bacteria in the influent solutions was 4.104 ± 2.104 CFU/mL. This density was reduced to an average of 5.103 ± 4.103 CFU/mL in the effluent solutions. The reduction rate of bacterial densities in nutrient solutions was clearly higher and more homogeneous with the bacteria-amended filters (90.3 ± 4.8% with the P-filter and 91.1 ± 3.8% with the B-filter) than with the C-filter (67.1 ± 17.5%). The C-filter reached its maximum reduction rate at the end of the cultivable season in September (88.1%) but lost efficacy in June and July (53.1 and 47.1%). P- and B-filters maintained high reduction levels throughout the cultivation season (85–97%) and were always more efficient than the C-filter according to the Kruskal-Wallis test (α = 0.05).
Bacterial Communities Analysis from their Metabolic and Molecular Fingerprints
MFA eigenvalues indicated that the first two axes, PC1 and PC2, explained 29% of the total data variance and gave a sufficient description of the two groups of active variables (Fig. 2). The first axis was more related to the structure group (68% of contribution), while the second axis was more related to the metabolism group (75% of contribution).
Distribution of variable groups on the principal plan designed by the multiple factor analysis. The two active groups (red triangle) metabolism and structure came respectively from the Biolog® GN profile (95 carbon sources) and the SSCP profile (650 scans) of the different microbial populations. Six descriptive variables (open triangle) are represented according to the sampling month (time), the sampling position before and after filtration (effluent), the filter type (filter), and the three correlations (time.filter, time.effluent, and filter.effluent). For example, the time.filter variable describes the microbial populations grouped by time and by filter. The total variation (%) explained by each principal component (PC) is given in brackets
All the time-related variables were significantly correlated with PC1, both for time, time.effluent (p = 0.001), and time.filter (p = 0.05). They were even more correlated with PC2 (p = 0.001). The distribution of time-correlated variables close to the metabolism group (Fig. 2) indicated that the time of sampling strongly and mainly defined the metabolic fingerprints of communities. Indeed, individuals grouped by sampling month (Fig. 3a) progressively shifted from positive coordinates (May, June, July) to negative coordinates (August, October, November) along the PC2 axis. May and November groups were very strongly correlated with this axis (p = 0.001). The close examination of carbon sources describing this dimension (Table 2) showed that, in the first month of cultivation season, microbial populations had a metabolic profile mainly oriented towards carbohydrates (55%), i.e., xylitol, d-sorbitol, α-d-lactose, d-fructose, d-raffinose, and l-rhamnose, whereas, at the end, they metabolized especially carboxylic acids (35%), i.e., citric acid, succinic acid, d-gluconic acid, d,l-lactic acid, α-ketoglutaric acid, cis-aconitic acid, and quinic acid, or amino acids (25%), i.e., γ-amino butyric acid, l-serine, l-asparagine, l-alanine, and l-aspartic acid.
Distribution of individual groups on the principal plan designed by the multiple factor analysis. Points represent microbial community profiles. Boxes indicate average positions of sample groups, and ellipses surround the confidence interval (p = 0.05). Samples are grouped according to the time (a) and time.effluent (b) variables. The second graph discriminates samples upstream of filters (full line) from samples downstream (dotted line)
The molecular structure was also time-dependent, but no linear shift appeared during the cultivation season (Fig. 3a). For example, SSCP patterns of May and November were positively correlated to PC1 while June and August had a negative correlation on this axis (p = 0.05). These changes in the molecular structure at the community level were related to the presence of specific SSCP domains (Table 2). Individuals positively correlated to PC1 were characterized by SSCP scans all located in the first half of the entire profile, whereas samples with a negative correlation had SSCP scans mainly located (79%) in the second half.
The high coordinates of the time.effluent variable both on PC1 and PC2 supported that, on a given date, the molecular and metabolic fingerprints of bacterial populations depended also on the effluent type, i.e., nutrient solutions upstream or downstream filters (Fig. 2). Time.effluent was the best descriptive variable associated with PC1 (R 2 = 0.79). This observation supported that, on a given date, the filtration process changed the molecular structure of bacterial populations colonizing the nutritive solutions. The distribution of individuals grouped by sampling month and effluent type (Fig. 3b) confirmed this result: for each month, excluding July, the average profile of solutions after filtration had, on PC1, higher coordinate than solutions coming to filters, whatever the filter. Moreover, from July to November, the filter output solutions were significantly distinct from the input ones (p = 0.05).
The descriptive variable filter did not discriminate the bacterial communities (Fig. 2), but the time.filter variable was significantly more correlated with PC1 (R 2 = 0.70) and PC2 (R 2 = 0.85) than the time variable (respectively 0.57 and 0.77). Thus, on a fixed date, bacterial communities differed also according to the filter type (C-, P-, or B-filter). Mapping of sample groups according to the filter type did not reveal a clear trend about these differences (data not shown).
Analysis of the 16S rDNA Clone Libraries
Description of the Global Database
A total of 537 16S ribosomal RNA genes from the nutrient solutions collected upstream and downstream the three filters were sequenced and analyzed. They were grouped into 228 different phylotypes according to phylogenetic uniqueness of the closest describe relative. The majority of them (71%) were related to uncultured bacterial taxa (Supplementary Table 1).
Bacterial clones were successfully assigned to the phylum taxonomic level (percentage of similarities up to 82 to 100%). Twenty of the phyla described [42, 43] were detected and half were candidate divisions for which new etymology has been proposed [44,45,46]. The most represented phyla were Proteobacteria (35% of sequences), mainly α-Proteo (23%) and γ-Proteo (7%); Parcubacteria (OD1) (24%); Microgenomates (OP11) (12%); and Bacteroidetes (8%). The remaining minor phyla accounting each less than 5% of sequences were Verrucomicrobia, Actinobacteria, Saccharibacteria (TM7), Cyanobacteria, Dependentiae (TM6), Armatimonadetes (OP10), Chloroflexi, Planctobacteria, Acidobacteria, Firmicutes, Spirochaetes, Gracilibacteria (BD), and Latescibacteria (WS3) and candidate divisions SR1, WS6, and TG1.
Among sequences having up to 95% of similarity with a closest described relative, 92 were affiliated with a bacterial genus. The two most abundant genera present in the nutrient solution circulation systems of the three filters were identified as Sphingomonas (24 sequences) and Rhizobium (13 sequences) representatives isolated from plant roots [47,48,49]. The others minor genera affiliated were Bosea (8), Pseudomonas (8), Burkholderia (6), Mycobacterium (5), Flavobacterium (4), Aquaspirillum (2), Clostridium (2), Haliscomenobacter (2), Hyphomicrobium (2), Legionella (2), Mesorhizobium (2), Acidovorax, Caedibacter, Cytophaga, Chromobacterium, Dyadobacter, Fluviicola, Hydrogenophaga, Merismopedia, Micrococcus, Microvirga, Spirochaetes, and Zoogloea.
Comparison of the Different Libraries
Firstly, the percentage of similarity between libraries, as given by the Chao Sorensen abundance-based index, indicated that influent solutions of P- and B-filters were closer on each other than with the C-filter (61% against 52 and 48%, respectively) and the same observation was made for effluent solutions (51% against 45 and 32%) (Table 3).
Secondly, the bacteria-amended filters drastically reduced the biodiversity of effluents comparatively to influents: the phylotype diversity, as measured by the ChaoI phylotype richness estimator, was reduced by 81% in the P-filter and by 53% in the B-filter whereas biodiversity was increased by 14% in the control filter (Table 3).
Lastly, the phylogenetic distribution of libraries was clearly distinct in the bacteria-amended filters compared to those of the control filter (Fig. 4). Proteobacteria were more abundant in influent solutions of B- and P-filters than in the C-filter (40 and 41% against 30% of sequences). B- and P-filters reduced the proportion of Proteobacteria in effluent solutions (31 and 21%) whereas the C-filter increased it (42%), especially for α- and β-Proteobacteria. The proportion of candidate phyla without cultivated representatives (candidate division) was strongly enhanced in effluent solutions from bacteria-amended filters, (from 33 to 52% in the B-filter and from 37 to 62% in the P-filter) whereas it decreased in the effluent of the C-filter (from 39 to 28%). If proportions of candidate division OD1 were similar in influent of the three filters (from 27 to 29%), the importance of this phylum was increased in effluent solutions of B- and P-filters whereas it decreased in the C-filter (43 and 41% against 20%, respectively).
Discussion
In soilless cultivation, the disinfection of recycled water is crucial for the control of plant pathogens. Active sterilizing tools such as heat treatment, ozonization, ultraviolet radiation, or chlorination [5, 9, 50, 51] constitute effective methods but destroy both the target pathogens and the non-target microorganisms [2, 11]. On the contrary, several authors reported that slow filtration control targeted pathogens while preserving part of the endogenous microflora [22, 26]. A large part of the slow filtration efficiency was dependent on the formation of bacterial biofilms on filtering substrates [19]. Amendments of filtering columns with bacteria with antagonistic and plant growth-promoting traits were described as a useful method to increase the efficiency of the pathogenic fungi control [22]. However, if the preservation of a natural and non-pathogenic microflora in nutrient solutions is a key point to guarantee a safe microbial ecosystem in closed irrigation systems, little is known about the bacterial population status in these nutrient solutions. The present study is focused on the characterization and comparison of the whole bacterial populations colonizing the nutrient solutions flowing through natural and bacteria-amended filters.
This study confirmed the benefit of bacteria amendment of slow filters, mainly Pseudomonas amendments, for enhancing the plant pathogen elimination [22]. Plate counting of Pythium spp. and Fusarium oxysporum communities revealed their presence in nutrient solutions over the cultivation season. The low amount of Pythium spp. found in influent solution ecosystems (< 102 prop/L) is congruent with the literature [23, 52]. With respect to the control filter that reduced from 56 to 99% the Pythium spp. and Fusarium oxysporum communities, we proved the superior effectiveness of bacteria-amended filters (82–99%), especially at the beginning of filtration. As colonization of filtering media by bacterial populations is known to contribute to the removal of plant pathogens [19, 20], we assume that bacterial amendments stimulate a more rapid bacterial colonization of filtering columns that lead to a more rapid control of pathogenic fungi or oomycetes in the nutrient solutions.
In agreement with Déniel et al. [22], the total bacteria communities flowing in effluents were estimated in the present work at 3–4.104 CFU/mL. This amount of bacteria is relatively low, compared to the average of 105–106 CFU/mL reported in circulating solutions of other tomato-closed soilless systems [24, 25, 52]. In the present experiment, except for the first month of filtration where amended filters released large amounts of bacteria, we showed that the slow filtration reduced the density of the cultivable bacterial population from approximately 1 Log. In comparison with the control filter, bacteria-amended filters induced a stronger and more stable reduction of mesophyllic aerobic bacteria in effluent solutions (68 ± 16 against 91 ± 5%). A slow pozzolana filtration seemed to be more efficient in reducing bacterial communities from nutrient solution than a slow-sand filtration. In the latter, van Os and Postma [26] detected relatively similar numbers of aerobic bacteria communities in the influent and effluent solutions.
The community-level physiological profiling indicated a temporal shift of bacterial populations from nutrient solutions from the beginning to the end of the cultivation season (here a 7-month period). At the beginning of recycling, the metabolism of nutrient solutions was mainly oriented towards carbohydrates. It progressively shifted to degradation of several specific amino acids and carboxylic acids, several of whom are known to be components of root exudates: γ-amino butyric acid, l-alanine, l-aspartic acid, l-serine, l-asparagine, hydroxylproline, citric acid, succinic acid, and malonic acid [53]. In the literature, the addition of maize rhizodeposits in the soil was proved to shift the physiological structure of bacterial microflora [54]. In the same extent, we may assume that the physiological shift observed along months in our study is due to the accumulation in the nutrient solutions of some amino and carboxylic acids released by tomato roots. As Alsanius et al. [55] reported a strong metabolization of amino acids by Pythium spp., the existence of a time-increased competition for amino acids by microbial communities of nutrient solutions likely took place. Moreover, in agreement with van Os and Postma and Postma et al. [10, 26], we recorded distinctive carbon utilization profiles between effluent and influent solutions of slow filters. Our result underlined that these differences were time-dependent, i.e., stronger in autumn than in summer.
The bacterial diversity of nutrient solutions was studied by 16S rDNA PCR-SSCP, and a progressive temporal shift was also recorded in the SSCP patterns of microbial ecosystems. Bacterial populations also proved to be diverse. In their study, Calvo-Bado et al. [52] described that DGGE microbial profiles in nutrient solutions of a small-scale circulating tomato growing system did not vary significantly over their 2 months of analysis. As our study was conducted over a longer period of time (7 months), that can explain the substantial variations we observed.
Another major finding of this SSCP study was the shift between influent and effluent solutions of slow filters observed each month of sampling. Effluent microflora were all the more different from the influent ones when slow filters were bacteria-amended. Thus, bacterial populations of nutrient solution were not only more quantitatively reduced but also more modified in their SSCP structure for amended filters than for the control one.
The sequencing of the clone libraries constructed in this study illustrated the modification of the nutrient solution microflora provided by slow filtration. When solutions were filtered on a natural filter (C-filter), the proportion of Proteobacteria (α- and β-) increased but it decreased after filtration with the two bacteria-amended filters. Conversely, the proportion of uncultured candidate phyla, i.e., candidate division OD1, was reduced in the effluent solutions of the C-filter while it rose in those of P- and B-filters. These differences should be related to the bacterial communities colonizing the substrate of the biofilters. The bacterial communities colonizing the three filters used in this study have already been characterized [27]. The massive addition of bacteria in P- and B-filters was shown to cause a significant microbial shift leading to the development of bacterial populations very different comparing to the ones of the C-filter. Indeed, the microflora of the C-filter mainly consisted of Proteobacteria representatives (69% of sequences) whereas this phylum was considerably less present in P- and B-filters (22 and 22%, respectively). The bacteria-inoculated filters were mostly colonized by Firmicutes representatives, mainly anaerobic strains of Clostridium spp. [27]. We assumed that these differences in microflora between biofilters should have influenced the bacterial communities of nutrient solutions drained out of the filters. This assumption does not mean that the bacterial populations colonizing filters will systematically be present in effluent solution: indeed, if Proteobacteria were abundant both in the C-filter and its effluents, the taxonomic group of Firmicutes was not detected in the effluent of P- and B-filters. The latter were strongly colonized (45 and 48%, respectively) by the candidate division bacteria. The most abundant were OD1: they represented 28% of the bacteria we analyzed in the effluents of the control filter, and 62 and 52% were respectively isolated in the effluents of P- and B-filters. The ecological influence of candidate phylogenetic divisions on nutrient solution ecosystems is still difficult to assess because they have no cultivated members and their physiological properties is unknown.
In a previous study, Berkelmann et al. [24] have biochemically identified 160 bacterial strains isolated from nutrient solutions of a hydroponic system of tomatoes. They were distributed among five taxonomic groups: Actinobacteria, Bacteroidetes, α-Proteobacteria, β-Proteobacteria, and γ-Proteobacteria. We confirmed here the presence of all these phyla in the microflora colonizing the nutrient solutions from another soilless culture, but numerous additional taxonomic groups were identified: Acidobacteria, Chlorobi, Chloroflexi, Cyanobacteria, Firmicutes, Gemmatimonadetes, Lentisphaerae, Planctomycetes, δ-Proteobacteria, Spirochaetes, and Verrucomicrobia, and candidate divisions BRC1, OD1, OP10, OP11, OP3, SR1, TM6, TM7, and BD1-5. The bacterial diversity described by the authors [24] was certainly underestimated because of the use of culture media for bacteria isolation. Indeed, cultured microorganisms represented only a small fraction of the microbial diversity compared to the abundance of environmental species. Calvo-Bado et al. [20] pointed out that DGGE-dominating bands present in slow filter ecosystems represented less than 2% of the cultivable population. In the present study, 86% of sequences identified were affiliated to uncultured bacteria. The status of these microorganisms regarding the plants, either neutral, plant-protective, or plant-pathogen, is unknown. For these bacteria, their usefulness to create a suppressive microflora in nutrient solutions over time is unknown too, but the absence of disease on the root of tomatoes suggests they are not pathogenic and may be involved in the protection of root systems. Further studies aimed at studying this key point for plant protection in soilless culture has to be carried out.
To conclude, the occurrence of a diverse microflora in the nutrient-solution effluents of biofilters raises the question of their impact on the microbial ecosystems of the tomato rhizosphere. As the solutions irrigate the root systems, part of their bacterial microflora has the opportunity to colonize the roots; consequently, shifts in the microflora of the circulating solutions may induce shifts in the rhizospheric microflora. As suggested by Vallance et al. [56], a relationship between microflora from either roots or solutions exists even if the links between the two are difficult to determine. Regarding the suppressiveness activity of this microflora, the shift in the bacterial communities over time suggests that its management is difficult to achieve by using biofilters. For instance, it will be not be possible to promote a specific bacterial species, e.g., Pseudomonas, by using biofiltration. However, as the diversity of the bacterial species in the effluent is high, the presumably non-pathogenic activity of most of them and the interactions that occur between them suggest that it will be difficult for a plant pathogenic bacteria or fungi to settle and become predominant in the effluent solutions. In that sense, the bacterial diversity observed in the various effluents is likely useful for the management of a potentially suppressive microflora.
References
Stanghellini ME, Rasmussen SL (1994) Hydroponics, a solution for zoosporic pathogens. Plant Dis. 78:1129–1138
Vallance J, Déniel F, Le Floch G, Guérin-Dubrana L, Blancard D, Rey P (2011) Potentially pathogenic and beneficial microorganisms in soilless cultures. A review. Agron. Sustain. Dev. 31:191–203
Davies JML (1980) Diseases in NFT. Acta Hort 98:299–305
Evans SG (1979) Susceptibility of plants to fungal pathogens when grown by the nutrient-film technique (NFT). Plant Pathol. 28:45–48
Runia WT (1995) A review of possibilities for disinfection of recirculation water from soilless cultures. Acta Hort 382:221–229
Paulitz TC (1997) Biological control of root pathogens in soilless and hydroponic systems. Hortscience 32:193–196
McPherson GM, Harriman MR, Pattison D (1995) The potential for spread of root diseases in recirculating hydroponic systems and their control with disinfection. Meded Fac Landbouwwet Univ Gent 60/2b:371–379
Van OEA (1999) Closed soilless growing systems: a sustainable solution for Dutch greenhouse horticulture. Water Sci. Technol. 39:105–112
Ehret DL, Alsanius B, Wohanka W, Menzies JG, Utkhede R (2001) Disinfestation of recirculating nutrient solutions in greenhouse horticulture. Agronomie 21:323–339
Postma J, Willemsen de Klein MJEIM, van Elsas FD (2000) Effect of the indigenous microflora on the development of root and crown rot caused by Pythium aphanidermatum in cucumber grown on rockwool. Phytopathology 90:125–133
Zhang W, JC T (2000) Effect of ultraviolet disinfection of hydroponic solutions on Pythium root rot and non-target bacteria. Eur. J. Plant Pathol. 106:415–421
Tu JC, Papadopoulos AP, Hao X, Zheng J (1999) The relationship of Pythium root rot and rhizosphere microorganisms in a closed circulating and an open system in rockwool culture of tomato. Acta Hort 481:577–583
Chave M, Dabert P, Brun R, Godon JJ, Poncet C (2008) Dynamics of rhizoplane bacterial communities subjected to physicochemical treatments in hydroponic crops. Crop Prot. 27:418–426
Postma J, Geraats BPJ, Pastoor R, van Elsas JD (2005) Characterization of the microbial community involved in the suppression of Pythium aphanidermatum in cucumber grown on rockwool. Phytopathology 95:808–818
Ellis KV (1985) Slow sand filtration. Crit Rev Environ Contr 15:315–354
Wohanka W (1995) Disinfection of recirculating nutrient solutions by slow sand filtration. Acta Hort 382:246–255
Rey P, Picard K, Déniel F, Benhamou N, Tirilly Y (1999) Development of an IPM sytem in soilless culture by using slow sand filtration and a biocontrol fungus, Pythium oligandrum. IOBC Bulletin 22:205–208
Van OEA, Amsing JJ, van Kuik AJ, Willers H (1999) Slow filtration: a potential method for the elimination of pathogens and nematodes in recirculating nutrient solutions from glasshouse-grown crops. Acta Hort 481:519–526
Brand T, Wohanka W (2001) Importance and characterization of the biological component in slow filters. Acta Hort 554:313–321
Calvo-Bado LA, Pettitt TR, Parsons N, Petch GM, Morgan JAW, Whipps JM (2003) Spatial and temporal analysis of the microbial community in slow sand filters used for treating horticultural irrigation water. Appl. Environ. Microbiol. 69:2116–2125
Renault D, Tirilly Y, Benizri E, Sohier D, Barbier G, Rey P (2007) Characterization of Bacillus and Pseudomonas strains with suppressive traits isolated from tomato hydroponic-slow filtration unit. Can. J. Microbiol. 53:784–797
Déniel F, Rey P, Chérif M, Guillou A, Tirilly Y (2004) Indigenous bacteria with antagonistic and plant-growth-promoting activities improve slow-filtration efficiency in soilless cultivation. Can. J. Microbiol. 50:499–508
Déniel F, Renault D, Tirilly Y, Barbier RP (2006) Dynamic filtration in tomato soilless greenhouse: evolution of microbial communities on filtering media and control of potentially suppressive and pathogenic microorganisms. Agron. Sustain. Dev. 26:185–193
Berkelmann B, Wohanka W, Wolf GA (1994) Characterization of the bacterial flora in circulating nutrient solutions of a hydroponic system with rockwool. Acta Hort 361:372–381
Koohakan P, Ikeda H, Jeanaksorn T, Tojo M, Kusakari SI, Okada K, Sato S (2004) Evaluation of the indigenous microorganisms in soilless culture: occurrence and quantitative characteristics in the different growing systems. Sci. Hortic. 101:179–188
Van OEA, Postma J (2000) Prevention of root diseases in closed soilless growing systems by microbial optimisation and slow sand filtration. Acta Hort 532:97–102
Renault D, Vallance J, Déniel F, Wery N, Godon JJ, Barbier G, Rey P (2012) Diversity of bacterial communities that colonize the filter units used for controlling plant pathogens in soilless cultures. Microb Ecol 63:170–187
Jeffers SN, Martin SB (1986) Comparison of two media selective for Phytophthora and Pythium species. Plant Dis. 70:1038–1043
Komada H (1975) Development of a selective medium for quantitative isolation of Fusarium oxysporum from natural soils. Rev Plant Prot Res 8:114–125
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 57:2351–2359
Garland JL (1996) Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol. Biochem. 28:213–221
Grove JA, Kautola H, Javadpour S, Moo-Young M, Anderson WA (2004) Assessment of changes in the microorganism community in a biofilter. Biochem. Eng. J. 18:111–114
Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R (1997) Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802–2813
Zemb O, Haegeman B, Delgenes JP, Lebaron P, Godon JJ (2007) SAFUM: statistical analysis of SSCP fingerprints using PCA projections, dendrograms and diversity estimators. Mol. Ecol. Notes 7:767–770
Fox J (2005) The R Commander: a basic statistics graphical user interface to R. J Stat Softw 14(9):1–42
Ewing B, Hillier L, Wendl M, Green P (1998) Basecalling of automated sequencer traces using phred. Genome Res. 8:175–194
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J. Mol. Biol. 215:403–410
Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, Ludwig W, Glöckner FO, Roselló-Móra R (2008) The all-species living tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31:241–250
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Colwell RK (2005) EstimateS version 7.5: statistical estimation of species richness and shared species from samples; user’s guide and application published at http://purl.oclc.org/estimates
Chao A, Chazdon RL, Colwell RK, Shen TJ (2005) A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 8:148–159
Rappé MS, Giovannoni SJ (2003) The uncultured microbial majority. Ann Rev Microbiol 57:369–394
Harris JK, Kelley ST, Pace NR (2004) New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845–849
Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437
Ferrari B, Winsley T, Ji M, Neilan B (2014) Insights into the distribution and abundance of the ubiquitous Candidatus Saccharibacteria phylum following tag pyrosequencing. Sci. Rep. 4(3957):1–9
Yeoh YK, Sekiguchi Y, Parks DH, Hugenholtz P (2015) Comparative genomics of candidate phylum TM6 suggests that parasitism is widespread and ancestral in this lineage. Mol Biol Evol 33:915–927
Oyaizu H, Matsumoto S, Minamisawa K, Gamou T (1993) Distribution of rhizobia in leguminous plants surveyed by phylogenetic identification. J. Gen. Appl. Microbiol. 39:339–354
Gao J, Terefework Z, Chen W, Lindström K (2001) Genetic diversity of rhizobia from Astragalus adsurgens growing in different geographical regions of China. J. Biotechnol. 91:155–168
Ogita N, Hashidoko Y, Limin SH, Tahara S (2006) Linear 3-hydroxybutyrate tetramer (HB4) produced by Sphingomonas sp. is characterized as a growth promoting factor for some rhizomicrofloral composers. Biosci. Biotechnol. Biochem. 70(9):2325–2329
Steinberg C, Moulin F, Gaillard P, Gautheron N, Stawiecki K, Bremeersch P, Alabouvette C (1994) Disinfection of drain water in greenhouses using a wet condensation heater. Agronomie 14:627–635
Rey P, Déniel F, Vasseur V, Benhamou N, Tirilly Y (2001) Evolution of Pythium spp. populations in soilless cultures and their control by active disinfecting methods. Acta Hort 554:341–348
Calvo-Bado LA, Petch G, Parsons NR, Morgan JAW, Pettitt TR, Whipps JM (2006) Microbial community responses associated with the development of oomycete plant pathogens on tomato roots in soilless growing systems. J. Appl. Microbiol. 100:1194–1207
Campbell CD, Grayston SJ, Hirst DJ (1997) Use of rhizosphere carbon sources in sole carbon source tests to discriminate soil microbial communities. J. Microbiol. Methods 30:33–41
Benizri E, Dedourge O, Dibattista-Leboeuf C, Piutti S, Nguyen C, Guckert A (2002) Effect of maize rhizodeposits on soil microbial community structure. Appl. Soil Ecol. 21:261–265
Alsanius BW, Khalil S, Hultberg M (1998) Biochemical and chemical characterization of Pythium ultimum. Meded Fac Landbouwwet Univ Gent 63/3a:891–897
Vallance J, Déniel F, Barbier G, Guérin-Dubrana L, Benhamou N, Rey P (2012) Influence of Pythium oligandrum on the bacterial communities that colonize the nutrient solutions and the rhizosphere of tomato plants. Can. J. Microbiol. 58:1124–1134
Acknowledgements
This project was supported by the Regional Councils of Brittany and Pays de la Loire and by the French Ministry of Research.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Supplementary Table 1
Abundance of 16S–ribosomal RNA gene phylotypes in the clone libraries constructed from nutrient solutions sampled from influent and effluent of C-filter (iClib, eClib), B-filter (iBlib, eBlib) and P-filter (iPlib, ePlib). For each phylotype affiliation, the closest described relative from the GenBank database with its accession number (in brackets) and the percentage of similarity as given by the Blast program are provided. (DOCX 172 kb)
Rights and permissions
About this article
Cite this article
Renault, D., Déniel, F., Vallance, J. et al. Bacterial Shifts in Nutrient Solutions Flowing Through Biofilters Used in Tomato Soilless Culture. Microb Ecol 76, 169–181 (2018). https://doi.org/10.1007/s00248-017-1117-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-017-1117-5