Abstract
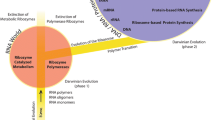
The ability to encode and convert heritable information into molecular function is a defining feature of life as we know it. The conversion of information into molecular function is performed by the translation process, in which triplets of nucleotides in a nucleic acid polymer (mRNA) encode specific amino acids in a protein polymer that folds into a three-dimensional structure. The folded protein then performs one or more molecular activities, often as one part of a complex and coordinated physiological network. Prebiotic systems, lacking the ability to explicitly translate information between genotype and phenotype, would have depended upon either chemosynthetic pathways to generate its components—constraining its complexity and evolvability— or on the ambivalence of RNA as both carrier of information and of catalytic functions—a possibility which is still supported by a very limited set of catalytic RNAs. Thus, the emergence of translation during early evolutionary history may have allowed life to unmoor from the setting of its origin. The origin of translation machinery also represents an entirely novel and distinct threshold of behavior for which there is no abiotic counterpart—it could be the only known example of computing that emerged naturally at the chemical level. Here we describe translation machinery’s decoding system as the basis of cellular translation’s information-processing capabilities, and the four operation types that find parallels in computer systems engineering that this biological machinery exhibits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The ability to encode and convert heritable information into molecular function is a defining feature of life as we know it (Schrödinger 1944; Joyce 1994). The conversion of information into molecular function is performed by the translation process, in which triplets of nucleotides in a nucleic acid polymer (mRNA) encode specific amino acids in a protein polymer that folds into a three-dimensional structure. The folded protein then performs one or more molecular activities, often as one part of a complex and coordinated physiological network (Goldman et al. 2012; Cuevas-Zuviría et al. 2023). Prebiotic systems, lacking the ability to explicitly translate information between genotype and phenotype, would have depended upon either chemosynthetic pathways to generate its components—constraining its complexity and evolvability (Vasas et al. 2010; Tessera 2018)—or on the ambivalence of RNA as both carrier of information and of catalytic functions—a possibility which is still supported by a very limited set of catalytic RNAs (Bernhardt 2012, Goldman et al. 2021). Thus, the emergence of translation during early evolutionary history may have allowed life to unmoor from the setting of its origin (Wolf and Koonin 2007; Goldman et al. 2016).
The emergence of translation machinery during early evolution also represents an entirely novel and distinct threshold of behavior for which there is no abiotic counterpart–it could be the only known example of computing that emerged naturally at the chemical level. The translation machinery’s decoding system is the basis of cellular translation’s information-processing capabilities, and it exhibits at least four operation types that find parallels in computer systems engineering.
First, this system performs an explicit mapping operation between one set of molecules, nucleic acids, and another (very different) set of molecules, proteins. This mapping operation, or ‘the genetic code,’ is arguably the most fundamental logical relationship of all living systems, establishing a semantic connection between two different kinds of polymers, each with orthogonal functions in the cell. Second, this operation is followed by the folding of the resulting string of amino acids, a process that we could consider like compiling a set of instructions. Recent research has shown that the folding process requires a very delicate kinetic control of the translation pace (Jiang et al. 2022). Third, translation has flow-control operations, i.e., using “riboswitches,” to debug and regulate its elemental operations. Translation can be stopped, re-started, stalled, and rescued by different processes that ensure that it continues to operate under diverse contingencies (Starosta et al. 2014; Breaker 2018). Fourth and finally, translation’s accuracy is afforded by non-linear process controls between different translation components and assemblies, with different kinetic partitioning checkpoints that favor forward-process steps for correct substrates and disfavor or delay these steps for the entry of poor substrates (Milón and Rodnina 2012). No other chemical system, either natural or synthetic, shows so many information-processing features bundled together as part of a single, multi-layered apparatus.
The principal of the correspondence between discrete nucleic acid and protein sequences is so inherently informatic that it can be analyzed in such terms without explicit reference to the underlying chemistry (Shannon 1948; Itzkovitz and Alon 2007; Adami 2012; Wills et al. 2015, Sonnerbon 1965). The information changes from a storage format into a functional format by decoding a set of three nucleotide monomers (a triplet codon) into one amino acid monomer along a growing peptide chain. The use of three bits of four RNA letters (64 possibilities) to specify one bit of amino acid information (20 possibilities), at first glance, may seem wasteful. But the excess capacity of the mapping assignment scheme, ‘code degeneracy,’ remarkably connects two very disparate needs of life: the consistency of information processing with the variability of novelty (Haig and Hurst 1991; Koonin and Novozhilov 2016; Błażej et al. 2018).
Degeneracy affords, for example, multiple and adjacent code assignments for amino acids with similar chemical properties (impacting functional divergence), variable translation speed (affecting folding), multiple stop codons limiting frameshifting or read-throughs (avoiding the waste of resources) (Nirenberg et al. 1966; Taylor and Coates 1989; Lehmann and Libchaber 2008, Liu et al. 2020, Křížek and Křížek 2012), and codon content and preference variability across different organisms (Shiba et al. 1997; Yacoubi et al. 2012; Fujishima and Kanai 2014; Pust et al. 2022, Novoa et al. 2012). Synonymous codon usage patterns (Labella et al. 2019), a ribosome’s interaction partners, and ribosomal assembly processes may not strictly map across related organisms (Timsit et al. 2021) but code degeneracy in translation as a fundamental informatic attribute is universal and conserved. For all these reasons, code degeneracy is better viewed as a hedge against (inevitable) error and a springboard of potential novelty than a wasteful use of computing resources. It is a feature, not a bug.
The information-processing capabilities of translation are distributed among the many components and subprocesses of the translation machinery, but the process of reading and decoding of sequence information at the molecular scale is nevertheless incredibly efficient. For each round of translation, the ribosome expends around four GTP molecules through translation factors and one ATP during the aminocylation of tRNAs. Despite its multiple layers of control, the nominal cellular replication process is still orders of magnitude more efficient on a per-bit basis when compared to even the most idealized conceivable electronic computers; this is also within an order of magnitude of theorized universal minimal bounds for information processing of any architecture (Zhirnov and Cavin 2013; Kempes et al. 2017).
Translation represents a uniquely evolved form of chemical computation with no known naturally occurring equivalent. This chemical intelligence is reliably repetitive, and at the same time robust to perturbation, but it is also not monolithic. The many components that take part in the translation system have intertwined, yet potentially distinct histories (Fournier et al. 2011; Fournier and Alm 2015; Petrov et al. 2015; Pouplana 2020; Fer et al. 2022). As its components co-evolved, the translation system would likely have had changing informatic capabilities, which in turn would restrict or expand the possible functions that it could have conducted. Understanding the origin of translation, from the perspective of its emerging information-processing attributes, will lead to profound new insights into the conditions in which biochemistry emerged from geochemistry.
References
Adami C (2012) The use of information theory in evolutionary biology. Ann Ny Acad Sci 1256:49–65. https://doi.org/10.1111/j.1749-6632.2011.06422.x
Bernhardt HS (2012) The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others)a. Biol Direct 7:23–23. https://doi.org/10.1186/1745-6150-7-23
Błażej P, Wnętrzak M, Mackiewicz D, Mackiewicz P (2018) Optimization of the standard genetic code according to three codon positions using an evolutionary algorithm. Plos One 13:e0201715. https://doi.org/10.1371/journal.pone.0201715
Breaker RR (2018) Riboswitches and translation control. Csh Perspect Biol 10:a032797. https://doi.org/10.1101/cshperspect.a032797
Cuevas-Zuviría B, Fer E, Adam ZR, Kaçar B (2023) The modular biochemical reaction network structure of cellular translation. Biorxiv. https://doi.org/10.1101/2023.01.21.524914
de Pouplana LR (2020) The evolution of aminoacyl-tRNA synthetases: From dawn to LUCA. Enzym 48:11–37. https://doi.org/10.1016/bs.enz.2020.08.001
Fer E, McGrath KM, Guy L et al (2022) Early divergence of translation initiation and elongation factors. Protein Sci 31:e4393. https://doi.org/10.1002/pro.4393
Fournier GP, Alm EJ (2015) Ancestral reconstruction of a Pre-LUCA Aminoacyl-tRNA synthetase ancestor supports the late addition of Trp to the genetic code. J Mol Evol 80:171–185. https://doi.org/10.1007/s00239-015-9672-1
Fournier GP, Andam CP, Alm EJ, Gogarten JP (2011) Molecular evolution of aminoacyl trna synthetase proteins in the early history of life. Origins Life Evol B 41:621–632. https://doi.org/10.1007/s11084-011-9261-2
Fujishima K, Kanai A (2014) tRNA gene diversity in the three domains of life. Front Genet 5:142. https://doi.org/10.3389/fgene.2014.00142
Goldman AD, Horst JA, Hung L-H, Samudrala R (2012) Evolution of the Protein Repertoire. In: Meyers R (ed) Systems Biology. Wiley-VCH, Wienheim, Germany, pp 207–237. https://doi.org/10.1002/3527600906.mcb.200400157.pub2
Goldman AD, Beatty JT, Landweber LF (2016) The TIM barrel architecture facilitated the early evolution of protein-mediated metabolism. J Mol Evol 82:17–26. https://doi.org/10.1007/s00239-015-9722-8
Goldman AD, Kacar B (2021) Cofactors are remnants of life’s origin and early evolution. J Mol Evol 89:127–133
Haig D, Hurst LD (1991) A quantitative measure of error minimization in the genetic code. J Mol Evol 33:412–417. https://doi.org/10.1007/bf02103132
Itzkovitz S, Alon U (2007) The genetic code is nearly optimal for allowing additional information within protein-coding sequences. Genome Res 17:405–412. https://doi.org/10.1101/gr.5987307
Jiang Y, Neti SS, Sitarik I et al (2022) How synonymous mutations alter enzyme structure and function over long timescales. Nat Chem. https://doi.org/10.1038/s41557-022-01091-z
Joyce (1994) In Origins of life: The central concepts. In: Fleischaker GR (ed) Deamer DW. Jones and Bartlett, Boston, pp xi–xii
Kempes CP, Wolpert D, Cohen Z, Prez-Mercader J (2017) The thermodynamic efficiency of computations made in cells across the range of life. Philosoph Trans Royal Soc Math Phys Eng Sci 375:20160343. https://doi.org/10.1098/rsta.2016.0343
Koonin EV, Novozhilov AS (2016) Origin and evolution of the universal genetic code. Annu Rev Genet 51:1–18. https://doi.org/10.1146/annurev-genet-120116-024713
Křížek M, Křížek P (2012) Why has nature invented three stop codons of DNA and only one start codon? J Theor Biology 304:183–187. https://doi.org/10.1016/j.jtbi.2012.03.026
Labella AL, Opulente DA, Steenwyk JL, Hittinger CT, Rokas A (2019) Variation and selection on codon usage bias across an entire subphylum. PLoS Genet 15(7):e1008304
Lehmann J, Libchaber A (2008) Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon. RNA 14:1264–1269. https://doi.org/10.1261/rna.1029808
Liu Y (2020) A code within the genetic code: codon usage regulates co-translational protein folding. Cell Commun Signal 18:145. https://doi.org/10.1186/s12964-020-00642-6
Milón P, Rodnina MV (2012) Kinetic control of translation initiation in bacteria. Crit Rev Biochem Mol 47:334–348. https://doi.org/10.3109/10409238.2012.678284
Nirenberg M, Caskey T, Marshall R et al (1966) The RNA code and protein synthesis. Cold Spring Harb Sym 31:11–24. https://doi.org/10.1101/sqb.1966.031.01.008
Novoa EM, de Pouplana LR (2012) Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet 28:574–581. https://doi.org/10.1016/j.tig.2012.07.006
Petrov AS, Gulen B, Norris AM et al (2015) History of the ribosome and the origin of translation. Proc National Acad Sci 112:15396–15401. https://doi.org/10.1073/pnas.1509761112
Pust M, Timmis KN, Tümmler B (2022) Bacterial tRNA landscape revisited. Environ Microbiol 24:2890–2894. https://doi.org/10.1111/1462-2920.16033
Rodnina MV, Wintermeyer W (2001) Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and Structural Mechanisms. Annu Rev Biochem 70:415–435. https://doi.org/10.1146/annurev.biochem.70.1.415
Schrödinger E (1944) What is Life? The Physical Aspect of the Living Cell. Cambridge University Press. https://doi.org/10.1017/cbo9781139644129
Shannon CE (1948) A mathematical theory of communication. Bell Syst Technical J 27:379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x
Shiba K, Motegi H, Schmimmel P (1997) Maintaining genetic code through adaptations of tRNA synthetases to taxonomic domains. Trends Biochem Sci 22:453–457. https://doi.org/10.1016/s0968-0004(97)01135-3
Sonnerbon TM (1965) Evolving Genes and Proteins. Academic Press. https://doi.org/10.1016/b978-1-4832-2734-4.50034-6
Starosta AL, Lassak J, Jung K, Wilson DN (2014) The bacterial translation stress response. Fems Microbiol Rev 38:1172–1201. https://doi.org/10.1111/1574-6976.12083
Taylor FJR, Coates D (1989) The code within the codons. Biosystems 22:177–187. https://doi.org/10.1016/0303-2647(89)90059-2
Tessera M (2018) Is pre-Darwinian evolution plausible? Biol Direct 13:18. https://doi.org/10.1186/s13062-018-0216-7
Timsit Y, Sergeant-Perthuis G, Bennequin D (2021) Evolution of ribosomal protein network architectures. Sci Rep-Uk 11:625. https://doi.org/10.1038/s41598-020-80194-4
Vasas V, Szathmáry E, Santos M (2010) Lack of evolvability in self-sustaining autocatalytic networks constraints metabolism-first scenarios for the origin of life. Proc National Acad Sci 107:1470–1475. https://doi.org/10.1073/pnas.0912628107
Wills PR, Nieselt K, McCaskill JS (2015) Emergence of Coding and its Specificity as a physico-informatic problem. Origins Life Evol B 45:249–255. https://doi.org/10.1007/s11084-015-9434-5
Wolf YI, Koonin EV (2007) On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol Direct 2:14–14. https://doi.org/10.1186/1745-6150-2-14
Yacoubi BE, Bailly M, de Crécy-Lagard V (2012) Biosynthesis and Function of Posttranscriptional Modifications of Transfer RNAs. Genetics 46:69–95. https://doi.org/10.1146/annurev-genet-110711-155641
Zhirnov VV, Cavin RK (2013) Future Microsystems for Information Processing: Limits and Lessons From the Living Systems. Ieee J Electron Devi 1:29–47. https://doi.org/10.1109/jeds.2013.2258631
Acknowledgements
Funding was provided by the John Templeton Foundation Grant #61926 and Margarita Salas Fellowship (Recualifica program) of the Universidad Politécnica de Madrid, funded by the NextGenerationEU, European Commission.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: David Liberles.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cuevas-Zuviría, B., Adam, Z.R., Goldman, A.D. et al. Informatic Capabilities of Translation and Its Implications for the Origins of Life. J Mol Evol 91, 567–569 (2023). https://doi.org/10.1007/s00239-023-10125-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-023-10125-0