Abstract
Natural occurring groundwater with abnormally high ammonium concentrations was discovered in the aquifer-aquitard system in the Pearl River Delta, South China. The community composition and abundance of aerobic/anaerobic ammonia/ammonium-oxidizing microorganisms (AOM) in the aquitard were investigated in this study. The alpha subunit of ammonia monooxygenase gene (amoA) was used as the biomarker for the detection of aerobic ammonia-oxidizing archaea (AOA) and bacteria (AOB), and also partial 16S rRNA gene for Plantomycetes and anaerobic ammonium-oxidizing (anammox) bacteria. Phylogenetic analysis showed that AOA in this aquitard were affiliated with those from water columns and wastewater treatment plants; and AOB were dominated by sequences among the Nitrosomonas marina/Nitrosomonas oligotropha lineage, which were affiliated with environmental sequences from coastal eutrophic bay and subtropical estuary. The richness and diversity of both AOA and AOB communities had very little variations with the depth. Candidatus Scalindua-related sequences dominated the anammox bacterial community. AOB amoA gene abundances were always higher than those of AOA at different depths in this aquitard. The Pearson moment correlation analysis showed that AOA amoA gene abundance positively correlated with pH and ammonium concentration, whereas AOB amoA gene abundance negatively correlated with C/N ratio. This is the first report that highlights the presence with low diversity of AOM communities in natural aquitard of rich ammonium.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Microbial nitrification plays an important role in the biogeochemical nitrogen cycle, which is a key process for ammonia oxidization to nitrate via nitrite. Ammonia-oxidizing bacteria (AOB) was once considered as the only contributor responsible for ammonia oxidation in the environment [1] and there were two groups of AOB, β-proteobacteria such as the genera Nitrosomonas and Nitrosospira and γ-proteobacteria like genus Nitrosococcus [2]. A number of studies showed that β-AOB were predominant in both the wastewater treatment plants and natural environments among the AOB communities [3]. Recently, the discoveries of both aerobic ammonia-oxidizing archaea (AOA) and anaerobic ammonium oxidizing (anammox) bacteria have updated our knowledge about the microbial nitrogen cycle [4, 5].
Some archaea in the Crenarchaeota were discovered to be able to carry out chemoautotrophic ammonia oxidation [6] and the first autotrophic ammonia-oxidizing marine archaeon, Nitrosopumilus maritimus SCM1 clone was isolated [4]. Soon after, AOA were found to be widely distributed and detected in various habitats, including estuary, coast wetlands, marine, soils, lake sediments, hot springs, wastewater treatment plants, and forest soils [7–23]. According to the genome studies of both AOA and AOB, ammonia-oxidizing genes (amo) were found to encode the ammonia monooxygenase responsible for ammonia-oxidation in all strains of both AOA and AOB [4, 24]. As a result, the corresponding α subunit of amo gene (amoA) in AOA and AOB was used as a molecular biomarker for the detection of AOA and AOB communities in environmental samples.
The relative copy numbers and dominance of bacterial and archaeal amoA genes in specific environments are still debatable. Many reports found that the copy number of archaeal amoA genes were higher than that of bacterial amoA genes in different environments like fertilized soils, sandy loam, rhizosphere paddy soil, low-oxygen fresh and brackish estuary sediment and semiarid soils [7, 25–30]. However, others showed that the archaeal amoA gene copy numbers were lower than that of bacterial amoA in various environments including estuary sediment, soils under stands of red alder, lake sediments and activated sludge bioreactor [8, 14, 31–33]. As a result, more investigations are required to elucidate if AOA is really the dominant ammonia-oxidizing archaea and bacteria in soils.
Anammox bacteria were first discovered in 1995 from the wastewater treatment plants where ammonium and nitrite disappeared and nitrogen gas appeared which could not be accounted for through denitrification [34, 35]. The anammox reaction is a chemolithotrophic process in which ammonium is coupled with nitrite to yield nitrogen gas in the absence of molecular oxygen [36]. Subsequently, five genera of the anammox bacteria were reported including Candidatus Kuenenia [37], Ca. Scalindua, Ca. Brocadia [38], Ca. Anammoxoglobus [39], and Ca. Jettenia [40]. Anammox bacteria have been detected in marine, coastal and estuarine sediments [41, 42], mangroves [43, 44], permafrost soil [45], oxygen minimum zones and anoxic basins [46], sea-ice [47], freshwater lakes and rivers [48, 49], and agricultural soils [50].
Pearl River Delta Estuary of China is one of the most complex large-scale estuarine systems, which consists of three sub-estuary bays, the Lingdingyang, the Modaomen and the Huangmaohai, and a reticulated network system [51]. The delta is surrounded by three major tributaries: Xijiang, Beijiang and Dongjiang, and started to form since late Pleistocene with annual average river freshwater discharge of 3.26 × 1011 m3 yr−1 [52]. But the current delta network system was the result of middle Holocene and with the effect of world-wide transgression [53]. The average sedimentation rate of constructing the estuary land was at around 1.8 mm annually and hence the formation of the estuary required a long historic evolutional process via different geographic effects [53, 54].
In a recent study, in order to understand the spatial distribution of the ammonium in the aquifer and the mechanism of abnormally high ammonium groundwater in the Pearl River Delta, a total of 40 boreholes were drilled and core samples of the aquitard and groundwater samples in the basal aquifer were collected [55]. The results showed that the aquifer-aquitard system contains an exceptionally high ammonium concentration. This ammonium was naturally originated in the overlying organic-rich Holocene-Pleistocene aquitard [55]. Hence, it is interesting to explore the status of ammonium-related microorganisms in this special habitat. In the present study, aerobic/anaerobic ammonia/ammonium-oxidizing microorganisms (AOM, including AOA, AOB and anammox bacteria) in the high nitrogen environment in the aquitard were for the first time investigated to understand these microorganisms in such environment.
Materials and Methods
Sample Collection
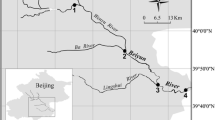
The sediment samples were collected from the cores of the borehole SDZK14 (N 22° 44′ 41″, E 113° 21′ 59″) drilled in the aquifer-aquitard to the north of Zhongshan, Guandong, China (Fig. 1). The borehole was ca. 50 m deep, drilled by the Department of Earth Sciences, The University of Hong Kong in January of 2008. The samples were immediately put onto ice bags in a heat-insulated cooler after collection and transported to the laboratory. Three sediment depths at 6.9, 23.5 and 37.4 m from the ground level were sampled for this study. Each sample was homogenized and split into three equal parts for chemical analysis, DNA extraction and stored at −20 °C.
The distribution of the ammonium-rich groundwater with NH4 + >10 mg/L (in brown) and the location of SDZK14 (N 22° 44′ 41″, E 113° 21′ 59″) in the Pearl River Delta. The white area is a fluvial and alluvial platform about 5–10 m in elevation and the grey areas are mountain areas with higher elevation (simplified from Jiao et al., 2010)
Environmental Physicochemical Analyses
Depth and pH of the sediments were measured in situ at various sediment depths with microelectrodes using the modified protocol at boreholes [56]. Five grams of each sediment sample was extracted with 2.0 M KCl by incubation at 150 rpm at 20 °C on an incubation shaker (Innova 4340, New Brunswick Scientific) overnight. Parameters including ammonium-N, nitrate-N, and nitrite-N were measured with an autoanalyzer (QuikChem, Milwaukee, WI) using standard flow injection analysis (FIA) technique according to the standard extraction procedures.
DNA Extraction and PCR Amplification
Total genomic DNA was extracted from 100 to 200 mg of sediment samples using the SoilMasterTM DNA Extraction Kit, according to the manufacturer’s protocol (Epicentre Biotechnologies, Madison, WI). Soil DNA was amplified with PCR primers targeting different genes (primers listed in Table 1).
In order to amplify the anammox 16S rRNA gene using the first pair of PCR primer set (Brod541F/Amx820R), the initial PCR amplification was performed with Pla46F/1037R which corresponded to one of the Planctomycetes 16S rRNA genes and 23S rRNA genes, respectively (Table 1) with GoTaq® DNA Polymerase (Promega, Madison, WI) in a 25-μl volume containing 10 μl of 5× colorless GoTaq® Flexi Buffer, 25 mM of MgCl2, 10 mM of each deoxyribonucleoside triphosphate, 2 μM of each primer, 1.25 U GoTaq® DNA Polymerase, 40 ng ml−1 BSA and 3 μl of DNA as templates (10–100 ng). When amplifying the anammox 16S rRNA gene using the second pair of PCR primer set (Amx368F/Amx820R), the initial PCR amplification was performed with Pla46F/1037R primers. The nested PCR was performed with the anammox-specific primers, Amx368F/Amx820R. The nested PCR condition was identical to that for Pla46F/1037R primers, except 1 min of extension at 72 °C and the cycle number increased to 36. The compositions of archaeal amoA and bacterial amoA PCR in 25 μl reaction mixture were identical to that of anammox 16S rDNA, except using different PCR primer pairs. The amplifying protocols are listed in Table 1.
Clone Library Construction and Phylogenetic Analysis
Purified DNA fragments were ligated into a pMD18-T vector (TaKaRa, Japan) and transformed into competent Escherichia coli DH5α cells. The library clones were screened directly by PCR for the presence of inserts using two M13 universal primers, M13F/M13R. The positive clones from each library were randomly selected and the PCR products were purified using a PCR Purification Kit (Qiagen, USA). The PCR products were sequenced by Tech Dragon Ltd (Hong Kong). DNA sequences were examined and edited using BioEdit (Tom Hall, North Carolina State University, NC) and MEGA, version 5.2 [60]. NCBI BLAST (http://www.ncbi.nih.gov) was used to find the closest related 16S rRNA gene and amoA gene sequences in the GenBank. The multiple alignments of partial 16S rRNA gene and amoA gene sequences were done and neighbor-joining phylogenetic trees with bootstrap values based on 1000 replications [61] were produced using MEGA program, version 5.2.
Quantitative PCR Assay
Quantitative PCR (q-PCR) was performed on ABI 7000 Real Time PCR System (Applied Biosystems, USA) to estimate the abundance of ammonia oxidizing archaea and bacteria in all three samples. The quantification was based on the fluorescent dye SYBR-Green I, which binds to double-stranded DNA during PCR amplification. In the process, bacterial and archaeal amoA gene were amplified by the same PCR primer as above. Archaeal and bacterial amoA gene fragments were cloned into pMD18-T plasmids (TaKaRa) and were extracted with QIAprep® Spin Miniprep Plasmid Kit (Qiagen, USA) according to the manufacturer’s protocols. The plasmid concentration was detected by UV-Visible Biophotometer (Eppendorf, Germany).
Tenfold serial dilution was carried out with the known copy number of plasmid of the archaeal and bacterial amoA genes and generated the standard curve over six orders of magnitude. The 25 μl archaeal amoA q-PCR reaction mixture contained 1–10 ng of DNA, 40 ng ml−1 BSA, 0.2 μM of each primer and 12.5 μl of FastStart Universal SYBR Green Master (ROX) (Roche). The modified q-PCR protocol was as follows: 3 min at 94 °C, followed by 50 cycles of 30 s at 94 °C, 1 min at 53 °C with the extension step of 1 min at 72 °C with fluorescence detection at the end of each cycle [13]. Bacterial amoA q-PCR was carried out in 25 μl reaction mixture with 1–10 ng of DNA, 40 ng ml−1 BSA, 0.2 μM of each primer and 12.5 μl of FastStart Universal SYBR Green Master (ROX) (Roche). The modified protocol was as follows: 3 min at 94 °C, followed by 50 cycles of 30 s at 94 °C, 30 s at 55 °C with 45 s extensions at 72 °C with fluorescence detection at the end of each cycle [59]. The intensity of fluorescence was measured at 83 °C and the melting curve analysis was carried out to confirm the specificity of the q-PCR products. The standard curves for q-PCR assays and data analysis were generated with ABI Prism 7000 SDS software (version 1.1).
Statistical Analysis
The DOTUR (Distance-Based OTU and Richness) program was employed to compare diversity for each anammox 16S rRNA and amoA sequence from each sample [62]. Operational taxonomic units (OTU) for community analysis were defined by a 3 % sequence variation in detecting anammox bacteria, AOA and AOB. DOTUR was also used to generate diversity analyses such as coverage, Chao1, Simpson index (D) and Shannon index (H) for each site. The H and D indices were calculated as the diversity indices by the program [62, 63]. The estimated coverage of the constructed anammox related 16S rRNA, archaeal amoA gene and bacterial amoA gene libraries were calculated using the formula of C = [1-(n1/N)] × 100, where n1 represents the number of OTUs detected in one clone library and N stands for the total number of clones in that particular library. This coverage estimates the probability that all the unique sequences present in a given sample were represented at least once in the library [64, 65]. Correlation analysis between the abundance of archaeal and bacterial amoA genes and environmental variables were conducted using Microsoft Excel program.
Nucleotide Sequence Accession Numbers
All the amoA genes sequences of ammonia oxidizing bacteria and archaea, and 16S rRNA gene sequences of anammox bacteria determined in this study were deposited in GenBank under accession numbers HM537238 to HM537467.
Results
Physicochemical Characteristics of Sediment Samples
The stratigraphy of the sampling site consisted of over 30 m of Quaternary aquitard of silty sediments. Detailed nutrient concentrations, C/N ratios and other chemical parameters of the three samples from SDZK14 are listed in Table 2. Samples from the site were enriched in ammonium (NH4 +), ranged from 144.8 to 568.6 mg kg−1 dry weight. The pH values of the three sediment samples were between neutral to slightly alkaline (from 6.98 to 7.79). The salinity of the groundwater in the aquifer below 40 m depth was 15.9‰ which was defined as intermediate salinity within the range of the subterranean estuary brackish waters [26]. The nitrite concentration remained low among the three samples with a range from 6.69 to less than 0.1 mg kg−1 dry weight which is even under the detection limit and did not show any characteristic trend. However, the concentrations of nitrate were relatively much higher from 124.3 to 142.1 mg kg−1 dry weight, showing a decreasing trend with the depths.
Effect of Depth on Phylogenetic Distribution and Diversity of AOA, AOB and Anammox
The operational taxonomic unit (OTU) was defined based on a 3 % sequence difference cut-off to represent the phylotype diversity of anammox bacteria, AOA and AOB (Table 3). The estimated coverage of the clone libraries was high enough to represent the majority of AOM in the samples. The diversity of AOA in the high ammonium aquitard sediment samples was generally low (Fig. 2a), all archaeal amoA gene sequences fell into the sub-cluster AI which is affiliated with the Water column/sediment sequences [13, 66]. A few archaeal amoA sequences were grouped together and formed a sub-cluster AII and three representatives of culturable archaeal isolates formed the soil/sediment clade (cluster B), defined by two previous studies [66, 67]. All cluster classifications were well supported by high bootstrap value (>96 %). Majority of the clone sequences (84 %) in sub-cluster AI were related to the environmental clone (GQ414591) found from natural drinking water column of Dongjiang River of the Pearl River Delta and the rest 16 % clone sequences were all from 6.9 m depth where the closest representative relative was Ca. Nitrosopumilus maritimus (EU239959) from tropical marine aquarium tank belonging to the ubiquitous marine group 1 Crenarchaeota [4]. Another closely related clone was an environmental archaeal amoA clone (EU860282) recovered from wastewater treatment plants [68]. Surprisingly, none of the clones was closely related with terrestrial clones but all were the same or similar to AOA detected in water related environment.
Phylogenetic relationship of the major phylogenetic groups of AOA (a), AOB (b), and anammox bacteria (c). The numbers at the bracket are the clone number of the related phylotype. The numbers at the nodes are percentages that indicate the levels of bootstrap support based on 1000 resample data sets (only values greater than 50 % are shown). Branch lengths correspond to sequence differences as indicated by the scale bar. For Fig. 2a, the clustering reference was defined elsewhere [13, 66, 67]. For Fig. 2c, sequences amplified by Brod541F/Amx820R primers are labeled with filled triangle and those by Amx368F/Amx820R primers are labeled with filled squares correspondingly
Evenly distributed and low diversity of AOB in the high ammonium aquitard sediment was detected (Fig. 2b). The distribution of AOB communities was uniform in general among the three depths based on the results in the three AOB clone libraries constructed. All sequences belonged to Nitrosomonas marina/Nitrosomonas oligotropha lineage [69, 70] instead of Nitrosospira group which was only found in acidic soils with between pH values from 4 to 6 [71]. Previous studies reported that Nitrosomonas marina/Nitrosomonas oligotropha lineage was dominant in wastewater treatment sludge and bioreactors [31, 72–74], in wastewater contaminated soil and freshwater sediments [70, 75]. Some species in this lineage adapted to the environment with high ammonium concentrations [76–79] as they have high ammonium affinity with Km values between 0.075 and 0.03 mM [79]. It was divided into two sub-clusters among the defined lineage, with 29 % of all sequenced clones (30 out of the 104 clones) in sub-cluster B closely associated with environmental clones from coastal eutrophication bay (EU244543) (unpublished data) and eutrophic subtropical estuary [63]. But 71 % of those (74 out of the 104 clones) were closely related to Nitrosomonas marina (AAG37813) [70] which was firstly retrieved from sea water, South Pacific [77] in cluster A. Both sub-clusters A and B provided evidence that AOB communities in these samples were related to the eutrophic environment and the libraries were dominated by sequences related to Nitrosomonas, which supports high ammonia concentration has a greater benefit for the Nitrosomonas species growth than that of Nitrosospira [80, 81].
From each individual clone library constructed, there were 1 and 2–3 anammox bacterial 16S rDNA OTUs using PCR primers Brod541F/Amx820R and Amx368F/Amx820R, respectively; 1–2 archaeal amoA OTUs and 2 bacterial amoA OTUs. Among the anammox bacteria clone libraries, the highest diversity of anammox bacteria was found at 23.5 and 37.4 m depths using both anammox PCR primer sets. At 6.9 m depth, anammox bacteria were below detection using the PCR primer set Brod541F/Amx820R; however, two anammox OTUs were found using the PCR primer set Amx368F/Amx820R. On the other hand, the highest diversity of archaeal amoA was detected at depth 6.9 m and evenly distributed bacterial amoA phylotypes among different depths, while the lowest diversity of anammox 16S rRNA genes was at 6.9 m depth (Table 3).
The phylogenetic study of the anammox bacterial community was based on PCR amplification of anammox related 16S rDNA in the samples after extraction. Initial PCR amplification did not result in any sequence with the primers Pla46F/1037R. However, in the subsequently nested PCR amplification reactions, PCR products with a size of 452 bp were obtained from all three samples using primers Amx368F/Amx820R. By amplifying anammox 16S rRNA gene using primers Brod541F/Amx820R, for sample at 6.9 m depth none of the sequences retrieved was related to any of the known anammox bacteria but all the amplicons fell into the Actinobacteria phylum; for samples at depths of 23.5 and 37.4 m, all sequences were affiliated with one anammox cluster, Ca. Scalindua group associated with the clones found in the marsh and river sediment.
From the phylogenetic tree constructed with 16S rDNA sequences recovered using primers Amx368F/Amx820R and Brod541F/Amx820R (Fig. 2c), two clusters amplified by Amx368F/Amx820R were detected in the three clone libraries. All clone sequences in the three samples fell into the two clades, Ca. Scalindua brodae/sorokinii clade and Ca. Scalindua sea sediment-like sequences clade. The closely related cultured relative to the sequenced clones was the annmmox bacterium isolate Ca. Scalindua brodae Strain EN 8 [38] with a high similarity from 93 to 99 %.
Effect of Depth on Abundance of AOA, AOB
The abundances of AOA and AOB were estimated by quantifying the amoA gene copy numbers. The highest archaeal amoA gene copy number was detected in depth of 23.5 m at 1.30 × 105 copies g−1 of dry soil and the lowest archaeal amoA gene copy number was found in depth of 37.4 m, which was eight times lower than that of 23.5 m depth (Fig. 3). The highest bacterial amoA gene copy numbers was found in depth of 23.5 m at 2.89 × 105 copies per gram of dry sediment and the depth of 6.9 m had the lowest bacterial amoA gene copy numbers with two times lower than that of depth of 23.5 m (Fig. 3). Surprisingly, the copy numbers of bacterial amoA gene in all samples ranged from 1.33 × 105 to 2.89 × 105 copies g−1 of dry sediment were higher than those of archaeal amoA copy numbers ranged from 1.62 × 104 to 1.30 × 105 copies g−1 of dry sediment. The AOA/AOB amoA gene ratios in 6.9, 23.5 and 37.4 m were 0.34, 0.45 and 0.07, respectively.
Archaeal amoA was much more abundant than bacterial amoA in many different environments, for example, semiarid soil, pristine and agricultural soils in climatic zone and sandy soil, and rhizosphere [25, 28–30, 66]. However, in this study the bacterial amoA, rather than archaeal amoA, was predominant in the high ammonium sediment. This is in the consistency with some other reports that AOB amoA copy numbers were greater than AOA amoA in some habitats [8, 14, 26, 32, 82]. AOB was thought to be obligate aerobes [83] before studies showed that Nitrosomonas species in AOB could survive under anoxic conditions [84, 85]. Though AOA appeared to be capable of survival in suboxic condition [9, 10], the role and effectiveness of AOA in nitrifier denitrification under low aeration environment are still unclear [86].
Effect of Environmental Variables on Diversity and Abundance of AOA, AOB and Anammox
The archaeal amoA abundance had a significant positive relationship with pH (Pearson correlation r = 0.97, p < 0.05) and ammonium (Pearson correlation r = 0.96, p < 0.05) (Table 4). The bacterial amoA abundance had a significant negative relationship with C/N ratio (Pearson correlation r = −0.96, p < 0.05). The ratios of AOA to AOB ranged from 0.07 to 0.45 and showed a significant positive correlation to the pH (Pearson correlation r = 0.97, p < 0.05).
Oxygen concentration might affect the sizes of the ammonia-oxidizer communities when low oxygen was available across the depth of sediment [87]. The effect of depth to the copy number of AOA amoA gene did not change significantly comparing to that of AOB amoA gene in either fertilized or unfertilized soils [28]. These previous findings might support higher amoA gene abundance of AOB over AOA detected in anoxic sediments in this study with the evidence of better utilization of ammonia by AOB under anoxic conditions.
The bacterial amoA gene copy numbers had a slightly positive correlation with the concentration of ammonia (Pearson correlation r = 0.82, p < 0.05), but a negative correlation with nitrite concentration (Pearson correlation r = 0.92, p = 0.01). It suggested that ammonia input could be a key factor determining the AOB abundance in natural soil environment, consistent with the results found in another study [27].
The inverse correlation of AOB bacterial amoA gene copy number with nitrite concentration in this study agreed with the study of activated sludge bioreactor published recently [31]. It was previously reported that a higher level of nitrite accumulation than this study would inhibit the ammonia-oxidizing activity of AOB by causing the loss of AMO activity under both aerobic and anaerobic conditions and more activity was lost under alkaline than under acidic conditions [31, 88, 89].
The AOB amoA copy number negatively correlated to the C/N ratios (Pearson correlation r = 0.96, p < 0.05), consistent with the results found in an estuary bay [32], freshwater sediment [67] and semiarid soils [25]. It was suggested that the negative relationship between AOB and soil C/N ratio was due to heterotrophs and AOB have high demand of nitrogen and face competition of AOA for available substrates under high C/N conditions [25].
Conclusion
Lower amoA gene abundance of AOA than that of AOB was detected, and anammox bacteria of low diversity at this site were dominated by Ca. Scalindua-related phylotypes. Among the AOA community, all archaeal amoA gene sequences were affiliated with the water column/sediment sequences. For the AOB community, only Nitrosomonas-like sequences with very low diversity were detected, all grouped into the Nitrosomonas marina/Nitrosomonas oligotropha lineage. The archaeal amoA abundance had a significant positive relationship with pH and ammonium, while the bacterial amoA abundance had a significant negative relationship with C/N ratio. The ammonia-oxidizing microorganism communities might cooperate with each other forming a stable dynamic partnership under the stresses of rich ammonia and low oxygen conditions in the aquitard.
References
Otte S, Schalk J, Kuenen J, Jetten M (1999) Hydroxylamine oxidation and subsequent nitrous oxide production by the heterotrophic ammonia oxidizer Alcaligenes faecalis. Appl Microbiol Biotechnol 51(2):255–261
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55(1):485–529. doi:10.1146/annurev.micro.55.1.485
Nold SC, Zhou J, Devol AH, Tiedje JM (2000) Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the β subdivision of the Proteobacteria. Appl Environ Microbiol 66(10):4532–4535. doi:10.1128/aem.66.10.4532-4535.2000
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437(7058):543–546
Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MSM (1999) Missing lithotroph identified as new planctomycete. Nature 400(6743):446–449
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y-H, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304(5667):66–74. doi:10.1126/science.1093857
Chen X-P, Zhu Y-G, Xia Y, Shen J-P, He J-Z (2008) Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ Microbiol 10(8):1978–1987
Jiang H, Dong H, Yu B, Lv G, Deng S, Berzins N, Dai M (2009) Diversity and abundance of ammonia-oxidizing archaea and bacteria in Qinghai Lake, Northwestern China. Geomicrobiol J 26(3):199–211
Wang Y-F, Gu J-D (2013) Higher diversity of ammonia/ammonium-oxidizing prokaryotes in constructed freshwater wetland than natural coastal marine wetland. Appl Microbiol Biotechnol 97(15):7015–7033. doi:10.1007/s00253-012-4430-4
Wang Y-F, Feng Y-Y, Ma X, Gu J-D (2013) Seasonal dynamics of ammonia/ammonium-oxidizing prokaryotes in oxic and anoxic wetland sediments of subtropical coastal mangrove. Appl Microbiol Biotechnol 97(17):7919–7934. doi:10.1007/s00253-012-4510-5
Laverock B, Tait K, Gilbert JA, Osborn AM, Widdicombe S (2014) Impacts of bioturbation on temporal variation in bacterial and archaeal nitrogen-cycling gene abundance in coastal sediments. Environ Microbiol Rep 6(1):113–121. doi:10.1111/1758-2229.12115
Restrepo-Ortiz CX, Auguet J-C, Casamayor EO (2014) Targeting spatiotemporal dynamics of planktonic SAGMGC-1 and segregation of ammonia-oxidizing thaumarchaeota ecotypes by newly designed primers and quantitative polymerase chain reaction. Environ Microbiol 16(3):689–700
Francis CA, Roberts KJ, Beman JM, Alyson ES, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102(41):14683–14688
Zheng Y, Hou L, Liu M, Lu M, Zhao H, Yin G, Zhou J (2013) Diversity, abundance, and activity of ammonia-oxidizing bacteria and archaea in Chongming eastern intertidal sediments. Appl Microbiol Biotechnol 97(18):8351–8363. doi:10.1007/s00253-012-4512-3
Wang J, Wang W, Gu J-D (2014) Conversion from soybean to rice paddy cultivation on community structure and abundance of ammonia-oxidizing archaea and bacteria in Baijiang soil of Northern China. Appl Microbiol Biotechnol. 98(6):2765–2778. doi:10.1007/s00253-013-5213-2
Sher Y, Zaady E, Nejidat A (2013) Spatial and temporal diversity and abundance of ammonia oxidizers in semi-arid and arid soils: indications for a differential seasonal effect on archaeal and bacterial ammonia oxidizers. FEMS Microbiol Ecol 86(3):544–556. doi:10.1111/1574-6941.12180
Qin H, Yuan H, Zhang H, Zhu Y, Yin C, Tan Z, Wu J, Wei W (2013) Ammonia-oxidizing archaea are more important than ammonia-oxidizing bacteria in nitrification and NO3 −-N loss in acidic soil of sloped land. Biol Fertil Soils 49(6):767–776. doi:10.1007/s00374-012-0767-1
Cao H, Auguet J-C, Gu J-D (2013) Global ecological pattern of ammonia-oxidizing archaea. PLoS One 8(2), e52853
Alves RJE, Wanek W, Zappe A, Richter A, Svenning MM, Schleper C, Urich T (2013) Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J 7:1620–1631. doi:10.1038/ismej.2013.35
Zhang L-M, Hu H-W, Shen J-P, He J-Z (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6(5):1032–1045
Sauder LA, Peterse F, Schouten S, Neufeld JD (2012) Low-ammonia niche of ammonia-oxidizing archaea in rotating biological contactors of a municipal wastewater treatment plant. Environ Microbiol 14(9):2589–2600. doi:10.1111/j.1462-2920.2012.02786.x
Zhang F-Q, Pan W, Gu J-D, Xu B, Zhang W-H, Zhu B-Z, Wang Y-X, Wang Y-F (2016) Dominance of ammonia-oxidizing archaea community induced by land use change from Masson pine to eucalypt plantation in subtropical China. Appl Microbiol Biotechnol. doi:10.1007/s00253-016-7506-8
Gan X-H, Zhang F-Q, Gu J-D, Guo Y-D, Li Z-Q, Zhang W-Q, Xu X-Y, Zhou Y, Wen X-Y, Xie G-G, Wang Y-F (2016) Differential distribution patterns of ammonia-oxidizing archaea and bacteria in acidic soils of Nanling National Nature Reserve forests in subtropical China. Anton Leeuw 109(2):237–251
Hanks JH, Weintraub RL (1936) The pure culture isolation of ammonia-oxidizing bacteria. J Bacteriol 32(6):653
Adair KL, Schwartz E (2008) Evidence that ammonia-oxidizing archaea are more abundant than ammonia-oxidizing bacteria in semiarid soils of northern Arizona, USA. Microb Ecol 56(3):420–426. doi:10.1007/s00248-007-9360-9
Santoro AE, Francis CA, de Sieyes NR, Boehm AB (2008) Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ Microbiol 10(4):1068–1079
Shen J-p, L-m Z, Zhu Y-g, J-b Z, He J-z (2008) Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ Microbiol 10(6):1601–1611
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442(7104):806–809
Strauss SL, Reardon CL, Mazzola M (2014) The response of ammonia-oxidizer activity and community structure to fertilizer amendment of orchard soils. Soil Biol Biochem 68:410–418
Hernández M, Dumont MG, Calabi M, Basualto D, Conrad R (2014) Ammonia oxidizers are pioneer microorganisms in the colonization of new acidic volcanic soils from South of Chile. Environ Microbiol Rep 6(1):70–79. doi:10.1111/1758-2229.12109
Wells GF, Park H-D, Yeung C-H, Eggleston B, Francis CA, Criddle CS (2009) Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ Microbiol 11(9):2310–2328
Mosier AC, Francis CA (2008) Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol 10(11):3002–3016
Boyle-Yarwood SA, Bottomley PJ, Myrold DD (2008) Community composition of ammonia-oxidizing bacteria and archaea in soils under stands of red alder and Douglas fir in Oregon. Environ Microbiol 10(11):2956–2965
Mulder A, van de Graaf AA, Robertson LA, Kuenen JG (1995) Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol Ecol 16(3):177–183
van de Graaf A, Mulder A, de Bruijn P, Jetten M, Robertson L, Kuenen J (1995) Anaerobic oxidation of ammonium is a biologically mediated process. Appl Environ Microbiol 61(4):1246–1251
Broda E (1977) Two kinds of lithotrophs missing in nature. Z Allg Mikrobiol 17(6):491–493
Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, Metzger JW, Schleifer K-H, Wagner M (2000) Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol 23(1):93–106. doi:10.1016/s0723-2020(00)80050-8
Schmid M, Walsh K, Webb R, Rijpstra WI, van de Pas-Schoonen K, Verbruggen MJ, Hill T, Moffett B, Fuerst J, Schouten S, Damsté JSS, Harris J, Shaw P, Jetten M, Strous M (2003) Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 26(4):529–538
Kartal B, Rattray J, van Niftrik LA, van de Vossenberg J, Schmid MC, Webb RI, Schouten S, Fuerst JA, Damsté Jaap S, Jetten MSM, Strous M (2007) Candidatus “Anammoxoglobus propionicus” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 30(1):39–49
Quan Z-X, Rhee S-K, Zuo J-E, Yang Y, Bae J-W, Park JR, Lee S-T, Park Y-H (2008) Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environ Microbiol 10(11):3130–3139
Song B, Buckner CT, Hembury DJ, Mills RA, Palmer MR (2014) Impact of volcanic ash on anammox communities in deep sea sediments. Environ Microbiol Rep 6(2):159–166. doi:10.1111/1758-2229.12137
Hou L, Zheng Y, Liu M, Gong J, Zhang X, Yin G, You L (2013) Anaerobic ammonium oxidation (anammox) bacterial diversity, abundance, and activity in marsh sediments of the Yangtze Estuary. J Geophys Res Biogeosci 118(3):1237–1246. doi:10.1002/jgrg.20108
Wang Y-F, Li X-Y, Gu J-D (2014) Differential responses of ammonia/ammonium-oxidizing prokaryotes in mangrove sediment to amendment of acetate and leaf litter. Appl Microbiol Biotechnol 98(7):3165–3180. doi:10.1007/s00253-013-5318-7
Wang Y-F, Gu J-D (2014) Effects of allylthiourea, salinity and pH on ammonia/ammonium-oxidizing prokaryotes in mangrove sediment incubated in laboratory microcosms. Appl Microbiol Biotechnol 98(7):3257–3274. doi:10.1007/s00253-013-5399-3
Penton CR, Devol AH, Tiedje JM (2006) Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl Environ Microbiol 72(10):6829–6832. doi:10.1128/aem.01254-06
Kuypers MMM, Sliekers AO, Lavik G, Schmid M, Jorgensen BB, Kuenen JG, Sinninghe Damsté JS, Strous M, Jetten MSM (2003) Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422(6932):608–611
Rysgaard S, Glud RN (2004) Anaerobic N2 production in Arctic sea ice. Limnol Oceanogr 49(1):86–94
Schubert CJ, Durisch-Kaiser E, Wehrli B, Thamdrup B, Lam P, Kuypers MM (2006) Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika). Environ Microbiol 8(10):1857–1863
Sun W, Xu MY, Wu WM, Guo J, Xia CY, Sun GP, Wang AJ (2014) Molecular diversity and distribution of anammox community in sediments of the Dongjiang River, a drinking water source of Hong Kong. J Appl Microbiol 116(2):464–476. doi:10.1111/jam.12367
Long A, Heitman J, Tobias C, Philips R, Song B (2013) Co-occurring anammox, denitrification, and codenitrification in agricultural soils. Appl Environ Microbiol 79(1):168–176. doi:10.1128/aem.02520-12
Dai M, Wang L, Guo X, Zhai W, Li Q, He B, Kao S-J (2008) Nitrification and inorganic nitrogen distribution in a large perturbed river/estuarine system: the Pearl River Estuary, China. Biogeosci Disc 5 (2)
Harrison PJ, Yin K, Lee J, Gan J, Liu H (2008) Physical–biological coupling in the Pearl River Estuary. Cont Shelf Res 28(12):1405–1415
Li P, Qiao P (1982) The model of evolution of the Pearl River Delta duing last 6,000 years. J Sediment Res 3:3
Wu C, BAO Y, REN J, Shi H, LEI Y A study on the Pearl Rive Delta in the last 6000 years-a long-term modeling approach. In: International conference on tidal dynamics and environment (TIDALITE 2002), 2002. Hangzhou
Jiao JJ, Wang Y, Cherry JA, Wang X, Zhi B, Du H, Wen D (2010) Abnormally high ammonium of natural origin in a coastal aquifer-aquitard system in the Pearl River Delta, China. Environ Sci Technol 44(19):7470–7475. doi:10.1021/es1021697
Conkling B, Blanchar R (1989) Glass microelectrode techniques for in situ pH measurements. Soil Sci Soc Am J 53(1):58–62
Neef A, Amann R, Schlesner H, Schleifer K-H (1998) Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144(12):3257–3266. doi:10.1099/00221287-144-12-3257
Francis CA, Beman JM, Kuypers MMM (2007) New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J 1(1):19–27
Okano Y, Hristova KR, Leutenegger CM, Jackson LE, Denison RF, Gebreyesus B, Lebauer D, Scow KM (2004) Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Appl Environ Microbiol 70(2):1008–1016. doi:10.1128/aem.70.2.1008-1016.2004
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. doi:10.1093/molbev/msr121
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71(3):1501–1506. doi:10.1128/aem.71.3.1501-1506.2005
Beman JM, Francis CA (2006) Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahía del Tóbari, Mexico. Appl Environ Microbiol 72(12):7767–7777. doi:10.1128/aem.00946-06
Dong L-h, Yang J-s, Yuan H-l (2008) Research advances in molecular ecology of ammonia oxidizing bacteria. Chin J Appl Ecol 19(6):1381–1388
Mullins TD, Britschgi TB, Krest RL, Giovannoni SJ (1995) Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr 40(1):148–158
Herrmann M, Saunders AM, Schramm A (2008) Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl Environ Microbiol 74(10):3279–3283. doi:10.1128/aem.02802-07
Herrmann M, Saunders AM, Schramm A (2009) Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl Environ Microbiol 75(10):3127–3136. doi:10.1128/aem.02806-08
Zhang T, Jin T, Yan Q, Shao M, Wells G, Criddle C, P Fang H (2009) Occurrence of ammonia‐oxidizing Archaea in activated sludges of a laboratory scale reactor and two wastewater treatment plants. J Appl Microbiol 107(3):970–977
Purkhold U, Wagner M, Timmermann G, Pommerening-Röser A, Koops H-P (2003) 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int J Syst Evol Microbiol 53(5):1485–1494
Purkhold U, Pommerening-Röser A, Juretschko S, Schmid MC, Koops H-P, Wagner M (2000) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66(12):5368–5382. doi:10.1128/aem.66.12.5368-5382.2000
Koops H-P, Pommerening-Röser A (2001) Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol Ecol 37(1):1–9
Hallin S, Lydmark P, Kokalj S, Hermansson M, Sörensson F, Jarvis Å, Lindgren PE (2005) Community survey of ammonia‐oxidizing bacteria in full‐scale activated sludge processes with different solids retention time. J Appl Microbiol 99(3):629–640
Dionisi HM, Layton AC, Harms G, Gregory IR, Robinson KG, Sayler GS (2002) Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl Environ Microbiol 68(1):245–253
Whang L-M, Chien I, Yuan S-L, Wu Y-J (2009) Nitrifying community structures and nitrification performance of full-scale municipal and swine wastewater treatment plants. Chemosphere 75(2):234–242
Cébron A, Berthe T, Garnier J (2003) Nitrification and nitrifying bacteria in the lower Seine River and estuary (France). Appl Environ Microbiol 69(12):7091–7100
Burrell PC, Phalen CM, Hovanec TA (2001) Identification of bacteria responsible for ammonia oxidation in freshwater aquaria. Appl Environ Microbiol 67(12):5791–5800. doi:10.1128/aem.67.12.5791-5800.2001
Koops H, Böttcher B, Möller U, Pommerening-Röser A, Stehr G (1991) Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J Gen Microbiol 137(7):1689–1699
Regan JM, Harrington GW, Noguera DR (2002) Ammonia- and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system. Appl Environ Microbiol 68(1):73–81. doi:10.1128/aem.68.1.73-81.2002
Stehr G, Böttcher B, Dittberner P, Rath G, Koops H-P (1995) The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol Ecol 17(3):177–186
Bollmann A, Schmidt I, Saunders AM, Nicolaisen MH (2005) Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl Environ Microbiol 71(3):1276–1282
Bollmann A, Bär-Gilissen M-J, Laanbroek HJ (2002) Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl Environ Microbiol 68(10):4751–4757. doi:10.1128/aem.68.10.4751-4757.2002
Lam P, Jensen MM, Lavik G, McGinnis DF, Müller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MMM (2007) Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci U S A 104(17):7104–7109. doi:10.1073/pnas.0611081104
Watson S, Bock E, Harms H, Koops H-P, Hooper A (1989) Nitrifying bacteria. In: Staley J, Bryant M, Pfennif N, Holt J (eds) Bergey’s manual of systematic microbiology, 3rd edn. Williams & Wilkins, Baltimore, pp 1808–1834
Casciotti KL, Ward BB (2001) Dissimilatory nitrite reductase genes from autotrophic ammonia-oxidizing bacteria. Appl Environ Microbiol 67(5):2213–2221. doi:10.1128/aem.67.5.2213-2221.2001
Diab S, Kochba M, Mires D, Avnimelech Y (1992) Combined intensive-extensive (CIE) pond system A: inorganic nitrogen transformations. Aquaculture 101(1):33–39
You J, Das A, Dolan EM, Hu Z (2009) Ammonia-oxidizing archaea involved in nitrogen removal. Water Res 43(7):1801–1809
Wang Y, Ke X, Wu L, Lu Y (2009) Community composition of ammonia-oxidizing bacteria and archaea in rice field soil as affected by nitrogen fertilization. Syst Appl Microbiol 32(1):27–36
Stein LY, Arp DJ (1998) Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl Environ Microbiol 64(10):4098–4102
Anthonisen AC, Loehr RC, Prakasam TBS, Srinath EG (1976) Inhibition of nitrification by ammonia and nitrous acid. J Water Pollut Control Fed 48(5):835–852
Acknowledgments
This study was supported financially by the General Research Fund of the Research Grants Council, the Hong Kong Special Administrative Region, China (HKU 702707P) and Guangdong Geological Survey.
Author information
Authors and Affiliations
Corresponding author
Additional information
Kwok-Ho Lee and Yong-Feng Wang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Lee, KH., Wang, YF., Wang, Y. et al. Abundance and Diversity of Aerobic/Anaerobic Ammonia/Ammonium-Oxidizing Microorganisms in an Ammonium-Rich Aquitard in the Pearl River Delta of South China. Microb Ecol 76, 81–91 (2018). https://doi.org/10.1007/s00248-016-0815-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0815-8