Abstract
We provide a kinetic characterization of (Na+, K+)-ATPase activity in a posterior gill microsomal fraction from the grapsid crab Goniopsis cruentata. (Na+, K+)-ATPase activity constitutes 95% of total ATPase activity, and sucrose density centrifugation reveals an ATPase activity peak between 25 and 35% sucrose, distributed into two, partially separated protein fractions. The (Na+, K+)-ATPase α-subunit is localized throughout the ionocyte cytoplasm and has an Mr of ≈ 10 kDa and hydrolyzes ATP obeying cooperative kinetics. Low (VM = 186.0 ± 9.3 nmol Pi min−1 mg−1 protein and K0.5 = 0.085 ± 0.004 mmol L−1) and high (VM = 153.4 ± 7.7 nmol Pi min−1 mg−1 protein and K0.5 = 0.013 ± 0.0006 mmol L−1) affinity ATP binding sites were characterized. At low ATP concentrations, excess Mg2+ stimulates the enzyme, triggering exposure of a high-affinity binding site that accounts for 50% of (Na+, K+)-ATPase activity. Stimulation by Mg2+ (VM = 425.9 ± 25.5 nmol Pi min−1 mg−1 protein, K0.5 = 0.16 ± 0.01 mmol L−1), K+ (VM = 485.3 ± 24.3 nmol Pi min−1 mg−1 protein, K0.5 = 0.9 ± 0.05 mmol L−1), Na+ (VM = 425.0 ± 23.4 nmol Pi min−1 mg−1 protein, K0.5 = 5.1 ± 0.3 mmol L−1) and NH4+ (VM = 497.9 ± 24.9 nmol Pi min−1 mg−1 protein, K0.5 = 9.7 ± 0.5 mmol L−1) obeys cooperative kinetics. Ouabain inhibits up to 95% of ATPase activity with KI = 196.6 ± 9.8 µmol L−1. This first kinetic characterization of the gill (Na+, K+)-ATPase in Goniopsis cruentata enables better comprehension of the biochemical underpinnings of osmoregulatory ability in this semi-terrestrial mangrove crab.
Graphic Abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Some semi-terrestrial crustaceans are among the few animal species that can respire in both water and in air since they can, respectively, employ their gills or lung-like branchial chambers for gas exchange in these media (Wolvenkamp and Waterman 1960). Despite such respiratory adaptations, variable salinity environments also represent a significant physiological challenge, and movements among habitats along salinity gradients demand considerable osmoregulatory adjustment (Rahi et al. 2018; Betancur-R et al. 2012). Mangroves constitute a steep transitional zone between marine and inland ecosystems and are characterized by a depositional coastal environment, including fine sediments with a high organic content, and a tidal regimen (Donato et al. 2011). The invertebrate fauna of the mangrove ecosystem consists primarily of decapod crustaceans, which are exposed to a variable tidal salinity regime, elevated temperatures, and desiccation (Donato et al. 2011).
Since fully amphibious species must survive both in air and in water, their evolution from aquatic ancestors likely passed through a transitional phase (Morris 2001; Hartnoll 1988). During this transition, recognized adaptations enabling the use of water and/or air for gas exchange include a reduction in gill number and surface area compared to aquatic crabs (Cameron 1981), an enlarged branchial chamber (Diaz and Rodriguez 1977), branchiostegites possessing a cutaneous lung-like respiratory epithelium (Innes and Taylor 1986) and water retention in the branchial chambers (Wood and Randall 1981). The branchial chambers of both aquatic and terrestrial crabs are ventilated by scaphognathites that can pump either water or air (Wilkens and Young 1992).
Osmoregulation constitutes the main physiological process by which crustaceans maintain ionic gradients between their body fluids and the surrounding medium (Mantel and Farmer 1983; Freire et al. 2008; Morris 2001). The ability to regulate hemolymph osmotic and ionic concentrations (Péqueux 1995) is accomplished mainly by the multi-functional gills (Freire et al. 2008), together with the antennal glands (McNamara et al. 2015) and the intestine (McNamara et al. 2005). The gill epithelial cells also play a central role in hemolymph acid–base regulation and in the excretion of nitrogenous metabolic end products (Péqueux, 1995; Lucu and Towle 2003; Weihrauch et al. 2004, 2017). Considering that semi-terrestrial and terrestrial crabs often inhabit burrows whose terminal chambers lie above the water table, water availability constitutes a limiting factor, driving a suite of morphological, physiological, and behavioral adaptations that underpin both water acquisition and retention (Cameron 1981). Thus, maintaining salt and water equilibria likely represents a further physiological challenge to semi-terrestrial species in addition to less effective gas exchange compared to fully aquatic species (Gannon et al. 2000; Burggren 1992).
The phyllobranchiate gills of brachyuran crustaceans are arranged in gill chambers formed between the thoracic wall and the branchiostegites and are termed pleurobranchiae owing to their insertion into the thoracic pleura, where they receive afferent and efferent vessels (Mantel and Farmer 1983). Each gill consists of a series of numerous flattened lamellae (Taylor and Taylor 1992), and different gills exhibit notable structural and functional differences: the 4–5 anterior gill pairs are the main sites of gas exchange while the 3–4 posterior pairs constitute sites of ion transport in hyper- and/or hypo-osmoregulating species (Freire et al. 2008; Péqueux 1995; Taylor and Taylor 1992). The lamellar epithelium of the posterior gills of osmoregulating marine and brackish water crabs is symmetrically thickened and consists of ionocytes and pillar cells (Taylor and Taylor 1992). This epithelium exhibits features characteristic of ion-transporting tissues such as a numerous mitochondria associated with basal membrane invaginations (Mantel and Farmer 1983; Onken and McNamara 2002), apical evaginations (Copeland and Fitzjarrell 1968; Péqueux 1995), and elevated (Na+, K+)-ATPase (Towle et al. 1976) and carbonic anhydrase activities (Henry and Cameron 1982).
The gill-based osmoregulatory mechanisms of decapods from distinct osmotic niches have been studied intensively (Péqueux 1995; Onken and McNamara 2002; Weihrauch et al. 2004; Freire et al. 2008; McNamara and Faria 2012). While enzymes such as the (Na+, K+)-ATPase, V(H+)-ATPase and carbonic anhydrase, the Cl–/HCO3− and Na+/H+ exchangers and the Na+/K+/2Cl− symporter are involved in ion transport, their roles in osmoregulatory mechanisms are still unclear (Onken and Putzenlechner 1995; Tsai and Lin 2007; Freire et al. 2008; McNamara and Faria 2012; Henry et al. 2012).
The (Na+, K+)-ATPase is a PIIc-type integral membrane ATPase (Chourasia and Sastry 2012) that exchanges three cytosolic Na+ for two extracellular K+ across cell membranes per molecule of ATP hydrolyzed (Jorgensen et al. 2003; Martin 2005). The resulting ionic gradients guarantee the secondary transport of other ions and molecules important for various cellular functions such as salt uptake and secretion and NH4+ excretion across the gill epithelium in crustaceans (McNamara and Faria 2012). The (Na+, K+)-ATPase consists of a ≈ 110-kDa catalytic α-subunit that possesses ten transmembrane segments and connecting loops, which contain the binding sites for Na+, K+ and ATP, the D369 aspartate residue and the inhibitory ouabain-binding site (Palmgren and Nissen 2011). The enzyme also possesses a ≈ 55-kDa β-subunit involved in K+ occlusion and in the regulation of Na+ and K+ affinities, which plays an important role in the correct folding, membrane insertion, and delivery of the catalytically active α-subunits to the plasma membrane (Kaplan 2002; Durr et al. 2009). In many cell types, including crustacean gill epithelia (Silva et al. 2012), the αβ-subunit complex interacts with the gamma subunit, a small regulatory transmembrane peptide of about 7 kDa belonging to the FXYD protein family (Geering 2008).
The (Na+, K+)-ATPase occurs in two main conformations, E1 and E2, both phosphorylated at the D369 aspartate residue. During the transition from the E1P conformational state, showing high affinity for intracellular Na+, to the E2P state, characterized by high affinity for extracellular K+, the binding of K+ accelerates dephosphorylation of the E2P form (Morth et al. 2009; Clausen et al. 2017). The crystal structure of E1·AlF4−·ADP·3Na+ from pig kidney suggests that coupling between the ion- and nucleotide-binding sites is mediated by helix M5 (Kanai et al. 2013). Crystallographic studies also show that the N-domain exhibits either low- or high-affinity ATP binding sites, depending on conformational state (Morth et al. 2007; Shinoda et al. 2009; Chourasia and Sastry 2012; Nyblom et al. 2013). In the presence of Na+, the E1 conformation binds ATP with high affinity (Kanai et al. 2013; Nyblom et al. 2013) while in the presence of K+, the E2 conformation binds ATP with low affinity (Morth et al. 2007; Shinoda et al. 2009). The high-affinity ATP binding site can be induced by Na+-like substances such as Tris+ (Middleton et al. 2015; Jiang et al. 2017). However, while these ions promote nucleotide binding with similar efficiency, the enzyme does not assume the Na+-like E1 form, low K+ leading to the dissociation of bound ADP (Middleton et al. 2015).
Magnesium ions are an essential cofactor for the (Na+, K+)-ATPase (Apell et al. 2017), and while not transported, Mg2+ is essential for activity; no phosphorylation occurs in its absence (Glynn 1985; Karlish 2003; Leone et al. 2017; Apell et al. 2017). Free Mg2+ concentrations above 3 mmol L−1 apparently shift the E1–E2 equilibrium toward the E2 conformation (Tribuzy et al. 2002). Under physiological conditions, Mg2+ (≤ 2 mmol L−1) is both a modulator and substrate of the (Na+, K+)-ATPase (Glynn 1985; Karlish 2003; Apell et al. 2017). The binding of ATP as a Mg2+ complex to the ATP binding site at the cytoplasmic N-domain of the (Na+, K+)-ATPase allows phosphorylation of the aspartate residue in the P domain (Glynn 1985; Karlish 2003; Apell et al. 2017).
The red mangrove crab Goniopsis cruentata Latreille 1803, known as the “maria mulata” or “aratu vermelho” in Brazil, is a semi-terrestrial brachyuran widely distributed throughout the western Atlantic Ocean, from southern Florida in the United States of America to southern Brazil, where it occurs from Pará to Santa Catarina States (Melo 1996). The crabs are found in salinities ranging from 6 to 33‰S, and inhabit cracks, logs, fallen tree trunks and roots, and are unable to survive for more than 48 h in or out of water (Zanders 1978). The gill epithelium consists of ionocytes and pillar cells that traverse the gill lamellae at regular intervals (Martelo and Zanders 1986). The basal region of the ionocytes is characterized by extensive invaginations associated with mitochondria which increase in crabs acclimated to low salinity. Epithelial ultrastructure does not differ appreciably in crabs acclimated to 53‰S compared to 35‰S, although the apical evaginations increase both in number and in height (Martelo and Zanders 1986).
Semi-terrestrial crabs capable of living in air or water rely on their gills for gas exchange when in water but can switch to the cutaneous epithelium of the lung-like branchiostegite when in air (Maitland 1990). However, both water and air likely pass over the gills of G. cruentata, since a well-developed respiratory epithelium lining the branchial chamber appears to be absent (Little 1983). Although considered a transitional crab, G. cruentata is found mainly in the lower, wetter parts of mangroves, in tree roots or burrows containing some water and, like the sympatric blue land crab Cardisoma guanhumi (Wood and Randall 1981), can retain water in the branchial chamber when active in air. The variable salinity conditions of the mangrove habitat and the scarce information available on the biochemical processes underlying ion transport by G. cruentata gills make this semi-terrestrial species an attractive model in which to characterize the gill (Na+, K+)-ATPase.
In this study, we provide an extensive kinetic characterization of (Na+, K+)-ATPase activity in a posterior gill microsomal fraction from fresh caught G. cruentata. We also investigate the enzyme distribution in the microsomal fraction and its cellular localization in the posterior gill lamellae. This is the first study to characterize the gill (Na+, K+)-ATPase in the red mangrove crab G. cruentata.
Materials and Methods
Material
All solutions were prepared using Millipore MilliQ ultrapure, apyrogenic water. Tris (hydroxymethyl) aminomethane, ATP di-Tris salt, NADH, NAD+, pyruvate kinase (PK), phosphoenolpyruvate (PEP), imidazole, N-(2-hydroxyethyl) piperazine-N′-ethane sulfonic acid (HEPES), lactate dehydrogenase (LDH), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ouabain, phosphoglycerate kinase (PGK), 4′,6-diamidino-2-phenylindole (DAPI), nitroblue tetrazolium (NBT), 5-bromo-4-chloro-3-indole phosphate (BCIP), alamethicin, bovine serum albumin, theophylline, thapsigargin, aurovertin, ethacrynic acid, ethylene glycol tetraacetic acid (EGTA), ethylene diamine tetraacetic acid (EDTA), bafilomycin A1, sodium orthovanadate, and anti-mouse IgG alkaline phosphatase conjugate were purchased from the Sigma Chemical Company (Saint Louis, USA). Ethanol, dimethyl sulfoxide (DMSO) and triethanolamine (TEA) were from Merck (Darmstadt, Germany). The protease inhibitor cocktail (1 mmol L−1 benzamidine, 5 µmol L−1 antipain, 5 µmol L−1 leupeptin, 1 µmol L−1 pepstatin A and 5 µmol L−1 phenyl-methane-sulfonyl-fluoride) was from Calbiochem (Darmstadt, Germany). Mouse monoclonal IgG α-5 antibody raised against chicken (Na+, K+)-ATPase α-subunit was from the Development Studies Hybridoma Bank (Iowa, USA). Spectra Multicolor Broad Range Protein Ladder (10–260 kDa) was purchased from Thermo Scientific (Rockford, USA). Alexa-fluor 488 conjugated donkey anti-mouse IgG was from Invitrogen (Carlsbad, USA). Optimal Cutting Temperature Compound was from Sakura Tissue-Tek (Torrance, USA); Fluoromount-G and p-formaldehyde were purchased from Electron Microscopy Sciences (Hatfield, USA). Polyvinylidene difluoride (PVDF) membranes were from BioRad (Hercules, USA). Ammonium sulfate-depleted PK, LDH, GAPDH, and PGK suspensions and stock solution of ATP were prepared according to Leone et al. (2015). When necessary, enzyme solutions were concentrated on YM-10 Amicon Ultra filters. All cations were used as chloride salts.
Crabs and Gill Dissection
Adult Goniopsis cruentata were caught using baited traps from Barra Seca mangrove (23° 24′ 58.9″ S, 45° 3′ 2.9″ W), Ubatuba, São Paulo State, Brazil. The crabs were transported to the laboratory in individual plastic boxes (20 × 20 cm2) containing a 1-cm layer of mangrove water (21‰S) and then held in glass aquaria containing a 1-cm layer of mangrove water with free access to a dry surface for 1–2 days, at ≈ 25 °C, under a natural photoperiod of 14-h daylight until preparation of the microsomal fraction. Crabs were fed on alternate days with shrimp tails, and the uneaten fragments were removed the following morning.
For each homogenate prepared, 5–10 intermolt specimens of about 3 cm carapace width were anesthetized by chilling in crushed ice for 5 min and then killed by sectioning the cerebral and thoracic ganglia. The four posterior most gill pairs (approximately 1.5 g wet weight) were rapidly excised and placed in ice-cold homogenization buffer (20 mmol L−1 imidazole buffer, pH 6.8, containing 250 mmol L−1 sucrose, 6 mmol L−1 EDTA, Tris.HCl 6 mmol L−1 and a protease inhibitor cocktail).
Preparation of the Gill Microsomal Fraction
The gills were rapidly diced and homogenized in a Potter homogenizer (600 rpm) in homogenization buffer (20 mL buffer/g wet gill tissue). After centrifuging the crude extract at 20,000×g for 35 min at 4 °C, the supernatant was placed on crushed ice and the pellet was resuspended in an equal volume of homogenization buffer. After a second centrifugation as above, the two supernatants were gently pooled and centrifuged at 100,000×g for 90 min at 4 °C. The resulting pellet containing the microsomal fraction was homogenized in homogenization buffer (15 mL buffer/g). Finally, 0.5-mL aliquots were rapidly frozen in liquid nitrogen and stored at − 20 °C. No appreciable loss of (Na+, K+)-ATPase activity was seen after 4 months’ storage of the microsomal preparation. All experiments were performed using the microsomal preparation held in a crushed ice bath for no longer than 4 h.
Measurement of Protein Concentration
Protein concentration was estimated according to Read and Northcote (1981) using bovine serum albumin as the standard.
Continuous-Density Sucrose Gradient Centrifugation
To accompany the distribution of the (Na+, K+)-ATPase in the microsomal preparation, an aliquot (2.5 mg protein) of the ATPase-rich gill microsomal preparation was layered into a 10 to 50% (w/w) continuous-density sucrose gradient in 20 mmol L−1 imidazole buffer, pH 6.8 and centrifuged at 180,000×g and 4 °C, for 3 h, using a Hitachi PV50T2 vertical rotor. Fractions (0.5 mL) collected from the bottom of the gradient were then assayed for total ATPase activity, ouabain-insensitive ATPase activity, protein concentration, and refractive index.
SDS-PAGE and Western Blotting Analyses
SDS-PAGE of the microsomal preparation from the posterior gill tissue was performed in 5–20% gels as described by Laemmli (1970), using 4 and 50 μg protein/lane for protein staining and blotting analyses, respectively. After electrophoresis, the gel was split, one half being stained with silver nitrate and the other electro-blotted in a Hoefer SE200 system using a nitrocellulose membrane (Towbin et al. 1979). The membrane was then blocked with 10% bovine serum albumin, freshly prepared in TBS-Tween buffer (50 mmol L−1 Tris–HCl buffer, pH 8.0, containing 150 mmol L−1 NaCl and 0.1% Tween 20) for 90 min under constant agitation. After washing, the membrane was incubated overnight at 4 °C in a 1:150 dilution of the α-5 monoclonal antibody, rewashed, and incubated with an alkaline phosphatase conjugated, anti-IgG mouse secondary antibody. Specific antibody binding was developed in 100 mmol L−1 Tris–HCl buffer, pH 9.5, containing 100 mmol L−1 NaCl, 5 mmol L−1 MgCl2, 0.2 mmol L−1 NBT, and 0.8 mmol L−1 BCIP. Western blotting was also performed in 6–12% SDS-PAGE gels using 50 μg protein/lane. All analyses were repeated three times using different microsomal preparations.
Immunolocalization of the Gill (Na+, K+)-ATPase
Seventh, right-side posterior gills were dissected and incubated in a fixative solution containing 2% p-formaldehyde in a phosphate-buffered saline (PBS, 10 mmol L−1 Na2HPO4, 2 mmol L−1 KH2PO4, 137 mmol L−1 NaCl, 2.7 mmol L−1 KCl, 290 mOsm kg−1 H2O), pH 7.4, for 1 h, then embedded in Optimal Cutting Temperature Compound Tissue-Tek® (Sakura Finetek Inc., Torrance, CA). Briefly, 15-μm thick cryosections were taken parallel to the gill long axis at − 20 °C using a Microm HM 505E Cryostatic Microtome (Walldorf, Germany). The sections were pre-incubated for 20 min with 100 mmol L−1 glycine in PBS and then for 10 min in a blocking solution containing 1% bovine serum albumin plus 0.1% gelatin in PBS. The (Na+, K+)-ATPase α-subunit was immunolocalized using a primary monoclonal mouse IgG α-5 anti-chicken (Na+, K+)-ATPase antibody and a secondary donkey anti-mouse IgG antibody conjugated with Alexa-fluor 488 according to Pinto et al. (2016). To locate nuclei, sections were stained for 20 min with DAPI, diluted 1:200 in PBS. Sections were mounted with Fluoromount-G slide mounting medium on Knittel Starfrost slides with coverslips (Bielefeld, Germany). Confocal images were obtained with a Leica TCS SP5 confocal laser scanning microscope (Leica Microsystems, Mannheim, Germany), using an argon laser for Alexa-fluor 488 staining or a diode laser for DAPI staining.
Measurement of (Na+, K+)-ATPase Activity
Total ATPase activity was assayed at 25 °C using a PK/LDH coupling system in which ATP hydrolysis was coupled to NADH oxidation (Leone et al. 2015). In this system, the phosphate released during ATP hydrolysis by the (Na+, K+)-ATPase is converted to ATP while phosphoenolpyruvate is converted to pyruvate by pyruvate kinase. Pyruvate is converted to lactate concomitant to NADH oxidation. The rate of ATP hydrolysis by the (Na+, K+)-ATPase can be estimated by monitoring NADH oxidation. The oxidation of NADH was monitored at 340 nm (ε340nm, pH7.5 = 6200 M−1 cm−1) in a Shimadzu UV-1800 spectrophotometer equipped with thermostated cell holders. Standard conditions were as follows: 50 mmol L−1 HEPES buffer, pH 7.5, containing 1 mmol L−1 ATP, 3 mmol L−1 MgCl2, 50 mmol L−1 NaCl, 10 mmol L−1 KCl, 10 µg alamethicin, 0.21 mmol L−1 NADH, 3.18 mmol L−1 PEP, 82 mg PK (49 U), 110 mg LDH (94 U), plus the microsomal preparation (10–20 μL), in a final volume of 1 mL. The activity was also assayed under stoichiometric ATP and Mg2+ concentrations.
Alternatively, ATPase activity was estimated using a GAPDH/PGK-linked system coupled to the reduction of NAD+ at 340 nm (Leone et al. 2015). This system was used only to examine the effects of K+ and NH4+ since K+ is a cofactor for pyruvate kinase. Standard conditions were: 50 mmol L−1 HEPES buffer, pH 7.5, containing 1 mmol L−1 ATP, 3 mmol L−1 MgCl2, 50 mmol L−1 NaCl, 10 mmol L−1 KCl, 10 µg alamethicin, 4.28 mmol L−1 NAD+, 1 mmol L−1 sodium phosphate, 2 mmol L−1 G3P, 150 mg GAPDH (24 U), 20 mg PGK (9 U), and the microsomal preparation (10–20 μL), in a final volume of 1 mL. The two coupling systems gave equivalent results with a difference of less than 10%.
ATP hydrolysis was also estimated using 3 mmol L−1 ouabain to assess ouabain-insensitive activity. The difference in activity measured without (total ATPase activity) or with ouabain (ouabain-insensitive activity) represents the (Na+, K+)-ATPase activity. ATP hydrolysis was also estimated after 10 min pre-incubation with alamethicin (1 μg/mg microsomal protein) to verify the presence of sealed vesicles. Controls without added enzyme were also included in each experiment to quantify non-enzymatic substrate hydrolysis. Initial velocities were constant for at least 15 min provided that less than 5% of the total NADH (or NAD+) was oxidized (or reduced). Neither NADH, PEP, LDH, PK, NAD+, G3P, PGK nor GAPDH was rate-limiting over the initial course of the assay, and no activity could be measured without NADH or NAD+. (Na+, K+)-ATPase activity was checked for linearity between 10 and 50 mg total protein; total microsomal protein added to the cuvette always fell well within the linear range of the assay.
For each ATP, Mg2+, Na+, K+, NH4+ and ouabain concentration, the reaction rate was estimated in duplicate using identical aliquots from the same preparation. The mean values for the duplicates were used to fit the corresponding saturation curves, each of which was repeated three times using a different microsomal homogenate (N = 3). The curves provided in the figures were fitted using the mean values (± SD, N = 3) for each of the three different homogenates. One enzyme unit (U) is defined as the amount of enzyme that hydrolyzes 1.0 nmol of ATP per minute, at 25 °C, and the (Na+, K+)-ATPase specific activity (U mg−1) appearing in the figures represents nmol Pi min−1 mg−1 protein.
Synthesis of [γ-32P]-ATP
Synthesis of γ32P-ATP disodium salt was performed as described by Walseth and Johnson (1979) modified by Maia et al. (1983).
Phosphorylation/dephosphorylation of gill microsomes by [γ-32P]-ATP
Aliquots of the gill microsomal (Na+, K+)-ATPase containing 40 μg protein were incubated at 27 °C for 10 min in an assay medium consisting of 50 mmol L−1 HEPES buffer, pH 7.5, 0.03 mmol L−1 MgCl2 and 50 mmol L−1 NaCl. The phosphorylation reaction was started by adding 0.02 mL 0.75 mmol L−1 ATP/γ32P-ATP (specific activity of 1 × 105 cpm/nmol) in a final volume of 0.5 mL (control E-P). After standing for 3 min at 27 °C, dephosphorylation was initiated by adding 0.05 mL 50 mmol L−1 ADP (ADP-induced dephosphorylation) or 0.025 mL of 2 mol L−1 KCl (K+-induced dephosphorylation) to each phosphorylation medium, the reaction proceeding for 5 min.
After stopping the reaction by adding 2 mL 125 mmol L−1 perchloric acid containing 5 mmol L−1 phosphoric acid and 5 mmol L−1 sodium pyrophosphate, the resulting mixture containing the 32P-phosphorylated enzyme (E-P) was filtered on 0.45-μm HAWP Millipore filters. The filters were washed three times with 2 mL 125 mmol L−1 perchloric acid and four times with 50 mmol L−1 perchloric acid (Fontes et al. 1992) to reduce signal in the blanks, and dried. Radioactivity was counted using a Beckman LS6500 liquid scintillation counter. Controls were performed in triplicate using the enzyme denatured previously with perchloric acid before the addition of [γ-32P]-ATP. Each experiment was repeated three times using a different microsomal preparation (N = 3). Results are given as the mean ± SD.
Estimation of Kinetic Parameters
SigrafW (Leone et al. 2005) was used to calculate the kinetic parameters VM (maximum velocity), K0.5 (apparent dissociation constant), KM (Michaelis–Menten constant), and the nH (Hill coefficient) values for ATP hydrolysis under the different assay conditions. The apparent dissociation constant, KI, of the enzyme-inhibitor complex was estimated as described by Leone et al. (2015).
The kinetic parameters furnished in the tables are calculated values and represent the mean ± SD derived from three different microsomal preparations (N = 3). The data were submitted to descriptive statistical analysis for parametric representation. SigrafW software can be freely obtained from http://portal.ffclrp.usp.br/sites/fdaleone/downloads.
Calculation of Free ATP, Mg2+, and ATP-Mg Species in Solution
The concentrations of free species of ATP, Mg2+, and ATP-Mg in solution were calculated according to Fabiato (1988) in congruence with the following equilibria:

in which pK1 = 6.53, pK2 = 4.06, pK3 = 1.00, pK4 = 1.00, pK5 = 4.22, pK6 = 2.24, pK7 = 1.58 and Me = Mg2+.
Results
Immunolocalization of the Gill (Na+, K+)-ATPase
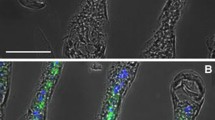
Fluorescent immunolabeling revealed that the (Na+, K+)-ATPase is distributed evenly throughout the gill lamellae, most epidermal cells showing intense signal (Fig. 1). The lamellae consist of a single-layered epithelium overlain by cuticle. Occasional paired pillar cells interspersed among the ionocytes project from either side of the lamella, narrowing at their opposing basal regions in the lamella center. The spaces between opposing pillar cell pairs form a system of irregular lacunae through which the hemolymph flows. Detailed immunolabeling showed that the (Na+, K+)-ATPase is distributed throughout the entire epithelial cell cytoplasm.
Subcellular immunolocalization of the (Na+, K+)-ATPase in posterior gill lamellae from Goniopsis cruentata. Phase contrast/confocal microscopy of the gill epithelium in a cryosection taken along the longitudinal gill axis revealing the lamellar architecture of the seventh gill. c Cuticle, pc pillar cells, h hemolymph lacunae. (Na+, K+)-ATPase α-subunit immunolabeling (green) is intense throughout the epidermal cell cytoplasm. Nuclei (blue) are stained with DAPI. Immunolabeling with mouse monoclonal IgG α-5 antibody against chicken (Na+, K+)-ATPase α-subunit. Fluorescent signal revealed with donkey anti-mouse IgG secondary antibody conjugated with Alexa-fluor 488 (495/519 nm). Scale bar 50 μm
Continuous-Density Sucrose Gradient Centrifugation Analysis
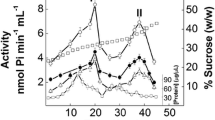
Centrifugation of the (Na+, K+)-ATPase microsomal preparation from the posterior gills of freshly caught crabs revealed an ATPase activity peak with a heavy shoulder lying between 25 and 35% sucrose density and apparently distributed into two membrane fractions partially separated along the gradient (Fig. 2). The fraction appearing at the lower sucrose concentrations showed a protein content twofold less than that at the higher concentrations. The ouabain-insensitive ATPase activity measured along the gradient suggests that the microsomal fraction contains ATP hydrolyzing enzymes other than the (Na+, K+)-ATPase.
Sucrose density gradient centrifugation of a microsomal fraction from the posterior gill tissue of Goniopsis cruentata. An aliquot containing 2.5 mg protein was layered into a 10–50% (w/w) continuous sucrose density gradient. Fractions (0.5 mL) were collected from the bottom of the gradient and were analyzed in duplicate for total ATPase activity (filled circle); ouabain-insensitive ATPase activity (filled square); (Na+, K+)-ATPase activity (open circle); protein concentration (open triangle) and sucrose concentration (open square)
SDS-PAGE and Western Blotting Analyses
SDS-PAGE of the microsomal fraction revealed protein profile characteristic of membrane protein (Fig. 3a). Western blotting analyses identified a broad immunoreactive band of Mr ≈ 110–115 kDa, corresponding to the (Na+, K+)-ATPase α-subunit (Fig. 3b). This broad band appears to be characteristic of the G. cruentata enzyme since SDS-PAGE performed in a 6–12% gels also revealed only a single immunoreactive band (Fig. 3c).
SDS-PAGE and Western blot analyses of microsomal fractions from the posterior gill tissue of Goniopsis cruentata. a Silver nitrate-stained SDS-PAGE (5–20%) using 4 µg gill tissue microsomal protein. b Western blotting (SDS-PAGE 5–20%) against the (Na+, K+)-ATPase α-subunit, revealed with an anti-mouse IgG alkaline phosphatase conjugate, using 50 µg gill tissue microsomal protein. c Western blotting (SDS-PAGE 6–12%) against the (Na+, K+)-ATPase α-subunit, revealed with an anti-mouse IgG alkaline phosphatase conjugate, using 50 µg gill tissue microsomal protein identifying an intensely staining band of ≈ 110 kDa. d Spectra Multicolor Broad Range Protein Ladder (10–180 kDa) standard used for 6–12% SDS-PAGE
(Na+, K+)-ATPase Activity in the Posterior Gill Microsomal Preparation
Maximum activities estimated with or without alamethicin were 512.9 ± 25.6 nmol Pi min−1 mg−1 protein and 419.4 ± 20.9 nmol Pi min−1 mg−1 protein, respectively, revealing that ≈ 20% of the ATPase is occluded in sealed vesicles. Ouabain inhibition revealed that the (Na+, K+)-ATPase represents up to 95% of the total ATPase activity of the microsomal preparation.
Increasing ATP concentrations from 10−7 to 2 × 10−3 mol L−1 stimulated (Na+, K+)-ATPase activity to a maximum of ≈ 480 nmol Pi min−1 mg−1 protein following a well-defined, biphasic saturation curve (Fig. 4a). Under saturating Mg2+ (3 mmol L−1), K+ (10 mmol L−1), and Na+ (50 mmol L−1) concentrations, the low-affinity ATP binding site showed VM = 186.0 ± 9.3 nmol Pi min−1 mg−1 protein and K0.5 = 0.085 ± 0.004 mmol L−1 while the high-affinity ATP binding site, accounting for 50% of (Na+, K+)-ATPase activity, showed VM = 153.4 ± 7.7 nmol Pi min−1 mg−1 protein and K0.5 = 0.013 ± 0.001 mmol L−1 (Table 1). ATP hydrolysis obeyed cooperative kinetics (Table 1). These data show that the increase in (Na+, K+)-ATPase activity correlates with the increase in Mg-ATP concentration, and not with free ATP, since for each total ATP concentration assayed, Mg-ATP concentration is ≈ 100-fold greater than that of free ATP (Fig. 4b). A maximum ouabain-insensitive ATPase activity of ≈ 30 nmol Pi min−1 mg−1 protein was measured over the same ATP concentration range (inset to Fig. 4a). A significant (Na+, K+)-ATPase activity of ≈ 140 nmol Pi min−1 mg−1 protein was found at ATP concentrations as low as 10−7 mmol L−1.
Effect of ATP concentration on (Na+, K+)-ATPase activity from the posterior gill tissue of Goniopsis cruentata. Activity was assayed continuously at 25 °C in 50 mmol L−1 HEPES buffer, pH 7.5, containing 3 mmol L−1 MgCl2, 50 mmol L−1 NaCI, 10 mmol L−1 KCl, 10 µg alamethicin, 0.21 mmol L−1 NADH, 3.18 mmol L−1 PEP, 49U PK and 94 U LDH, using duplicate aliquots from the same microsomal preparation (about 44 µg protein) as described in the Materials and Methods. Mean values (± SD) for the duplicates were used to fit each corresponding curve, which was repeated utilizing three different microsomal preparations (N = 3). a Modulation by ATP. Inset: total ATPase activity (filled circle), ouabain-insensitive ATPase activity (open circle). b Calculated concentrations of free species in solution: ATP (filled circle), Mg2+ (open circle) and Mg·ATP (open square)
Under stoichiometric concentrations of ATP and Mg2+, a single saturation curve was revealed as Mg-ATP concentration increased from 10−8 to 7 × 10−4 mol L−1 (Fig. 5a). The substrate was hydrolyzed following site–site interactions with VM = 443.0 ± 22.1 nmol Pi min−1 mg−1 protein and K0.5 = 0.060 ± 0.003 mmol L−1 (Table 1). The ouabain-insensitive ATPase activity (30 nmol Pi min−1 mg−1 protein) also was stimulated up to 60 nmol Pi min−1 mg−1 protein over the same Mg-ATP concentration range (inset to Fig. 5a). Under stoichiometric ATP and Mg2+ concentrations, steady-state analysis of (Na+, K+)-ATPase activity showed ≈ 20 nmol Pi min−1 mg−1 protein for ATP concentrations as low as 10−8 mol L−1 compared to ≈ 140 nmol Pi min−1 mg−1 protein seen under excess Mg2+ (see Fig. 4a).
(Na+, K+)-ATPase activity from the posterior gill tissue of Goniopsis cruentata assayed under stoichiometric ATP and MgCl2 concentrations. Activity was assayed continuously at 25 °C in 50 mmol L−1 HEPES buffer, pH 7.5, containing 50 mmol L−1 NaCI, 10 mmol L−1 KCl, 10 µg alamethicin, 0.21 mmol L−1 NADH, 3.18 mmol L−1 PEP, 49U PK and 94U LDH, using duplicate aliquots from the same microsomal preparation (about 44 µg protein) as described in Materials and Methods. Mean values (± SD) for the duplicates were used to fit each corresponding curve, which was repeated utilizing three different microsomal preparations (N = 3). a Modulation by Mg-ATP. Inset: total ATPase activity (filled circle), ouabain-insensitive ATPase activity (open circle). b Calculated concentrations of free species in solution: ATP (filled circle filled circle), Mg2+ (open square), and Mg-ATP (open circle)
In the absence of Mg2+, no (Na+, K+)-ATPase activity was found. Under saturating ATP (1 mmol L−1), K+ (10 mmol L−1) and Na+ (50 mmol L−1) concentrations, increasing Mg2+ concentrations from 10−5 to 2 × 10−3 mol L−1 stimulated (Na+, K+)-ATPase activity up to 425.9 ± 25.5 nmol Pi min−1 mg−1 protein, following cooperative kinetics (nH = 2.6) with K0.5 = 0.16 ± 0.01 mmol L−1 (Fig. 6a and Table 1). Low (Na+, K+)-ATPase activity (less than 40 nmol Pi min−1 mg−1 protein) was registered at Mg2+ concentrations below 10−5 mol L−1. Stimulation of ouabain-insensitive ATPase activity over the same Mg2+ concentration range (inset to Fig. 6a) represented less than 10% of the total ATPase activity. Analysis of the free species of ATP, Mg2+, and Mg·ATP in solution confirmed that the stimulation of (Na+, K+)-ATPase activity is due to Mg·ATP and not to free Mg2+ (Fig. 6b). At 10−5 mol L−1 MgCl2, Mg·ATP concentration is almost tenfold greater than that of free Mg2+.
Effect of magnesium ion concentration on (Na+, K+)-ATPase activity from the posterior gill tissue of Goniopsis cruentata. Activity was assayed continuously at 25 °C in 50 mmol L−1 HEPES buffer, pH 7.5, containing 1 mmol L−1 ATP, 50 mmol L−1 NaCl, 10 mmol L−1 KCl, 0.21 mmol L−1 NADH, 10 µg alamethicin, 3.18 mmol L−1 PEP, 49U PK, and 94U LDH, using duplicate aliquots from the same microsomal preparation (about 44 μg protein) as described in Materials and Methods. Mean (± SD) values for the duplicates were used to fit each corresponding curve, which was repeated utilizing three different microsomal preparations (N = 3). a Modulation by MgCl2—Inset: total ATPase activity (filled circle), ouabain-insensitive ATPase activity (open circle). b Calculated concentrations of free species in solution: ATP (filled triangle), Mg2+ (open circle), and Mg·ATP (filled circle)
Under saturating ATP (1 mmol L−1), Mg2+ (3 mmol L−1), and K+ (10 mmol L−1) concentrations, the Na+-dependent (Na+, K+)-ATPase activity showed a saturation curve obeying cooperative kinetics with VM = 425.0 ± 23.4 nmol Pi min−1 mg−1 protein and K0.5 = 5.1 ± 0.3 mmol L−1 (Fig. 7 and Table 1). A ouabain-insensitive ATPase activity of around 10% was found over the same Na+ concentration range (inset to Fig. 7) although there was no stimulation by increasing Na+ concentration.
Effect of sodium ion concentration on (Na+, K+)-ATPase activity from the posterior gill tissue of Goniopsis cruentata. Activity was assayed continuously at 25 °C in 50 mmol L−1 HEPES buffer, pH 7.5, containing 1 mmol L−1 ATP, 3 mmol L−1 MgCl2, 10 mmol L−1 KCl, 10 µg alamethicin, 0.21 mmol L−1 NADH, 3.18 mmol L−1 PEP, 49U PK and 94U LDH, using duplicate aliquots from the same microsomal preparation (about 44 μg protein) as given in Materials and Methods. Mean (± SD) values for the duplicates were used to fit each corresponding curve, which was repeated utilizing three different microsomal preparations (N = 3). Inset: total ATPase activity (filled circle), ouabain-insensitive ATPase activity (open circle)
Individually, both K+ and NH4+ stimulated (Na+, K+)-ATPase activity (Fig. 8). In the absence of NH4+, under saturating ATP (1 mmol L−1), Mg2+ (3 mmol L−1), and Na+ (50 mmol L−1) concentrations, K+ from 10−6 to 3 × 10−2 mol L−1 stimulated the enzyme following Michaelis–Menten kinetics (nH = 1.0), reaching VM = 485.3 ± 24.3 nmol Pi min−1 mg−1 protein with K0.5 = 0.9 ± 0.05 mmol L−1 (Fig. 8a and Table 1). The ouabain-insensitive ATPase activity was unaffected over this K+ concentration range (inset to Fig. 8a). In the absence of K+, NH4+ between 10−5 mol L−1 and 7 × 10−2 mol L−1 also stimulated enzyme activity, obeying cooperative kinetics (Fig. 8b). Under the above saturating conditions, VM was 497.9 ± 24.9 nmol Pi min−1 mg−1 protein with K0.5 = 9.7 ± 0.50 mmol L−1 (Table 1). Ouabain-insensitive ATPase activity was unaltered over this NH4+ concentration range (inset to Fig. 8b). Further, NH4+ plus K+ only stimulated (Na+, K+)-ATPase activity slightly (< 10%, data not shown).
Effect of potassium and ammonium ion concentrations on (Na+, K+)-ATPase activity from the posterior gill tissue of Goniopsis cruentata crab. Activity was assayed continuously at 25 °C in 50 mmol L−1 HEPES buffer, pH 7.5, containing 1 mmol L−1 ATP, 3 mmol L−1 MgCl2, 50 mmol L−1 NaCl, 10 µg alamethicin, 4.28 mmol L−1 NAD+, 1 mmol L−1 sodium phosphate, 2 mmol L−1 G3P, 12U GAPDH and 9U PGK using duplicate aliquots from the same microsomal preparation (about 44 μg protein) as described in Materials and Methods. Mean values (± SD) for the duplicates were used to fit each corresponding curve, which was repeated utilizing three different microsomal preparations (N = 3). a Modulation by K+ in the absence of NH4+. b Modulation by NH4+ in the absence of K+ Insets: total ATPase activity (filled circle), ouabain-insensitive ATPase activity (open circle)
Analysis of Phosphoenzyme Formation/Decomposition
Phosphoenzyme formation/decomposition was analyzed as a function of Mg2+ concentration to better understand why the high-affinity ATP binding site was not exposed at stoichiometric ATP and Mg2+ concentrations (Table 2). Phosphorylation with 0.03 mmol L−1 ATP and 0.03 mmol L−1 Mg2+ showed a maximum titer of phosphorylated enzyme of 0.283 ± 0.002 nmol Pi mg−1 protein, considered to be the control titer for E-P formation. K+-induced (100 mmol L−1) dephosphorylation was 0.247 ± 0.009 nmol Pi mg−1 (≈ 87% control titer), suggesting that only 13% of the total phosphorylated enzyme was dephosphorylated at 2.244 × 10−5 mol L−1 Mg2+. In contrast, at 5 mmol L−1 ADP, just 0.078 ± 0.004 nmol L−1 (≈ 27% control titer) of phosphorylated enzyme was measured, suggesting that ≈ 73% of the phosphorylated enzyme was dephosphorylated.
Inhibition by Ouabain
The monophasic inhibition curve seen using 10−5 to 5 × 10−3 mol L−1 ouabain suggests a single ouabain-binding site (Fig. 9). The decrease in activity to ≈ 50 nmol Pi min−1 mg−1 protein suggests that ≈ 95% of the microsomal ATPase activity corresponds to the (Na+, K+)-ATPase. The calculated KI value for ouabain inhibition was 196.6 ± 9.8 μmol L−1 (inset to Fig. 9, Table 1).
Inhibition by ouabain of (Na+, K+)-ATPase activity from the posterior gill tissue of Goniopsis cruentata. Activity was assayed continuously at 25 °C in 50 mmol L−1 HEPES buffer, pH 7.5, containing 1 mmol L−1 ATP, 50 mmol L−1 NaCI, 3 mmol L−1 MgCl2, 10 mmol L−1 KCl, 10 µg alamethicin, 0.21 mmol L−1 NADH, 3.18 mmol L−1 PEP, 49U PK and 94U LDH, using duplicate aliquots from the same microsomal preparation (about 44 μg protein) as described in Materials and Methods. Mean values (± SD) for the duplicates were used to fit each corresponding curve, which was repeated utilizing three different microsomal preparations (N = 3). Inset: Dixon plot for estimation of KI in which vc is the reaction rate corresponding to (Na+, K+)-ATPase activity alone
Discussion
We provide an extensive kinetic characterization of the (Na+, K+)-ATPase from the posterior ion-transporting gills of the red mangrove crab Goniopsis cruentata. The (Na+, K+)-ATPase constitutes up to 95% of total gill ATPase activity in the microsomal preparation, is distributed evenly throughout the gill epithelium, and has an Mr of ≈ 110 kDa. At very low ATP concentrations and under excess Mg2+, a high-affinity ATP binding site accounting for 50% of total (Na+, K+)-ATPase activity was revealed. The enzyme is stimulated by NH4+; however, synergistic stimulation by K+ plus NH4+ was not seen.
The epithelial architecture of the phyllobranchiate gill lamellae of G. cruentata conforms to the typical crab plan (Freire et al. 2008). The lamellae consist of a single-layered epithelium overlain by cuticle, sustained by occasional paired pillar cells. The (Na+, K+)-ATPase is located throughout all the epidermal cells, as seen in Cardisoma guanhumi (Farias et al. 2017) but contrasting with Callinectes sapidus and Carcinus maenas in which the enzyme is located in the basal invaginations of the epithelial ionocytes among the pillar cells (Towle and Kays 1986).
The single (Na+, K+)-ATPase peak with a heavy shoulder (see Fig. 2) revealed by sucrose gradient centrifugation is similar to the euryhaline blue crab Callinectes danae (Masui et al. 2002, 2009) but contrasts with findings for the semi-terrestrial mangrove crab Cardisoma guanhumi(Farias et al. 2017), the freshwater crab Dilocarcinus pagei (Furriel et al. 2010) and the blue crab Callinectes ornatus (Garçon et al. 2007), all of which show two distinct (Na+, K+)-ATPase peaks. Gill homogenization may produce membrane fragments derived from different regions of the gill epithelium or with distinct lipid to protein ratios (Furriel et al. 2010; Lucena et al. 2012), which might account for membrane fragments of different densities. However, whether these differences are species-specific or are artifacts resulting from the homogenization process remains to be established.
The broad immunoreactive band seen by Western blot (see Fig. 3b, c) suggests that only a single (Na+, K+)-ATPase α-subunit isoform is present in the microsomal fraction as seen in several crustacean species (Masui et al. 2002; Garçon et al. 2007; Leone et al. 2014, 2015). The existence of just a single isoform is corroborated by the single ouabain titration curve (see Fig. 9).
Compared to the sympatric semi-terrestrial crab C. guanhumi (Farias et al. 2017), both the high-affinity ATP binding site and the elevated basal (Na+, K+)-ATPase activity of ≈ 140 nmol Pi min−1 mg−1 protein seen at ATP concentrations as low as 10−7 mmol L−1 represent novel kinetic characteristics of the G. cruentata enzyme. This high basal (Na+, K+)-ATPase activity is not due to ATPase activities other than the (Na+, K+)-ATPase since the ouabain-insensitive ATPase activity is only 30 nmol Pi min−1 mg−1 protein (see Fig. 4a) and is stimulated up to 60 nmol Pi min−1 mg−1 protein over the same Mg·ATP concentration range (see inset to Fig. 4b). Rather, such activity is a consequence of excess free Mg2+ that triggers the exposure of a high-affinity ATP binding site (see Fig. 5 and Table 2).
High- and low-affinity ATP binding sites on the (Na+, K+)-ATPase are well known (Glynn 1985; Ward and Cavieres 1998). However, despite the plethora of crystallographic data suggesting a binding site within the (Na+, K+)-ATPase N-domain (Kanai et al. 2013; Nyblom et al. 2013; Chourasia and Sastry 2012; Shinoda et al. 2009; Morth et al. 2007), its exact localization on the enzyme molecule remains an open question (Morth et al. 2007). Experiments employing the fluorescent dye RH-421 and techniques like fluorescence energy transfer, molecular docking and site-directed mutagenesis on the (Na+, K+)-ATPase C45 loop fragment that contains the N and P domains, have identified the Phe482 residue as the center of the nucleotide-binding pocket responsible for orienting the adenine moiety during ATP binding (Kubala et al. 2003; Huliciak et al. 2017). Molecular dynamics studies (Tejral et al. 2017) postulate new cytoplasmic domain subconformations in which the N-domain alternates between “open,” “semi-open,” and “closed” conformations. The “open” and “semi-open” conformations (E2-related forms, with or without occluded K+) may bind two ATP molecules simultaneously (GROMACS data). However, in the “closed” conformation (E1 and Na+-related form) only a single ATP molecule can bind and be hydrolyzed with subsequent transphosphorylation of the Asp369 residue. These findings are similar to those for crystal structures with a single interchangeable ATP site. The high-affinity ATP binding site of the G. cruentata enzyme accounts for ≈ 60% of (Na+, K+)-ATPase activity, in contrast to just 10% for the mammalian and some crustacean (Na+, K+)-ATPases (Leone et al. 2017). With these exceptions, the kinetic characteristics of the low- and high-affinity binding sites are similar to those of Ucides cordatus (Leone et al. 2020), C. danae (Masui et al. 2002), Clibanarius vittatus (Gonçalves et al. 2006; Lucena et al. 2012), and Macrobrachium amazonicum (Santos et al. 2007).
The allosteric regulation of P-type ATPases by Mg2+ is well established (see Apell et al. 2017). Also, free Mg2+ is required for (Na+, K+)-ATPase phosphorylation by ATP or Pi, and for the conversion of the ADP-sensitive phosphoenzyme conformation E1P to the K+-sensitive phosphoenzyme conformation E2P (Fukushima and Post 1978; Hegyvary and Jorgensen 1981). For G. cruentata, exposure of the high-affinity ATP binding site induced by excess Mg2+ may be interpreted as follows. Between 10−8 and 10−5 mol L−1 Mg·ATP, the high-affinity ATP binding site is not exposed since free Mg2+ concentrations range between 9.854 × 10−7 and 2.244 × 10−5 mol L−1 (see Fig. 5b). These concentrations are very low compared to the free Mg2+ concentration required to trigger exposure of this ATP binding site (2.970 × 10−3 mol L−1, Fig. 4b). Thus, occlusion of the high-affinity ATP binding sites under stoichiometric ATP and Mg2+ concentrations results from the very low availability of free Mg2+ that cannot sustain the E1P to E2P transition, causing a drastic reduction in ATP hydrolysis. Data from Table 2 corroborate this interpretation since 100 mmol L−1 K+ induced only 13% dephosphorylation while the addition of 5 mmol L−1 ADP induced 73% dephosphorylation. At 0.03 mmol L−1 Mg2+ and 0.03 mmol L−1 ATP, the free Mg2+ concentration (2.244 × 10−5 mol L−1, see Fig. 5b) lies far below the 2.970 × 10−3 mol L−1 necessary to expose the high-affinity ATP binding site (see Fig. 4b). Thus, the 13% dephosphorylation seen with 100 mmol L−1 K+ (see Table 2) prevails since at this concentration (2.244 × 10−5 mol L−1 free Mg2+), the enzyme is in its E1 conformation that cannot be dephosphorylated by K+. Further, the high dephosphorylation titer seen with 5 mmol L−1 ADP (see Table 2) corroborates this finding for K+, suggesting that with 2.244 × 10−5 mol L−1 Mg2+ the enzyme is in its E1 conformation, and the E1P to E2P transformation cannot proceed.
Measurements using fluorescein and eosin to detect E1P to E2P transitions when performed with increasing free Mg2+ concentrations show that the K0.5 for Mg2+ related to the E2P transition lies in the range of 0.15 to 0.7 mmol L−1 (Hegyvary and Jorgensen 1981; Montes et al. 2015), a value similar to those estimated here (see Table 1). These data reveal unequivocally that the high-affinity ATP binding sites are not exposed under stoichiometric ATP and Mg2+ concentrations due to the lack of free Mg2+ since only 2.244 × 10−5 mol L−1 Mg2+ rather than 2.970 × 10−3 mol L−1 is available.
In the absence of Mg2+, the G. cruentata gill enzyme shows no (Na+, K+)-ATPase activity, corroborating previous findings (Masui et al. 2002; Furriel et al. 2000; Glynn, 1985). However, the inhibition seen above 3 mmol L−1 Mg2+, like the C. guanhumi enzyme (Farias et al. 2017), appears to result from Mg2+ binding to a second binding site causing an inhibitory effect (Pedemonte and Beaugé 1983). Enzyme affinity for Mg2+ seems to be both habitat- and species-specific and its affinity in G. cruentata lies in the range of marine species such C. danae (Masui et al. 2002) and C. ornatus (Garçon et al. 2007), seabob Xiphopenaeus kroyeri (Leone et al. 2015), striped hermit crab C. vittatus (Gonçalves et al. 2006) and mangrove crab C. guanhumi (Farias et al. 2017). The K0.5 for Mg2+ is lower under stoichiometric conditions (K0.5 = 0.06 ± 0.003 mmol L−1 and Table 1) compared to increasing Mg2+ concentrations (K0.5 = 0.16 ± 0.01 mmol L−1 and Table 1), similar to C. guanhumi (Farias et al. 2017).
The isoforms of the (Na+, K+)-ATPase α-subunit exhibit different apparent affinities for Na+ and K+ when ATP is used as a substrate (Blanco and Mercer 1998; Lopez et al. 2002), depending on the organism, tissue, membrane factors, and post-translational modifications (Therien et al. 1996; Beaugé et al. 1997; Geering 2001). The apparent affinity of the G. cruentata enzyme for Na+ (K0.5 = 5.1 ± 0.30 mmol L−1) lies within the range for freshwater, estuarine, and semi-terrestrial decapods (4 to 6.5 mmol L−1) (Furriel et al. 2000; Masui et al. 2002; Gonçalves et al. 2006; Farias et al. 2017) but is lower compared to marine crabs like Carcinus maenas, Cancer pagurus and Macropipus puber (Lucu and Towle 2003). Overall, the apparent affinity for Na+ varies markedly and has been correlated with habitat salinity (Lucu and Towle 2003). However, in our hands, the apparent Na+ affinities of gill (Na+, K+)-ATPases of crustaceans from different habitats are similar (Leone et al. 2015) and are unaltered by salinity acclimation to dilute medium (Garçon et al. 2009).
The K+ affinity of the G. cruentata gill (Na+, K+)-ATPase is comparable to those of various marine crabs such as C. danae (Masui et al. 2002, 2009), C. ornatus (Garçon et al. 2007), C. vittatus (Gonçalves et al. 2006; Lucena et al. 2012), and the freshwater crab Potamon potamios (Tentes and Stratakis 1991). However, compared to the semi-terrestrial crab C. guanhumi, the apparent affinity is about six-fold higher, suggesting that a different enzyme conformation or isoform may be expressed in the gill tissue of G. cruentata. In general, K+ affinity does not correlate with salinity acclimation in crustaceans since the potassium-binding site of the pump faces the hemolymph and not the external environment (Lucu and Towle 2003).
The K0.5 value for NH4+ stimulation of G. cruentata (Na+, K+)-ATPase activity is ≈ 15-fold higher compared to that of the semi-terrestrial C. guanhumi (Farias et al. 2017) but comparable to marine crabs like C. ornatus (Garçon et al. 2007, 2009), C. vittatus (Gonçalves et al. 2006; Lucena et al. 2012), and C. danae (Masui et al. 2002, 2009). The slight stimulation of (Na+, K+)-ATPase activity by NH4+ plus K+ (< 10%), similarly to C. guanhumi (Farias et al. 2017), seems to be characteristic of semi-terrestrial crabs. NH4+ can replace K+ in sustaining ATP hydrolysis by the crustacean gill (Na+, K+)-ATPase (Leone et al. 2015, 2017) and can substitute for K+ in Na+ transport in C. sapidus (Towle and Holleland 1987). Considering that G. cruentata is a benthic mangrove species that burrows into sediments under trees and rocks, the high K0.5 value (9.7 ± 0.5 mmol L−1) and the slight synergistic stimulation by NH4+ plus K+ do not appear to enable a rapid response to increased hemolymph NH4+ against toxic ammonia accumulation.
The (Na+,K+)-ATPase activity estimated in the gill tissue of G. cruentata (≈ 95%) is very similar to C. danae (Masui et al. 2002) but contrasts with various marine crustaceans (80%) (Gonçalves et al. 2006; Garçon et al. 2009; Lucena et al. 2012; Farias et al. 2017). The KI for ouabain inhibition also lies within the range of many crustaceans inhabiting freshwater, brackish, and marine habitats (Neufeld et al. 1980; Lucu and Towle 2003; Gonçalves et al. 2006; Garçon et al. 2009; Furriel et al. 2010; Lucena et al. 2012; Leone et al. 2015; Farias et al. 2017). This ample variation in KI (10–500 µmol L−1) seen among the Crustacea probably is due to differences in membrane lipid fluidity (Lucu and Towle 2003). The existence of different phosphorylated intermediates that occur between the E1P and E2P forms of the enzyme may explain these differences in KI values (Yoda and Yoda 1982), possibly involving phosphorylated E2P enzyme subconformations (Fedosova et al. 1998).
The gill (Na+, K+)-ATPase plays a major role in crustacean osmoregulation (Weihrauch et al. 2009; Leone et al. 2017). Our findings for the gill enzyme of the red mangrove crab, G. cruentata, particularly the elevated activity of the high-affinity ATP binding site, further characterize the biochemical adaptations of decapod crustaceans to their specific biotopes characterized by different salinity regimes and degrees of terrestrially.
References
Apell HJ, Hitzler T, Schreiber G (2017) Modulation of the Na, K-ATPase by magnesium ions. Biochemistry 56:1005–1016
Beaugé LA, Gadsby DC, Garrahan PJ (1997) (Na+, K+)-ATPase and related transport ATPases Structure, mechanism and regulation. Ann NY Acad Sci 834:1–694
Betancur-R R, Orti G, Stein AM, Marceniuk AP, Pyron RA (2012) Apparent signal of competition limiting diversification after ecological transition from marine to freshwater habitats. Ecol Lett 15:822–830
Blanco G, Mercer RW (1998) Isozymes of the (Na+, K+)-ATPase: heterogeneity in structure, diversity in function. Am J Physiol 44:F633–F650
Burggren WW (1992) Respiration and circulation in land crabs: novel variations on the marine design. Am Zool 32:417–427
Cameron JN (1981) Brief introduction to the land crabs of the Palau Islands: stages in the transition to air breathing. J Exp Zool 218:1–5
Chourasia M, Sastry N (2012) The nucleotide, inhibitor, and cation binding sites of P-type II ATPases. Chem Biol Drug Design 79:617–627
Clausen MV, Hilbers F, Poulsen H (2017) The structure and function of the Na, K-ATPase isoforms in health and disease. Front Physiol 8:371
Copeland DE, Fitzjarrell AT (1968) The salt absorbing cells in the gills of the blue crab (Callinectes sapidus, Rathbun) with notes on modified mitochondria. Z Zellforsch 92:1–22
Diaz H, Rodriguez G (1977) The branchial chamber in terrestrial crabs: a comparative study. Biol Bull 153:485–505
Donato DC, Kauffman JB, Murdiyarso D, Kurnianto S, Stidham M, Kanninem M (2011) Mangroves among the most carbon-rich forests in the tropics. Nature Geosci 4:293–297
Durr KL, Tavraz NN, Dempski RE, Bamberg E, Friedrich T (2009) Functional significance of E2 state stabilization by specific α/β-subunit interactions of Na, K- and H, K-ATPase. J Biol Chem 284:3842–3854
Fabiato A (1988) Computer programs for calculating total from specified free or free from specified total ionic concentrations in aqueous solution containing multiple metals and ligands. Methods Enzymol 157:378–417
Farias DL, Lucena MN, Garçon DP, Mantelatto FL, McNamara JC, Leone FA (2017) A kinetic characterization of the gill (Na+, K+)-ATPase from the semi-terrestrial mangrove crab Cardisoma guanhumi Latreille 1825 (Decapoda, Brachyura). J Membr Biol 250:517–534
Fedosova NU, Cornelius F, Klodos I (1998) E2P phosphoforms of (Na+, K+)-ATPase I: comparison of phosphointermediates formed from ATP e Pi by their reactivity toward hydroxylamine e vanadate. Biochem 37:13634–13642
Fontes CFL, Barrabin H, Scofano HM, Norby JG (1992) The role of Mg2+ and K+ in the phosphorylation of Na+, K+-ATPase by ATP in the presence of dimethyl sulfoxide but in the absence of Na+. Biochim Biophys Acta 1104:215–225
Freire CA, Onken H, McNamara JC (2008) A structure–function analysis of ion transport in crustacean gills and excretory organs. Comp Biochem Physiol 151A:272–304
Fukushima Y, Post RL (1978) Binding of divalent cation to phosphoenzyme of sodium-potassium-transport adenosine triphosphatase. J Biol Chem 253:6853–6862
Furriel RPM, McNamara JC, Leone FA (2000) Characterization of (Na+, K+)-ATPase in gill microsomes of the freshwater shrimp Macrobrachium olfersii. Comp Biochem Physiol 126B:303–315
Furriel RPM, Firmino KCS, Masui DC, Faleiros RO, Torres AH, McNamara JC (2010) Structural and biochemical correlates of (Na+, K+)-ATPase driven ion uptake across the gills of the true freshwater crab, Dilocarcinus pagei Brachyura, Trichodactylidae. J Exp Zool 313A:508–523
Gannon AT, Arunakul N, Henry RP (2000) Respiratory, cardiovascular, and hemolymph acid-base changes in the amphibious crab, Cardisoma guanhumi, during immersion and emersion. Mar Freshw Behav Physiol 34:73–92
Garçon DP, Masui DC, Mantelatto FLM, McNamara JC, Furriel RPM, Leone FA (2007) K+ and NH4+ modulate gill (Na+, K+)-ATPase activity in the blue crab, Callinectes ornatus: fine tuning of ammonia excretion. Comp Biochem Physiol 147A:145–155
Garçon DP, Masui DC, Mantelatto FLM, McNamara JC, Furriel RPM, Leone FA (2009) Hemolymph ionic regulation and adjustments in gill (Na+, K+)-ATPase activity during salinity acclimation in the swimming crab Callinectes ornatus (Decapoda, Brachyura). Comp Biochem Physiol 154A:44–55
Geering K (2001) The functional role of beta subunits in oligomeric P-type ATPases. J Bioenergy Biomemb 33:425–438
Geering K (2008) Functional roles of Na, K-ATPase subunits. Curr Opin Nephrol Hypert 17:526–532
Glynn IM (1985) The (Na+, K+)-transporting adenosine triphosphatase. In: Martonosi AN (ed) The enzymes of biological membranes, vol 3. Plenum Press, New York, pp 35–114
Gonçalves RR, Masui DC, McNamara JC, Mantelatto FLM, Garçon DP, Furriel RPM, Leone FA (2006) A kinetic study of the gill (Na+, K+)-ATPase, and its role in ammonia excretion in the intertidal hermit crab, Clibanarius vittatus. Comp Biochem Physiol 145A:346–356
Hartnoll RG (1988) Evolution, systematic and geographical distribution. In: Burggren WW, McMahon BR (eds) Biology of land crabs. Cambridge University Press, New York, pp 6–51
Hegyvary C, Jorgensen PL (1981) Conformational changes of renal sodium plus potassium ion transport adenosine triphosphatase labeled with fluorescein. J Biol Chem 256:6296–6303
Henry RP, Cameron JN (1982) The distribution and partial characterization of carbonic anhydrase in selected aquatic and terrestrial decapod crustaceans. J Exp Zool 221:309–321
Henry RP, Lucu C, Onken H, Weihrauch D (2012) Multiple functions of the crustacean gill: osmotic/ionic regulation, acid-base balance, ammonia excretion, and bioaccumulation of toxic metals. Front Physiol 3:1–33
Huličiak M, Bazgierb V, Berkab K, Kubala M (2017) RH421 binds into the ATP-binding site on the Na+/K+ -ATPase. Biochim Biophys Acta 1859:2113–2122
Innes AJ, Taylor EW (1986) The evolution of air-breathing in crustaceans: a functional analysis of branchial, cutaneous and pulmonary gas exchange. Comp Biochem Physiol 85A:621–637
Jiang Q, Garcia A, Han M, Cornelius F, Apell HJ, Khandelia H, Clarke RJ (2017) Electrostatic stabilization plays a central role in autoinhibitory regulation of the N+, K+ -ATPase. Biophys J 112:288–299
Jorgensen PL, Hakansson KO, Karlish SJD (2003) Structure and mechanism of functional sites and their interactions. Annu Rev Physiol 65:817–849
Kanai R, Ogawa H, Vilsen B, Cornelius F, Toyoshima C (2013) Crystal structure of a Na+-bound Na+, K+-ATPase preceding the E1 state. Nature 502:201–207
Kaplan JH (2002) Biochemistry of Na, K-ATPase. Annu Rev Biochem 71:511–535
Karlish SJ (2003) Investigating the energy transduction mechanism of P-type ATPases with Fe2+- catalyzed oxidative cleavage. Ann NY Acad Sci 986:39–49
Kubala M, Teisinger J, Ettrich R, Hofbauerova K, Kopecky V, Baumruk Krumscheid R, Plasek J, Schoner W, Amler E (2003) Eight amino acids form the ATP recognition site of Na+/K+-ATPase. Biochemistry 42:6446–6452
Laemmli UK (1970) Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature (London) 227:680–685
Leone FA, Baranauskas JA, Furriel RPM, Borin IA (2005) SigrafW: an easy-to-use program for fitting enzyme kinetic data. Biochem Mol Biol Educ 33:399–403
Leone FA, Bezerra TM, Garçon DP, Lucena MN, Pinto MR, Fontes CF, McNamara JC (2014) Modulation by K+ Plus NH4+ of microsomal (Na+, K+)-ATPase activity in selected ontogenetic stages of the diadromous river shrimp Macrobrachium amazonicum (Decapoda, Palaemonidae). PLoS ONE 9:e89625
Leone FA, Lucena MN, Rezende LA, Garçon DP, Pinto MR, Mantelatto FLM, McNamara JC (2015) Kinetic characterization and immunolocalization of (Na+, K+)-ATPase activity from gills of the marine seabob shrimp Xiphopenaeus kroyeri (Decapoda, Penaeidae). J Membr Biol 248:257–272
Leone FA, Lucena MN, Garçon DP, Pinto MR, McNamara JC (2017) Gill ion transport ATPases and ammonia excretion in aquatic crustaceans. In: Weihrauch D, O’Donnell MJ (eds) Acid-base balance and nitrogen excretion in invertebrates. Springer, New York, pp 61–107
Leone FA, Lucena MN, Fabri LM, Garçon DP, Fontes CFL, Faleiros RO, Moraes CM, McNamara JC (2020) Effect of salinity on modulation by ATP, protein kinases and FXYD2 peptide of gill (Na+, K+)-ATPase activity in the swamp ghost crab Ucides cordatus (Brachyura, Ocypodidae). https://doi.org/10.1101/2020.04.24.058297
Little C (1983) The colonization of land: origins and adaptations of terrestrial animals. Cambridge University Press, Cambridge, pp 92–93
Lopez LB, Quintas LEM, Noel F (2002) Influence of development on Na+/K+-ATPase expression: isoform- and tissue-dependency. Comp Biochem Physiol 131A:323–333
Lucena MN, Garçon DP, Mantelatto FLM, Pinto MR, McNamara JC, Leone FA (2012) Hemolymph ion regulation and kinetic characteristics of the gill (Na+, K+)-ATPase in the hermit crab Clibanarius vittatus (Decapoda, Anomura) acclimated to high salinity. Comp Biochem Physiol 161B:380–391
Lucu C, Towle DW (2003) (Na+, K+)-ATPase in gills of aquatic crustacea. Comp Biochem Physiol 135A:195–214
Maia JCC, Gomes SL, Juliani MH, Morel CM (1983) Laboratory manual proceedings. In: Morel CM (ed) Genes and Antigens of Parasites. FIOCRUZ, Rio de Janeiro, pp 144–157
Maitland DP (1990) Aerial respiration in the semaphore crab Holoecius cordiformis, with or without branchial water. Comp Biochem Physiol 95A:267–274
Mantel LH, Farmer LL (1983) Osmotic and ionic regulation. In: Bliss DE (ed) The biology of Crustacea: internal anatomy and physiological regulation, vol 5. Academic Press, New York, pp 53–159
Martelo MJ, Zanders IP (1986) Modifications of gill ultrastructure and ionic composition in the crab Goniopsis cruentata acclimated to various salinities. Comp Biochem Physiol 84A:383–389
Martin DW (2005) Structure–function relationships in the (Na+, K+)-Pump. Seminars Nephrol 198:282–291
Masui DC, Furriel RPM, McNamara JC, Mantelatto FLM, Leone FA (2002) Modulation by ammonium ions of gill microsomal (Na+, K+)-ATPase in the swimming crab Callinectes danae: a possible mechanism for regulation of ammonia excretion. Comp Biochem Physiol 132C:471–482
Masui DC, Mantelatto FLM, McNamara JC, Furriel RPM, Leone FA (2009) (Na+, K+)-ATPase activity in gill microsomes from the blue crab, Callinectes danae, acclimated to low salinity: novel perspectives on ammonia excretion. Comp Biochem Physiol 153A:141–148
McNamara JC, Faria SC (2012) Evolution of osmoregulatory patterns and gill ion transport mechanisms in the decapod Crustacea: a review. J Comp Physiol 182B:997–1014
McNamara JC, Zanotto FP, Onken H (2005) Adaptation to hyposmotic challenge in brachyuran crabs: a microanatomical and electrophysiological characterization of the intestinal epithelia. J Exp Zool 303A:880–893
McNamara JC, Freire CA, Torres AH, Faria SC (2015) The conquest of fresh water by the palaemonid shrimps: an evolutionary history scripted in the osmoregulatory epithelia of the gills and antennal glands. Biol J Linn Soc 114:673–688
Melo GAS (1996) Manual de Identificação dos Brachyura (Caranguejos e Siris) do Litoral Brasileiro. São Paulo FAPESP, Pleiade, p 604
Middleton DA, Fedesova NU, Esmann M (2015) Long-range effects of Na+ binding in Na, K-ATPase reported by ATP. Biochemistry 54:7041–7047
Montes MR, Ferreira-Gomes MS, Centeno M, Rossi RC (2015) The E2P-like state induced by magnesium fluoride complexes in the Na, K-ATPase. Kinetics of formation and interaction with Rb(+). Biochim Biophys Acta 1848:1514–1523
Morris S (2001) Neuroendocrine regulation of osmoregulation and the evolution of air-breathing in decapod crustaceans. J Exp Biol 204:979–989
Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TLM, Petersen J, Eersen JP, Vilsen B, Nissen P (2007) Crystal structure of the sodium–potassium pump. Nature 450:1043–1050
Morth JP, Poulsen H, Toustrup-Jensen MS, Shack VR, Egebjerg J, Andersen JP, Vilsen B, Nissen P (2009) The structure of the Na+, K+-ATPase and mapping of isoform differences and disease-related mutations. Phil Trans R Soc 364B:217–227
Neufeld GJ, Holliday CW, Pritchard JB (1980) Salinity adaptation of gill (Na+, K+)-ATPase in the blue crab, Callinectes sapidus. J Exp Zool 211:215–224
Nyblom M, Poulsen H, Gourdon P, Reinhard I, Andersson M (2013) Crystal structure of Na+, K+-ATPase in the Na+-bound state. Science 342:123–127
Onken H, McNamara JC (2002) Hyperosmoregulation in the red freshwater crab Dilocarcinus pagei (Brachyura, Trichodactylidae): structural and functional asymmetries of the posterior gills. J Exp Biol 205:167–175
Onken H, Putzenlechner M (1995) A V-ATPase drives active, electrogenic and Na+-independent Cl− absorption across the gills of Eriocheir sinensis. J Exp Biol 198:767–774
Palmgren MG, Nissen P (2011) P-Type ATPases. Annu Rev Biophys 40:243–266
Pedemonte CH, Beaugé L (1983) Inhibition of (Na+, K+)-ATPase by magnesium-ions and inorganic-phosphate and release of these ligands in the cycles of ATP hydrolysis. Biochim Biophys Acta 748:245–253
Péqueux A (1995) Osmotic regulation in crustaceans. J Crust Biol 15:1–60
Pinto MR, Lucena MN, Faleiros RO, Almeida EA, McNamara JC, Leone FA (2016) Effects of ammonium stress in the Amazon River shrimp Macrobrachium amazonicum (Decapoda, Palaemonidae). Aquat Toxicol 170:13–23
Rahi L, Moshtaghi A, Mather PB, Hurwood DA (2018) Osmoregulation in decapod crustaceans: physiological and genomic perspectives. Hydrobiology 825:177–188
Read SM, Northcote DH (1981) Minimization of variation in the response to different proteins of the Coomassie blue-G dye-binding assay for protein. Analyt Biochem 116:53–64
Santos LCF, Belli NM, Augusto A, Masui DC, Leone FA, McNamara JC, Furriel RPM (2007) Gill (Na+, K+)-ATPase in diadromous, freshwater palaemonid shrimps: species-specific kinetic characteristics and α-subunit expression. Comp Biochem Physiol 148A:178–188
Shinoda T, Ogawa H, Cornelius F, Toyoshima C (2009) Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature 459:446–450
Silva ECC, Masui DC, Furriel RP, McNamara JC, Barrabin H, Scofano HM, Perales J, Teixeira-Ferreira A, Leone FA, Fontes CFL (2012) Identification of a crab gill FXYD2 protein and regulation of crab microsomal (Na+, K+)-ATPase activity by mammalian FXYD2 peptide. Biochim Biophys Acta 1818:2588–2597
Taylor HH, Taylor EW (1992) Microscopic anatomy of invertebrates. In: Harrison FW, Humas AG (eds) Decapod Crustacea, vol 10. Wiley-Liss, New York, pp 203–293
Tejral G, Sopko B, Necas A, Schoner W, Amler E (2017) Computer modelling reveals new conformers of the ATP binding loop of Na+/K+-ATPase involved in the transphosphorylation process of the sodium pump. Peer J. https://doi.org/10.7717/peerj.3087
Tentes I, Stratakis E (1991) Partial purification and properties of (Na+, K+)-ATPase from Potamon potamios. Comp Biochem Physiol 100C:619–624
Therien AG, Nestor NB, Ball WJ, Blostein R (1996) Tissue-specific versus isoform-specific differences in cation activation kinetics of the Na, K-ATPase. J Biol Chem 271:7104–7112
Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets – procedure and some applications. Proc Natl Acad Sci USA 76:4350–4354
Towle DW, Holleland T (1987) Ammonium ion substitutes for K+ in ATP dependent Na+ transport by basolateral membrane vesicles. Am J Physiol 252R:427–489
Towle DW, Kays WT (1986) Basolateral localization of Na++K+-ATPase in gill epithelium of two osmoregulating crabs, Callinectes sapidus and Carcinus maenas. J Exp Zool 239:311–318
Towle DW, Palmer G, Harris J (1976) Role of gill Na++ K+ -dependent ATPase in acclimation of blue crabs (Callinectes sapidus) to low salinity. J ExpZool 196:315–322
Tribuzy AVB, Fontes CFL, Norby JG, Barrabin H (2002) Dimethyl sulfoxide-induced conformational state of Na/K-ATPase studied by proteolytic cleavage. Arch Biochem Biophys 399:89–95
Tsai JR, Lin HC (2007) V-type H1-ATPase e Na+, K+-ATPase in the gills of 13 euryhaline crabs during salinity acclimation. J Exp Biol 210:620–627
Walseth TF, Johnson RA (1979) The enzymatic preparation of alpha [32P] nucleoside triphosphates, cyclic [32P]AMP and cyclic [32P]GMP. Biochim Biophys Acta 562:11–31
Ward DG, Cavieres JD (1998) Affinity labeling of two nucleotide sites on (Na+, K+)-ATPase using 2′(3′)-O-(2,4,6-trinitrophenyl) 8-azidoadenosine 5′-[alpha-P-32] diphosphate (TNP-8N(3)-[alpha-P-32]ADP) as a photoactivatable probe Label incorporation before and after blocking the high affinity ATP site with fluorescein isothiocyanate. J Biol Chem 273:33759–33765
Weihrauch D, McNamara JC, Towle DW, Onken H (2004) Ion-motive ATPases and active, transbranchial NaCl uptake in the red freshwater crab, Dilocarcinus pagei (Decapoda, Trichodactylidae). J Exp Biol 207:4623–4631
Weihrauch D, Wilkie MP, Walsh PJ (2009) Ammonia and urea transporters in gills of fish and aquatic crustaceans. J Exp Biol 212:1716–1730
Weihrauch D, Fehsenfeld S, Quijada-Rodriguez AR (2017) Nitrogen excretion in aquatic crustaceans. In: Weihrauch D, O’Donnell MJ (eds) Acid-base balance and nitrogen excretion in invertebrates. Springer, New York, pp 1–24
Wilkens JL, Young RE (1992) Regulation of pulmonary blood flow and of blood pressure in a mangrove crab (Goniopsis cruentata). J Exp Biol 163:297–316
Wolvenkamp HP, Waterman TH (1960) The physiology of Crustacea, vol 1. Academic Press, New York, pp 35–100
Wood CM, Randall DJ (1981) Oxygen and carbon dioxide exchange during exercise in the land crab (Cardisoma carnifex). J Exp Zool 218:7–22
Yoda A, Yoda S (1982) Interaction between ouabain and the phosphorylated intermediate of (Na+, K+)-ATPase. Mol Pharmacol 22:700–705
Zanders IP (1978) Ionic regulation in the amgrove crab Goniopsis cruentata. Comp Biochem Physiol 60:293–302
Acknowledgements
The authors thank the Instituto Chico Mendes de Conservação da Biodiversidade, Ministério do Meio Ambiente (ICMBio/MMA) for collecting permit #29594-9 to JCM, INCT-ADAPTA (Instituto Nacional de Ciência e Tecnologia para Adaptações da Biota Aquática da Amazônia, ADAPTA-II) with which this laboratory (FAL) is integrated, and the Amazon Shrimp Network (Rede de Camarão da Amazônia). We are grateful to Ana Pitteri for kindly allowing use of her original photograph of Goniopsis cruentata.
Funding
This investigation was financed by research grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2013/22625-1 and 2016/25336-0), Conselho de Desenvolvimento Científico e Tecnológico (CNPq 445078/2014-6) and in part by INCT-ADAPTA-II (Grant No. 465540/2014-7) and the Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM 062.1187/2017). MNL received a post-doctoral scholarship from FAPESP (2013/24252-9). FAL (Grant No. 302776/2011-7), CFLF (Grant No. 308847/2014-8), DPG (Grant No. 458246/2014-0) and JCM (Grant No. 303613/2017-3) received Excellence in Research scholarships from CNPq. LMF received a scholarship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Finance Code 001).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Preparation of biological material, data collection, and analysis were performed by Cintya M. Moraes, Leonardo M. Fabri, Marcelo R. Pinto, Rogério O. Faleiros, John C. McNamara and Carlos F.L. Fontes. The first draft of the manuscript was written by Francisco A. Leone, John C. McNamara, Malson N. Lucena and Daniela P. Garçon, and all authors actively participated in subsequent versions. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflicts of interest were disclosed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Moraes, C.M., Lucena, M.N., Garçon, D.P. et al. Biochemical Characterization and Allosteric Modulation by Magnesium of (Na+, K+)-ATPase Activity in the Gills of the Red Mangrove Crab Goniopsis cruentata (Brachyura, Grapsidae). J Membrane Biol 253, 229–245 (2020). https://doi.org/10.1007/s00232-020-00120-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-020-00120-7