Abstract
The aim of this study was to obtain high-purity galacto-oligosaccharide (GOS) by combining Lactobacillus-derived β-galactosidase enzymatic synthesis with fermentation and purification. GOS can be enzymatically synthesized by β-galactosidase from Lactobacillus plantarum CICC 22186. Temperature, pH and lactose concentration are important parameters during enzymatic synthesis. The optimal production process (35 °C, pH 7.0, 400 g/L) was obtained by optimization. The glycosidic bond structures of GOS were β-D-Galp-(1 → 6)-D-Lac, β-D-Galp-(1 → 3)-D-Lac, and β-D-Galp-(1 → 6)-D-Glc by GC–MS. The soluble solid content (SSC) and inoculum concentration of Kluyveromyces lactis 1772 both influenced purification, and the GOS content increased under optimal conditions (10%, 75 g/L). To elucidate the purification mechanism of Kluyveromyces lactis 1772, the metabolic changes during purification were analyzed by investigating related enzyme activities and transcriptomics. The results showed that the synthesis of macromolecules, such as proteins in yeast cells, was blocked, but cell growth was resumed by increasing the processes of glucose metabolism and substance transport. As a result, high-purity GOS was obtained.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Galacto-oligosaccharides (GOS) are important components of human milk and have potential for use as prebiotics in the food industry [10]. GOS stimulates the proliferation of gut microbiota, such as Bifidobacterium, and contributes greatly to intestinal health [2, 5]. It has been reported to exert important physiological effects, including promoting mineral absorption and lipid metabolism [3, 26].
GOS can be obtained through various means and sources, including chemical synthesis, microbial, and enzymatic synthesis. The chemical synthesis of GOS requires many reaction steps, and the environmental impact of unwanted harmful substances is greater than that of enzymatic synthesis [9]. Enzymatic synthesis is more feasible, safe, and less costly than chemical synthesis [11]. In the enzymatic synthesis of GOS, GOS is mainly synthesized by the transposition of β-galactosidase enzyme. β-Galactosidase enzyme is responsible for simultaneous lactose hydrolysis and rearrangement in larger molecules [22]. Sources of commercial β-galactosidase enzymes used in the GOS synthesis process include specific microorganisms, such as Aspergillus spp. and Kluyveromyces spp. Lactic acid bacteria (LAB) are recognized as food-grade and safe microorganisms; they also multiply very quickly. In general, the glycosidic bonds of most commercial GOS are β-(1,4) and β-(1,6), while some GOS synthesized by the LAB-derived β-galactosidase enzymatic method have β-D-Galp-(1 → 6)-D-Lac and β-D-Galp-(1 → 3)-D-Lac. They had a better effect of promoting the proliferation of Bifidobacterium [13, 20]. Therefore, LAB have the potential to be a rich source of galactosidases.
The reaction conditions were optimized to obtain the highest yield of GOS with the expected composition. The production yield of GOS is dependent on the various parameters involved in the reaction, including temperature, pH, and lactose concentration. In a previous study, González-Delgado et al. found that the optimal conditions for high-yield GOS production (12.18%) was at 40 °C, 5 U/mL enzyme concentration, and pH 7.0 [8]. Some of the by-products produced during the enzymatic synthesis of GOS, such as glucose, galactose, and unreacted lactose, affect the physiological activity of GOS and restrict its applications in commerce. Therefore, the purification of raw GOS is essential to enhance its commercial use and applications. However, few yeast species have the ability to ferment lactose, of which the foremost is Kluyveromyces lactis.
Therefore, the objectives of this study were to: (1) explore the synthesis of GOS by the Lactobacillus plantarum-derived β-galactosidase enzymatic method; (2) optimize the conditions of enzymatic synthesis of GOS; (3) investigate the purification mechanism involved in Kluyveromyces lactis.
Materials and methods
Bacterial strains
Lactobacillus plantrum CICC 22186, Lactobacillus acidophilus NCFM, Lactobacillus plantrum Lp39, Lactobacillus plantrum C8-1, Lactobacillus acidophilus CICC 6075, Lactobacillus plantrum B7, and Kluyveromyces lactis 1772 were obtained from China Industrial Microbial Culture Preservation Management Center (CICC). Lactobacillus strains were cultured in De Man, Rogosa, and Sharpe (MRS) medium for 18 h at 37 °C. Viability was determined using the standard count methods in MRS agar at 37 °C for 48 h. Kluyveromyces lactis 1772 was incubated in yeast peptone dextrose (YPD) medium for 24 h at 30 °C.
Analysis of Lactobacillus-derived β-galactosidase
Extraction of crude β-galactosidase
MRS solid medium containing X-Gal (30 mg/L) was used as the preliminary screening medium. The β-galactosidase-producing strains were selected by observing any blue matter around the colony in the medium.
To obtain crude β-galactosidase extracts, the strains were amplified in a fluid lactose medium, Cells were collected by centrifugation at 8000×g for 5 min at 4 °C. Next, 20 mL of 1 mol/L phosphatic buffer solution (PBS) was added to the analysis system. The cell pellets were washed twice with sterilized saline water, and then fragmentized using an ultrasonic cell disruption system (Scientz Biotechnology Co., Ltd., Ningbo, China) at an amplitude of 40 Hz for 15 min (time interval was three seconds). Lastly, the cells were centrifuged (8000×g, 20 min, 4 °C), and the supernatants were used for subsequent applications.
β-galactosidase activity assay
In the assay of β-galactosidase activity, the synthetic substrate o-nitrophenyl-β-D-galactopyranoside (ONPG) was used to evaluate the hydrolytic activity of β-galactosidase. Briefly, 0.5 mL of 20 mmol/L ONPG was added to 0.5 mL of crude β-galactosidase. After incubation in a water bath for 30 min at 37 °C, 4 mL of 0.5 mol/L Na2CO3 solution was added. The absorbance was measured using a UV–Vis spectrophotometer (Purkinje General Instrument Co., Ltd., Beijing, China) at 420 nm. An o-nitrophenol (ONP) standard curve was plotted to quantify the enzymatic production (expressed in mmol/L). The β-galactosidase activity assay was defined as the quantity necessary for the enzyme to release 1 μmol ONP per minute during hydrolysis. β-galactosidase enzymatic activity was calculated using the following equation:
XNL denotes β-galactosidase enzymatic activity in U/mL, where C is the concentration of ONP from the standard curve in mmol/L, V is the final volume of the reaction mixture in mL, f is the dilution factor, T is the reaction time in minutes, m represents the sample mass or volume in g/mL, and 1.3 is the unit definition coefficient of neutral lactase activity in μmol/min.
Transglycosylation activity assay
Lactose (0.5 mol/L) solution was prepared in 1 mol/L PBS (pH 6.0). A total of 400 μL lactose solution was added to 200 μL of crude β-galactosidase extracts in a reaction system at 35 °C for 4 h. The analysis system was kept in boiling water for 5 min, followed by the addition of an equal volume of acetonitrile solution. The GOS content was determined by high-performance liquid chromatography (HPLC) (LC-20AT; Shimadzu Co., Ltd., Japan) equipped with a Sugar-D column (4.6 × 250 mm; Cosmosil). Each sample (20 μL) was injected into the HPLC system combined with a RID-20 differential detector. The mobile phases were composed of acetonitrile (A) and water (B) (75:25, v/v) at a flow rate of 1.0 mL/min. Glucose, galactose, and lactose standards were used to determine the retention time by HPLC. GOS was quantified using the normalization method.
Identification of β-galactosidase by SDS-PAGE and LC–MS
Crude β-galactosidase was identified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The samples were boiled for 3–5 min, followed by the preparation of a 10% polyacrylamide gel. After applying the corresponding protein markers and samples to the gel, SDS-PAGE was performed at a constant voltage of 120 V. The proteins were stained with Coomassie blue after fixation.
The isolated peptides were analyzed using liquid chromatography and mass spectrometry (LC–MS). The peptides were subjected to LC separation equipped with a column (Acclaim PepMap RSLC C18, 5 μm, 100 A, 300 μm × 5 mm) and a column (Acclaim PepMap C18, 3 μm, 100 A, 75 μm × 150 mm, nanoViper). The mobile phases were composed of 0.1% v/v formic acid in water (A) and 80% v/v CAN (B). A 40-min LC gradient was run as follows: 5–50% B in 20 min, 50–90% B in 10 min, and 90–5% B in 10 min. The flow rate was set to 300 nL/min. On-line MS measurement (Thermo Scientific Q Exactive) was used to further analyze the samples. Data were recorded alternating 0.04-s scans over an m/z range of 350–1800. A database search was performed using the MaxQuant database.
Enzymatic synthesis and identification of GOS
Effect of temperature, pH, and lactose concentration on enzymatic synthesis
To determine the effect of temperature on enzymatic synthesis, a mixture of 300 μL of lactose solution (400 g/L, pH 6.0) and 150 μL of β-galactosidase from Lactobacillus plantarum CICC 22186 was heated at 30, 32, 35, 37, and 40 °C, respectively. Samples were taken at 0, 2, 4, 6, 8, and 10 h. The samples were kept in boiling water for 5 min and then analyzed by HPLC.
To determine the effect of pH on enzymatic synthesis, 400 g/L lactose solution was prepared in phosphate buffer at different pH values. The pH of the phosphate buffer was adjusted to 5.0, 6.0, 7.0, and 8.0. Then, lactose solutions at different pH values and β-galactosidase concentrations from Lactobacillus plantarum CICC 22186 were mixed. Samples were taken at 0, 2, 4, 6, 8, and 10 h and analyzed by HPLC.
To determine the effect of lactose concentration on enzymatic synthesis, different concentrations (100, 200, 300, 400, and 500 g/L) of lactose solution were prepared in phosphate buffer (pH 6.0). Then, lactose solution at different concentrations and β-galactosidase from Lactobacillus plantarum CICC 22186 were mixed. The samples were taken at 0, 2, 4, 6, 8, and 10 h and analyzed by HPLC.
Identification of GOS
Initially, GOS was derivatized with hexamethyldisilazane (HMDS), and then injected into a gas chromatography–mass spectrometry (GC–MS) system (QP2010 ultra; Shimadzu Co., Ltd., Japan). An Agilent J&W DB-5 ms capillary column (30 m × 0.25 mm i.d., 0.25-μm film thickness) was selected for analyte separation. The carrier gas was helium at a flow rate of 1 mL/min. The temperature program was as follows: held at 200 °C for 15 min, increased from 200 to 260 °C at 3 °C/min, from 260 to 280 °C at 1 °C /min, and from 280 to 320 °C at 5 °C /min, then held for 10 min. The MS source was set at 220 °C and MS Quad at 280 °C. Data were recorded at scans over an m/z range of 45–500.
Purification mechanism of Kluyveromyces lactis 1772
GOS was purified by fermenting and purifying Kluyveromyces lactis 1772.
Effect of Kluyveromyces lactis 1772 inoculum concentration and SSC on purification
To prepare the simulated GOS solution, lactose and galactose were added to a commercial GOS solution. The concentration was adjusted to the same concentration as that synthesized by Lactobacillus plantarum CICC 22186-derived β-galactosidase enzymatic synthesis.
To determine the effect of the Kluyveromyces lactis 1772 inoculum concentration on purification, the yeast inoculum concentration was adjusted to 25, 50, 75, and 100 g/L, respectively. The cells were incubated in 20% solid content of simulated GOS solution, followed by culturing at 24 °C and measured every 2 h. After treatment, the cells were collected by centrifugation at 9000×g for 5 min and the samples were analyzed by HPLC.
To determine the effect of SSC on purification, the SSC of the simulated GOS solution was adjusted to 5, 10, 15, and 20%, respectively. The cells were incubated in simulated GOS solution, cultured at 24 °C, and measured every 2 h. After treatment, the cells were collected by centrifugation at 9000×g for 5 min and the samples were analyzed by HPLC.
Finally, to assess the purification effect of Kluyveromyces lactis 1772, the bacterial cells were added to the GOS solution for Lactobacillus plantarum CICC 22186-derived β-galactosidase enzymatic synthesis under optimal conditions. Sugar utilization was detected by HPLC.
Related enzyme activity assay
Cells were collected by centrifugation at 8000×g for 5 min at 4 °C. First, the pellets were washed twice with sterilized saline water. Then, GOS was purified under optimal conditions. Finally, the cells were centrifuged (8000×g, 5 min, 4 °C). Samples were obtained using a one-step bacterial active protein extraction kit, and the protein content was determined using a BCA protein concentration assay kit (Sangon Biotech Co., Ltd., Shanghai, China) according to the manufacturer’s instructions.
According to the operating instructions, the hexokinase (HK) and pyruvate kinase (PK) enzymatic activity were determined using a hexokinase activity assay kit and pyruvate kinase activity assay kit, respectively (Comin Biotechnology Co., Ltd., Jiangsu, China). HK enzyme activity (U/mg) was defined as the production of 1 nmol NADPH per minute and per mg of histone. PK enzyme activity (U/mg) was defined as the production of 1 nmol NADH per minute and per mg of histone.
The assay method for β-galactosidase was the same as that of the former method in the study.
Analysis of Kluyveromyces lactis 1772 gene expression during purification
The transcriptome method was used to investigate the gene expression of Kluyveromyces lactis 1772 during purification. Total RNA was extracted from Kluyveromyces lactis 1772 cells using the TRIzol kit (Sangon Biotech Shanghai Co., Ltd., Shanghai, China). RNA quality was detected by agarose gel electrophoresis using NanoDrop T6 visible spectrophotometer (PGENERAL, Co., Ltd., Beijing, China), Qubit, and Agilent 2100 (Agilent Technologies, Santa Clara, CA) instruments.
Ribosomal RNA was obtained from the samples using Ribo-Zero Magnetic Kit (MRZB12424; Epicentre Biotechnologies, Wisconsin, USA). First-strand cDNA was synthesized using random hexamer primers, while second-strand cDNA synthesis was performed using dNTPs, DNA polymerase I, and RNase H. Then, the purified and amplified fragments were used to create the final cDNA library. cDNA sequencing was performed using a high-throughput sequencing platform (Illumina HiSeq 2000; Illumina, California, USA).
RNA-Seq bioinformatics analysis was performed to sequence the mRNA of the Kluyveromyces lactis 1772 transcriptome during the purification process. CASAVA base recognition analysis was used to convert raw reads into clean reads [24]. Clean reads were matched to the reference genome using HISAT and Bowtie2. RSEM (RNA-Seq by Expectation Maximization) was used to calculate the gene expression levels. DEGseq was applied to differential expression analysis using a screening criteria of p < 0.05. The absolute value of log2 (fold-change) was greater than or equal to 1.
Based on the standard classification of DEGs and the annotated results in the Gene Ontology (GO) database, the GO functions or biological pathways of the DEGs were classified. A significantly enriched term in high-level expressed genes was identified by comparison to the entire genome.
Statistical analysis
All experiments and analyses were performed in triplicate. The results are presented as the mean values. Statistical Package for Social Science (SPSS) (version 17.0) and Microsoft Office Excel 2010 action supplement were used for data analysis. One-way analysis of variance (ANOVA) and the Independent Samples t test were used for statistical analysis.
Results
Analysis of Lactobacillus-derived β-galactosidase
Based on the color change of the colony in the preliminary screening medium, Lactobacillus acidophilus CICC 6075, Lactobacillus plantarum C8-1, and Lactobacillus plantarum CICC 22186 were selected for further activity assays. As shown in Table 1, the β-galactosidase activity of the three strains ranged from 16.35 to 17.55 U/mL, and the value of transglycosyl activity ranged from 25 to 33%. The β-galactosidase activity and transglycosylation activity of Lactobacillus plantarum CICC 22186 were higher than those of the other two strains, with a β-galactosidase activity of 17.55 U/mL and a glycosyltransferase activity of 33%. Therefore, Lactobacillus plantarum CICC 22186 was chosen for further study.
Lactobacillus plantarum CICC 22186-derived β-galactosidase was separated by SDS-PAGE and identified by LC–MS. According to the electrophoretic results (Fig. 1a), two distinct protein bands with molecular weights between 35 and 70 kDa were observed. Five unique peptides of β-galactosidase were retrieved in band 2 with a theoretical molecular weight of 35.025 kDa by searching the MaxQuant database, which matched the theoretical masses.
Effect of temperature, pH, and lactose concentration on enzymatic synthesis
The effect of temperature on enzymatic synthesis is shown in Fig. 2a. GOS production and temperature were positively correlated at five different temperatures between 0 and 6 h. However, a downward trend was observed after 6 h. At 35 °C, the production of GOS was up to 29.37%, which was a relatively high value.
As shown in Fig. 2b, GOS production and pH also showed a positive correlation at five different pH values between 0 and 8 h. However, when the reaction time increased above 8 h, GOS production decreased. In addition, GOS production at was relatively low at pH 5 and 6. The maximum production of GOS (34.62%) was observed at pH 7.
The effect of lactose concentration on enzymatic synthesis is shown in Fig. 2c. At a lactose concentration ranging from 200 to 400 g/L, the production of GOS increased with an increasing lactose concentration. The maximum production of GOS increased from 20.54 to 30.63%. However, GOS production was found to decrease as the lactose concentration rose above 400 g/L.
Structural identification of GOS by GC–MS analysis
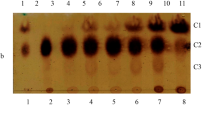
According to Fig. 1a, b and c, total of eight disaccharide and four oligotrisaccharides were identified. Their main glycosidic bond structures were β-D-Galp-(1 → 6)-D-Lac, β-D-Galp-(1 → 3)-D-Lac, and β-D-Galp-(1 → 6)-D-Glc.
Effect of Kluyveromyces lactis 1772 inoculum concentration and SSC on purification
As shown in Fig. 3a, the value of GOS production was the highest when the inoculum concentration of Kluyveromyces lactis 1772 was 75 g/L. At reaction times above 12 h, the production value was over 80%. While the inoculation of yeast was adjusted to 25 g/L, the value was at the minimum, which was lower than 70%.
The effect of SSC on purification is shown in Fig. 3b. As indicated by the concave curve, the purification speed was relatively high at the beginning and the purification speed was gradually reduced. As the SSC increased, the purification time was elongated. At an SSC of 20%, the GOS value reached its minimum. An increase in the concentration resulted in an increased viscosity, while the reaction mass transfer efficiency was decreased.
HPLC was used to determine the changes in the sugar content of the purified solution during the purification process. As a result, only lactose, glucose, and galactose were found to be consumed, and not GOS, proving the feasibility of purification by Kluyveromyces lactis 1772. The proportion of GOS increased from 30 to 75% under optimal conditions (75 g/L, 10%) after 8 h of purification.
Purification mechanism of Kluyveromyces lactis 1772
Enzyme activity during purification
As shown in Fig. 4, the levels of hexokinase activity first decreased and then increased slowly, reaching 57.33 U/mg. For pyruvate kinase, a slow upwards trend from 14.95 to 20.44 U/mg was observed. By contrast, β-galactosidase activity decreased initially, and was then maintained at a certain level, ranging from 57.57 to 39.64 U/mg.
Transcriptomic analysis
According to the comparative transcriptomic results (Fig. 5), significant changes occurred at the mRNA level during purification (p < 0.05). A total of 1287 genes in group (4 vs. 0 h) were differentially expressed, of which 573 DEGs were up-regulated and 714 DEGs were down-regulated. In group (8 vs. 4 h), 141 genes were differentially expressed, 99 genes were up-regulated, and 42 genes were down-regulated. In group (12 vs. 8 h), 18 genes were differentially expressed, 6 genes were up-regulated, and 12 genes were down-regulated.
The functional categories of DEGs were annotated using GO enrichment analysis (Table 2). DEGs were classified into three categories: biological process (BP), cellular component (CC), and molecular function (MF).
In the BP annotation of group (4 vs. 0 h), the annotated down-regulated genes were located in gene translation, amide synthesis, peptide biosynthesis, cellular macromolecular biosynthesis, protein metabolism, and RNA processing and modification, among others. The up-regulated genes played roles in transmembrane transport effect and oxidation–reduction. Within the CC category, the down-regulated DEGs were located in ribosome, protein complex, cytoplasm, and organelles. Upregulated genes were mainly concentrated in membrane and membrane parts. In the MF category, the down-regulated genes annotated were located in RNA binding, nucleic acid binding, polymerase activity, and transferase activity. These results indicate that the material transport was accelerated by up-regulating related genes, such as membrane and transport function, at the beginning of purification.
Within the BP annotation of group (8 vs. 4 h), the annotated down-regulated genes were located in transmembrane transport and amino acid catabolism. In the CC annotation, the down-regulated DEGs were located in membrane function. In the MF category, the annotated down-regulated genes were located in RNA binding, nucleic acid binding, polymerase, and transferase activity. The up-regulated genes were mainly concentrated in the membrane and membrane parts. In the MF category, the annotated down-regulated genes were located in translocator activity. Overall, the up-regulated genes in the three categories mainly exerted roles in gene expression, RNA modification and metabolism, ribosomal protein complex, rRNA metabolism, and nucleic acid metabolism.
In group (12 vs. 8 h), the annotated down-regulated genes were located in oxidoreductase activity and oxidation–reduction process, while up-regulated genes were mainly concentrated in carbohydrate metabolism, carbohydrate binding, and galactose and monosaccharide binding. In the middle and later stages of purification, the cells showed slower material transport and enhanced gene expression, transcription, and replication.
To systematically analyze the function of DEGs further, KEGG enrichment analysis was used to categorize DEGs into different pathways based on bioinformatics databases. As shown in Table 3, down-regulated pathways were related to the ribosomal and RNA pathways compared with the results at 0 h. By contrast, the up-regulated pathways were mainly involved in the pathways for fatty acid metabolism, amino acid metabolism, and carbohydrate metabolism. These results demonstrate that genetic transcription and translation were inhibited at the beginning of purification, and that the growth of cells was maintained by up-regulating material metabolism. At 4 h, up-regulated pathways were related to the ribosomal pathway. Compared with 8 h, the cells up-regulated glycometabolism to maintain cell viability and translation, while the sugar metabolism was accelerated to ensure cell vitality during the middle and later stages of purification.
Discussion
β-Galactosidases are glycosidases with both transglycosylation and hydrolytic activities [16]. In transglycosylation activity, β-galactosidases catalyze the transfer of sugar residues to acceptors, resulting in the formation of new glycosidic bonds. The hydrolysis and transglycoside activities of β-galactosidases from different sources were significantly different [27]. Sanaullah et al. found that β-galactosidase from Lactobacillus plantarum WCFS1 consisted of a small subunit of 35 kDa and a large subunit of 72 kDa, similar to our findings [14].
Temperature plays an important role in enzymatic reaction efficiency. When the temperature increases, the mass transfer effect of the substrate improves and molecular collision is enhanced, which is beneficial to the production of GOS. However, when the reaction temperature exceeds the optimum temperature of the enzyme, enzyme activity is reduced or even deactivated, which may lead to a decrease in the reaction speed [28]. Therefore, it is of great significance to select the appropriate reaction temperature when other reaction conditions are certain for a rapid enzymatic reaction.
Each enzyme has a pH range and an optimum pH value. For example, β-galactosidase is an alkalescent enzyme and can remain stable in an alkalescent range, but is easily deactivated in an acidic pH range [21]. At its optimal pH value, enzyme activity is high and GOS is produced in abundance.
In the reaction system, the lactose hydrolysis reaction and transgalactosylation reaction run at the same time. The reaction tends to hydrolyze at low lactose concentrations. By contrast, transgalactosylation reactions will tend to occur under high lactose concentrations, increasing the chance of galactosyl bond to glycosyl conversion, which is helpful for production [6]. In a separate study, Warmerdam found that a high initial lactose concentration had a large positive effect on enzyme activity and stability [25]. However, crystallization can occur at a certain temperature and concentration, promoting a hydrolysis reaction.
As a result of the enzymatic synthesis of GOS, undesired molecules are also produced, the properties of which are similar. As a result, the structural identification of GOS is difficult. Owing to the different glycoside bond types found in GOS, the strength of the characteristic peak ions was also different. However, GC–MS data of GOS with different glycosylated bond types and retention times can be used to analyze and identify GOS precisely [1]. As a result, Lactobacillus plantarum CICC 22186-derived β-galactosidase was found to synthesize GOS of β-(1 → 6) and β-(1 → 3) bonds. According to Sanaullah et al., Lactobacillus-derived β-galactosidase showed a strong preference for the formation of β-(1 → 6) linkages in its transgalactosylation mode, while β-(1 → 3)-linked products were formed to a lesser extent [13]. Maischberger reported that a maximum yield of 31% GOS of total sugars was obtained at 78% lactose conversion. The enzyme showed a strong preference for the formation of β-(1 → 3) and β-(1 → 6) linkages [12].
By-products can affect the purity of GOS during its enzymatic synthesis. Purification can be carried out using various techniques, including chromatography columns, membranes, enzymes, and fermentation [4]. Fermentation has the advantage of being simple and low cost, as well as having a good separation effect. Sun et al. found that Kluyveromyces lactis could be used to obtain GOS with a purity of over 95% of [19]. During purification, the concentration of the inoculum and the SSC are important factors. Santibáñez et al. [15] reported that the total carbohydrate concentration (10–50%) has an effect on the efficiency of producing high-purity GOS.
Despite the diversity of the substrates and metabolic modes displayed by yeasts, the glycolytic pathway is found in many species of yeast. This pathway consists of many enzymes, including hexokinase, phosphorylates, glucose, and fructose, at the expense of ATP. A second phosphoryl transfer reaction occurs to form pyruvate and a second ATP molecule, catalyzed by pyruvate kinase. A lack of nutrient substances would result in a decline in the growth rate of cells. The activity of the rate-limiting enzyme (hexokinase) in the glycolytic pathway was found to decrease at the initial stage of purification. With an acceleration of the glycolysis pathway, the activity of hexokinase and pyruvate kinase activity increased gradually. In addition, Suleau et al. [17] found that genes of the yeast Kluyveromyces lactis involved in pyruvate metabolism were more highly expressed in the presence of lactose than glucose. Although lactose was the only carbon source, cells preferentially utilized glucose and galactose at the beginning of purification, which resulted in a decrease in β-galactosidase activity. The cells then hydrolyzed lactose to maintain normal growth, allowing β-galactosidase activity to remain stable [7].
The defining trait of Kluyveromyces lactis, a food yeast, is its capacity to grow on lactose as a sole carbon source. Although Kluyveromyces lactis is known to be able to grow with carbon sources alone, the effects of this condition on the physiology of this yeast has yet to investigated extensively. However, differential gene expression is expected when cells are grown in the absence of a nitrogen source, with the majority of DEGs associated with protein. This demonstrates that the protein function of Kluyveromyces lactis 1772 is significantly affected during purification when nitrogen sources are lacking from the GOS solution. Varela et al. [23] found that two genes, LAC12 (lactose permease) and LAC4 (lactase), were sufficient for lactose uptake and hydrolysis to maintain normal growth during lactose fermentation in Kluyveromyces lactis. Their analysis of Kluyveromyces lactis 1772-related enzyme activities and transcriptomics demonstrated that although the synthesis of macromolecules, such as proteins, in cells was blocked, cell growth resumed by increasing the processes of glucose metabolism and substance transport. In the present study, Kluyveromyces lactis 1772 was found to successfully generate high-purity GOS. Suleau et al. [18] reported extensive differences in gene expression between Kluyveromyces lactis with regards to sugar transport, glucose repression, ethanol metabolism, and amino acid import. These differences can be partly attributed to repression by glucose and other unknown regulation mechanisms.
Conclusion
In this study, GOS was synthesized using a Lactobacillus plantarum CICC 22186-derived β-galactosidase enzymatic method. The main structural formula of the GOS produced was β-D-Galp-(1 → 6)-DLac, β-D-Galp-(1 → 3)-D-Lac, and β-D-Galp-(1 → 6)-D-Glc. Based on the fact that various factors, including temperature, pH, and lactose concentration, are involved in the synthesis of GOS, we optimized the conditions for the enzymatic of synthesis (35 °C, pH 7.0, and 400 g/L). Using this method, a GOS content of 30% was achieved. Under the optimal purification process conditions, (SSC of 10% and the addition of 75 g/L yeast), the GOS content was increased from the initial 30 to 75%. Thereafter, the purification mechanisms of Kluyveromyces lactis 1772 were explored, and the analysis of Kluyveromyces lactis 1772-related enzyme activities and transcriptomics demonstrated that cell growth was resumed by increasing the processes of glucose metabolism and substance transport. Thus, by combining the process of enzymatic synthesis with the fermentation of Kluyveromyces lactis 1772, GOS with a high degree of purity was obtained.
References
Cardelle-Cobas A, Martínez-Villaluenga C, Sanz ML, Montilla A (2009) Gas chromatographic–mass spectrometric analysis of galactosyl derivatives obtained by the action of two different β-galactosidases. Food Chem 114(3):1099–1105. https://doi.org/10.1016/j.foodchem.2008.10.057
Davis LMG, Martínez I, Walter J, Hutkins R (2010) A dose dependent impact of prebiotic galactooligosaccharides on the intestinal microbiota of healthy adults. Int J Food Microbiol 144(2):285–292. https://doi.org/10.1016/j.ijfoodmicro.2010.10.007
Djouzi Z, Andlueux C (1997) Compared effects of three oligosaccharides on metabolism of intestinal microflora in rats inoculated with a human faecal flora. Br J Nutr 78(2):313–324. https://doi.org/10.1079/BJN19970149
Feng YM, Chang XL, Wang WH, Ma RY (2009) Separation of galacto-oligosaccharides mixture by nanofiltration. J Taiwan Inst Chem Eng 40(3):326–332. https://doi.org/10.1016/j.jtice.2008.12.003
Gopal PK, Sullivan PA, Smart JB (2001) Utilisation of galacto-oligosaccharides as selective substrates for growth by lactic acid bacteria including Bifidobacterium lactis DR10 and Lactobacillus rhamnosus DR20. Int Dairy J 11(1):19–25. https://doi.org/10.1016/S0958-6946(01)00026-7
Guerrero C, Vera C, Conejeros R, Illanes A (2015) Transgalactosylation and hydrolytic activities of commercial preparations of β-Galactosidase for the synthesis of prebiotic carbohydrates. Enzyme Microbial Technol 70:9–17. https://doi.org/10.1016/j.enzmictec.2014.12.006
Gondium MDB, Souza CG, Simões DA, Morais MA (2002) The β-galactosidase activity in Kluyveromyces marixanus decrease by high concentration of galactose. Curr Microbiol 44:379–382. https://doi.org/10.1007/s00284-001-0052-2
González-Delgado I, López-Muñoz MJ, Morales G, Segura Y (2016) Optimisation of the synthesis of high galacto-oligosaccharides (GOS) from lactose with β-galactosidase from Kluyveromyces lactis. Int Dairy J 61:211–219. https://doi.org/10.1016/j.idairyj.2016.06.007
Hansson T, Kaper T, Oost JVD, Vos WMD, Adlercreutz P (2001) Improved oligosaccharide synthesis by protein engineering of β-galactosidase CelB from hyperthermophilic Pyrococcus furiosus. Biotechnol Bioengr 73:203–210. https://doi.org/10.1002/bit.1052
Knol J, Scholtens P, Kafka C, Steenbakkers J, Gro S, Helm K, Klarczyk M, Schopfer H, Bockler HM, Wells J (2005) Colon Microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J Pediatr Gastroenterol Nutr 40(1):36–42. https://doi.org/10.1097/00005176-200501000-00007
Liao XY, Huang JJ, Zhou QL, Guo LQ, Lin JF, You LF, Liu S, Yang JX (2017) Designing of a novel β-galactosidase for production of functional oligosaccharides. Eur Food Res Technol 243(6):979–986. https://doi.org/10.1007/s00217-016-2813-y
Maischberger T, Leitner E, Nitisinprasert S, Juajun O, Yamabhai M, Nguyen T, Haltrich D (2010) β-Galactosidase from Lactobacillus pentosus: purification, characterization and formation of galacto-oligosaccharides. J Biotechnol 5:838–847. https://doi.org/10.1002/biot.201000126
Sanaullah I, Thu-Ha N, Hoang Anh N, Tien Thanh N, Thomas M, Roman K, Haltrich D (2011) Characterization of a heterodimeric GH2 β-Galactosidase from lactobacillus sakei Lb790 and formation of prebiotic galacto-oligosaccharides. J Agric Food Chem 59(8):3803–3811. https://doi.org/10.1021/jf103832q
Sanaullah I, Thu-Ha N, Tien Thanh N, Thomas M, Dietmar H (2010) β-Galactosidase from Lactobacillus plantarumWCFS1: biochemical characterization and formation of prebiotic galacto-oligosaccharides. Carbohydrate Res 345(10):1408–1416. https://doi.org/10.1016/j.carres.2010.03.028
Santibánez L, Fernández-Arrojo L, Guerrero C, Plou FJ, Illanes A (2016) Removal of lactose in crude galacto-oligosaccharides by β-galactosidase from Kluyveromyces lactis. J Mol Catal B Enzym 133:85–91. https://doi.org/10.1016/j.molcatb.2016.07.014
Silvério SC, Macedo EA, Teixeira JA, Rodrigues LR (2018) New β-galactosidase producers with potential for prebiotic synthesis. Bioresour Technol 250:131–139. https://doi.org/10.1016/j.biortech.2017.11.045
Suleau A, Jacques N, Reitz-Ausseur J, Casaregola S (2005) Intraspecific gene expression variability in the yeast Kluyveromyces lactis revealed by micro-array analysis. FEMS Yeast Res 5(6–7):595–604. https://doi.org/10.1016/j.femsyr.2004.11.009
Suleau A, Gourdon P, Reitz-Ausseur J, Casaregola S (2006) Transcriptomic analysis of extensive changes in metabolic regulation in Kluyveromyces lactis strains. Eukaryot Cell 5(8):1360–1370. https://doi.org/10.1128/EC.00087-06
Sun H, You S, Wang M, Qi W, Su R, He Z (2016) Recyclable strategy for the production of high-purity galacto-oligosaccharides by Kluyveromyces lactis. J Agric Food Chem 64(28):5679–5685. https://doi.org/10.1021/acs.jafc.6b01531
Thongaram T, Hoeflinger JL, Chow JM, Miller M (2017) Prebiotic Galactooligosaccharide Metabolism by Probiotic Lactobacilli and Bifidobacteria. J Agric Food Chem 65(20):4184–4192. https://doi.org/10.1021/acs.jafc.7b00851
Torres DPM, Gonçalves MPF, Teixeira JA, Rodrigues LR (2010) Galacto-oligosaccharides: production, properties, applications, and significance as prebiotics. Compr Rev Food Sci Food Safety 9(5):438–454. https://doi.org/10.1111/j.1541-4337.2010.00119.x
Urrutia P, Rodriguez-Colinas B, Fernandez-Arrojo L, Ballesteros AO, Wilson L, Illanes A, Plou FJ (2013) Detailed analysis of galacto-oligosaccharides synthesis with β-Galactosidase from Aspergillus oryzae. J Agric Food Chem 61(5):1081–1087. https://doi.org/10.1021/jf304354u
Varela JA, Puricelli M, Ortiz-Merino A, Giacomobono R, Morrissey JP (2019) Origin of lactose fermentation in Kluyveromyces lactis by interspecies transfer of a neo-functionalized gene cluster during domestication. Curr Biol 29(24):4284–4290. https://doi.org/10.1016/j.cub.2019.10.044
Wang L, Yue T, Yuan Y, Wang ZL, Ye MQ, Cai R (2015) A new insight into the adsorption mechanism of patulin by the heat inactive lactic acid bacteria cells. Food Control 50:104–110. https://doi.org/10.1016/j.foodcont.2014.08.041
Warmerdam A, Wang J, Boom RM, Janssen AEM (2013) Effects of carbohydrates on the ONPG converting activity of β-galactosidases. J Agric Food Chem 61(26):6458–6464. https://doi.org/10.1021/jf4008554
Weaver CM, Martin BR, Nakatsu CH, Armstrong AP, Clavijo A, Linda D, McCabe LD, McCabe GP, Duignan S, Schoterman MHC, van den Heuvel EGHM (2011) Galactooligosaccharides improve mineral absorption and bone properties in growing rats through gut fermentation. J Agric Food Chem 59(12):6501–6510. https://doi.org/10.1021/jf2009777
Yin H, Bultema JB, Dijkhuizen L, Van Leeuwen SS (2017) Reaction kinetics and galactooligosaccharide product profiles of the β-galactosidases from Bacillus circulans, Kluyveromyces lactis and Aspergillus oryzae. Food Chem 225:230–238. https://doi.org/10.1016/j.foodchem.2017.01.030
Zarate S, Lopez-Leiva MH (1990) Oligosaccharide formation during enzymatic lactose hydrolysis. J Food Protect 53:262–268. https://doi.org/10.1016/0168-1656(90)90083-N
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Compliance with ethics requirements
This study does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, X., Yao, C., Wang, T. et al. Production of high-purity galacto-oligosaccharides (GOS) by Lactobacillus-derived β-galactosidase. Eur Food Res Technol 247, 1501–1510 (2021). https://doi.org/10.1007/s00217-021-03727-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-021-03727-9