Abstract
Nitrogen assimilation is a highly regulated process requiring metabolic coordination of enzymes and pathways in the cytosol, chloroplast, and mitochondria. Previous studies of prasinophyte genomes revealed that genes encoding nitrate and ammonium transporters have a complex evolutionary history involving both vertical and horizontal transmission. Here we examine the evolutionary history of well-conserved nitrogen-assimilating enzymes to determine if a similar complex history is observed. Phylogenetic analyses suggest that genes encoding glutamine synthetase (GS) III in the prasinophytes evolved by horizontal gene transfer from a member of the heterokonts. In contrast, genes encoding GSIIE, a canonical vascular plant and green algal enzyme, were found in the Micromonas genomes but have been lost from Ostreococcus. Phylogenetic analyses placed the Micromonas GSIIs in a larger chlorophyte/vascular plant clade; a similar topology was observed for ferredoxin-dependent nitrite reductase (Fd-NiR), indicating the genes encoding GSII and Fd-NiR in these prasinophytes evolved via vertical transmission. Our results show that genes encoding the nitrogen-assimilating enzymes in Micromonas and Ostreococcus have been differentially lost and as well as recruited from different evolutionary lineages, suggesting that the regulation of nitrogen assimilation in prasinophytes will differ from other green algae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary productivity in marine ecosystems is often regulated by the availability of nitrogen in the surrounding environment. Various cellular transporters and enzymes regulate nitrogen uptake and assimilation, for which the biochemical pathways are well conserved among photosynthetic eukaryotes. Membrane-localized transporters allow for the regulated uptake of nitrate and ammonium while enzymes in the cytosol, chloroplast, and mitochondria catalyze reduction and condensation reactions that allow for the regulated assimilation of inorganic nitrogen into organic molecules. The expression and activity of the enzymes are highly regulated, allowing for the coordination of nitrogen and carbon assimilatory pathways with cellular energy demands.
Nitrate and ammonium are the principle forms of inorganic nitrogen assimilated by photosynthetic eukaryotes. In general, nitrate is reduced to ammonium via the sequential reactions catalyzed by nitrate reductase (NR) and nitrite reductase (NiR). The ammonium that is produced is assimilated into the amino acid glutamine and glutamate through the enzymatic activity of glutamine synthetase (GS) and glutamate synthase and represents the major route for the entry of inorganic nitrogen into organic compounds. The nitrogen-assimilating enzymes have been well characterized in green algae and vascular plants, and while the pathways are well conserved, the enzymes within the pathways show differences in their evolutionary histories. For example, multiple GSII isoforms are observed in vascular plants and green algae. Phylogenetic analyses have shown that the GSII isoenzymes in vascular plants evolved by gene duplication, while in contrast, chlorophytes (Chlorella, Chlamydomonas, and Volvox) and bryophytes (Physcomitrella) share one GSII (GSIIE) that is homologous to that of vascular plants while a second GSII (GSIIB) evolved by horizontal gene transfer (HGT) from eubacteria (Ghoshroy et al. 2010).
Prasinophytes are morphologically diverse, free-living, unicellular green algae found predominately in marine environments, with a few representatives found in freshwater. Phylogenetic analyses have shown prasinophytes to be a paraphyletic grouping of early diverging lineages within the Chlorophyta (Turmel et al. 2009; Leliaert et al. 2011, 2012). Currently, ten major lineages are recognized within the prasinophytes and there are additional groups that have yet to be placed in the phylogenetic tree with confidence (Leliaert et al. 2011, 2012).
The Mamiellophyceae is the largest clade of prasinophytes and unites members of the Mamiellales, Dolichomastigales, and Monomastigales (Leliaert et al. 2012). Within this group, two genera within the Mamiellales, Ostreococcus and Micromonas, are globally distributed, marine phytoplankton with relatively small genomes (Derelle et al. 2006; Palenik et al. 2007; Worden et al. 2009). Although small in size, Ostreococcus and Micromonas are dominant members of picoeukaryotic communities and contribute significantly to oceanic primary productivity, food webs, and biogeochemical cycling.
Nitrogen availability in marine ecosystems varies across several spatial and temporal scales. Previous studies have shown that the number of genes encoding nitrate (NRT) and ammonium (AMT) transporters varies among species of Micromonas and Ostreococcus and that genes encoding multiple AMTs evolved via prokaryotic-to-eukaryotic and eukaryote-to-eukaryote HGT events (McDonald et al. 2010). The expansion of the transporter gene family by HGT may provide a selective advantage for these organisms that are often found in low-nitrogen environments or environments in which nitrogen source and supply varies (Derelle et al. 2006; Worden et al. 2009).
Given the evidence for variable copy numbers of nitrogen transporters, the HGT origin of AMTs in Ostreococcus and Micromonas, and the evidence of the HGT of GSII in other lineages of chlorophytes, we were interested in exploring the evolutionary history of several key enzymes involved in the regulated reduction and assimilation of nitrate and ammonium in Micromonas and Ostreococcus. We present phylogenetic analyses of GS (both GSIII and GSII), Fd-NiR, and NR, and provide evidence of multiple evolutionary sources for these genes within prasinophytes. In particular, we present evidence that genes encoding GSIII in prasinophytes may have evolved by HGT from a heterokont ancestor while genes encoding Fd-NiR and GSIIE evolved by vertical transmission. The evolutionary history of the NR encoding genes was unresolved. Fd-NiRs of prasinophytes are shown to have additional protein domains that may facilitate reduction of nitrite in low light or the dark. Our results indicate that the regulation of nitrogen assimilation in marine prasinophytes differs between genera of prasinophytes and from other green algal lineages and thus merits further investigation.
Materials and Methods
Phylogenetic Analyses
Amino acid sequences for four nitrogen assimilatory enzymes, GSIII, GSII, NR and Fd-NiR, were retrieved from NCBI and from the complete genomes at JGI DOE by the following methods. Both databases were searched using the complete enzyme names as keywords. In addition, BLAST searches were performed in NCBI using the following sequences from the diatom Thalassiosira pseudonana: GSIII (XP_002295274), GSII (EED87725), NR (XP_002294410) and Fd-NiR (XP_002289265). For some taxa, complete in-frame translations of the protein sequences were done in silico from EST contigs using CodonCode Aligner (CodonCode Corporation, Dedham, MA, USA). Where possible, taxa were selected to provide broad taxonomic sampling and an even distribution of taxa across the tree. Accession numbers and protein identifiers for sequences as well as the taxa used in the present study are provided in the Supplementary Tables 1–4. (Supplementary Materials online). GSIII sequences from Perkinsus marinus and Trichomonas vaginalis were not included in the current study as our previous analyses established the bacteria-to-eukaryote HGT origin of these sequences (Ghoshroy and Robertson 2012).
GSIII, GSII, NR, and Fd-NiR data matrices were constructed using amino acid sequences aligned with MAFFT v.6 (http://mafft.cbrc.jp/alignment/software; Katoh et al. 2002) followed by manual adjustment in MacClade 4.08 (Maddison and Maddison 2000). Functional domains within the enzymes were well conserved and aligned readily. More variable regions, including N- and C-terminal regions and indels were excluded from the phylogenetic analyses.
Bayesian analyses of the protein alignments were done using MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) with the same parameters for each dataset. These included two parallel runs, each with four chains (three heated and one cold) for 106 generations. The evolutionary models were explored using the mixed amino acid model available in MrBayes and the WAG model was identified as the best amino acid replacement model. Rate variation across sites was approximated using a gamma distribution with the proportion of invariable sites estimated from the data. Trees were sampled every 100 generations. Trees remaining (10,000) after a “burnin” of 5,001 for each run were used to compute the 50 % majority rule consensus trees.
Maximum likelihood (ML) analyses were performed for each dataset with RAxML v. 7.2.8 (http://phylobench.vital-it.ch/raxml-bb/; Stamatakis et al. 2008) using the GAMMA model for rate heterogeneity and the WAG amino acid substitution matrix; 100 rapid bootstrap replicates were performed. The number of taxa and characters included for each dataset in both Bayesian and ML analyses are presented in Table 1.
Predictions of Cellular Localization
In silico predictions of the cellular localization of GSIII, GSII, and Fd-NiR from Micromonas and Ostreococcus were performed using Target P 1.1(Emanuelsson et al. 2007), MitoProtII v.1.101 (Claros and Vincens 1996) and Predotar v. 1.03 (Small et al. 2004; Emanuelsson et al. 2007). For all proteins analyzed, we searched for N-terminal extensions upstream of conserved amino acid domains, and for the presence of any additional, upstream methionine (M) codons that were in-frame with the predicted open reading frame. If additional N-terminal amino acids were identified, they were included in the in silico targeting prediction analyses.
Domain Predictions for Fd-NiR
Domain predictions for Fd-NiR from O. lucimarinus CCE9901 (XP_001419920), O. tauri (XP_003081527), Micromonas sp. RCC299 (XP_002507511), M. pusilla CCMP1545 (XP_003057941), and Bathycoccus prasinos (CCO14961) were done using the Pfam 27.0 sequence search feature (http://pfam.sanger.ac.uk; Punta et al. 2012).
Results
GS in Micromonas and Ostreococcus
GS (E.C. 6.3.1.2) is an essential enzyme catalyzing the ATP-dependent condensation of ammonium and glutamate producing glutamine. The GS superfamily of enzymes has three classes, GSI, GSII, and GSIII, which are broadly distributed across prokaryotic and eukaryotic domains of life. In photosynthetic eukaryotes, GS isoforms are found predominantly in the cytosol and chloroplast, however recent studies have shown that GS is targeted to the mitochondria in some vascular plants and diatoms (Taira et al. 2004; Allen et al. 2006; Ghoshroy and Robertson 2012). Although GSII is considered the canonical eukaryotic form of GS, GSIII has been described in several eukaryotic lineages, but it has not been previously characterized in green algae or plants.
We uncovered differences in the distribution of GSII and GSIII genes within the genomes of Micromonas and Ostreococcus (Table 2). One gene encoding GSII and two genes encoding GSIII were identified in the genomes of M. pusilla CCMP1545 and Micromonas sp. RCC299. In contrast, the Ostreococcus genomes (Ostreococcus sp. RCC809, O. lucimarinus, and O. tauri) yielded a single GSIII gene and there were no genes encoding GSII.
In our phylogenetic analyses, GSIII from Micromonas and Ostreococcus formed a well-supported clade (BPP > 0.95 and ML = 99) in which Micromonas GSIIIs were found in two groups, with one group forming a close sister association with the GSIII sequences from Ostreococcus (Fig. 1). These results suggest that the GSIII genes in M. pusilla CCMP1545 and Micromonas sp. RCC299 arose via a gene duplication event that preceded the divergence of Micromonas and Ostreococcus and was followed by the loss of one gene copy in the latter genus (Fig. 1).
Evolutionary relationships among eukaryote and eubacterial GSIII protein sequences. Phylogenetic analyses included 68 GSIII sequences. An unrooted 50 % majority rule consensus tree from the Bayesian analysis is shown as inferred from 10,000 trees as described in the “Materials and Methods” section. Nodes with BPP support >0.95 are represented with thick lines. RAxML bootstrap values are indicated for most eukaryotic nodes. The collapsed eubacterial node represents 30 taxa. A complete taxa list for GSIII protein analyses is presented in SI Table 1. Target sequences identified in the N-terminal end of the prasinophyte sequences are marked as TS. “D” indicates the node of a putative gene duplication within the prasinophytes
The chromosomal locations for the GS genes in Micromonas (GSII and GSIII) and Ostreococcus (GSIII) were determined from genome data (Table 2). The GSIII sequences in the three species of Ostreococcus were located on chromosome 1. In Micromonas sp. RCC299, genes encoding GSIII, which arose via gene duplication, were located on chromosome 1 (JGI protein id [JPI]: 112708) and chromosome 3 (JPI: 113822), whereas the gene encoding GSII (JPI: 58286) was located on chromosome 5. The chromosomes have not been assembled for M. pusilla, however, each of the GS genes were found on different scaffolds, suggesting they are located in different regions of the genome.
ESTs were retrieved for all GSIII sequences from both genera, but transcripts from the GSII genes in Micromonas spp. were not detected. This could reflect an underrepresentation of the transcripts in the EST studies or may indicate that these genes were not transcribed under the conditions used to generate the ESTs.
Molecular Evolution of GSIII
As GSIII enzymes have not been reported in other green algal lineages, phylogenetic analyses were used to explore the evolutionary origin of GSIII in prasinophytes. Phylogenetic analyses of GSIII protein sequences were performed using two datasets: one with sequences with complete open reading frames and the other incorporating both complete and partial protein sequences. In these analyses, eukaryotic GSIII sequences were well resolved from prokaryotic sequences (Figs. 1 and 2). The eukaryote clade included sequences from members of the Amoebozoa, Excavata (Euglena gracilis), cryptophytes, haptophytes, heterokonts, dinoflagellates, and the prasinophytes. A consistent grouping of Heterokont + Prasinophyte GSIII sequences was recovered in all our phylogenetic analyses with strong to moderate support (Figs. 1 and 2). When the dataset containing complete protein sequences was used, prasinophyte (Micromonas and Ostreococcus) GSIII enzymes were placed as a sister clade to heterokont sequences (Thalassiosira pseudonana, Chaetoceros compressus, Phaeodactylum tricornutum, Fragilariopsis cylindrus, and Aureococcus anophagefferens) with strong support (BPP > 0.95; MLBS = 84; Fig. 1). Phylogenetic analyses using the extended taxa dataset, which included partial sequences from four additional eukaryotes (Pyramimonas gelidicola [prasinophyte], Ochromonas danica [heterokont], Prymnesium parvum [haptophyte], and Karlodinium micrum [dinoflagellate]), resulted in tree topologies where the prasinophyte clade (including the P. gelidicola sequence) was nested within the heterokont clade (Fig. 2); O. danica GSIII was placed in association with the prasinophyte clade with moderate to low support (BPP = 0.92; MLBS = 52; Fig. 2). Although the support for the Heterokont + Prasinophyte clade decreased when partial GSIII sequences were included, the clade was consistently recovered in our analyses.
Evolutionary relationships among eukaryote and eubacterial GSIII protein sequences as inferred from the extended dataset. Phylogenetic analyses included 72 GSIII sequences. An unrooted 50 % majority rule consensus tree from the Bayesian analysis is shown as inferred from 10,000 trees as described in the “Materials and Methods” section. Nodes with BPP support > 0.95 are represented with thick lines. The node containing Ochromonas danica and the prasinophytes had a BBP value of 0.92 as indicated with an arrow toward this node. RAxML bootstrap values are indicated for most nodes. The prasinophytes + heterokonts node and the prasinophytes + chromists node are also indicated with arrows
As opposed to the consistent grouping of Heterokont + Prasinophyte, the placement of the remaining eukaryotic taxa varied among the methods of analyses and the datasets analyzed. A single heterokont sequence from the parasitic Blastocystis hominis nested within the Amoebozoa in both ML and Bayesian analyses (Figs. 1, 2). A single representative sequence from Excavata (E. gracilis) nested within the chromists (sensu Cavalier-Smith 1986 which includes cryptophytes, haptophytes and heterokonts) and dinoflagellate sequences, but the placement of this sequence varied among analyses. The affinity of the E. gracilis plastid genome with prasinophyte lineages has been described (Turmel et al. 2009); however, based on our phylogenetic analyses, the GSIII in E. gracilis does not appear to have been derived from the prasinophyte endosymbiont.
Molecular Evolution of GSII
As in previous analyses (Ghoshroy et al. 2010; Nedelcu et al. 2009; Robertson and Tartar 2006), eubacterial (GSIIB) and eukaryotic (GSIIE) GSIIs formed two distinct clades in all phylogenetic analyses (Fig. 3). The GSIIE sequences from the two Micromonas species (RCC299 and CCMP 1545) were placed within a group that contained chlorophytes, mosses, and plants and were sister to the moss and vascular plant sequences (BPP > 0.95; MLBS = 80). GSIIE sequences from the remaining chlorophytes formed a separate well-supported clade (BPP > 0.95; MLBS = 98) that was distinct from the Micromonas sequences.
Evolutionary relationships among eukaryote and eubacterial GSII protein sequences. Phylogenetic analyses included 144 GSII protein sequences. An unrooted 50 % majority rule consensus tree from the Bayesian analysis is shown, as inferred from 10,000 trees as described in the “Materials and Methods” section. The tree is rooted at the branch leading toward the eubacterial and eukaryotic GSIIB sequences. Nodes with BPP support > 0.95 are represented with thick lines. RAxML bootstrap values are indicated for major nodes. The collapsed clade representing fungal sequences contains 13 taxa, metazoan collapsed clade has 11 taxa, “eubacteria others” are represented by 30 taxa, and Actinobacteria are represented by 12 taxa. A complete taxa list for GSII protein analyses is presented in SI Table 2. The prasinophytes are shaded for ease of identification in the tree
Previous analyses identified GSIIB protein sequences in chlorophytes and streptophytes, which were determined phylogenetically to be of eubacterial origin (Ghoshroy et al. 2010). In the current analyses, the GSIIB sequences formed a well-supported clade (BPP > 0.95; MLBS 100) nested within the larger eubacterial clade and distinct from the GSIIE sequences. Homologs of the GSIIB genes were not found in the genomes of either Micromonas or Ostreococcus, suggesting either the loss of this gene from these genera or that the HGT of GSIIB from eubacteria to eukaryotes occurred following the divergence of prasinophytes from other chlorophytes and plants.
Cellular Location of GS
In silico predictions of cellular localizations of the seven prasinophyte GSIII enzymes were performed as described in the Materials and Methods. Four GSIII sequences did not have transit peptides (TP), while three sequences (O. lucimarinus [JPI: 39960], O. tauri [JPI: 15060], and M. pusilla CCMP 1545 [JPI: 55724]), were predicted to have TP by all algorithms. O. lucimarinus (JPI: 39960) and O. tauri (JPI: 15060) were unanimously predicted to have mitochondrial TP while predictions for M. pusilla CCMP 1545 (JPI: 55724) GSIII varied between mitochondria and chloroplast localization (Table 3). The variability in the predictions may reflect the difficulty in recognizing chloroplast TP in green algae (Franzén et al. 1990) or indicate that the GSIII in O. lucimarinus and O. tauri, functions in the mitochondria or may be dual-targeted to both the chloroplast and mitochondria in these prasinophytes.
The M. pusilla CCMP 1545 GSII protein (JPI: 4228) lacked the N-terminal methionine in the annotated genome. We identified a putative in-frame start codon upstream of the ORF identified in the JGI database and added the following amino acid sequence (MSYAATGSQEGSGHGGARALDRL) to the N-terminal region of the annotated sequence. Even with the additional amino acid residues, TP were not predicted for CCMP 1545 or RCC 299 GSII sequences, suggesting the enzymes function in the cytosol.
Molecular Evolution of NR
Assimilatory NAD(P)H:NRs (E.C. 1.7.1.1-3) belong to the family of molybdenum-cofactor containing enzymes and catalyze the reduction of nitrate to nitrite. In plants and algae, NADH-specific forms are common while NAD(P)H-specific and bi-specific forms are also observed in eukaryotes (Fischer et al. 2005). The eukaryotic NR enzymes appear to have evolved from sulfite oxidases and are not homologous to prokaryotic NRs (Stolz and Basu 2002).
The prasinophyte genomes each contained a single gene encoding NR (Micromonas sp. RCC299 [NCBI: XP_002507512; JPI: 113975], M. pusilla CCMP1545 [NCBI: XP_003058321; JPI: 70878], O. lucimarinus [NCBI: XP_001420098; JPI: 37938], and O. tauri [NCBI: XP_003081526; JPI: 35072]); there was no evidence of a TP in the prasinophyte NRs indicating that the enzymes function in the cytosol.
In our phylogenetic analyses, NR sequences from the chlorophytes (excluding sequences from Micromonas and Ostreococcus) formed a well-supported clade (BPP > 0.95; MLBS = 91; Fig. 4). NR sequences from Micromonas and Ostreococcus were excluded from the chlorophyte clade and were associated with a larger group of sequences, the majority of which represented sequences from heterokont lineages (diatoms, raphidophytes, brown algae, oomycetes, and pelagophytes) and red algae (Gracilaria tenuistipitata, Porphyra yezoensis, and Cyanidioschyzon merolae). Diatom NR sequences formed a well-supported clade (BPP > 0.95; MLBS = 100) but deeper nodes in the tree were not supported (Fig. 4). Although Micromonas +Ostreococcus NR sequences were never recovered associated with other chlorophyte or moss + vascular plant sequences, the evolutionary history of the NR genes in prasinophytes remains unresolved.
Evolutionary relationships among NR protein sequences from eukaryotes. Phylogenetic analyses included 76 NR protein sequences. An unrooted 50 % majority rule consensus tree from the Bayesian analysis is shown, as inferred from 10,000 trees as described in the “Materials and Methods” section. Nodes with BPP support >0.95 are represented with thick lines. RAxML bootstrap values are indicated for major nodes. A complete taxa list for NR protein analyses is presented in SI Table 3. Clade A and B indicate sequences from heterokont lineages. The prasinophytes are shaded for ease of identification in the tree
Fd-NiR in Micromonas and Ostreococcus
Fd-NiR (E.C. 1.7.7.1) catalyzes the 6-electron reduction of nitrite to ammonium. Reduced ferredoxin is the physiological electron donor for this reaction in photosynthetic organisms, which in the chloroplast is reduced via photosynthetic electron transport (Privalle et al. 1985). In non-photosynthetic tissues, redox pathways that include NADPH, ferredoxin: NADP(H) oxidoreductase (FNR), and ferredoxin provide reduced ferredoxin to ferredoxin-dependent enzymes, such as NiR (Green et al. 1991; Hanke et al. 2005). Fd-NiR is found in cyanobacteria and photosynthetic eukaryotes and, in the present study, five prasinophyte Fd-NiR sequences (B. prasinos, Micromonas sp. RCC299, M. pusilla CCMP1545, O. lucimarinus and O. tauri) were retrieved from public databases for inclusion in phylogenetic analyses and analyses of domain structures.
Molecular Evolution of Fd-NiR
Fd-NiR sequences were resolved into three major clades in both Bayesian and ML analyses (Fig. 5). Among the three clades, the cyanobacteria were monophyletic and received strong support in both analyses (BPP > 0.95; MLBS = 100). Chlorophyte Fd-NiR sequences were not monophyletic. Sequences from Prasinophyceae showed a well-supported sister association with moss and vascular plants (BPP > 0.95; MLBS = 100), while the Chlorophyceae were grouped with the remaining eukaryotes, including sequences from rhodophyte, heterokonts, and a chlorarachniophyte. The remaining chlorophyte sequences showed a sister relation with Porphyra yezoensis with moderate support (BPP > 0.95; MLBS = 83). These results are consistent with an endosymbiotic origin of Fd-NiR in photosynthetic eukaryotes.
Evolutionary relationships among Fd-NiR protein sequences from photosynthetic eukaryotes and cyanobacteria. Phylogenetic analyses included 45 Fd-NiR protein sequences. An unrooted 50 % majority rule consensus tree from the Bayesian analysis is shown, as inferred from 10,000 trees as described in the “Materials and Methods” section. Nodes with BPP support >0.95 are represented with thick lines. RAxML bootstrap values >70 are indicated for nodes. A complete taxa list for Fd-NiR protein analyses is presented in SI Table 4. Target sequences identified in the N-terminal end of the prasinophyte sequences are marked as TS. The prasinophytes are shaded for ease of identification in the tree
Cellular Location and Domain Structure of Fd-NiR in Prasinophytes
In plants, Fd-NiR is localized to the chloroplast and catalyzes the reduction of nitrite to ammonium using electrons supplied from ferredoxin reduced by photosynthetic electron flow. McDonald et al. (2010) identified chloroplast TP in Fd-NiR from Micromonas RCC299 and CCMP1545 and we identified similar sequences in Fd-NiR of B. prasinos, O. lucimarinus, and O. tauri (Table 4). Predictions of localization varied between the chloroplasts and mitochondria, as did the location of the putative cleavage site. As observed with GSIII, these results may reflect the difficulty in recognizing chloroplast TP in green algae (Franzén et al. 1990) or indicate that the Fd-NiRs in prasinophytes are targeted to both the chloroplast and mitochondrion.
Fd-NiR enzymes have two well-conserved nitrite/sulfite ferredoxin like half domains (Pfam: PF3460.12) and two nitrite/sulfite reductase 4Fe-4S domains (Pfam: PF01077.17), which were present in the five prasinophyte Fd-NiR sequences (Fig. 6). In addition, three conserved domains, a rubredoxin domain (PF00301.15), an oxidoreductase FAD-binding domain (Pfam: FAD_binding_6: PF00970.19), and an oxidoreductase NAD-binding domain (Pfam: NAD_binding_1: PF00175.16), were identified at the C-terminal end of the prasinophyte Fd-NiR, as previously reported for O. tauri, Micromonas sp. RCC299, and M. pusilla 1545 (Derelle et al. 2006; McDonald et al. 2010). The annotated genome sequence from O. lucimarinus Fd-NiR lacked the C-terminal domains; however, extension of the open reading frame to the first, in-frame stop codon resulted in the addition of 358 amino acids, which contained the domains (Fig. 6). The three domains present in the C-terminal region of the prasinophyte Fd-NiRs share similarity with cytochrome b5 and the ferredoxin: NADP(H) oxidoreductase (FNR) family of enzymes. As observed in non-photosynthetic tissues in vascular plants, this region of the prasinophyte Fd-NiR may allow for NADPH or NADH to provide reduced ferredoxin for nitrite reduction independent of photosynthetic electron transport.
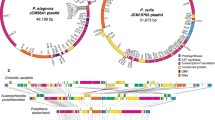
Domain structures in Fd-NiR. A comparison of domains found in green algal lineages and additional domains reported for Fd-NiR protein sequences in prasinophytes. a Shows the respective domains present in Fd-NiR of Chlorella variabilis and Micromonas pusilla, the domain name, protein family (PF) number and the amino acid location in the respective proteins for the domain location is identified. b Shows schematic of multiple alignment of Fd-NiR protein sequences from chlorophyte taxa used in the present study. C-terminal extensions are noted for prasinophyte lineages
Discussion
This is the first detailed analysis of genes encoding a member of the GSIII family in the green algae. The presence of GSIII in the prasinophytes is surprising as this gene is absent from genomes and EST’s of other members of the Archaeplastida. Although GSIII is present in some cyanobacteria, the cyanobacterial GSIII sequences are distantly related to the eukaryotic sequences, negating the possibility of a chloroplast-endosymbiotic origin in eukaryotes. Alternatively, Ghoshroy and Robertson (2012) proposed that GSIII in eukaryotes arose either through a mitochondrial endosymbiotic gene transfer or genes encoding GSIII were present in the nucleus of the ancestral eukaryote prior to the acquisition of mitochondria and chloroplasts.
In our phylogenetic analyses, the prasinophyte GSIIIs grouped with cryptophyte, haptophytes, and heterokonts and were found to be nested within or sister to the heterokonts (Figs. 1 and 2). If genes encoding GSIII were widespread early in the evolution of eukaryotes and evolved by vertical transmission, the prasinophyte GSIII sequences should branch outside of the cryptophyte-haptophyte-heterokont clade. Thus, our phylogenetic results are consistent with an HGT origin of GSIII in prasinophytes and propose that the transfer was from the heterokonts to the prasinophytes.
Previous studies have identified genes in heterokonts that have a phylogenetic affinity with genes in the green algae. Using an automated phylogenomic pipeline, Moustafa et al. (2009) identified over 1,700 genes with green algal affinity within the genomes of the diatoms, Thalassiosira pseudonana and P. tricornutum, and 500 of these genes grouped with prasinophytes. Based on these results, Moustafa et al. (2009) proposed that these genes arose via endosymbiotic gene transfers (EGT) from a cryptic green algal endosymbiont. However, revaluation of the dataset indicated that the majority of these genes more likely evolved via EGT from the red algal ancestor of chloroplasts thus reducing the likelihood of a cryptic green algal endosymbiont event in the evolution of heterokonts (Dorrell and Smith 2011; Burki et al. 2012; Deschamps and Moreira 2012; Keeling 2013). As GSIII is not observed in other Archaeplastida lineages and the reduced likelihood of a green algal endosymbiont in the evolutionary history of the heterokonts, we propose that the HGT of GSIII occurred from heterokonts to the ancestor of the Mamiellales and Pyramimonadales lineages. This hypothesis is strongly supported by our phylogenetic analyses and it requires only one gain of GSIII prior to the divergence of Pyramimonadales (prasinophyte clade I) + Mamiellophyceae (prasinophyte clade II) node in the consensus tree of Leliaert et al. (2012). This hypothesis also predicts that GSIII will be present in members of the Dolichomastigales and Monomastigales, which can be tested with broader taxon sampling.
Evidence of HGT events in eukaryotes is increasing as more prokaryotic and eukaryotic genomes become available (e.g., Keeling and Palmer, 2008; Yue et al. 2012; Huang and Yue 2013; Huang 2013). There appears a bias to which genes are retained and fixed after the horizontal transfer, with genes encoding enzymes in metabolic pathways and transport proteins showing high levels of retention (Schönknecht et al. 2014). Schönknecht et al. (2014) proposed that HGT may be advantageous to populations of different species sharing similar environments in which the HGT acquisition of transport proteins or enzymes rapidly produces novel phenotypes. Previous studies characterized regulatory and kinetic differences among the GS isoenzymes (Robertson and Alberte 1996; Reyes et al. 1997; Bernard and Habash 2009; Seabra et al. 2013). The acquisition of paralogous enzymes via HGT, as proposed here for GSIII, may provide organisms with a “step change” in enzyme kinetics, substrate affinity, or regulation that confers a selective advantage and thus favors gene retention and fixation.
Marine viruses are important regulators of oceanic primary productivity, influencing the size and duration of phytoplankton blooms and mediating nutrient cycles. Interestingly, the prasinophytes included in our analyses have documented viruses whose complete genomes have been sequenced (Grimsley et al. 2012). A recent study of phosphate transporters (pho4) in marine phytoplankton and their viruses found that pho4 genes from pelagophytes, haptophytes and their infecting viruses were closely related to homologues in prasinophytes, suggesting the possibility of viral mediated HGT (Monier et al. 2012). Prasinoviruses have been found harboring genes encoding enzymes involved in amino acid synthesis and GSIIE genes have been identified in marine viruses (Allen et al. 2012; Grimsley et al. 2012). While many marine viruses are species specific (Clerissi et al. 2013), some may be vehicles for HGT among marine eukaryotes, similar to what has been observed in planktonic cyanobacteria (e.g., Sullivan et al. 2005; Suttle 2007), and may contribute to the high number of HGTs observed in marine prasinophytes.
The number of GS genes varied among the prasinophyte genomes. Each of the Ostreococcus species studied here had a single GS (GSIII) gene, which had a TP that was predicted to localize the enzyme to the chloroplast or mitochondrion. In contrast, both Micromonas species had three GS sequences (two GSIIIs and one GSII). The GSIII genes appear to have arisen via a gene duplication event, with subsequent loss in Ostreococcus spp. Neither Micromonas nor Ostreococcus had a GSIIB homolog, a gene present in other chlorophytes and early evolving plants (Ghoshroy et al. 2010). As GSIIB appears to have evolved via a eubacteria-eukaryote HGT, the absence of GSIIB in Micromonas and Ostreococcus may be the result of gene loss or may indicate that the HGT occurred after the divergence of these lineages from other green algae. This question can be explored by further sampling examination of GSII in the prasinophytes.
The reduction in the number of GS genes in Ostreococcus relative to Micromonas may be a reflection of the extremely reduced genome in Ostreococcus (12–13 Mbp compared to the 21 Mbp genome of Micromonas). Alternatively, as Micromonas is more broadly distributed than Ostreococcus (Worden et al. 2009) and flagellated, the multiple GS isoenzymes observed in Micromonas may provide a selective advantage to cells experiencing more variable nutrient environments associated with a larger distribution or as a result of increased motility.
In contrast to GSIII, our phylogenetic analyses support a cyanobacterial-endosymbiotic origin of Fd-NiR in photosynthetic eukaryotes. However, as observed with GSIIE, Fd-NiR from green algae were not monophyletic; the prasinophyte sequences were associated with the moss and vascular plant clade while the remainder chlorophytes associated with a rhodophyte (Porphyra yezoensis) sequence. The observed phylogeny may reflect an ancient gene duplication event, followed by gene loss, resulting in Chlorophyte and Prasinophyte paralogs being represented in the phylogenetic tree.
Organellar targeting sequences were identified in the prasinophyte GSIII and Fd-NiR protein sequences. The dual predictions (chloroplast and mitochondria) in prasinophytes may reflect the ambiguous nature of TP in green algae, where chloroplast TP were found to be similar to mitochondrial TP (Franzén et al. 1990). In our analyses, Ostreococcus enzymes with TP were predicted more often to mitochondria than Micromonas TP. Alternatively, the enzymes may be targeted to both organelles. This is more likely for GSIII than Fd-NiR as dual targeting has been reported for GS in Arabidopsis thaliana (Taira et al. 2004). GS is involved in the assimilation of ammonium from nitrite reduction in the chloroplast as well as ammonium produced during photorespiration. The localization of GS to the mitochondria may help in the recovery of ammonium produced during photorespiration in photosynthetic eukaryotes (Taira et al. 2004). As in diatoms, genes encoding urea transporters, as well as enzymes involved in urea catabolism and the urea cycle, have been reported in the genomes of Micromonas sp RCC299, O. tauri and O. lucimarinus (Solomon et al. 2010; Derelle et al. 2006). The prediction of mitochondrial-localized GSIII enzymes in both diatoms and prasinophytes may reflect of a role for this enzyme in urea metabolism (Allen et al. 2011) as well as in the assimilation of ammonium produced during photorespiration.
Fd-NiR protein sequences from the Mamiellales (B. prasinos, Ostreococcus, and Micromonas) had additional domains in the C-terminal region of the enzyme that are not present in other photosynthetic eukaryotes and may represent an evolutionary innovation in this group. The domains are shared among members of the cytochrome B5 reductase and ferredoxin: NADP(H) oxidoreductase-like superfamilies. These additional domains in the Fd-NiR enzymes may allow for non-photosynthetic reduction of ferredoxin for use in nitrite reduction (Hanke et al. 2005). The use of NADPH or NADH to reduce ferredoxin may provide the physiological capacity to assimilate nitrate in low-light environments and possibly the dark respiration of nitrate, similar to what is observed in diatoms (Kamp et al. 2011).
NR catalyzes the rate-limiting step in the assimilation of nitrate and eukaryotic NRs have a well-conserved domain structure (reviewed in Campbell 1999). However in our analyses, the deeper nodes in the phylogeny were not resolved and thus the evolutionary history of NR genes in prasinophytes is currently unresolved.
Nitrogen Assimilatory Pathways in Prasinophytes
While the major pathways for assimilating inorganic nitrogen into organic molecules are conserved among photosynthetic eukaryotes, this study demonstrates the chimeric nature of the nitrate assimilatory pathway in Micromonas and Ostreococcus, summarized in Fig. 7. Genes encoding ammonium transporters in Micromonas and Ostreococcus evolved via prokaryotic-to-eukaryotic and eukaryotic-to-eukaryotic gene transfer events (McDonald et al. 2010). Our phylogenetic analyses suggest that genes encoding GSIII arose in the prasinophytes through eukaryotic-to-eukaryotic HGT. Genes encoding other Fd-NiR and GSIIE enzymes in the prasinophytes appear to have evolved via vertical transmission and in the case of Fd-NiR, following endosymbiotic acquisition.
Diagram summarizing the diversity and predicted evolutionary history of key nitrogen-assimilating enzymes in chlorophytes. The results from the phylogenetic analyses presented in the text are summarized. As discussed in the text, enzymes shown as chloroplast localized may also be targeted to the mitochondria. The distribution of NRTs and AMTs are based on data presented by McDonald et al. (2010)
The incorporation of horizontally transferred genes into genomes would require the evolution of the mechanisms that regulate gene expression and enzyme activity, suggesting that the regulation of nitrogen assimilation in the prasinophytes is likely to differ from other lineages of green algae. In addition, the number of nitrate and ammonium transporters (McDonald et al. 2010) and the number and cellular distribution of GS isoenzymes varies among the prasinophytes, suggesting there will be physiological and regulatory differences in nitrogen assimilation within this group of early diverging green algae (Fig. 7).
The Prasinophytes are one of the most abundant groups of marine photosynthetic eukaryotes (Not et al. 2005; Worden et al. 2009). In photosynthetic eukaryotes assimilation of inorganic nitrogen into organic molecules is a key factor regulating population growth and primary productivity. The mixed evolutionary history uncovered for key nitrogen-assimilating enzymes and transporters in the prasinophytes indicates the capacity of these organisms to acquire and maintain genes from disparate sources. This capacity may promote evolutionary innovation of physiological processes. Future studies examining the regulation of gene expression and enzyme activity may provide insight as to how genes from different evolutionary lineages can be integrated into highly regulated and coordinated metabolic pathways.
References
Allen A, Vardi A, Bowler C (2006) An ecological and evolutionary context for integrated nitrogen metabolism and related signaling pathways in marine diatoms. Curr Opin Plant Biol 9:264–273
Allen A, Dupont C, Oborník M, Horák A, Nunes-Nesi A, McCrow J, Zheng H, Johnson D, Hu H, Fernie A, Bowler C (2011) Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473:203–207. doi:10.1038/nature10074
Allen LZ, Allen E, Badger J, McCrow J, Paulsen I, Elbourne L, Thiagarajan M, Rusch D, Nealson K, Williamson S, Venter J, Allen A (2012) Influence of nutrients and currents on the genomic composition of microbes across an upwelling mosaic. ISME J 6:1403–1414. doi:10.1038/ismej.2011.201
Bernard SM, Habash DZ (2009) The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol 182:608–620
Burki F, Flegontov P, Oborník M, Cihlář J, Pain A, Lukeš J, Keeling PJ (2012) Re-evaluating the green versus red signal in eukaryotes with secondary plastid of red algal origin. Genome Biol Evol 4:738–747
Campbell WH (1999) Nitrate reductase structure, function and regulation: bridging the gap between biochemistry and physiology. Annu Rev Plant Physiol Plant Mol Biol 50:277–303
Cavalier-Smith T (1986) The kingdom chromista: origin and systematics. Progr Phycol Res 4:309–347
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786
Clerissi C, Grimsley N, Desdevises Y (2013) Genetic exchanges of inteins between prasinoviruses (Phycodnaviridae). Evolution 67:18–33
Derelle E, Ferraz C, Rombauts S, Rouze P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynie S, Cooke R, Saeys Y, Wuyts J, Jabbari K, Bowler C, Panaud O, Piegu B, Ball SG, Ral JP, Bouget FY, Piganeau G, De Baets B, Picard A, Delseny M, Demaille J, Van de Peer Y, Moreau H (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103:11647–11652
Deschamps P, Moreira D (2012) Reevaluating the green contribution to diatom genomes.Genome Biol Evol 4:683–688. doi: 10.1093/gbe/evs053
Dorrell RG, Smith AG (2011) Do red and green make brown?: perspectives on plastid acquisitions within chromalveolates. Eukaryot Cell 10:856–868
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc 2:953–971
Fischer K, Barbier GG, Hecht H-J, Mendel RR, Campbell WH, Schwarz G (2005) Structural basis of eukaryotic nitrate reduction: crystal structures of the nitrate reductase active site. Plant Cell 17:1167–1179
Franzén L-G, Rochaix J-D, von Heijne G (1990) Chloroplast transit peptides from the green alga Chlamydomonas reinhardtii share features with both mitochondrial and higher plant chloroplast presequences. FEBS Lett 260:169–172
Ghoshroy S, Robertson DL (2012) Molecular evolution of glutamine synthetase II and III in the chromalveolates. J Phycol 48:768–783
Ghoshroy S, Binder M, Tartar A, Robertson DL (2010) Molecular evolution of glutamine synthetase II: phylogenetic evidence of a non-endosymbiotic gene transfer event early in plant evolution. BMC Evol Biol 10:198
Green LS, Yee BC, Buchanan BB, Kamide K, Sanada Y, Wada K (1991) Ferredoxin and ferredoxin-NADP reductase from photosynthetic and nonphotosynthetic tissues of tomato. Plant Physiol 96:1207–1213
Grimsley NH, Thomas R, Kegely JU, Jacquetz S, Moreau H, Desdevises Y (2012) Genomics of algal host–virus interactions. Chapter 9. Advances in Botanical Research, Volume 64. Elsevier Ltd. ISSN 0065-2296. http://dx.doi.org/10.1016/B978-0-12-391499-6.00009-8
Hanke GT, Okutani S, Satomi Y, Takao T, Suzuki A, Hase T (2005) Multiple iso-proteins of FNR in Arabidopsis: evidence for different contributions to chloroplast function and nitrogen assimilation. Plant, Cell Environ 28:1146–1157. doi:10.1111/j.1365-3040.2005.01352.x
Huang J (2013) Horizontal gene transfer in eukaryotes: the weak-link model. BioEssays 35:868–875
Huang J, Yue J (2013) Horizontal gene transfer in the evolution of photosynthetic eukaryotes. J Syst Evol 51:13–29
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Kamp A, de Beer D, Nitsch J, Lavik G, Stief P (2011) Diatoms respire nitrate to survive dark and anoxic conditions. Proc Natl Acad Sci USA 108:5649–5654. doi:10.1073/pnas.1015744108
Katoh K, Misawa K, Kuma K-I, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res 30:3059–3066. doi:10.1093/nar/gkf436
Keeling PJ (2013) The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol 64:583–607
Keeling PJ, Palmer JD (2008) Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9:605–618. doi:10.1038/nrg2386
Leliaert F, Verbruggen H, Zechman FW (2011) Into the deep: new discoveries at the base of the green plant phylogeny. BioEssays 33:683–684
Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O (2012) Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci 31:1–46
Maddison W, Maddison D (2000) MacClade 4: analysis of phylogeny and character evolution. Sinauer Associates Sunderland, MA
McDonald SM, Plant JN, Worden AZ (2010) The mixed lineage nature of nitrogen transport and assimilation in marine eukaryotic phytoplankton: a case study of Micromonas. Mol Biol Evol 27:2268–2283
Monier A, Welsh RM, Gentemann C, Weinstock G, Sodergren E, Armbrust EV, Eisen JA, Worden AZ (2012) Phosphate transporters in marine phytoplankton and their viruses: cross-domain commonalities in viral-host gene exchanges. Environ Microbiol 14:162–176. doi:10.1111/j.1462-2920.2011.02576.x
Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D (2009) Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324:1724–1726
Nedelcu AM, Blakney AJC, Logue KD (2009) Functional replacement of a primary metabolic pathway via multiple independent eukaryote-to-eukaryote gene transfers and selective retention. J Evol Biol 22:1882–1894. doi:10.1111/j.1420-9101.2009.01797.x
Not F, Massana R, Latasa M, Marie D, Colson C, Eikrem W, Pedros-Alio C, Vaulot D, Simon N (2005) Late summer community composition and abundance of photosynthetic picoeukaryotes in Norwegian and Barents Seas. Limnol Oceanogr 50:1677–1686
Palenik B, Grimwood J, Aerts A, Rouze P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, Zhou K, Otillar R, Merchant SS, Podell S, Gaasterland T, Napoli C, Gendler K, Manuell A, Tai V, Vallon O, Piganeau G, Jancek S, Heijde M, Jabbari K, Bowler C, Lohr M, Robbens S, Werner G, Dubchak I, Pazour GJ, Ren Q, Paulsen I, Delwiche C, Schmutz J, Rokhsar D, Van de Peer Y, Moreau H, Grigoriev IV (2007) The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA 104:7705–7710. doi:10.1073/pnas.0611046104
Privalle LS, Privalle CT, Leonardy NJ, Kamin H (1985) Interactions between spinach ferredoxin-nitrite reductase and its substrates. Evidence for the specificity of ferredoxin. J Biol Chem 260:14344–14350
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD (2012) The Pfam protein families database. Nucl Acids Res 40:D290–D301
Reyes JC, Muro-Pastor MI, Florencio FJ (1997) Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J Bacteriol 179:2678
Robertson DL, Alberte RS (1996) Isolation and characterization of glutamine synthetase from the marine diatom Skeletonema costatum. Plant Physiol 111:1169–1175
Robertson DL, Tartar A (2006) Evolution of glutamine synthetase in heterokonts: evidence for endosymbiotic gene transfer and the early evolution of photosynthesis. Mol Biol Evol 23:1048–1055
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Schönknecht G, Weber APM, Lercher MJ (2014) Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. BioEssays 36:9–20
Seabra A, Silva L, Carvalho H (2013) Novel aspects of glutamine synthetase (GS) regulation revealed by a detailed expression analysis of the entire GS gene family of Medicago truncatula under different physiological conditions. BMC Plant Biol 13:137
Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4:1581–1590
Solomon CM, Collier JL, Berg GM, Glibert PM (2010) Role of urea in microbial metabolism in acquatic systems: a biochemical and molecular review. Aquat Microb Ecol 59:67–88. doi:10.3354/ame01390
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771
Stolz JF, Basu P (2002) Evolution of nitrate reductase: molecular and structural variations on a common function. ChemBioChem 3:198–206
Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW (2005) Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3:e144. doi:10.1371/journal.pbio.0030144
Suttle CA (2007) Marine viruses-major players in the global ecosystem. Nat Rev Microbiol 5:801–812
Taira M, Valtersson U, Burkhardt B, Ludwig RA (2004) Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell 16:2048–2058
Turmel M, Gagnon MC, O’Kelly CJ, Otis C, Lemieux C (2009) The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol Biol Evol 26:631–648
Worden AZ, Lee JH, Mock T, Rouze P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, Foulon E, Grimwood J, Gundlach H, Henrissat B, Napoli C, McDonald SM, Parker MS, Rombauts S, Salamov A, Von Dassow P, Badger JH, Coutinho PM, Demir E, Dubchak I, Gentemann C, Eikrem W, Gready JE, John U, Lanier W, Lindquist EA, Lucas S, Mayer KF, Moreau H, Not F, Otillar R, Panaud O, Pangilinan J, Paulsen I, Piegu B, Poliakov A, Robbens S, Schmutz J, Toulza E, Wyss T, Zelensky A, Zhou K, Armbrust EV, Bhattacharya D, Goodenough UW, Van de Peer Y, Grigoriev IV (2009) Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324:268–272
Yue J, Hu X, Sun H, Yang Y, Huang J (2012) Widespread impact of horizontal gene transfer on plant colonization of land. Nat Commun 3:1152. doi:10.1038/ncomms2148
Acknowledgments
This research was supported by a Grant (IOS105207) from the National Science Foundation (United States) to D. L. R. The authors thank Alfred Justo and Romina Gazis for their helpful comments on the manuscript. We also thank two anonymous reviewers for their constructive comments and suggestions.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ghoshroy, S., Robertson, D.L. Molecular Evolution of Nitrogen Assimilatory Enzymes in Marine Prasinophytes. J Mol Evol 80, 65–80 (2015). https://doi.org/10.1007/s00239-014-9659-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-014-9659-3