Abstract
The flat oyster (Ostrea edulis) is one of the most appreciated molluscs in Europe, but natural beds have been greatly reduced due to harvesting and the effects of the parasite Bonamia ostreae. Characterization of current wild populations is required to develop long-term bed restoration programmes by enhancing genetic diversity and tolerance to bonamiosis. Oysters from different locations corresponding to the main natural beds from Denmark, The Netherlands, England, Ireland, France and Spain, including two different cohorts per location were sampled in 2011 and 2013. Sixteen microsatellite loci were used to study temporal and geographical genetic structure. Temporal variation was low, although sometimes significant probably due to high variance in reproductive success among individuals. Conversely, samples from different countries showed much higher genetic divergence, and Ireland and France presented differences among locations within country. Clustering analyses grouped samples into three main geographical regions, associated with oceanic fronts: one group constituted by The Netherlands and Denmark; another by France, Ireland and England; and a third one exclusively by Spain. Effective population sizes (Ne) within regions were high (>1000), which reflects population stability and low levels of genetic drift. The presence of regional genetic structure shows the potential for local adaptation of O. edulis and suggests caution when transplanting individuals, especially between distant geographical regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The European flat oyster (Ostrea edulis) is a sessile, protandrous hermaphroditic, filter-feeding bivalve mollusc with a wild distribution ranging from Norway to Morocco in the Atlantic Ocean, and across all the Mediterranean Sea up to the Black Sea (Jaziri 1990; Lallias et al. 2010a). O. edulis is a highly appreciated aquaculture species in Europe and, due to its economic potential, has been introduced in other parts of the world such as USA, Canada, Japan and South Africa (Launey et al. 2002; FAO 2015). This mollusc species has had a historical importance as part of the human diet for centuries, being reared since ancient civilizations such as Greek and Roman (Gonzalez-Serrano 2001). However, during the first half of the twentieth century overharvest and habitat destruction have caused severe decline of flat oyster populations throughout its native distribution (Laing et al. 2006). Further, two diseases (caused by the Protozoa Marteilia refringens and Bonamia ostreae), spread in the early 1970s and 1980s, drastically reduced the production of O. edulis in the majority of European traditional rearing areas (Lallias et al. 2007). Despite improved management practices and intensive enhancement programmes, the production of O. edulis has remained low since then. Thus, oyster production declined from the 10,000–30,000 tons/year in the 1960s to c. 2000 in 2012 (FAO 2015). Selective breeding programmes, mainly for resistance to B. ostreae, were successfully developed in different European countries (Naciri-Graven et al. 1998; Culloty et al. 2004; da Silva et al. 2005) which indicates a substantial genetic variance underlying this trait. Thus, restoring O. edulis wild populations or improving production using pathogen-resistant seed could be feasible.
Genetic characterization of natural and cultured beds is required to develop long-term bed restoration and to identify the raw material for enhancing tolerance to bonamiosis in breeding programmes. Moreover, native oyster reefs provide many ecosystem services including water filtration, food and habitat for many animals, shoreline stabilization, coastal defence and fisheries (Beck et al. 2011), being essential for habitat management and conservation. The OSPAR (from Oslo and Paris conventions) Commission, which is in charge of the protection and conservation of the North-East Atlantic resources, has listed O. edulis as one of the threatened species in the region. This commission has also provided management recommendations for wild oyster beds inside Marine Protected Areas (OSPAR 2008). Pilot experiments using oyster spat transplantation to re-establish degraded areas have been recently performed and demonstrated higher settlement success in non-harrowed areas (Bromley et al. 2016).
Over the last three decades, genetic diversity and geographical structure of flat oyster wild populations have been analysed using different molecular markers such as allozymes (Blanc et al. 1986; Jaziri 1990; Saavedra et al. 1993, 1995), mitochondrial DNA (mtDNA) (Diaz-Almela et al. 2004) and microsatellites (Launey et al. 2002; Sobolewska and Beaumont 2005; Beaumont et al. 2006; Lallias et al. 2010a). Based on allozyme data from a limited number of sample locations, Jaziri (1990) suggested that, after the last glacial Pleistocene episode, Mediterranean oysters colonized the Atlantic areas. Saavedra et al. (1993, 1995) analysed a larger number of locations (19) encompassing the entire natural range of the species (excluding the Black Sea) and suggested the presence of two population units allopatrically differentiated and associated with each basin (i.e. Atlantic and Mediterranean), which would have merged in the Strait of Gibraltar since the last glaciation. Further studies across the distribution range including the Black Sea did not detect such transition between areas using either microsatellite loci (Launey et al. 2002) or mtDNA (Diaz-Almela et al. 2004). High genetic diversity has been reported in oyster populations, being generally lower in the Atlantic than in the Mediterranean area. This has been associated with shorter favourable periods for reproduction and stronger effects of parasites in the Atlantic region (Launey et al. 2002), which would have caused a reduction of the effective population size (N e). Data also suggest that the genetic structure of flat oyster follows an isolation by distance model (Saavedra et al. 1995; Launey et al. 2002; Diaz-Almela et al. 2004). In summary, available information suggests that O. edulis does not constitute a single panmictic unit throughout its natural range despite the high gene flow mediated by pelagic larvae (8–10 days pelagic larval duration; Launey et al. 2002). In the Atlantic area, Sobolewska and Beaumont (2005) using four microsatellite loci found weak population differentiation among locations, and argued that the Atlantic region would constitute a single genetic resource relevant for conservation. However, Beaumont et al. (2006), using six loci, identified genetically isolated groups within the Atlantic region and suggested that they should be taken into account for management and that movements of live oysters within this region should be avoided. Genetic variability in wild and pond-produced locations, as well as in hatchery-produced stocks (four locations for each group, 12 locations in total), was estimated using five microsatellite loci (Lallias et al. 2010a). Hatchery stocks showed lower genetic diversity, high differentiation from wild locations and signs of inbreeding. Thus, Lallias et al. (2010a) argued that restocking of wild populations with hatchery stocks could be detrimental for their conservation. However, pond production systems could represent a valuable alternative source for restocking wild O. edulis populations because pond production seems to maintain genetic diversity better than other culture procedures (Lallias et al. 2010a).
The differences observed in allele frequencies and genetic diversity (i.e. number of alleles) among local stocks may reflect a certain isolation and determine variation in effective population size (N e), a key parameter to evaluate population viability and conservation (Sobolewska and Beaumont 2005; Beaumont et al. 2006; Vercaemer et al. 2006). The effective number of breeders (N b) within populations can be low as observed in two French flat oyster populations (N b = 10–20 individuals), supporting the hypothesis of sweepstake reproductive success in the species (Hedgecock et al. 2007). The lowest N b estimates were observed in hatchery stocks; pond-produced oysters had intermediate estimates; and wild populations showed significantly higher N b estimates (Lallias et al. 2010a). However, variance in reproductive success was observed in both hatchery and wild conditions (Lallias et al. 2010b). Moreover, sequencing of mitochondrial DNA showed a reduced number of effective females (N e♀ ) within the species (Diaz-Almela et al. 2004) mainly due to the higher variance in female reproductive success (Boudry et al. 2002) and to the male biased sex ratio during reproductive season (3♂:1♀; Le Dantec and Marteil 1976). This bias could be caused by flat oyster being protandrous hermaphrodites and the higher incidence of B. ostreae on 2–3-year-old individuals, mostly females, further skewing the sex ratio (Culloty and Mulcahy 1996). Hence, differences in sex ratio between Bonamia-free and Bonamia-infected areas could be expected, an observation not documented to date; thus, further information is needed.
Despite the significant effort to elucidate genetic structure of flat oyster, no information is available on temporal genetic variance to date, essential to evaluate the reproductive bias whereby cohorts can substantially differ from the parental generation (Hedgecock et al. 2007). Moreover, the number of microsatellite loci used in previous studies was low (i.e. ≤6 microsatellite loci), which may have limited the potential to detect subtle population structure inside the Atlantic region. The aim of this study is to analyse the current genetic status of the main Atlantic remnant natural beds of the flat oyster (many of them infected by B. ostreae) with a larger number of microsatellite loci (16 markers) to describe both temporal and geographical structure related to oceanographic dynamics and connectivity. This information, including basic population parameters such as N e, and N em (i.e. number of effective migrants), is essential to assess the current status of flat oyster beds and to enable appropriate management and restoration (if necessary) of the wild resource which is also the raw material for breeding programmes.
Materials and methods
Biological material
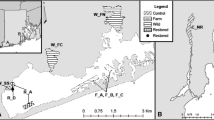
Tissue samples were collected from a total of 1595 oysters across 16 wild natural beds throughout the European Atlantic Coast (Fig. 1) and stored in 100 % ethanol. Roughly 50 oysters were sampled twice from each location in 2011 and 2013, resulting in a total sample of about 100 individuals per location (Table 1). To ensure that two different cohorts are included in the sampling design, adult individuals were collected in 2011 while spats were collected in 2013. All locations were affected by bonamiosis, except those in Denmark (DK-LHAVN and DK-BRED), where the parasite has not been reported (Flannery et al. 2014) .
Locations of Ostrea edulis analysed in the present study. Doted lines and roman numbers show the OSPAR regions. Location codes are those shown in Table 1
Molecular markers
Total DNA was extracted from gill tissue samples using Chelex-Proteinase K (Estoup et al. 1996), the eZN.A. E-96 mollusc DNA kit (Omega Bio-Tek) or standard phenol–chloroform procedures (Sambrook et al. 1989). A set of 13 anonymous microsatellite loci (Launey et al. 2002; Lallias et al. 2009) was selected based on their polymorphism and technical properties (e.g. low frequency of null alleles, high genotyping accuracy) using information from a pilot study (comprising 48 oysters, 24 from Ireland and 24 from Spain). Three additional recently developed microsatellite loci were included (OE-27, OE-03 and OE-11; Vera et al. 2015). The 16 loci were split into three PCR multiplexes and amplified using the Qiagen multiplex kit with labelled (FAM, NED, VIC and PET, Applied Biosystems) primers following manufacturer recommendations (detailed PCR multiplex conditions are shown in Table 2). The size of amplified fragments was analysed using GeneScan™ 500 LIZ® (Applied Biosystems) size standard on ABI 3730xl genetic analyzers (Applied Biosystems, Forest City, CA, USA) located in Queens University Belfast or in University of Santiago de Compostela (USC). Allele scoring was performed with GENEMARKER ver. 1.97 (Softgenetics, State College, PA, USA) at University College Cork (UCC) and with GENEMAPPER ver. 4.0 (Applied Biosystems, Forest City, CA, USA) at USC. Calibration of allele calling was performed between UCC and USC for all loci by genotyping in both platforms a reference panel of individuals composed of representative flat oysters across the sampled area in order to identify most of the allelic series per locus (reference individuals and genotypes for validation are available upon request to authors). Only individuals that showed successful amplification at ten or more loci were retained for further analyses.
Genetic data analyses
The software MICRO-CHECKER v. 2.2.3 (Van Oosterhout et al. 2004) was used to identify genotyping errors (stuttering, allele dropout and null alleles; 1000 randomizations). Allele frequencies, number of alleles, observed and expected heterozygosities, heterozygote excess and deficiency (exact test, 10,000 dememorizations and 5000 iterations) and tests for linkage disequilibrium (Fisher’s method, 10,000 dememorizations and 5000 iterations) were estimated with the programme GENEPOP v. 4.0.9 (Rousset 2008). Allelic richness among locations was calculated using the FSTAT v. 2.9.3 programme (Goudet 2001). Weir and Cockerham (1984) unbiased estimator of Wright’s (1951) F-statistics (F ST) was used as implemented in the software package Microsatellite Analyser (MSA, Dieringer and Schlötterer 2003) to assess the relative component of genetic differentiation among locations. Partitioning of the observed structure into local/regional and temporal variation was performed using the AMOVA approach as implemented in the ARLEQUIN v. 3.5.1.3 software package (Excoffier et al. 2005). Their significance was tested with 10,000 permutations. Alpha values corrections for multiple tests were adjusted using Bonferroni correction. The STRUCTURE v. 2.3.3 software (Pritchard et al. 2000; Falush et al. 2007) was employed to sort individuals into clusters (using the admixture and correlated allele frequency model with 100,000 replicates and a burn-in length of 10,000) using K (number of clusters) from 1 to 32 (the number of samples in the data) with five replicates for each K. The most likely K was assessed by plotting ln(PD) and implementing the ΔK method as in Evanno et al. (2005). Further, STRUCTURE analyses were performed applying the correction for null alleles in locations and loci for which MICRO-CHECKER indicated presence of null alleles (Falush et al. 2007) in an effort to avoid potential biases caused by null alleles. The CLUMPP v. 1.1.2 software (Jakobsson and Rosenberg 2007) was applied on STRUCTURE results to account for cluster label switching and to assign which clusters each run corresponded to (search options: greedy G′, using random input orders and 100 repeats). A neighbour-joining (NJ) tree was constructed from a matrix of Nei’s Da genetic distances between all pairs of populations (Nei 1987) using POPULATIONS 1.2.26 (Langella 2002) after pooling the weakly differentiated temporal replicates (see below). The robustness of the branches was tested with 1000 bootstrap replicates. The tree was visualized using TreeView version 3.2 (Page 1996). In order to detect isolation by distance (IBD) pattern, correlation among geographical (measured as the shortest coastline between two locations in Km) and genetic (measured as F ST/1−F ST; Rousset 1997) distances was evaluated with a Mantel test with 10,000 permutations using NTSYS v.2.1 (Rohlf 1993). The programme MIGRATE-N v. 3.1.6 (Beerli and Felsenstein 2001) was used to estimate theta (Θ), which is equal to 4N e μ where N e is the long-term (inbreeding) effective population size and μ is the mutation rate. From inspecting how the effective population size is estimated (N e = Θ/4μ), it is clear that the mutation rate has a large effect on estimates of N e. Mutation rates (μ) for microsatellites have been reported to range from 10−5 to 10−2 per locus and generation (Weber and Wong 1993). Hence, any estimate of N e is subjected to large variation depending on the specific mutation rate. However, mutation rates should be similar for each marker independently of the studied population. Therefore, no attempt was made to estimate actual effective population sizes and Θ estimates were instead used as proxies for measures of relative effective population sizes. In addition, MIGRATE-N was also used to estimate M, where M = m/μ, m is the migration rate per generation and μ is the mutation rate. The number of immigrants per generation, 4N em (for nuclear data), can be estimated by multiplying Θ by M. In addition to the long-term effective population size, genotype data were analysed using the programme LDNe (Waples and Do 2008) employing the default setting to estimate contemporary N e under random mating conditions. Genotype data from each individual sample were analysed with BOTTLENECK v. 1.2.02 [using the two-phase mutational model (TPM) with an IAM and SMM variance of 30 and 70 %, respectively] to assess whether any of the samples showed indications of having undergone genetic bottlenecks. Finally, the LOSITAN software (Beaumont and Nichols 1996; Antao et al. 2008) was used to detect evidence for selection at individual loci (using 50,000 simulations with the switches ‘“Neutral” mean Fst’ and ‘Force mean Fst’) under both the infinite alleles model (IAM) and stepwise mutation model (SMM).
Results
Genetic variability
After retaining individuals with at least ten genotyped loci, 1591 individuals were available out of 1595 for further analyses. Control for null alleles and technical artefacts using MICRO-CHECKER indicated the presence of null alleles at a total of 52 evaluations out of 512 (16 loci × 32 samples), representing twice more than the 5 % expected by chance. The spread of null alleles across loci was rather even and not particularly prominent for any locus (Supplementary Table 1), except for Oed325 (12 out of 32 samples affected), OeduJ12 (8), Oed202a (7) and Oedu-U2 (7). The locus Oed325 showed a geographical pattern of null allele occurrence with null alleles observed in all samples except the Danish and Dutch samples. However, while there was a lack of consistent null alleles across most loci and samples, and given the low expected effect of null alleles on F ST estimations and assignment tests accuracy (Carlsson 2008), analyses were done both with all loci included and with loci showing high frequency of null alleles removed. MICRO-CHECKER indicated stutter only at six occasions across the 512 tests (1 %), well within the 5 % expected by chance, and not associated with a specific locus or particular location. Further, MICRO-CHECKER did not indicate any large allele dropout. A total of 183 loci combinations of the 3840 tests (5 %) showed significant linkage (or gametic phase disequilibrium), a proportion very close to the expected false-positive rate of 5 %, with no particular pair of loci being linked in multiple locations (≤6 out of the 32 tested). Based on the relative low temporal variation (see below), temporal samples were pooled for descriptive statistical analyses (Supplementary Table 2). Number of analysed individuals per locus ranged from 61 at locus OE-11 in DK-LHAVN to 149 in UK-FAL at loci OE-03, Oed202a, Oed177a, Oed243 and OE-11. The number of alleles (Na) ranged from three at locus OE-03 in SP-PNTD to 39 at locus Oed202a in SP-OR (mean = 19.8 ± 8.4). Allelic richness (Ar) varied from 2.58 at locus OE-03 in SP-PNTD to 33.99 at locus Oed202b in NL-GREV (mean = 19.8 ± 8.4). Expected heterozygosity (He) ranged from 0.243 at locus OE-11 in DK-BRED to 0.966 at locus Oed202a in SP-NOI (mean = 0.839 ± 0.178). Mean Ar and He values per pooled location ranged from 16.50 in SP-FER to 19.30 in IE-LFOYL (mean = 18.0 ± 1.0) and from 0.819 in NL-GREV to 0.855 in FR-QUIV (mean = 0.838 ± 0.010), respectively, with similar diversity levels for all locations when analysing temporal replicates separately and pooled temporal replicates (Table 3).
A number of loci deviated significantly from Hardy–Weinberg expectations in some samples after sequential Bonferroni correction (initial α = 0.05/16, P = 0.0031). However, of the 41 observed deviations, 25 were associated with three loci. Locus OdeuJ12 deviated in six samples, OE-03 deviated in nine samples and Oed325 in ten samples. While deviations at OeduJ12 and Oed325 were associated with null alleles in eight and twelve samples, respectively, OE-03 showed no indications of null alleles. There was no apparent geographical pattern to deviations from Hardy–Weinberg expectations for any locus except Oed325 that showed heterozygosity deficiency in UK-FAL, IE-CLEWB and all samples from Spain and France; while no indications of heterozygote deficiency could be detected in UK-CHIC, IE-LFOYL or samples from the Netherlands and Denmark (Table 3, Supplementary Tables 1 and 2). Three loci showed indications of potentially being under directional selection. OE-27 and Oed325 showed footprints of directional selection under both IAM and SMM models, while Oed240 only showed footprints of positive selection under the SMM model after corrections for multiple tests using the false discovery rate (FDR) model included in LOSITAN software.
Temporal and geographical variation
Pairwise multilocus F ST between years within locations ranged from 0.0000 to 0.0090. Five of the 16 analysed locations (UK-CHIC, DK-LHAVN, NL-GREV, NL-HW and FR-QUIB; pairwise F ST values >0.0033) showed significant temporal variation after correction for multiple test (sequential Bonferroni correction, initial α = 0.05/16, P = 0.0031). However, AMOVA analysis (Table 4) indicated that the temporal variability within location was relatively low (F SC = 0.0016, P < 0.001) compared with the geographical variability among locations (F CT = 0.0061, P < 0.001). Consequently, temporal replicates where pooled for further analyses. Global population differentiation (F ST) was 0.0079 (P < 0.001; F ST = 0.0089, P < 0.001 when temporal replicates were separated). When loci affected by null alleles were removed (i.e. Oed 325, OeduJ12, Oed202a and Oedu-U2), global F ST was slightly higher (0.0084, P < 0.001; F ST = 0.0094, P < 0.001 when temporal replicates were separated), probably reflecting the effect of decreased heterozygosity on F ST caused by null alleles. All in all, in accordance with previous observations (Carlsson 2008) loci with null alleles did not cause significant distortion of data and were subsequently retained for downstream analyses.
The closest geographical samples (i.e. from each country) showed no significant multilocus F ST, while they showed significant values regarding those more distant after correction for multiple tests (sequential Bonferroni correction, initial α = 0.05/71, P = 0.0007; Supplementary Table 3). No within-country differentiation could be observed with the exceptions of the two locations within Ireland (F ST = 0.0053, P < 0.001) and within France (F ST = 0.0011, P < 0.001).
In spite of the low within-country differentiation, AMOVA analyses (Table 4) indicated significant variation between locations within countries (F SC = 0.0012, P < 0.001), but the variation among countries was nearly six times higher (F CT = 0.0068, P < 001). The STRUCTURE analysis assessed K from 1 to 32 (the number of samples) and the resulting ln(PD) plot gave very similar support for K from 1 to 14 (Supplementary Fig. 1). However, when using the ΔK approach by Evanno et al. (2005), K = 2 showed the highest support (Supplementary Fig. 2). Scenarios from K = 2 to K = 5 were further analysed using CLUMPP (Fig. 2). At K = 2 it was clear that the Spanish and Danish/Dutch samples formed separated clusters with the English/Irish/French samples constituting an intermediate/mixed cluster. At K = 3, the three regions suggested above were evident as independent clusters (cluster I: Danish/Dutch, cluster II: English/Irish/French and cluster III: Spanish samples), while with K = 4 and K = 5, neither further geographical structure and additional substructuring nor temporal differentiation could be detected (Fig. 2). As expected, the same results were rendered when correction for null alleles was applied for STRUCTURE analyses (Supplementary Fig. 3). As no further geographical or temporal structure could be observed after K = 3, we performed an AMOVA analysis using the clusters from this scenario (Table 4), which indicated a significant amount of variation within these three regions (F SC = 0.0017, P < 0.001), but with five times higher variation among them (F CT = 0.0074, P < 0.001). In spite of the low bootstrap values shown for the robustness of the branches, the NJ tree also corroborated the presence of three different clusters geographically grouped (Fig. 3). Finally, a significant correlation among geographical and genetic distance matrices was detected suggesting isolation by distance (r = 0.82439, P = 0.0001; Supplementary Fig. 4).
Demographics and gene flow
The BOTTLENECK analysis did not show indications of genetic bottlenecks in any sample when assuming the TMP model of mutation (Wilcoxon test resulting in nonsignificant probabilities after sequential Bonferroni correction for multiple tests initial α = 0.05/32, P = 0.0016). Estimates of effective population size per location obtained with LDNe (allowing for a minimum allele frequency of 1 %) generated negative N e for some samples which were interpreted as ∞, and hence, the LDNe method could not successfully produce estimates for these samples. In samples where LDNe was able to estimate N e the estimates ranged from c. 1000 to 17,000 (Supplementary Fig. 5). Most locations showed 95 % confidence intervals (CI) that included infinity, while the lower 95 % CIs ranged from 700 to 8400 (Supplementary Fig. 5). The regional subdivision with K = 3 suggested by STRUCTURE and NJ analyses was also analysed with LDNe. While LDNe could only generate a N e point estimate for the Danish/Dutch group (22,800), the programme generated 95 % CIs for all groups that ranged from 2600 to ∞ (Table 5). Similarly, the long-term relative inbreeding effective population size was analysed for these three groups with MIGRATE-N resulting in Θ estimates ranging from 0.072 in the Danish/Dutch group to 0.098 in the English/Irish/French group (Fig. 4). The scale and directionality of global gene flow was also estimated with MIGRATE-N showing that the Danish/Dutch group received more migrants than those generated, while gene flow was symmetric between the English/Irish/French and Spanish groups (Fig. 5).
Discussion
Genetic diversity and temporal stability
Preservation of genetic diversity is vital to allow natural populations to adapt to environmental changes and maintained population demographics rely on wild individuals to counterbalance depletion caused by overexploitation or diseases (Frankham et al. 2002). Moreover, maintaining natural genetic variability is also important for farmed species, like O. edulis, as this variability constitutes the raw material for broodstock trait variability and selective breeding (Lallias et al. 2010a). The current study reports similar genetic diversity within all analysed flat oyster populations (Ar > 16, He > 0.800) in accordance with previous reports from the Atlantic region (Sobolewska and Beaumont 2005; Beaumont et al. 2006; Lallias et al. 2010a).
The frequency of null alleles (i.e. non-amplified alleles by PCR due to mutations in primer annealing sites) across most loci was low except for Oed325 and OeduJ12, and their impact on population structure was negligible. This presence of null alleles in microsatellite loci has previously been reported for flat oyster (Beaumont et al. 2006) and in general for molluscs (McGoldrick et al. 2000; Hedgecock et al. 2004; Diz and Presa 2008; Pino-Querido et al. 2015). Single nucleotide polymorphisms (SNPs) are widespread in mollusc genomes, which increases the probability of null alleles because of primer annealing failure in adjacent regions. While SNP frequency in O. edulis range from one SNP every 47 bp in noncoding regions to one every 76 bp in coding regions (Harrang et al. 2013), it can be even higher for other oyster species. For example, one SNP every 60 bp in coding regions and each 40 bp in noncoding regions have been reported for Crassostrea gigas (Sauvage et al. 2007), and even higher in C. virginica (1 SNP every 20 bp; Zhang and Guo 2010). Despite the high frequency of SNP that can lead to inflated deviations from HW expectations, the effects of null alleles on F ST estimations and assignment tests are low and in line with data presented in Carlsson (2008). The current study corroborates this observation, and the inclusion of loci with moderate null allele frequencies can increase accuracy of estimated population parameters by diminishing inter-locus variance.
Effective population size (N e) is a key parameter for assessing the viability of populations for conservation (Frankham et al. 2002; Sobolewska and Beaumont 2005). The reported genetic differences among some temporal replicates in flat oyster (i.e. cohorts) may suggest variance in reproductive success among cohorts, thus reducing N e (Lallias et al. 2010b). Flat oyster has shown sex-biased ratio (3♂:1♀) at reproductive season (Le Dantec and Marteil 1976), although proportion of females in our samples varied between 20 and 100 % depending on local environmental factors which play an important role on sex (e.g. Orton 1927; Cole 1942; Korringa 1957). Controlled experiments of temperature and photoperiod showed that oysters are predominantly females at the beginning of spawning season when the water temperature is colder, while the frequency of males increase and are more frequent when the temperatures are warmer at the end of the spawning season (Joyce et al. 2013). Skewed sex ratio on populations can lead to decreasing N e (Wright 1931). Hedgecock et al. (2007) hypothesized sweepstakes on reproductive success (i.e. low number of progenitors succeeds in replacing an entire population, while the majority fails to procreate) with very low effective number of breeders (N b) estimates (10–20), hence increasing inbreeding. Moreover, N b has been shown to be reduced in hatchery stocks when compared to wild populations (Lallias et al. 2010a). Most of our temporal cohorts showed genetic temporal stability, which suggests that unequal spawning events are less frequent than previously suggested (Hedgecock et al. 2007; Lallias et al. 2010b). Additionally, although B. ostreae has drastically reduced O. edulis production during the twentieth century (Lallias et al. 2007), no signs of significant bottleneck have been detected. Estimates of N e per location were around or higher than 1000 in the most conservative estimates within regions, with N e values higher than 2500 by region. In conclusion, these results show low impact of genetic drift on flat oyster populations and suggest the viability of populations, since most adaptive potential has likely been retained (Frankham et al. 2002). Similarly, genetic studies of marine species suffering a mass mortality by pathogens (e.g. sea urchin, Diadema antillarum) or overfishing (e.g. European spiny lobster, Palinurus elephas) were unable to detect bottleneck signals, and despite the severe reductions in census population size, the species retained large and stable historical N e and high diversity levels (Lessios et al. 2001; Palero et al. 2011).
Population structure and connectivity at macro- and microgeographical scale
Despite the low level of temporal variation observed among flat oyster cohort, spatial differentiation was close to four times higher than temporal variation (F ST 0.0061 vs. 0.0016). Three consistent differentiated clusters (Danish/Dutch cluster, English/Irish/French cluster and Spanish cluster) were suggested by STRUCTURE analyses and supported by NJ tree and AMOVA. Isolation by distance (IBD) pattern has been suggested for O. edulis in its whole natural distribution range (e.g. Launey et al. 2002), with slight, but significant structure inside the Atlantic area (Beaumont et al. 2006). Our study confirmed structuring in the Atlantic region that follows isolation by distance (IBD)—a pattern not previously reported (Sobolewska and Beaumont 2005; Beaumont et al. 2006). The reason why the current study was able to detect IBD in the Atlantic may be related to the higher power associated with a high number of molecular markers and large number of sampling locations considered in our study. Beaumont et al. (2006) grouped locations from French Brittany and The Netherlands using Nei’s (1978) unbiased genetic distance (D) and reported a pairwise F ST between Norway and The Netherlands (0.051) that was lower than that observed between The Netherlands and France (0.056).The discrepancy with our data may be related to sampling effort between both studies. Beaumont et al. (2006) analysed northern Scottish populations in British Isles and three continental sites corresponding to Norway, The Netherlands and France, thus biasing the geographical representation and not incorporating representative samples for Europe.
Connectivity in marine species is assumed to be higher than in terrestrial species, due to lack of geographical barriers to gene flow expected in marine environments (Waples 1998). However, connectivity in the marine environment is often associated with oceanographic dynamics (e.g. oceanic currents, tides, bathymetry, upwelling systems, fronts, etc.), which can allow or prevent dispersal of pelagic forms and/or gametes (Palumbi 2004; Galarza et al. 2009). Oceanic fronts are narrow zones of enhanced horizontal gradient of water parameters (e.g. temperature, salinity, nutrients, etc.), often persistent over seasons, and separate broader areas with different water masses of different vertical structure (stratification) (Belkin 2002). Different fronts have been described inside the three large marine ecosystems (LME) included in this study: Iberian Coastal LME (Spanish locations), Celtic-Biscay Shelf LME (Irish, French and English locations) and North Sea LME (Dutch and Danish locations) (Belkin and Cornillon 2007; Belkin et al. 2009). These fronts represent major barriers to planktonic dispersal, and they have strong influence on population structure of littoral organisms with both inshore (<2 km) and offshore (>2 km) pelagic larvae distribution (Galarza et al. 2009). Therefore, although the potential for migration is presumably high, fronts in the studied region could drive the differentiation among the three population groups. Thus, the Shelf-Slope Front of the Iberian coast (Atlantic and Cantabrian shores) is a thermo-haline front which separates the saltier and mixed oceanic waters from the fresher and haline-stratified coastal waters (Fernandez et al. 1993). This front has been proposed as a retention mechanism for fish larvae (Fernandez et al. 1993; Gonzalez-Quiros et al. 2003, 2004; Rodriguez et al. 2009). This front could in effect isolate Spanish populations of flat oysters from others. The Irish Front, located in the Celtic Sea around the Saint George Channel and Bristol Channel, and the Ushant Front, located in front of French Brittany coast, are tidal mixing fronts which completely separate mixed waters columns over the coastal shallow zones and the stratified water column over the deeper oceanic zones (Belkin and Cornillon 2007). The Mid-Shelf Front, situated along Biscay Bay, western Irish coast and northern Scottish coast, is a shelf break front separating on-shelf coastal waters from off-shelf oceanic waters (Belkin and Cornillon 2007). These fronts are related with the distribution of nutrients and plankton in North-East Atlantic (O’Boyle and Raine 2007) and they would reduce the connectivity between the former locations and those from Ireland, England and France. Finally, the Frisian Front, located off the Dutch and German coasts, and the Danish Front, located in the western Jutland coast, are also tidal mixing fronts proposed to be the responsible of the differences in the macrofauna community structure (Kroencke et al. 2011), and they would further enforce the isolation of Dutch and Danish populations from the rest of European O. edulis beds (Belkin and Cornillon 2007). Interestingly, an oceanic salinity front caused by the freshwaters from the fjords and Skagerrak seas has been described along the Norwegian coast (Norwegian Coastal Current Front; Belkin et al. 2009), which could drive the flat oyster genetic differentiation previously reported for this region (Beaumont et al. 2006). Very similar population structure has been recently reported for the edible cockle (Cerastoderma edule), with the presence of a southern group from Iberian coast to France, a northern group with populations from Denmark to Russia and a third central group composed by Irish, British and southern North Sea populations (Martinez et al. 2015). Previous studies explained the grouping of population by marine currents and historical processes including population expansions and secondary contacts of southern and northern groups in the English Channel zone, which is suggested as a hybrid/transition zone for other marine species including fish (Vandamme et al. 2014) and mollusc (Smietanka et al. 2004; Krakau et al. 2012). It is likely that glaciations have shaped the current distribution of population groups in the northern hemisphere favouring secondary contacts among groups previously differentiated (Hewitt 2000). Thus, high genetic variability is common in southern regions (e.g. southern European peninsulas including Iberian Peninsula), considered as glacial refugia, whereas rapid postglacial colonization resulted in northern groups with lower genetic diversity (Hewitt 1996, 2000; Larmuseau et al. 2010; Albaina et al. 2012). However, northern cryptic refugia have been previously described for marine species (Smietanka et al. 2004; Nikula et al. 2007; Krakau et al. 2012) associated with ecologically productive polynyas (areas of persistent open water), likely located between the Scottish and the Fennoscandia ice sheets (Krakau et al. 2012). Phylogeographic analyses with mtDNA in O. edulis showed the presence of ancestral related haplotypes in the most distant areas of its natural distribution (i.e. Norway and Black Sea; see Diaz-Almela et al. 2004). Thus, to speculate, the mtDNA results and those in the present study (genetic diversity and differentiation) could agree with the presence of two ancestral groups, a southern group composed by Spanish locations, a northern group by Dutch and Danish locations and a central one, where the two previous groups merged. This would be in accordance with the results reported by Martinez et al. (2015) for C. edule showing similar F ST values to those described for O.edulis (F ST values higher than 0.010 among sampling sites for different groups, see Supplementary Table 3). Neutral markers provide information about the effect of gene flow and genetic drift on genetic variability and its distribution. Thus, in spite of all results presented in our study suggest the presence of different genetic groups of O. edulis in the Atlantic area, further analyses based on genomic screening of a huge number of molecular markers (i.e. SNPs) should be performed to confirm the existence of local adaptation and thus the biological significance of the proposed groups.
Biogeographical barriers can also preclude the dispersal of individuals in marine ecosystems. Cape Finisterre (NW Iberian Peninsula) has been defined as a biogeographical barrier for some marine individuals including molluscs (Piñeira et al. 2008) and fish (Abaunza et al. 2008; Larmuseau et al. 2009), while it does not act as a barrier for gene flow in other species (e.g. seahorses, Lopez et al. 2015). The survey carried out at microgeographical scale in the Galician coasts (NW Spain), with locations situated northwards (SP-EOA, SP-OR, SP-FER, SP-PNTD) and southwards (SP-NOI, SP-PON) of this discontinuity, did not show population differentiation among locations within the region. This result indicates the potential presence of a single panmictic population unit (i.e. high natural gene flow) in the region apparently unaffected by the Cape Finisterre discontinuity. However, Beaumont et al. (2006) performed genetic studies along the Scottish coasts on a similar geographical scale as the Galician coast and suggested that genetic homogeneity could be the consequence of human-mediated transplantation. Therefore, anthropogenic causes for the lack of clear population structure cannot be ruled out as the cause for the homogeneity detected among samples from the Galician coast, and also in the other two proposed clusters.
Management and conservation of wild flat oyster beds
The main objective for species conservation is the preservation of viable wild populations. This is particularly important for farmed species as wild populations are the source of genetic variation of pinnacle importance for breeding programmes. Flat oysters have been reared for many centuries in Europe, and human-mediated transplantations are well recorded including transplantation of seeds from geographically close regions but also from more distant ones (Magennis et al. 1983; Sobolewska and Beaumont 2005; Beaumont et al. 2006). These transplantations would tend to genetically homogenize the wild oyster beds. However, our results show that genetic differentiation of wild flat oyster populations along Atlantic region is still present, despite human-mediated transplantation since ancient civilizations, which could impact mainly at regional level as suggested previously by Beaumont et al. (2006). The OSPAR convention defines five main regions inside the North-East Atlantic region based on their biogeographical characteristics and exploitation uses, and O. edulis is present in region II (Greater North Sea), region III (Celtic Sea) and region IV (Bay of Biscay and Iberian coast) (Fig. 1). Hence, the three suggested genetic clusters in the present study fit with OSPAR regions, although English locations were genetically clustered with Irish ones while they belong to two different OSPAR regions (region II and III, respectively).
Another important issue for the O. edulis conservation is the control of the parasite Bonamia ostreae. Bonamiosis has drastically reduced the production of O. edulis in Europe during the second half of the twentieth century (Lallias et al. 2007). Limited population differentiation at neutral molecular markers (i.e. microsatellite loci) has been detected among the naïve (free of disease) Danish and Bonamia-affected Dutch oysters (pairwise F ST values ranged from 0.0025 to 0.0034), and accordingly these two regions were clustered in a single genetic group. However, differences among genomic regions under selective pressure driven by B. ostreae infection cannot be ruled out. Differences in bonamiosis resistance have been described for individuals collected in different regions (Greece, Galicia and Ireland), and families from populations long exposed to the parasite are more tolerant (da Silva et al. 2005). Also, breeding programmes for resistance to bonamiosis have been successful, thus demonstrating genetic variation underlying tolerance to the parasite (Naciri-Graven et al. 1998; Culloty et al. 2004). It is clear that populations which have not been in contact yet with the parasite will be more susceptible than those exposed for a long time which could have developed some resistance. Thus, it is important to avoid transplantations of individuals (mainly adults, see Culloty and Mulcahy 1996) as naïve populations might be more sensitive to the disease. In addition, transplantations should be avoided in an effort to limit the spread of the disease to new areas. As suggested above, it is possible that genetic screening with a larger number of markers (e.g. SNP-based methods) could detect footprints of selection related to resistance to bonamiosis with potential use in breeding programmes and conservation.
Despite the high gene flow among regions, the significant genetic differentiation detected in the present study suggests that there are differentiated gene pools with possible potential for local adaptation in O. edulis which should be confirmed by the identification of footprints of selection at specific markers (i.e. outliers). Moreover, these gene pools have relatively large effective population sizes. Thus, the findings from our study suggest that transplantations among countries should be avoided and that, to improve the wild bed conditions/environments, habitat restoration should be the focus of O. edulis restoration. In addition, it is essential to control the presence of B. ostreae to avoid an increase of parasite dispersion to regions free of the disease (e.g. Danish populations) (Laing et al. 2006; Beck et al. 2011; Bromley et al. 2016).
References
Abaunza P, Murta AG, Campbell N, Cimmaruta R, Comesana AS, Dahle G, Santamaria MTG, Gordo LS, Iversen SA, MacKenzie K, Magoulas A, Mattiucci S, Molloy J, Nascetti G, Pinto AL, Quinta R, Rarnos P, Sanjuan A, Santos AT, Stransky C, Zimmermann C (2008) Stock identity of horse mackerel (Trachurus trachurus) in the Northeast Atlantic and Mediterranean Sea: integrating the results from different stock identification approaches. Fish Res 89:196–209. doi:10.1016/j.fishres.2007.09.022
Albaina N, Olsen JL, Couceiro L, Miguel Ruiz J, Barreiro R (2012) Recent history of the European Nassarius nitidus (Gastropoda): phylogeographic evidence of glacial refugia and colonization pathways. Mar Biol 159:1871–1884. doi:10.1007/s00227-012-1975-9
Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G (2008) LOSITAN: a workbench to detect molecular adaptation based on a F(st)-outlier method. BMC Bioinformatics. doi:10.1186/1471-2105-9-323
Beaumont MA, Nichols RA (1996) Evaluating loci for use in the genetic analysis of population structure. Proc R Soc B-Biol Sci 263:1619–1626. doi:10.1098/rspb.1996.0237
Beaumont A, Garcia MT, Honig S, Low P (2006) Genetics of Scottish populations of the native oyster, Ostrea edulis: gene flow, human intervention and conservation. Aquat Living Resour 19:389–402. doi:10.1051/alr:2007008
Beck MW, Brumbaugh RD, Airoldi L, Carranza A, Coen LD, Crawford C, Defeo O, Edgar GJ, Hancock B, Kay MC, Lenihan HS, Luckenbach MW, Toropova CL, Zhang GF, Guo XM (2011) Oyster reefs at risk and recommendations for conservation, restoration, and management. Bioscience 61:107–116. doi:10.1525/bio.2011.61.2.5
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA 98:4563–4568. doi:10.1073/pnas.081068098
Belkin IM (2002) Front. In: Nybakken JW, Broenkow WW, Vallier TL (eds) Interdisciplinary Encyclopedia of marine sciences. Grolier, Danbury, pp 433–436
Belkin IM, Cornillon PC (2007) Fronts in the world ocean’s large marine ecosystems. ICES CM 2007/D:21, 33 pp
Belkin IM, Cornillon PC, Sherman K (2009) Fronts in large marine ecosystems. Prog Oceanogr 81:223–236. doi:10.1016/j.pocean.2009.04.015
Blanc F, Pichot P, Attard J (1986) Genetic variability in the European oyster, Ostrea edulis, geographic variation between local French stocks. Aquaculture 57:362–363
Boudry P, Collet B, Cornette F, Hervouet V, Bonhomme F (2002) High variance in reproductive success of the Pacific oyster (Crassostrea gigas, Thunberg) revealed by microsatellite-based parentage analysis of multifactorial crosses. Aquaculture 204:283–296
Bromley C, McGonigle C, Ashton E, Roberts D (2016) Restoring degraded European native oyster, Ostrea edulis, habitat: is there a case for harrowing? Hydrobiologia in press. doi:10.1007/s10750-015-2544-2
Carlsson J (2008) Effects of microsatellite null alleles on assignment testing. J Hered 99:616–623. doi:10.1093/jhered/esn048
Cole HA (1942) Primary sex-phases in Ostrea edulis. Q J Microsc Sci 2:317–356
Culloty SC, Mulcahy MF (1996) Season-, age-, and sex-related variation in the prevalence of bonamiosis in flat oysters (Ostrea edulis L.) on the south coast of Ireland. Aquaculture 144:53–63
Culloty SC, Cronin MA, Mulcahy MF (2004) Potential resistance of a number of populations of the oyster Ostrea edulis to the parasite Bonamia ostreae. Aquaculture 237:41–58. doi:10.1016/j.aquaculture.2004.04.007
da Silva P, Fuentes J, Villalba A (2005) Growth, mortality and disease susceptibility of oyster Ostrea edulis families obtained from brood stocks of different geographical origins, through on-growing in the Ria de Arousa (Galicia, NW, Spain). Mar Biol 147:965–977. doi:10.1007/s00227-005-1627-4
Diaz-Almela E, Boudry P, Launey S, Bonhomme F, Lapegue S (2004) Reduced female gene flow in the European flat oyster Ostrea edulis. J Hered 95:510–516. doi:10.1093/jhered/esh073
Dieringer D, Schlötterer C (2003) Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169. doi:10.1046/j.1471-8286.2003.00351.x
Diz AP, Presa P (2008) Regional patterns of microsatellite variation in Mytilus galloprovincialis from the Iberian Peninsula. Mar Biol 154:277–286. doi:10.1007/s00227-008-0921-3
Estoup A, Largiadier CR, Perrot E, Chourrout D (1996) Rapid one-tube DNA extraction for reliable PCR detection of fish polymorphic markers and transgenes. Mol Mar Biol Biotechnol 5:295–298
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578. doi:10.1111/j.1471-8286.2007.01758.x
FAO (2015) Global aquaculture production: quantities 1950–2012. http://www.fao.org/fishery/statistics/global-aquaculture-production/query. Accessed 21 Jan 2015
Fernandez E, Cabal J, Acuna JL, Bode A, Botas A, Garciasoto C (1993) Plankton distribution across a slope current-induced front in the southern Bay of Biscay. J Plankton Res 15:619–641. doi:10.1093/plankt/15.6.619
Flannery G, Lynch SA, Carlsson J, Cross TF, Culloty SC (2014) Assessment of the impact of a pathogen, Bonamia ostreae, on Ostrea edulis oyster stocks with different histories of exposure to the parasite in Ireland. Aquaculture 432:243–251. doi:10.1016/j.aquaculture.2014.04.038
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, Turner GF, Rico C (2009) The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci USA 106:1473–1478. doi:10.1073/pnas.0806804106
Gonzalez-Quiros R, Cabal J, Alvarez-Marques F, Isla A (2003) Ichthyoplankton distribution and plankton production related to the shelf break front at the Avile’s Canyon. ICES J Mar Sci 60:198–210. doi:10.1016/s1054-3139(03)00009-2
Gonzalez-Quiros R, Pascual A, Gomis D, Anadon R (2004) Influence of mesoscale physical forcing on trophic pathways and fish larvae retention in the central Cantabrian Sea. Fish Oceanogr 13:351–364. doi:10.1111/j.1365-2419.2004.00295.x
Gonzalez-Serrano JL (2001) Evolución histórica y situación actual de la acuicultura en el mundo y en España. In: Técnica SG (ed) Comercialización y distribución de productos pesqueros. Ministerio de Agricultura, Pesca y Alimentación, Madrid, pp 89–160
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). http://www.unil.ch/izea/softwares/fstat.html
Harrang E, Lapegue S, Morga B, Bierne N (2013) A high load of non-neutral amino-acid polymorphisms explains high protein diversity despite moderate effective population size in a marine bivalve with sweepstakes reproduction. G3-Genes Genomes Genet. doi:10.1534/g3.112.005181
Hedgecock D, Li G, Hubert S, Bucklin K, Ribes V (2004) Widespread null alleles and poor cross-species amplification of microsatellite DNA loci cloned from the Pacific oyster, Crassostrea gigas. J Shellfish Res 23:379–385
Hedgecock D, Launey S, Pudovkin AI, Naciri Y, Lapegue S, Bonhomme F (2007) Small effective number of parents (Nb) inferred for a naturally spawned cohort of juvenile European flat oysters Ostrea edulis. Mar Biol 150:1173–1182. doi:10.1007/s00227-006-0441-y
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. doi:10.1093/bioinformatics/btm233
Jaziri H (1990) Variations génétiques et structuration biogéographique chez un bivalve marin: l’huître plate Ostrea edulis (L.). Science et Techniques du Languedoc, Montpellier
Joyce A, Holthuis TD, Charrier G, Lindegarth S (2013) Experimental effects of temperature and photoperiod on synchrony of gametogenesis and sex ratio in the european oyster Ostrea edulis (Linnaeus). J Shellfish Res 32:447–458. doi:10.2983/035.032.0225
Korringa P (1957) Water temperature and breeding throughout the geographical range of Ostrea edulis. Ann Biol Paris 33:1–17
Krakau M, Jacobsen S, Jensen KT, Reise K (2012) The cockle Cerastoderma edule at Northeast Atlantic shores: genetic signatures of glacial refugia. Mar Biol 159:221–230. doi:10.1007/s00227-011-1802-8
Kroencke I, Reiss H, Eggleton JD, Aldridge J, Bergman MJN, Cochrane S, Craeymeersch JA, Degraer S, Desroy N, Dewarumez J-M, Duineveld GCA, Essink K, Hillewaert H, Lavaleye MSS, Moll A, Nehring S, Newell R, Oug E, Pohlmann T, Rachor E, Robertson M, Rumohr H, Schratzberger M, Smith R, Berghe EV, van Dalfsen J, van Hoey G, Vincx M, Willems W, Rees HL (2011) Changes in North Sea macrofauna communities and species distribution between 1986 and 2000. Estuar Coast Shelf Sci 94:1–15. doi:10.1016/j.ecss.2011.04.008
Laing I, Walker P, Areal F (2006) Return of the native—is European oyster (Ostrea edulis) stock restoration in the UK feasible? Aquat Living Resour 19:283–287. doi:10.1051/alr:2006029
Lallias D, Lapegue S, Haley C, Heurtebise S, Boudry P, Beaumont A (2007) A preliminary genetic linkage map for the European flat oyster Ostrea edulis. Aquaculture 272:S281–S282. doi:10.1016/j.aquaculture.2007.07.117
Lallias D, Stockdale R, Boudry P, Beaumont AR, Lapegue S (2009) Characterization of 27 microsatellite loci in the European flat oyster Ostrea edulis. Mol Ecol Resour 9:1276. doi:10.1111/j.1755-0998.2009.02722.x
Lallias D, Boudry P, Lapegue S, King JW, Beaumont AR (2010a) Strategies for the retention of high genetic variability in European flat oyster (Ostrea edulis) restoration programmes. Conserv Genet 11:1899–1910. doi:10.1007/s10592-010-0081-0
Lallias D, Taris N, Boudry P, Bonhomme F, Lapegue S (2010b) Variance in the reproductive success of flat oyster Ostrea edulis L. assessed by parentage analyses in natural and experimental conditions. Genet Res 92:175–187. doi:10.1017/s0016672310000248
Langella O (2002) POPULATIONS 1.2.26. Logiciel de Génétique de populations. Laboratoire populations, génétique et évolution. CNRS UPR 9034. Gif-sur-Yvette. France. http://www.bioinformatics.org/project/?group_id=84
Larmuseau MHD, Van Houdt JKJ, Guelinckx J, Hellemans B, Volckaert FAM (2009) Distributional and demographic consequences of Pleistocene climate fluctuations for a marine demersal fish in the north-eastern Atlantic. J Biogeogr 36:1138–1151. doi:10.1111/j.1365-2699.2008.02072.x
Larmuseau MHD, Raeymaekers JAM, Hellemans B, Van Houdt JKJ, Volckaert FAM (2010) Mito-nuclear discordance in the degree of population differentiation in a marine goby. Heredity 105:532–542. doi:10.1038/hdy.2010.9
Launey S, Ledu C, Boudry P, Bonhomme F, Naciri-Graven Y (2002) Geographic structure in the European flat oyster (Ostrea edulis L.) as revealed by microsatellite polymorphism. J Hered 93:331–338
Le Dantec J, Marteil L (1976) La reproduction des huitres. Revue des Travaux de l’Institut des Peches Marines 40:233–256
Lessios HA, Garrido MJ, Kessing BD (2001) Demographic history of Diadema antillarum, a keystone herbivore on the Caribbean reefs. Proc R Soc Lond Ser B Biol Sci 268:2347–2353. doi:10.1098/rspb.2001.1806
Lopez A, Vera M, Planas M, Bouza C (2015) Conservation genetics of threatened Hippocampus guttulatus in vulnerable in NW Spain: temporal and spatial stability of wild populations with flexible polygamous mating system in captivity. PLoS One 10:e0117538. doi:10.1371/journal.pone.0117538
Magennis BA, Gosling E, Wilkins NP (1983) Irish oyster populations: a historical and genetic study. Proc R Irish Acad Sec B-Biol Geol Chem Sci 83:291–299
Martinez L, Freire R, Arias-Perez A, Mendez J, Insua A (2015) Patterns of genetic variation across the distribution range of the cockle Cerastoderma edule inferred from microsatellites and mitochondrial DNA. Mar Biol 162:1393–1406. doi:10.1007/s00227-015-2676-y
McGoldrick DJ, Hedgecock D, English LJ, Baoprasertkul P, Ward RD (2000) The transmission of microsatellite alleles in Australian and North American stocks of the Pacific oyster (Crassostrea gigas): selection and null alleles. J Shellfish Res 19:779–788
Naciri-Graven Y, Martin AG, Baud JP, Renault T, Gerard A (1998) Selecting the flat oyster Ostrea edulis (L.) for survival when infected with the parasite Bonamia ostreae. J Exp Mar Biol Ecol 224:91–107. doi:10.1016/s0022-0981(97)00171-8
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nikula R, Strelkov P, Vainola R (2007) Diversity and trans-Arctic invasion history of mitochondrial lineages in the North Atlantic Macoma balthica complex (Bivalvia:Tellinidae). Evolution 61:928–941. doi:10.1111/j.1558-5646.2007.00066.x
O’Boyle S, Raine R (2007) The influence of local and regional oceanographic processes on phytoplankton distribution in continental shelf waters off north-western Ireland. Biol Environ-Proc R Irish Acad 107B:95–109. doi:10.3318/bioe.2007.107.2.95
Orton JH (1927) Observations and experiments on sex-change in the European oyster (O. edulis). J Mar Biol Assoc UK 14:967–1045
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Palero F, Abello P, Macpherson E, Beaumont M, Pascual M (2011) Effect of oceanographic barriers and overfishing on the population genetic structure of the European spiny lobster (Palinurus elephas). Biol J Linn Soc 104:407–418. doi:10.1111/j.1095-8312.2011.01728.x
Palumbi SR (2004) Marine reserves and ocean neighborhoods: the spatial scale of marine populations and their management. Annu Rev Environ Resour 29:31–68. doi:10.1146/annurev.energy.29.062403.102254
Piñeira J, Quesada H, Rolan-Alvarez E, Caballero A (2008) Genetic discontinuity associated with an environmentally induced barrier to gene exchange in the marine snail Littorina saxatilis. Mar Ecol Prog Ser 357:175–184. doi:10.3354/meps07278
Pino-Querido A, Alvarez-Castro JM, Vera M, Pardo BG, Fuentes J, Martinez P (2015) A molecular tool for parentage analysis in the Mediterranean mussel (Mytilus galloprovincialis). Aquac Res 46:1721–1735. doi:10.1111/are.12329
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rodriguez JM, Gonzalez-Nuevo G, Gonzalez-Pola C, Cabal J (2009) The ichthyoplankton assemblage and the environmental variables off the NW and N Iberian Peninsula coasts, in early spring. Cont Shelf Res 29:1145–1156. doi:10.1016/j.csr.2008.11.020
Rohlf FJ (1993) NTSYS-pc. Numerical taxonomy and multivariate analysis system, Version 2.1. Setauket, New York
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rousset F (2008) GENEPOP ‘ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106. doi:10.1111/j.1471-8286.2007.01931.x
Saavedra C, Zapata C, Guerra A, Alvarez G (1993) Allozyme variation in european populations of the oyster Ostrea edulis. Mar Biol 115:85–95. doi:10.1007/bf00349389
Saavedra C, Zapata C, Alvarez G (1995) Geographical patterns of variability at allozyme loci in the European oyster Ostrea edulis. Mar Biol 122:95–104. doi:10.1007/bf00349282
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. New York
Sauvage C, Bierne N, Lapegue S, Boudry P (2007) Single nucleotide polymorphisms and their relationship to codon usage bias in the Pacific oyster Crassostrea gigas. Gene 406:13–22
Smietanka B, Zbawicka M, Wolowicz M, Wenne R (2004) Mitochondrial DNA lineages in the European populations of mussels (Mytilus spp.). Mar Biol 146:79–92. doi:10.1007/s00227-004-1418-3
Sobolewska H, Beaumont AR (2005) Genetic variation at microsatellite loci in northern populations of the European flat oyster (Ostrea edulis). J Mar Biol Assoc UK 85:955–960
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. doi:10.1111/j.1471-8286.2004.00684.x
Vandamme SG, Maes GE, Raeymaekers JAM, Cottenie K, Imsland AK, Hellemans B, Lacroix G, Mac Aoidh E, Martinsohn JT, Martinez P, Robbens J, Vilas R, Volckaert FAM (2014) Regional environmental pressure influences population differentiation in turbot (Scophthalmus maximus). Mol Ecol 23:618–636. doi:10.1111/mec.12628
Vera M, Bello X, Álvarez-Dios JA, Pardo B, Sánchez L, Carlsson J, Carlsson J, Bartolomé C, Maside X, Martinez P (2015) Screening of repetitive motifs inside the genome of the flat oyster (Ostrea edulis): transposable elements and short tandem repeats. Mar Genom 24:335–341. doi:10.1016/j.margen.2015.08.006
Vercaemer B, Spence KR, Herbinger CM, Lapegue S, Kenchington EL (2006) Genetic diversity of the European oyster (Ostrea edulis L.) in Nova Scotia: comparison with other parts of Canada, Maine and Europe and implications for broodstock management. J Shellfish Res 25:543–551
Waples RS (1998) Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J Hered 89:438–450. doi:10.1093/jhered/89.5.438
Waples RS, Do C (2008) LDNe: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756. doi:10.1111/j.1755-0998.2007.02061.x
Weber JL, Wong C (1993) Mutation of human short tandem repeats. Hum Mol Genet 2:1123–1128. doi:10.1093/hmg/2.8.1123
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Zhang L, Guo X (2010) Development and validation of single nucleotide polymorphism markers in the eastern oyster Crassostrea virginica Gmelin by mining ESTs and resequencing. Aquaculture 302:124–129. doi:10.1016/j.aquaculture.2010.02.012
Acknowledgments
Authors wish to thank S. Sanchez-Darriba, L. Insua and S. Gomez for their technical support and D. Iglesias, A. van den Brink, E. Brummelhuis, A. van Gool, C. Fomsgard, J. K. Petersen and M. Longshaw for providing wild samples. The authors are also grateful to Dr. O. Puebla and four anonymous reviewers for their constructive comments on the earlier version of this manuscript. This study has been supported by the OYSTERECOVER project (FP7-SM3-2008-2-243583) from the European Community’s Seventh Framework Programme.
Funding
This study was funded by the OYSTERECOVER project from the European Community’s Seventh Framework Programme (FP7-SM3-2008-2-243583).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national and/or institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
Additional information
Responsible Editor: O. Puebla.
Reviewed by undisclosed experts.
M. Vera and J. Carlsson have contributed equally to this work and are first co-authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vera, M., Carlsson, J., Carlsson, J.E. et al. Current genetic status, temporal stability and structure of the remnant wild European flat oyster populations: conservation and restoring implications. Mar Biol 163, 239 (2016). https://doi.org/10.1007/s00227-016-3012-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-016-3012-x