Abstract
While amphipols have been proven useful for refolding of seven transmembrane helical (7-TM) proteins including G-protein-coupled receptors (GPCRs) and it could be shown that an amphipol environment is in principle suitable for NMR structural studies of the embedded protein, high-resolution NMR insights into amphipol refolded and isotopically labeled GPCRs are still very limited. Here we report on the recent progress toward NMR structural studies of the melanocortin-2 and -4 receptors, two class A GPCRs which so far have not been reported to be incorporated into an amphipol environment. Making use of the established 7-TM protein bacteriorhodopsin (BR) we initially tested and optimized amphipol refolding conditions. Most promising conditions were transferred to the refolding of the two melanocortin receptors. Analytical-scale refolding experiments on the melanocortin-2 receptor show very similar behavior to the results obtained on BR. Using cell-free protein expression we could generate sufficient amounts of isotopically labeled bacteriorhodopsin as well as melanocortin-2 and -4 receptors for an initial NMR analysis. Upscaling of the amphipol refolding protocol to protein amounts needed for NMR structural studies was, however, not straightforward and impeded detailed NMR insights for the two GPCRs. While well-resolved and dispersed NMR spectra could only be obtained for bacteriorhodopsin, a comparison of NMR data recorded on the melanocortin-4 receptor in SDS and in an amphipol environment indicates that amphipol refolding induces larger structural modifications in the receptor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amphipathic polymers have demonstrated great potential as suitable membrane substitutes (Zoonens and Popot 2014). Among the reasons that favor amphipols over conventional detergent-based surfactants are their ability to increase stability of the embedded membrane protein in certain cases as well as their good refolding properties in particular for seven transmembrane helical (7-TM) proteins (Dahmane et al. 2009; Pocanschi et al. 2006). These properties make amphipols very promising for the investigation of 7-TM proteins including G-protein-coupled receptors (GPCRs) (Baneres et al. 2011; Mary et al. 2014). In addition, it could be demonstrated that amphipol-stabilized membrane proteins including β-barrel (Zoonens et al. 2005) and 7-TM proteins (Etzkorn et al. 2013) are in general accessible by solution-state NMR techniques (Planchard et al. 2014).
In regard to GPCRs, amphipols were used to determine the structure of leukotriene B4 (LTB4), a small molecule ligand, bound to an amphipol-refolded and stabilized BLT2 receptor (Catoire et al. 2010). Ligand-binding capabilities of amphipol-refolded GPCRs could be demonstrated and high-resolution insights of the LTB4 ligand while bound to perdeuterated BLT2 receptor could be obtained using homonuclear 1H spectroscopy of the ligand. However, NMR spectra of the respective GPCR in amphipols still suffer from limited sensitivity, resolution, and dispersion (Catoire et al. 2010).
Here we report on the initial results obtained using amphipols for the structural investigation of the melanocortin-2 and the melanocortin-4 receptors (MC2R and MC4R, respectively). The signaling pathways of the MC4R are of great pharmaceutical relevance due to their role in the control of body weight and appetite, regulation of blood pressure, and the inhibition of inflammation (Breit et al. 2011; Tao 2010). The native melanocortin agonists comprise the adrenocorticotropic hormone (ACTH) as well as the α-, β-, and γ-melanocyte-stimulating hormones (MSHs). Current efforts have mainly focused on ligand-binding studies, see e.g., (Fani et al. 2013; Liang et al. 2013) for recent reviews. However, no experimental high-resolution structural information is available for these two receptors so far.
In the following we provide a biophysical characterization of the MC2R and MC4R receptors in an amphipol environment. These initial results will guide our efforts toward the characterization of structural details underlying hormone–receptor communication. NMR sample preparation including refolding strategies was optimized using the well-established 7-TM protein bacteriorhodopsin (BR).
Materials and Methods
Cell-Free Protein Expression
The bacterioopsin and melanocortin receptors were expressed using an E.coli-based cell-free expression system following the established procedures (Klammt et al. 2007; Schwarz et al. 2007). The wt-MC2R and wt-MC4R sequence was cloned into a pIVEX2.4d expression vector containing an N-terminal His10-tag followed by a Factor Xa cleavage site. Dialysis mode reactions were carried out at 28 °C in the absence of ligands and surfactants. After 12–16 h, the reaction mix was centrifuged for 10 min at 12,000×g. The resulting pellet was stored at −20 °C or directly refolded. Pellets for NMR sample preparation were additionally washed with buffer (10 mM Tris–acetate (pH 8.2), 14 mM Mg2+ acetate, and 0.6 mM K+ acetate). Note that residual Mg2+ could lead to aggregation of amphipols and the addition of EDTA prior to refolding could be beneficial (Picard et al. 2006).
Amphipol Refolding
Amphipols were synthesized following the published procedures for A8-35 (Gohon et al. 2004; Tribet et al. 2009). Note that NMR spectra of the amphipol batch used in this study show an increase in the ratio of free carboxyl to isopropylamine compared to A8-35. Based on our NMR data, the amphipol side-chain composition of the batch was determined to be 57 % free carboxyls, 12 % isopropylamine, and 31 % octylamine. Refolding into amphipols was done by initially resuspending the protein pellets in SDS buffer (50 mM sodium phosphate (pH 7.5) and 20 mM SDS). For BO refolding, no additional purification step was carried out prior to refolding with amphipols; for the GPCRs, cell-free protein pellet was purified using immobilized metal affinity chromatography (IMAC) in SDS buffer before refolding. For BR we could not observe differences in the refolding yield when refolding was carried out before or after IMAC purification. To induce folding, four different strategies for SDS removal were tested:
KCl Precipitation
Amphipols (to 2.2 % w/v) and (unless otherwise stated) the ligand (fivefold molar excess) were added, the mixture was kept at room temperature for 15–30 min, and SDS was precipitated by the addition of KCl to a final concentration of 150 mM and kept at room temperature with occasional shaking for additional 1–2 h. Residual SDS was not removed using a dialysis step, instead the refolded protein was directly purified using a Ni–NTA agarose column. Buffer A (50 mM sodium phosphate buffer (pH 8) and 150 mM NaCl) supplemented with 0.08 % w/v amphipols and 20 mM imidazole was used for washing (3 steps of two column volumes), and buffer A supplemented with 0.15 % amphipols and 250 mM imidazole was used for elution (5 steps of one column volume). Protein-containing fractions were pooled and centrifuged for 5 min at 12,000×g. 100 μl of the supernatant was directly analyzed by analytical gel filtration using a Superdex 200 10/300 GL (GE) column equilibrated in amphipol-free buffer A using a flow rate of 0.5 ml/min at 4 °C.
Dilution
Amphipols (to 2.2 % w/v) and the ligand (fivefold molar excess) were added; the mixture was diluted 1:10 by fast addition of SDS-free buffer A and kept at room temperature for 1–2 h. Ni–NTA purification and gel filtration were carried out as described above.
Bio-Bead Preparations
Amphipols (to 2.2 % w/v) and the ligand (5- to 10-fold access) were added. Washed and buffer-equilibrated Bio-Beads (Bio-Beads SM-2; Biorad) were added (up to 80 % w/v) and kept at room temperature for 8–14 h under constant shaking. Bio-Beads were removed using a centrifugation step. Ni–NTA purification and gel filtration were done as described above.
Refolding on Ni–NTA Agarose Matrix
SDS-solubilized protein was loaded onto Ni–NTA beads prior to the addition of amphipols or ligands. After protein was bound to the Ni–NTA matrix (15–30 min at room temperature) refolding by SDS removal using KCl precipitation, dilution or Bio-Beads was carried out as described above.
No residual SDS was detected in the BR–amphipol NMR samples by 1D and 2D 1H correlation spectroscopy after refolding into amphipols followed by IMAC and SEC (data not shown). The ligands used in this study comprise all-trans retinal (Sigma-Aldrich) in 10 mM stock solution in ethanol for BR refolding as well as versions of the adrenocorticotropic hormone (ACTH 1-39) for MC2R and MC4R refolding. For MC4R also the high-affinity antagonist SHU 9119 (Tocris Bioscience) was used.
Small-scale refolding was typically carried out in 50–150 μl sample volumes while large-scale refolding was done in sample volumes between 1 and 6 ml. Protein concentrations during refolding and SEC were in the order of 10–20 μM. Purification of NMR samples as well as all GPCR preparations was carried out with protease inhibitor-supplemented buffer (Complete EDTA free, Roche).
NMR Sample Preparation and Experimental Setup
Isotope-labeled proteins were produced using double (2H and 15N) or triple (2H, 15N, and 13C) labeled ALGAL isotope mixtures (Cambridge Isotope Laboratories or Sigma Isotec). The ALGAL isotope mixtures contain all amino acids (in different concentrations) except for the four amino acids: Cys, Trp, Gln, and Asn. The missing amino acids were added in natural abundance. All NMR samples were expressed under >90 % D2O conditions. Typical BR concentrations were in the range of 100–300 μM, the melanocortin receptors’ NMR samples had protein concentrations between 40 and 80 μM. Measurements were carried out at 25 °C (melanocortin receptors) and 38 °C (BR), at proton resonance frequencies of 750 or 800 MHz. Duration of 2D experiments was in the order of 4–12 h for BR and 24–48 h for the melanocortin receptors.
Results and Discussion
Amphipol refolding of BR and the two melanocortin receptors MC2R and MC4R was studied using cell-free protein expression, size-exclusion chromatography (SEC), and nuclear magnetic resonance (NMR) spectroscopy.
All proteins used in this study were produced using an E. coli extract-based cell-free expression system in the absence of any membrane-mimicking surfactants. Figure 1a shows SDS-PAGE results of cell-free (CF) expressed bacterioopsin (BO). As reported before, the resulting CF pellet predominantly contains the expressed protein (Klammt et al. 2012; Schneider et al. 2010). Still Ni–NTA purification in SDS buffer clearly improves sample purity (Fig. 1a).
Biochemical and biophysical characterization of amphipol refolding of bacteriorhodopsin. a Coomassie blue stained SDS-PAGE showing cell-free expression, SDS purification, and amphipol solubilization. b–m Normalized size-exclusion chromatograms of different amphipol–bacteriorhodopsin preparations. b Unfolded bacterioopsin in SDS micelles, c amphipol-refolded BR purified without excess of free amphipols, d same as in c but after incubation in 1 % w/v amphipol buffer for 12 h, e–l amphipol refolding from 15 mM SDS using different strategies as summarized in Table 1. In short: e, f without retinal, g, h using KCl precipitation, i, j using dilution, k, l using Bio-Beads. m Reinjection of separated BR–amphipol peak after 48 h (see text and Table 1 for more details). Note that SEC results in SDS buffer b were obtained at room temperature, whereas all the other results were recorded at 4 °C
Protein Refolding into Amphipathic Polymers
We investigated refolding into amphipols in particular with regard to a subsequent solution-state NMR spectroscopic study, which, similar to crystallization attempts (Charvolin et al. 2014), should preferentially be carried out with a sample consisting of small and homogenous particles. Therefore, the refolding product was assessed using size-exclusion chromatography to optimize refolding with respect to homogeneity and particle size. To simplify efficient removal of the SDS, which is used to solubilize the cell-free expressed protein pellet, we reduced the SDS concentration from 0.8 % (w/v) as reported in (Dahmane et al. 2009) to 0.6 % (w/v).
Noteworthily, cell-free expression enables a well-defined starting condition which is not biased by the presence of coordinated lipids (Etzkorn et al. 2013) and also allows the easy production of ligand-free BO. Figure 1b shows the SEC profile of purified cell-free expressed BO in SDS. Figure 1c–m summarizes our SEC results for refolding of BO from SDS into amphipols. As reported before (Gohon et al. 2008; Zoonens et al. 2007), the amphipol-solubilized receptor can be diluted to a large extent with amphipol-free buffer; however, removing all non-protein attached amphipols, e.g., by washing amphipol-solubilized BR with amphipol-free buffer while being immobilized on a Ni–NTA column, leads to formation of very large protein–amphipol particles (Fig. 1c) (also see (Arunmanee et al. 2014)). In agreement with previous findings, these large particles can be converted into regular-sized particles to some extent by the addition of free amphipols (Fig. 1d).
Refolding of BO in the absence of retinal significantly reduces the amount of protein that is incorporated into amphipols while increasing the protein aggregation as visible by very strong absorbance in the SEC void volume (Fig. 1e). Our results on cell-free expressed BO, i.e., BO that never came into contact with its native lipids, are consistent with previous studies on BO which was expressed in Halobacterium salinarum and successively delipidated (Dahmane et al., 2013). Refolding in the absence of the ligand was improved when BO was immobilized on a Ni–NTA matrix during refolding (Fig. 1f). However, both SEC profiles reveal a rather heterogeneous size distribution when BO is refolded in the absence of retinal. Interestingly, in the presence of retinal the efficiency of refolding is not enhanced by immobilizing BR (Fig. 1g, h), suggesting that the ligand has a strong positive effect on the refolding of BR.
Refolding of proteins in general is accomplished by reducing the concentration of denaturant that keeps the protein of interest in an unfolded state. It is known that the method for reducing the denaturant concentration can influence refolding yields. In our study, refolding was originally initiated by precipitating the denaturant SDS as its potassium salt by the addition of KCl. In the following, we also tested different methods of SDS removal including the addition of Bio-Beads as well as dilution below the critical micelle concentration (CMC). For both methods we again also tested refolding of BR while being immobilized (see Table 1 for an overview of the different methods used). In general, all strategies produced correctly refolded BR (as indicated by the characteristic color of the samples, data not shown). Refolding of immobilized BR by dilution produced a very homogeneous SEC profile (Fig. 1j) offering an attractive alternative method to the precipitation of SDS by KCl. SDS removal using Bio-Beads results in a SEC profile (Fig. 1k) that is similar to the precipitation method (Fig. 1g). The simultaneous use of Bio-Beads and Ni–NTA beads is generally feasible (Fig. 1l); however, it makes sample handling more difficult and in our case resulted in significant sample loss. This may be partially improved by the use of magnetic beads. Note that the use of Bio-Beads may interfere with refolding in the presence of ligand due to the adsorption of ligand. For the BR–retinal system, this was clearly the case as evidenced by the yellow color of the Bio-Beads after refolding of BR in the presence of all-trans retinal.
We also tested reinjection of the fractions containing the regular-sized BR–amphipol particles. The respective elution fractions (i.e., elution volumes 12.5–15.5 ml of the conventional KCl precipitation shown in Fig. 1g) were pooled, concentrated, and stored at 5 °C for 48 h. The clean SEC profile (Fig. 1m) of this sample suggests that homogenous and stable particles can be generated and isolated.
The insight gained from the optimization of BR refolding was used to study the amphipol refolding of two human GPCRs. Cell-free expression enabled the expression of the melanocortin-2 and the melanocortin-4 receptors in sufficient yields for further biophysical characterization as well as potential NMR structural studies. Purification of SDS-solubilized receptor using a Ni–NTA column results in relatively pure unfolded receptor. In the following, we investigated the transfer of SDS-solubilized melanocortin receptor into amphipols. No protein bands are detected by SDS-PAGE in the insoluble pellet fraction after refolding into amphipols followed by 10-min centrifugation at 12,000×g (Fig. 2a, b) suggesting that the amphipols efficiently solubilize both melanocortin receptors. Figure 2c–f shows SEC profiles of different amphipol refolding strategies for the MC2 receptor. The behavior observed for the MC2 receptor under the different refolding conditions closely resembles the results obtained for BR. Note that the additional peak with an elution volume of approximately 19 ml matches the SEC results obtained on free ACTH (data not shown). In accordance with our observations for BO, refolding of the MC2 receptor can be improved when the receptor is immobilized on Ni–NTA beads (Fig. 2f). Due to the potential interference of Bio-Beads with the receptor ligands, the KCl precipitation method (in the presence of Ni–NTA beads) was selected as a straightforward method for refolding of larger quantities as required for NMR structural studies.
Biochemical and biophysical characterization of amphipol refolding of the two cell-free expressed melanocortin receptors MC2R and MC4R. a, b Coomassie blue stained SDS-PAGE results of expression, purification, and amphipol solubilization of MC2R (a) and MC4R (b). c–f SEC profiles of amphipol-incorporated MC2R using different transfer strategies from SDS: c Bio-Beads, d dilution, e, f KCl precipitation in the absence (e) and presence (f) of Ni–NTA beads
NMR Characterization of Amphipol-Refolded 7-TM Proteins
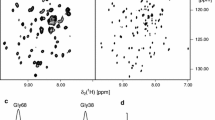
Figure 3 shows the resulting SEC profiles after upscaling of amphipol refolding for BR as well as the MC2 receptor (see Materials and Methods for more details). In the case of BR, linear upscaling resulted in the predominant occurrence of the desired (smaller-sized) population (Fig. 3a, BR–APOL). However, non-ideal preparations can also contain a sizable population of a larger fraction (Fig. 3a, BR APOL2). (In this preparation, IMAC was carried out with amphipol-free buffer (see Fig. 1c) and free amphipols were added after elution (see Fig. 1d)). Notably, this preparation allowed the comparison of NMR spectral quality of the smaller-sized and the larger-sized fractions (Fig. 3b, c, respectively). While the smaller-sized fraction gives rise to a very well-resolved and dispersed TROSY–HSQC spectrum (indicative for a well-structured and homogenous sample), the NMR data of the larger-sized fraction show very limited dispersion (indicative for random coil segments). Importantly, it is evident by the characteristic color of the samples that both fractions are well folded. Quantitative absorbance measurements of BR purified with and without excess of free amphipols show that the amount of well-folded BR is similar in both preparations although SEC profiles differ significantly (see supplementary material Fig. SI1). This strongly suggests that only the highly flexible parts of BR, including the protein termini, are visible in the spectrum of the larger-sized fraction (Fig. 3c).
Preparative-scale SEC profiles and NMR results of amphipol-refolded BR and MC2R. Black chromatogram in (a) shows optimal refolding of BR, blue line shows additional species after not optimal folding of BR. Red curve shows chromatogram of amphipol-refolded MC2R after upscaling. b–d 2D [15N,1H]-TROSY spectra of optimal refolded BR species (b), only the larger BR species (c) and of MC2R (d). Note that the spectra in (b) and in (c) both contain well-folded BR as indicated by the characteristic color of the sample (see also supplementary material Fig. Sl1)
Upscaling of amphipol refolding for the MC2 receptor results in a broad distribution of different sizes as shown by SEC (most of which are too large for high-resolution solution-state NMR detection). At this point, it is not clear why upscaling changes the MC2R SEC profile after refolding and why the MC2R behavior differs in this point from BR upscaling. In line with the broad SEC profile, the NMR spectrum of amphipol-refolded MC2R shows very limited dispersion and resolution (Fig. 3d).
The observed spectral properties of amphipol-refolded MC2R would be commonly attributed to a not appropriately folded receptor. However, as evident by our NMR data recorded on the larger-sized fraction of correctly folded BR (Fig. 3c), it is difficult to judge the quality of refolding based on a badly dispersed NMR spectrum.
In the next setup, we, therefore, first recorded a spectrum of the MC4 receptor solubilized in SDS to have a reference for its unfolded state (Fig. 4a). Transferring the receptor into an amphipol environment clearly changes the NMR spectrum (Fig. 4b). Although several well-resolved and partly dispersed peaks are visible it is clear that not all resonances expected for the MC4R are present. Based on the rather unconventional dispersion of the peaks as well as the presence of small residual impurities visible on the SDS gel, it cannot be fully excluded at this point that parts of the spectrum in Fig. 4b represent degradation products and/or signals not originating from the receptor. However, arguments in favor of an NMR spectrum representing the MC4R in amphipols include the fact that cell-free expression does not produce larger amounts of isotope-labeled impurities as well as that protein expression and purification were carried out in the presence of protease inhibitors. In addition, the spectrum of MC4R in amphipols was recorded after refolding the receptor directly from the MC4R-SDS sample which showed expected peak dispersions (spectrum in Fig. 4a). Hence, the major difference between Fig. 4a, b is the absence of SDS as well as the presence of amphipols and the ligand in Fig. 4b. Since both compounds, amphipols and the ligand (SHU9119), were added in relative large quantities we additionally recorded 2D TROSY–HSQC spectra of isolated amphipols and SHU9119 to exclude contributions of natural abundance signal in Fig. 4b (see supplementary material Fig. SI2).
Therefore, our NMR data would be most consistent with a partly folded receptor as well as with an amphipol–receptor complex that is too large for a detailed NMR spectroscopic study. The spectrum shown in Fig. 4b strongly suggests that replacing SDS with amphipols induces larger structural rearrangements in the receptor.
Conclusion
We investigated the effects of amphipol-assisted refolding of the 7-TM protein bacteriorhodopsin as well as the melanocortin-2 and melanocortin-4 receptors using size-exclusion chromatography and solution-state NMR. We measured biophysical properties and recorded NMR spectra of the proteins after they were solubilized by SDS and refolded using amphipathic polymers. We found that the choice of the method for removal of the SDS plays a critical role in obtaining a decent NMR spectrum. In addition, upscaling the refolding protocol for the required amounts to produce NMR samples was not always straightforward and the case of the two tested GPCRs largely interfered with the production of suitable NMR samples.
In general, all resulting NMR spectra are consistent with their underlying SEC profile, suggesting that sample optimization toward small, homogeneous particles is of fundamental importance for high-resolution NMR studies of amphipol-stabilized membrane proteins in solution. Amphipol refolding of MC4R induces clear changes in the NMR spectrum; however, only parts of the receptor are visible in the spectrum and further sample optimization toward smaller homogenous particle sizes has to be carried out to obtain high-resolution insights of MC2R and MC4R in an amphipol environment. For BR it became clear that amphipol refolding can be very efficient and in general permits high-resolution NMR insights into a fully folded 7-TM protein. On the other hand, our data of well-folded BR in larger amphipol particles (Fig. 3c) also show that badly dispersed NMR spectra of 7-TM proteins alone are not sufficient to exclude well-structured proteins.
Here we could optimize SEC profiles for amphipol-refolded bacteriorhodopsin as well as for small-scale refolding of the MC2 receptor. Refolding in the presence of the ligand and/or while attached to a Ni–NTA matrix can have significant positive effects on the refolding product.
We could also generate mg amounts of isotope-labeled MC2 and MC4 receptors for NMR investigation using a home-built cell-free expression system. Straightforward upscaling of the refolding protocol did not yield the homogenous and small particle distribution required for high-resolution NMR studies. Nevertheless, based on our small-scale refolding results, we anticipate that amphipol refolding of NMR quantities of cell-free expressed MC2 and MC4 receptors in respect to a smaller and more homogenous size distribution should be feasible. A thorough optimization of the upscaling conditions has to be carried out which naturally would strongly benefit from a reliable functional assay. Our current NMR comparison of the MC4 receptor in SDS and in an amphipol environment already provides initial evidence that amphipol refolding induces major structural rearrangements.
References
Arunmanee W, Harris JR, Lakey JH (2014) Outer membrane protein F stabilised with minimal amphipol forms linear arrays and LPS-dependent 2D crystals. J Membrane Biol. doi:10.1007/s00232-014-9640-5
Baneres JL, Popot JL, Mouillac B (2011) New advances in production and functional folding of G-protein-coupled receptors. Trends Biotechnol 29(7):314–322
Breit A, Buch TR, Boekhoff I, Solinski HJ, Damm E, Gudermann T (2011) Alternative G protein coupling and biased agonism: new insights into melanocortin-4 receptor signalling. Mol Cell Endocrinol 331:232–240
Catoire LJ, Damian M, Giusti F, Martin A, van Heijenoort C, Popot JL, Guittet E, Baneres JL (2010) Structure of a GPCR ligand in its receptor-bound state: leukotriene B4 adopts a highly constrained conformation when associated to human BLT2. J Am Chem Soc 132:9049–9057
Charvolin D, Picard M, Huang LS, Berry EA, and Popot JL (2014) Solution behavior and crystallization of cytochrome bc1 in the presence of amphipols. J Membrane Biol (under review)
Dahmane T, Damian M, Mary S, Popot JL, Baneres JL (2009) Amphipol-assisted in vitro folding of G protein-coupled receptors. Biochemistry 48:6516–6521
Dahmane T, Rappaport F, Popot JL (2013) Amphipol-assisted folding of bacteriorhodopsin in the presence or absence of lipids: functional consequences. Eur Biophys J 42:85–101
Etzkorn M, Raschle T, Hagn F, Gelev V, Rice AJ, Walz T, Wagner G (2013) Cell-free expressed bacteriorhodopsin in different soluble membrane mimetics: biophysical properties and NMR accessibility. Structure 21:394–401
Fani L, Bak S, Delhanty P, van Rossum EF, and van den Akker EL (2014) The melanocortin-4 receptor as target for obesity treatment: a systematic review of emerging pharmacological therapeutic options. Int J Obes (Lond) 38:163–169
Gohon Y, Pavlov G, Timmins P, Tribet C, Popot JL, Ebel C (2004) Partial specific volume and solvent interactions of amphipol A8-35. Anal Biochem 334:318–334
Gohon Y, Dahmane T, Ruigrok RW, Schuck P, Charvolin D, Rappaport F, Timmins P, Engelman DM, Tribet C, Popot JL et al (2008) Bacteriorhodopsin/amphipol complexes: structural and functional properties. Biophys J 94:3523–3537
Klammt C, Schwarz D, Dotsch V, Bernhard F (2007) Cell-free production of integral membrane proteins on a preparative scale. Methods Mol Biol 375:57–78
Klammt C, Maslennikov I, Bayrhuber M, Eichmann C, Vajpai N, Chiu EJ, Blain KY, Esquivies L, Kwon JH, Balana B et al (2012) Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat Methods 9:834–839
Liang L, Angleson JK, Dores RM (2013) Using the human melanocortin-2 receptor as a model for analyzing hormone/receptor interactions between a mammalian MC2 receptor and ACTH(1-24). Gen Comp Endocrinol 181:203–210
Mary S, Damian M, Rahmeh R, Marie J, Mouillac B, and Baneres JL (2014) Amphipols in G protein-coupled receptor pharmacology: What are they good for? J Membrane Biol. doi:10.1007/s00232-014-9665-9
Picard M, Dahmane T, Garrigos M, Gauron C, Giusti F, le Maire M, Popot JL, Champeil P (2006) Protective and inhibitory effects of various types of amphipols on the Ca2+-ATPase from sarcoplasmic reticulum: a comparative study. Biochemistry 45:1861–1869
Planchard N, Point E, Dahmane T, Giusti F, Renault M, Le Bon C, Durand G, Milon A, Guittet E, Zoonens M, et al. (2014) The use of amphipols for solution NMR studies of membrane proteins: advantages and constraints as compared to other solubilizing media. J Membrane Biol. doi:10.1007/s00232-014-9654-z
Pocanschi CL, Dahmane T, Gohon Y, Rappaport F, Apell HJ, Kleinschmidt JH, Popot JL (2006) Amphipathic polymers: tools to fold integral membrane proteins to their active form. Biochemistry 45:13954–13961
Schneider B, Junge F, Shirokov VA, Durst F, Schwarz D, Dotsch V, Bernhard F (2010) Membrane protein expression in cell-free systems. Methods Mol Biol 601:165–186
Schwarz D, Junge F, Durst F, Frolich N, Schneider B, Reckel S, Sobhanifar S, Dotsch V, Bernhard F (2007) Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nat Protoc 2:2945–2957
Tao YX (2010) The melanocortin-4 receptor: physiology, pharmacology, and pathophysiology. Endocr Rev 31:506–543
Tribet C, Diab C, Dahmane T, Zoonens M, Popot JL, Winnik FM (2009) Thermodynamic characterization of the exchange of detergents and amphipols at the surfaces of integral membrane proteins. Langmuir 25:12623–12634
Zoonens M, Popot JL (2014) Amphipols for each season. J Membrane Biol. doi:10.1007/s00232-014-9666-8
Zoonens M, Catoire LJ, Giusti F, Popot JL (2005) NMR study of a membrane protein in detergent-free aqueous solution. Proc Natl Acad Sci USA 102:8893–8898
Zoonens M, Giusti F, Zito F, Popot JL (2007) Dynamics of membrane protein/amphipol association studied by Forster resonance energy transfer: implications for in vitro studies of amphipol-stabilized membrane proteins. Biochemistry 46:10392–10404
Acknowledgments
This work was supported by grants from the German Academic Exchange Service (DAAD) and the DFG (ET 103/2-1) to M. E. as well as grants from the NIH (GM094608, GM075879, EB002026 and S10 RR029236).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Elter, S., Raschle, T., Arens, S. et al. The Use of Amphipols for NMR Structural Characterization of 7-TM Proteins. J Membrane Biol 247, 957–964 (2014). https://doi.org/10.1007/s00232-014-9669-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-014-9669-5