Abstract
Optimization of the thrombin inhibitory activities of different enzymatic hydrolysates was conducted, and then an optimal hydrolysis condition by trypsin (5000 u/g) was determined as follows, digested at 45 °C and pH 8.5 for 2 h with a protein concentration of 25 mg/mL. Thrombin inhibitory activity was proved to be 76.92 ± 4.66% under this condition. A total of 39 peptides were identified in the hydrolysate by UPLC-Q-TOF–MS/MS, and all the peptides were predicted to be nontoxic by in silico predictive approaches. Twenty-six peptides were predicted to be anticoagulant peptides by molecular docking method, and the peptide 26 (Lys-Asn-Ala-Glu-Asn-Glu-Leu-Gly-Glu-Val-Thr-Val-Arg) was predicted to be a better anticoagulant peptide through both structure–activity relationship and affinity activity to thrombin. The interactional positions between peptide and thrombin were also involved in the interaction site on the S1 pocket of thrombin and strongly promoted its thrombin inhibitory activity. The firmly non-bonded interactions made the bound of peptide and thrombin firmly. Eventually, the chemical identification and activity verification of synthetic peptide 26 were conducted, and the thrombin inhibitory activity was 89.96 ± 5.30% at the concentration of 9 mg/mL. This study optimized an enzymatic hydrolysis and a virtual screening method for predicting and verifying the anticoagulant peptide from Mytilus edulis, respectively, which provided a good theoretical basis and application method for the research and development of the anticoagulant peptides, especially from the seafood products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Marine bioactive peptides have been reported with a wide range of biological functions on account of their structural properties, amino acid composition and sequences [1,2,3]. Mytilus edulis is a kind of bivalve mollusks that belongs to Mollusca, Laaellibraachia, Anisomyaria, Mytilacea, Mytidea. It widely distributes in all the countries of the Atlantic coast. Mytilus edulis contains much higher essential amino acids than eggs, chickens, ducks, fish, shrimp and meat and so on [4,5,6].
Thrombin is the key enzyme of the coagulation system with the functions of activating of platelets, transforming of fibrinogen into a fibrin network and feedbacking amplification of coagulation. The essence of the blood clotting is conversion of soluble fibrinogen into insoluble fibrin in the blood plasma [7,8,9]. The anticoagulant peptide inhibition of thrombin is an important way to inhibit thrombosis. In the blood coagulation system, it is a complex process for thrombin to convert fibrinogen into insoluble fibrin. An antithrombotic peptide will inhibit the activity of thrombin, reduce the formation of fibrin aggregates, and then result in a decrease of absorbance of testing system. Thus, inhibition of thrombin can be calculated from the decrease in absorbance [10, 11]. It is becoming more and more important for anticoagulant peptides to prevent ischemic events in patients with cardiovascular diseases. Besides the natural anticoagulants identified in the coagulation cascade, series of natural sources have also been found to contain anticoagulant peptides [12].
It is well known that bioactive peptides generally contain 5–40 amino acid residues with specific structural properties and amino acid sequence [13]. To obtain bioactive peptides, the enzymatic hydrolysis method was used. At present, biological enzymes are chosen to break the protein molecule based on the restriction sites into peptides with smaller molecular weight, which proved to be a faster approach to obtain target peptides. Precursor proteins can be hydrolyzed with the enzymes such as alcalase, trypsin, neutrase, papain, and protamex [14].
Molecular docking method is generally used for the sake of electrostatic interaction, hydrogen bonding and hydrophobic effects between the ligand and receptor, which also can be used to analyze the interaction of anticoagulant peptides against thormbin [12]. The purpose of molecular docking is to predict the best mode by which a given compound will fit into a binding site of a macromolecular target. The method has been developed and coupled with suitable scoring functions [15]. Molecular docking works on the basis of the ligand and receptor function “lock-key principle”, simulation interaction between small molecular biological macromolecular ligand (peptides) and the receptor (thrombin). Depending on theoretical calculations of the interaction patterns and affinity against thrombin, virtual screening for anticoagulant peptides was carried out.
In the present study, the peptidome of M. edulis protein hydrolysates by three kinds of enzymes was analyzed by UPLC-Q-TOF–MS/MS. Moreover, the bioactive peptides were subjected to in silico prediction and peptides with potential antithrombotic activity were synthesized and the thrombin inhibitory activity was analyzed. These results would provide some basic instructions for production of target bioactive peptides from M. edulis protein by enzymatic technology.
Materials and methods
Materials and chemicals
Fresh mussels had an average shell width of about 5–6 cm, and an approximate weight of 13 g, with a breeding time of 1 year. Trypsin (EC 3.4.21.5, ≥200 u/mg, Beijing Boise Technology Co. Ltd, China); Fibrinogen (Nanjing Oddo’s Biological Technology Co., LTD, China); Pepsin and Neutrase (Ameresco, China); Molecular weight marker for peptides (Thermo Scientific, America).
Preparation of M. edulis protein hydrolysate
The experiments were operated at 4°C unless stated otherwise. M. edulis was washed three times with deionized water to remove salts and other contaminants. After removing the shells, 200 g of edible parts was minced and homogenized at 5000 rpm for 10 min, and the insoluble homogenates were removed by centrifugation at 8000 rpm for 15 min with a table-top low speed centrifuge (Hunan Xiangyi Instrument Development Co. Ltd, China). 5000 u/g of pepsin, trypsin, and neutrase were added to the supernatants, respectively. The temperature and pH were adjusted to 37 °C and pH 2.0 for pepsin, 37 °C and pH 8.0 for trypsin, 40 °C and pH 7.5 for neutrase, respectively. The hydrolysis was performed for 1–3 h at constant pH value maintained by the addition of 1 M HCl or 1 M NaOH. Inactivation of the enzyme was obtained by immersing it in boiling water bath for 10 min and then cooled immediately. The hydrolysates were then centrifuged at 8000 rpm for 15 min and the supernatants were stored at 4 °C [16].
Determination of the degree of hydrolysis
Reactions were monitored by measuring the extent of proteolytic degradation with the method of pH–stat described by Adler-Nissen [17,18,19]. The values for the degree of hydrolysis (DH) were determined using the following Eqs. (1) and (2):
where DH is the ratio of the numbers of peptide bonds cleaved (h) to the total number of peptide bonds in the substrate studied (h tot). The variable B is the amount of alkaline consumed to keep the pH constant during the reaction, α is the average degree of dissociation of α-NH2 groups released during hydrolysis, and N b is the normality of the alkali, M p is the mass of the substrate (protein, determined as N × 6.25) in the reaction, t is the temperature of the reaction environment, and K is the equilibrium constant.
Thrombin inhibitory activity determination
Thrombin inhibitory activity was determined according to previous reports at 37 °C [20]. Peptide, thrombin, and fibrinogen were separately dissolved in 50 mM Tris–HCl buffer (pH 7.4) containing 0.154 mM NaCl. A 0.1% fibrinogen solution (140 µL) and 40 µL sample solution at a protein concentration of 1 mg/mL were first injected into the plate wells and mixed, and then the absorbance at 405 nm was recorded (sample blank or control blank). 10 µL of thrombin solution (12 U/mL) was added to each well to initiate the reaction of thrombin-catalyzed fibrinogen coagulation. After 10 min of incubation, sample absorbance was detected again (sample or control). Rather than adding the sample solution, the control treatment contained 40 µL of Tris–HCl buffer (pH 7.4, 50 mM) as the control blank. Inhibition rate was calculated using Eq. (3):
where A C, A CB, A S, and A SB represent the absorbance of the control, the control blank, the sample and the sample blank, respectively [20, 21]. The thrombin inhibitory activity was determined by the inhibition of fibrin cross-linking which is responsible for the clotting activity.
Optimization of thrombin inhibitory activity
An orthogonal array design was used to reduce the total number of experiments for the L9 (3)4 orthogonal array design, there are 9 pretreatment combinations that were derived by altering the four independent variables in this experimental design: enzyme loading (2500–7500 u/g), protein concentration (20–33 mg/mL), pH (7.5–9.5) and temperature (37–45 °C). Thrombin inhibitory activity was determined to assess the effects of different enzymatic hydrolysis conditions [21].
Identification of peptides by UPLC-Q-TOF
A Cleanert S C18-SPE column (Agela Technologies Inc., USA) was used to desalt trypsin enzymatic hydrolysate as described with some modifications [22,23,24]. Briefly, samples were redissolved in 0.1% formic acid (FA) aqueous solution to a total volume of 1 mL. The C18 SPE column was preconditioned with 3 ml methyl alcohol for 3 min before it was rinsed with 3 × 1 mL 0.1% FA. Then, the sample was loaded on to the top of the SPE cartridge column followed by washing it with 3 × 1 mL 0.1% FA; finally, the peptides were eluted with 3 × 500 µL 80% methyl alcohol. The eluted samples were lyophilized before analysis. The desalted samples were then subjected to analysis on a UPLC-Q-TOF system coupled to a Synapt Mass Quadrupole Time-of-Flight Mass Spectrometer. 10 µL of sample was loaded onto an C18 column (150 mm × 3 mm, 3 µm), and the column was run using a linear gradient of water-acetic acid (100:0.5, v/v) (Elution solution A) and methanol (Elution solution B) as follows: 0–2 min, 10% B; 2–30 min, 35% B; 30–34 min, 35% B; 34–36 min, 10% B and, finally, the initial conditions were held for 4 min as a re-equilibration step; flow rate: 0.4 ml/min. The MS spectra were acquired under the positive electrostatic ionization. Collision energy in the range of 10–35 eV was tested for each preselected precursor ion and optimized based on the quality of ion fragmentation. The optimum values of the ESI–MS parameters were as follows: capillary voltage, +4.5 kV; drying gas temperature, 200 °C; drying gas flow, 6.0L/min; nebulizer gas pressure, 2.0 bar [25]. The top 20 most intense ions were obtained according to auto MS/MS after the fragmentation of protonated molecular ions. Each sample was analyzed in triplicate.
Automated spectral processing, peak list generation and database search for the identification of the peptides were performed using Mascot v1.4.0.38 software with analyzing ion peaks in the m/z range of 50–2200 m/z. Mass tolerance was 10 ppm for MS and 0.03 Da for MS/MS ions, and allowed for up to one missed proteolytic sites with trypsin for protein cleavage and the selected fixed and variable modification was oxidation. Identifications were validated by performing a decoy database search for the estimation of false discovery rate (FDR). The Swiss-Prot protein database was used to identify the peptides with an ions score threshold of 20, a significance threshold p < 0.05, FDR ≤ 1% [26]. The potential toxicity of the peptides in silico was detected using ToxinPred, available at http://www.imtech.res.in/raghava/toxinpred/ [27].
Molecular docking
Molecular docking of the estimated antithrombotic peptides and thrombin was performed using Discovery Studio 3.5 software according to the operation procedures [28]. The 3D structure of the human thrombin complex with the hirudin fragment, TYS, and 4CP was downloaded from the PDB database. The PDB code of the complex is 2BVR [29]. First, the ligands of hirudin, TYS, and 4CP were removed from the 3D structure of the complex, and then the interaction site was set at the original position of hirudin in the 2BVR complex for the docking of bioactive peptides and thrombin. The interaction mechanisms of some peptides with inhibitory activity against thrombin have been reported previously [30, 31], which were used as references for molecular docking. Additionally, the ligand of hirudin (fragment 54–65) was docked as a control, which was compared with all peptides tested in the present study.
Synthesis of thrombin inhibitory peptide
Peptide with high thrombin inhibitory activity identified in the hydrolysates was chemically synthesized by the conventional Fmoc solid-phase synthesis method (Hefei Semanno biotechnology co., LTD, China). The purity and sequence of the synthesized peptides were verified by HPLC and UPLC-Q-TOF, respectively. As for HPLC method, the synthetic peptide at a concentration of 1 mg/mL was performed on an Agilent HPLC system using an Inertsil ODS-SP column (5 µm, 4.6 mm × 250 mm) with a linear gradient of 0.1% trifluoroacetic acid in 100% acetonitrile (Elution solution A) and 0.1% trifluoroacetic acid in 100% water (Elution solution B) [32].
Statistical analysis
All measurements were replicated in triplicates. The results were analyzed by One-Way ANOVA using the Statistical Package for the Social Sciences SPSS 18.0 software and expressed as the mean ± SD. p < 0.05 was considered to be statistically significant.
Results and discussion
Effects of enzymatic hydrolysis on degree of hydrolysis
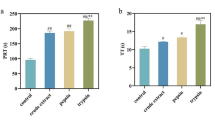
Enzymes have specific cleavage sites within polypeptide chains. To select suitable proteases, M. edulis protein was independently hydrolyzed with pepsin, trypsin, and neutrase. As shown in Fig. 1, the proteins were hydrolyzed in different degrees. With the extension of hydrolysis time, the degree of hydrolysis (DH) of all the hydrolysates kept increasing during the hydrolysis of 2 h. Compared to the other two enzymes, the effect of pepsin on the hydrolysis was the weakest. DH of the hydrolysate by pepsin, neutrase and trypsin at the time of 2 h was 2.31 ± 0.71, 12.76 ± 0.52 and 15.67 ± 0.40%, respectively.
Effects of enzymatic hydrolysis on the thrombin inhibitory activity of hydrolysates
So as to figure out the optimal hydrolysis parameters, the inhibitory activities against thrombin of the different hydrolysates of M. edulis proteins were compared. It is a complex process of thrombin to turn fibrinogen into insoluble fibrin. Thrombin cut fibrin peptide A and B from the Aα chain and N-terminal of Bβ chain, respectively, which activated the fibrinogen into insoluble fibrin and lead to thrombus. When the amount of fibrinogen is constant, the amount of fibrin polymer formed is linked to the activity of thrombin. The activity of thrombin is inhibited by adding a thrombin inhibitor, and the formation of fibrin aggregates is reduced, resulting in a decrease in absorbance. Thus, inhibition of thrombin can be calculated from the decrease in absorbance [10, 11]. As shown in Fig. 2, the inhibitory ability of M. edulis protein and its hydrolysates against thrombin were compared, and the activity had a significant increase in each hydrolysate compared with control group. The thrombin inhibitory activity of 57.62 ± 3.36% was observed in the hydrolysate by trypsin at 2 h, and followed by pepsin hydrolysate (31.89 ± 2.74%) and neutral hydrolysate (30.48 ± 4.17%). At the same time, trypsin hydrolysate also showed the highest DH, which indicated that there were more peptide bonds cleaved in trypsin hydrolysates of M. edulis. Hence, trypsin is better for preparing peptides with higher antithrombotic activity under the present conditions.
To optimize the enzymatic hydrolysis conditions with high thrombin inhibitory activity, an orthogonal design test (L9 (3)4) was carried out due to the better activity of the trypsin hydrolysates. In orthogonal experiment, K value is defined as the sum of the evaluation indexes of all levels in each factor and K − (mean value of K) is used to determine impact of factors on enzymatic hydrolysis of thrombin inhibitory activity. R is defined as the range between the maximum and minimum value of K, and is used to evaluate the importance of the factors [21, 33]. As shown in Table 1, the K value revealed that the optimal enzymatic hydrolysis conditions were enzyme application of 5000 u/g, pH 8.5, temperature 45°C and protein concentration 25 mg/mL. R value showed that the amount of enzyme showed the most significant effect on activity, and the order of importance that influenced thrombin inhibitory activity was found to be as follows: amount of enzyme > pH > temperature > protein concentration. A thrombin inhibitory activity of 76.92 ± 4.66% was obtained under the optimal conditions.
Identification of peptides by UPLC-Q-TOF
Trypsin hydrolysate was submitted to UPLC-Q-TOF–MS/MS analysis and the peptides involved were subsequently identified. As a result, a total of 39 peptides were identified based on the MS/MS data (Table 1). These peptides were sequenced using the Mascot v1.4.0.38 software and scored. Most of the identified peptides were paramyosin OS that belong to myofibril protein which have a large proportion of the M. edulis muscle. The proteins were identified and scored as shown in Table 2. Obviously, there were four kinds of proteins identified including astropomyosin OS, sperm-specific protein Phi-1 OS, paramyosin OS and heavy metal-binding protein HIP OS of M. edulis. The scores were much higher(>60 points),which indicated that these proteins identified were genuine and believable. Table 2 shows the peptides identified corresponding to the three kinds of proteins which belong to M. edulis. The peptides were selected from the Swiss-Prot database and showed compatibility with scores.
Prediction of toxicity of identified peptides
ToxinPred is a widely used website to identify toxic regions in a protein sequence based on the database of all possible overlapping peptides [27]. Researchers can easily identify highly toxic regions in a protein and obtain results. Results showed that all the peptides have no toxicity and the non-toxicity peptides are listed in Table 1. As a result, prediction of toxicity before their synthesis and other analysis is beneficial for developing peptide drugs or application in functional foods.
Molecular docking of peptides against thrombin
The interaction forces between the identified peptides and the thrombin were detected by Discovery Studio 3.5 software. The thrombin structure in Fig. 3a (PDB code, 2BVR) was cleaned firstly by removing the water molecules and hirudin, adding hydrogen atoms and then TYS18 ligand was removed after defining it as the active and is shown in Fig. 3b. Peptides identified were subsequently docked to the active site of the final thrombin. The scoring functions (“–CDOCKER_Energy”) were calculated in silico method [34] and listed in Table 3. These results indicated that 26 peptides showed better interaction effect than the contrast (hirudin) against thrombin. The interaction of anticoagulant peptide 1 (Leu-Thr-Gln-Glu-Asn-Phe-Asp-Leu-Gln-His-Gln-Val-Gln-Glu-Leu-Asp-Gly-Ala-Asn-Ala-Gly-Leu-Ala-Lys) had the highest affinity with an “–CDOCKER_Energy” of 307.670 kcal/mol compared with the control of 181.530 kcal/mol. Ligand–protein interactions of peptide 1 and thrombin are shown in Fig. 4b, and the interactional amino acids were marked with red color including THR74, TYR76, ARG67, CYS51, PRO111, LYS110, MET84, LEV65, GLN38, PRO37A and LYS36. The non-bonded interactions between the amino acid residues of receptor protein and the ligand docking poses included hydrogen bond; electrostatic and other hydrophobic categories including salt bridge, attractive charge, sulfur-X, hydrophobic alkyl, and pi-alkyl. Although there were few references about structure–activity relationship of anticoagulant peptide, on the one hand, there is also evidence that the charged amino acid in C-terminal of the peptide was considered to have effect on thrombin inhibition activity or participated in the clotting process at the same time [35]. Negatively charged C-terminal may play an important role in binding to positively charged thrombin for preventing coagulation process. At the same time, lysine (Lys) may also be a vital amino acid residue in performing anticoagulant activity [36, 37]. On the other hand, in order to research the activities of short peptides upon the request of economical and extensive applications with similar “–CDOCKER-_Energy” compared to the control at the same time, the anticoagulant peptide 26 (Lys–Asn-Ala-Glu-Asn-Glu-Leu-Gly-Glu-Val-Thr-Val-Arg) was within the scope of consideration and more suitable for this research since it had a similar and strong affinity with an “–CDOCKER-_Energy” of 190.077 kcal/mol compared with the control of hirudin. Ligand–protein interactions of peptide 26 and thrombin are shown in Fig. 4c. As a result, the interactional amino acids included LYS110, MET84, PRO111, SER83, LEU65, ILE82, GLN38, ARG67, THR74, and ARG73 which were also involved in the interaction site on the S1 pocket of thrombin and indicated that the combination between the peptide and the thrombin active site strongly promoted its thrombin inhibitory activity [38]. The non-bonded interactions between the amino acid residues of receptor protein and the ligand docking poses included one salt bridge, three attractive charges, twenty-seven hydrogen bonds and three hydrophobic Alkyls. These firmly non-bonded interactions made the bound firmly between the peptide and thrombin.
The thrombin structure (PDB code: 2BVR) and the modified forms. 2BVR was downloaded from PDB and was prepared for simulation by cleaning it. In a, the red dots are water molecules, and the short chain at the bottom is hirudin. The water molecules, 4CP ligand and hirudin in the 2BVR are removed. Then, hydrogen atoms were added and the status was shown. Then, TYS18 site was removed after defining it as the active center; the final results are shown in b
Activity determination of the synthetic thrombin inhibitory peptide
In accordance with the HPLC chromatogram, the peptide 26 was shown at the retain time of 9.961 min with a high purity of 99.21%. UPLC-Q-TOF was performed to verify the peptide sequence, and the secondary mass spectrogram at the m/z of 729.8801 Da for peptide 26 is shown in Fig. 5, and the molecular weight of the identified peptide was 1459.7602 Da, and the peptide score was up to 85.39 with a m/z error of −0.0009 Da which indicated that the results were reliable and can be used for the following analysis. The thrombin inhibitory activities of the synthetic peptide 26 at different concentrations were verified in Fig. 6 after chemical identification. The thrombin inhibitory activity was 67.21 ± 5.50% at the concentration of 1 mg/mL which showed a higher activity. The activity was increased with the increase of the peptide concentration and the thrombin inhibitory activity was achieved the highest of 89.96 ± 5.30% at the concentration of 9 mg/mL. The results showed that the peptide 26 had a stronger thrombin inhibitory activity both in prediction and practical methods and can be used as a thrombin inhibitor.
Conclusion
Anticoagulant hydrolysate of M. edulis can be chosen by optimization of thrombin inhibitory activity through enzymatic hydrolysis process. 26 of 39 peptides identified by UPLC-Q-TOF were predicted to be anticoagulant peptides by the molecular docking method. Peptides with better inhibitory activities can be predicted based on both structure–activity relationship and their affinity activity against thrombin by interaction analysis. The peptide sequence of Lys-Asn-Ala-Glu-Asn-Glu-Leu-Gly-Glu-Val-Thr-Val-Arg was finally predicted with higher inhibitory activity and synthesized. The experiment in vitro was verified to prove its thrombin inhibitory activity. Based on optimization and in silico analysis of antithrombotic peptides in proteins of M. edulis, trypsin was a suitable enzyme for produce peptides with a higher antithrombotic activity, and both the prediction and verification of the peptide were of great importance to clarify the process. The present study can be further developed to facilitate the rational production of bioactive peptides for their utilization in various functional food applications.
References
Lazcano-Pérez F, Román-González SA, Sánchez-Puig N, Arreguin-Espinosa R (2012) Bioactive peptides from marine organisms: a short overview [J]. Protein Pept Lett 19:700–707
Kim SK, Mendis E (2006) Bioactive compounds from marine processing byproducts—a review [J]. Food Res Int 39:383–393
Dutta PK, Tripathi S, Mehrotra GK, Joydeep D (2009) Perspectives for chitosan based antimicrobial films in food applications [J]. Food Chem 114:1173–1182
Najafian L, Babji AS (2012) A review of fish-derived antioxidant and antimicrobial peptides: their production, assessment, and applications [J]. Peptides 33:178–185
Betoret E, Betoret N, Vidal D, Fito P (2011) Functional foods development: trends and technologies [J]. Trends Food Sci Technol 22:498–508
Rajanbabu V, Chen JY (2011) Antiviral function of tilapia hepcidin 1–5 and its modulation of immune-related gene expressions against infectious pancreatic necrosis virus (IPNV) in Chinook salmon embryo (CHSE)-214 cells. Fish Shellfish Immunol 30:39–447
Mann KG, Lorand L (1992) Introduction: blood coagulation [J]. Methods Enzymol 222:1–10
Furie B, Furie BC (1992) Molecular and cellular biology of blood coagulation [J]. N Engl J Med 326:800–806
Davie EW (1995) Biochemical and molecular aspects of the coagulation cascade [J]. Thromb Haemost 74:1–610
Ricardo LH, Walter SH (1960) Proteolytic and polymerase activity of thrombin [J]. Am J Physisiol 198:173–179
Sidelmann JJ, Gram J, Jespersen J, Kluft C (2000) Fibrin clot formation and lysis: basic mechanisms [J]. Semin Thromb Hemost 26:605–618
Chen H, Lyne PD, Giordanetto F, Lovell T, Jin L (2006) On evaluating molecular-docking methods for pose prediction and enrichment factors [J]. J Chem Inf Model 46:401–415
Je JY, Park PJ, Byun HG, Jung WK, Kim SK (2005) Angiotensin I converting enzyme (ACE) inhibitory peptide derived from the sauce of fermented blue mussel, Mytilus edulis [J]. Biores Technol 96:1624–1629
Dai ZY, Zhang YP, Zhang H, Lu YB (2012) Preparation and characterization of mussel (Mytilus edulis) protein hydrolysates with angiotensin-I-converting enzyme (ACE) inhibitory activity by enzymatic hydrolysis [J]. J Food Biochem 36:66–74
Kumar JV, Chen WY, Tsai JJP, Hu WP, et al (2013) Molecular simulation methods for selecting thrombin-binding aptamers [J]. Inf Technol Converg 253:743–749 (Springer, Netherlands)
Wei JT, Chiang BH (2009) Bioactive peptide production by hydrolysis of porcine blood proteins in a continuous enzymatic membrane reactor [J]. J Sci Food Agric 89:372–37817
Adler-Nissen J (1982) Limited enzymic degradation of proteins: a new approach in the industrial application of hydrolases [J]. J Chem Technol Biotechnol 32:138–156
Guérard F, Dufossé L, Broise DDL, Binet A (2001) Enzymatic hydrolysis of proteins from yellowfin tuna (Thunnus albacares) wastes using Alcalase [J]. J Mol Catal B Enzym 11:1051–1059
Nasri R, Amor IB, Bougatef A, Nedjar-Arroume N, Dhulster P, Gargouri J et al (2012) Anticoagulant activities of goby muscle protein hydrolysates [J]. Food Chem 133:835–841
Rojas-Ronquillo R, Cruz-Guerrero A, Flores-Nájera A, Rodríguez-Serrano G, Gómez-Ruiz L, Reyes-Grajeda JP et al (2012) Antithrombotic and angiotensin-converting enzyme inhibitory properties of peptides released from bovine casein by Lactobacillus casei Shirota [J]. Int Dairy J 26:147–154
Yan Z, Li J, Li S et al (2015) Impact of lignin removal on the enzymatic hydrolysis of fermented sweet sorghum bagasse [J]. Appl Energy 160:641–647
Lassoued I, Mora L, Barkia A et al (2015) Bioactive peptides identified in thornback ray skin’s gelatin hydrolysates by proteases from Bacillus subtilis and Bacillus amyloliquefacien [J]. J Proteom 128:8–17
Zhou C, Zhong Q, Rhodes LV et al (2012) Proteomic analysis of acquired tamoxifen resistance in MCF-7 cells reveals expression signatures associated with enhanced migration [J]. Breast Cancer Res 14:R45
Burton LJ, Rivera M, Hawsawi O et al (2016) Muscadine grape skin extract induces an unfolded protein response-mediated autophagy in prostate cancer cells: a TMT-based quantitative proteomic analysis [J]. PLoS One 11:e0164115
Liu R, Zheng W, Li J, Wang L, Wu H, Wang X et al (2015) Rapid identification of bioactive peptides with antioxidant activity from the enzymatic hydrolysate of Mactra veneriformis by UHPLC–Q-TOF mass spectrometry [J]. Food Chem 167:484–489
Savitski MM, Wilhelm M, Hahne H et al (2015) A scalable approach for protein false discovery rate estimation in large proteomic data sets [J]. Mol Cell Proteom 14:2394–2404
Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Raghava GP (2013) In silico approach for predicting toxicity of peptides and proteins [J]. PLoS One 8:e73957
Rawendra RDS, Chang CI, Chen HH, Huang TC, Hsu JL (2013) A novel angiotensin converting enzyme inhibitory peptide derived from proteolytic digest of Chinese soft-shelled turtle egg white proteins [J]. J Proteom 94:359–36929
Neumann T, Junker HD, Keil O, Burkert K, Ottleben H, Gamer J, Metz G (2005) Discovery of thrombin inhibitor fragments from chemical microarray screening [J]. Lett Drug Des Discov 2:590–594
Corralrodríguez MÁ, Bock PE, Hernándezcarvajal E, Gutiérrezgallego R, Fuentesprior P (2011) Structural basis of thrombin-mediated factor V activation: the Glu666-Glu672 sequence is critical for processing at the heavy chain-B domain junction [J]. Blood 117:7164–7173
Koh CY, Kumar S, Kazimirova M, Nuttall PA, Radhakrishnan UP, Kim S et al (2011) Crystal structure of thrombin in complex with S-variegin: insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors [J]. PLoS One 6:e26367
He D, Gu D, Huang Y et al (2009) Separation and purification of phenolic acids and myricetin from black currant by high-speed countercurrent chromatography [J]. J Liq Chromatogr Relat Technol 32:3077–3088
Ling JY, Zhang GY, Cui ZJ et al (2007) Supercritical fluid extraction of quinolizidine alkaloids from Sophora flavescens Ait. and purification by high-speed counter-current chromatography [J]. J Chromatogr A 1145:123–127
Wu G, Robertson DH, Vieth M (2003) Detailed analysis of grid-based molecular docking: a case study of CDOCKER-A CHARMm-based MD docking algorithm [J]. J Comput Chem 24:1549–1562
Nasria R, Amorb IB, Bougatefa A, Nedjar-Arroume N, Dhulster P, Gargouri J (2012) Anticoagulant activities of goby muscle protein hydrolysates [J]. Food Chem 133(3):835–841
Dodt J, Seemuller U, Maschler R, Fritz H (1985) The complete covalent structure of hirudin. Localization of the disulfide bonds [J]. Bio Chem Hoppe-Seyler 366:379–385
Mao SJ, Yates MT, Owen TJ, Krstenansky JL (1988) Interaction hirudin with thrombin: identification of a minimal bindingdomain of hirudin that inhibits clotting activity. Biochem US 27(21):8170–8173
Biela A, Khayat M, Tan H, Kong J, Heine A, Hangauer D, Klebe G (2012) Impact of ligand and protein desolvation on ligand binding to the S1 pocket of throm-bin. J Mol Biol 418:350–366
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (31371805), the State key research and development plan “Modern Food Processing and Food Storage and Transportation Technology and Equipment (No. 2017YFD0400200). The authors appreciated the NeoTrident Technology Limited. (Beijing, China) for the guide of molecular docking.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors have no conflict of interest for this article.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Rights and permissions
About this article
Cite this article
Feng, L., Tu, M., Qiao, M. et al. Thrombin inhibitory peptides derived from Mytilus edulis proteins: identification, molecular docking and in silico prediction of toxicity. Eur Food Res Technol 244, 207–217 (2018). https://doi.org/10.1007/s00217-017-2946-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-017-2946-7