Abstract
Drug screening tests are mandatory in the search for drugs in forensic biological samples, and immunological methods and mass spectrometry (e.g., gas chromatography-mass spectrometry and liquid chromatography-tandem mass spectrometry) are commonly used for that purpose. However, these methods have some drawbacks, and developing new screening methods is required. In this study, we develop a rapid-fire drug screening method by probe electrospray ionization tandem mass spectrometry (PESI-MS/MS), which is an ambient ionization mass spectrometry method, for human urine, named RaDPi-U. RaDPi-U is carried out in three steps: (1) mixing urine with internal standard (IS) solution and ethanol, followed by vortexing; (2) pipetting the mixture onto a sample plate for PESI; and (3) rapid-fire analysis by PESI-MS/MS. RaDPi-U targets 40 forensically important drugs, which include illegal drugs, hypnotics, and psychoactive substances. The analytical results were obtained within 3 min because of the above-mentioned simple workflow of RaDPi-U. The calibration curves of each analyte were constructed using the IS method, and they were quantitatively valid, resulting in good linearity (0.972–0.999) with a satisfactory lower limit of detection and lower limit of quantitation (0.01–7.1 ng/mL and 0.02–21 ng/mL, respectively). Further, both trueness and precisions were 28% or less, demonstrating the high reliability and repeatability of the method. Finally, we applied RaDPi-U to three postmortem urine specimens and successfully detected different drugs in each urine sample. The practicality of the method is proven, and RaDPi-U will be a strong tool as a rapid-fire drug screening method not only in forensic toxicology but also in clinical toxicology.
Graphical abstract

Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Drug analysis is mandatory to determine the cause of acute intoxication, and to evaluate toxicity levels by the quantification of drug concentrations in blood, though it is unusual to get detailed information on ingested drugs of abusers and/or victims. Thus, a drug screening test should be efficient for searching for drugs in forensic biological samples. Immunological methods are commonly used for on-site drug screening tests because of their high operability [1, 2], whereas mass spectrometric screening methods are generalized for their high selectivity and quantitativity [2,3,4,5,6,7,8,9,10,11,12,13]. Immunological drug screening tests can provide results in a short time, though targeted drugs are considerably limited and the tests often show false-positive results because of their cross-reactivity [14]. Thus, confirmation tests by more selective analytical methods, such as mass spectrometry, are indispensable for determining what drugs are used. In contrast, mass spectrometric screening tests using gas chromatography-mass spectrometry (GC-MS) [2,3,4] and liquid chromatography-tandem mass spectrometry (LC-MS/MS) [5,6,7,8,9,10,11,12,13] provide more specific results than immunological tests. However, sample pretreatment, such as extraction, before the mass spectrometry-based screening tests is somewhat tedious for analysts.
To overcome such issues, ambient ionization mass spectrometry (AIMS) has been used for drug screening tests [15,16,17,18,19,20,21,22,23,24,25,26,27,28,29]. The AIMS can directly analyze targeted compounds in biological samples under open-air conditions, with little or no sample preparation [15]. In other words, the AIMS allows the direct detection of analytes in a biological sample with minimum sample pretreatment. For instance, direct analysis in real-time tandem mass spectrometry (DART-MS/MS) is used for the screening analysis of illegal drugs in urine and blood [18, 26]. Additionally, paper spray ionization (PSI)- and swab touch spray ionization-MS/MS are applied to the screening tests of illegal drugs and psychoactive substances in blood, urine, or oral fluid [19, 20, 23, 24, 27].
To date, our group has reported many studies using probe electrospray ionization-MS/MS (PESI-MS/MS) [30,31,32,33,34,35,36]. The PESI is an ambient ionization technique [37], and our group applies the PESI-MS/MS to the direct analysis of metabolites and poisons [30,31,32,33,34]. PESI uses an ultra-fine metal probe needle (the tip diameter 700 nm) as the sampling and ionization unit, allowing the direct detection of analytes in biological samples, particularly in tissues. In particular, our group has reported the world’s first study for the combinational use of PESI-MS/MS for metabolome analysis, where we achieve not only direct metabolome analysis in mouse and human tissues but also the real-time monitoring of metabolites in a living mouse liver and brain [30,31,32, 34]. Moreover, our group has achieved the rapid quantification of extracellular neurotransmitters in microdialysates obtained from mouse brains [35] and the screening methods for cyanide and its metabolite in human blood by PESI-MS/MS [33]. Since the publication of our first report on PESI-MS/MS, this technique has been extended to other researchers’ studies. Usui et al. report quantification methods for acetaminophen and glyphosate by PESI-MS/MS [38, 39], and Griffeuille et al. successfully develop an analytical method for eight illegal drugs in saliva by PESI-MS/MS [40]. As described here, PESI-MS/MS has been applied to drug analysis in human biological samples, though there is no report for applying PESI-MS/MS to a simultaneous screening method for several dozen drugs.
Therefore, in this study, we develop a rapid-fire drug screening method by PESI-MS/MS for 40 forensically important drugs in urine, named RaDPi-U. To achieve a simultaneous drug screening method, we apply the scheduled-selected reaction monitoring (scheduled-SRM) mode technique, which is used in our previous metabolomics study [34], to this method. We target 40 drugs, including illegal drugs, hypnotics, and psychoactive substances, and construct each calibration curve using an internal standard (IS) method. Also, we evaluate the quantitativity of the method using quality control (QC) samples as a drug screening method. Finally, we apply the RaDPi-U to three postmortem urine specimens, proving its practicality.
Materials and methods
Materials
Authentic standards for drugs and diazepam-d5 were purchased from Merck (Darmstadt, Germany); Sumitomo Pharma Co., Ltd. (Osaka, Japan); Takeda Pharmaceutical Co., Ltd. (Tokyo, Japan); and Fujifilm Wako Chemicals (Osaka, Japan). All organic solvents were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). A blank human urine sample was purchased from Cosmo Bio Co., Ltd (Tokyo, Japan). A stock solution of the drug mixture was prepared at 10 μg/mL by dissolving each drug in methanol. An IS solution was prepared at 250 ng/mL by dissolving diazepam-d5 in distilled water. These solutions were stored at − 30 °C until analysis. Postmortem urine samples were obtained from three anonymous drug abusers by autopsy, though no personal information of the victims, including sex and age, was available for bioethical reasons and privacy protection. These samples were stored at − 30 °C until analysis. This work was approved by the Ethics Committee of Nagoya University Graduate School of Medicine (approval number: 2017-0175).
Analytical conditions
An LCMS-8060 triple quadrupole tandem mass spectrometer with a probe electrospray ionization source (DPiMS-8060, Shimadzu Corporation, Kyoto, Japan) was used. Figure 1 shows the details of the ionization source and exterior of the instrument.
A PESI probe needle (the tip diameter 700 nm) and PESI sample plate were manufactured by Shimadzu Corporation. The resolution of the mass spectrometer was set to unit resolution, and the scheduled-SRM mode was used according to our previous report [34]. Here, the SRM transitions and collision energies for each drug were optimized using authentic standards, and these are listed in Table 1. The dwell and pause times for each SRM transition were 1 ms. The frequency of the PESI probe movement was 3.3 Hz, where the ionization and sampling times were 150 and 30 ms, respectively. The applied voltages for the PESI positive and negative modes were + 2.5 and − 2.5 kV. The heat block and desolvation line temperatures were set at 50 °C and 300 °C, respectively.
Sample preparation
For preparing calibrants and QC samples, 10 μg/mL of a drug mixture solution was spiked with blank human urine, and their concentrations were adjusted to 25–500 ng/mL by dilution with blank human urine. For analysis, 10 μL of the spiked urine, 90 μL of the IS solution, and 100 μL of ethanol were mixed. After vortexing the mixture solution for 1 min, 10 μL of the solution was pipetted onto a PESI sample plate, and PESI-MS/MS was performed. Further, the postmortem urine samples were similarly analyzed: sample preparation and clean-up processes including pH adjustment were not performed.
Method validation
The calibration curves for each drug were constructed by analyzing five-level calibrants (25, 50, 100, 250, and 500 ng/mL). According to our previous reports [33, 41, 42], the lower limit of detection (LLOD) and the lower limit of quantitation (LLOQ) were calculated from the calibration curves using the following formulas: LLOD = 3.3 σ blank/S and LLOQ = 10 σ blank/S, where σ blank is the standard deviation of the blank and S is the slope of each calibration curve. The postmortem samples were automatically quantified via the built-in software (LabSolutions, Shimadzu). The method was validated using the QC samples (75, 150, and 300 ng/mL), and their intra- and inter-day truenesses and precisions were calculated.
Results and discussion
Analytical conditions of RaDPi-U
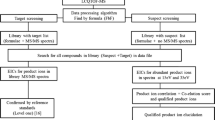
According to our previous report [31], the 40 targeted drugs were separated into 5 groups and assigned to each segment in the scheduled-SRM method (Table 1). Each segment included diazepam-d5 as the IS for the targeted drugs. Based on the pre-experiment results and our previous study [34], the surface adsorption of non-ionized contaminants onto the needle surface could cause the instability of the ionization of the targeted drugs when we used either polarity ionization mode. Our previous report clarified that the accumulation of such contaminants on the needle surface caused such instability of ionization, and it is necessary to incorporate positive and negative ionization modes in each segment to flash out the needle surface [34]. However, for RaDPi-U, all targeted drugs were ionized at positive ionization mode, and thus, a dummy segment with negative ionization mode was inserted in each segment (dummy transitions are not shown in Table 1). Additionally, in our previous reports [34, 36], we optimized segment time, resulting that 0.3 min per one segment showed the highest repeatability. Thus, we also set 0.3-min segment time in this study, and thus, the analysis time for 40 drugs was 1.5 min under the present analytical conditions. Figure 2 shows the workflow of the RaDPi-U, and it consists of just three steps: (1) mixing 10 μL of urine with 90 μL of IS solution and 100 μL of ethanol, followed by vortexing for 1 min; (2) pipetting 10 μL of the mixture onto a PESI sample plate; and (3) analyzing by PESI-MS/MS (1.5 min/run).
The analytical results were obtained within 3 min because of such a simple workflow. Compared to the previously reported screening methods using GC-MS and LC-MS/MS [2, 3, 5, 9, 12] (Table 2), the analysis of the RaDPi-U was considerably fast.
Sensitivity and quantitativity of RaDPi-U
The validation data for each drug are listed in Table 3. The calibration curves were constructed using calibrants of 25–500 ng/mL and R2 values range of 0.972–0.999, demonstrating good linearity. The LLOD and LLOQ values were 0.01–7.1 ng/mL and 0.02–21 ng/mL, respectively. In the previous studies using other AIMS methods (Table 4), the LLOD and LLOQ values were 0.1–2.5 ng/mL and 1.0–25 ng/mL for DART-MS/MS, whereas the LLOQ values were 0.7–41 ng/mL for PSI-MS/MS (LLOD was not shown for PSI-MS/MS) [18, 20]. Compared with these methods, our method achieved equivalent quantitativity and sensitivity. Also, there are several reports on drug screening method using other ambient ionization techniques (AITs). Gómez-Ríos and Pawliszyn reported coated blade spray ionization (CBS)-MS for analyzing 21 drugs, and their analytical process can be completed within 3 min [43]. Wiseman et al. reported direct analysis of dried blood spots (DBSs) using desorption electrospray ionization-MS for 4 drugs employed for therapeutic drug monitoring [28]. Moreover, Thirukumaran et al. reported solid-phase microextraction (SPME)-PESI for screening analysis of 8 drugs with the optimal desorption time (30 min) [44]. Compared to these reports using AITs, our method is equivalently rapid and do not require additional things such as DBSs. Furthermore, there is no need to extract drugs like CBS-MS and SPME-PESI in our method. Prior to the validation of the method, we evaluated the selectivity of the RaDPi-U by analyzing 10 independent blank urine samples, resulting in no specific noise for each drug in every urine sample. Moreover, good linearity of the calibration curves for each drug can be obtained because there were no specific noises for each drug in blank urine samples. As representative data, the chronograms of the blank urine and the spiked urine samples (25, 50, 100, 250, and 500 ng/mL of flurazepam) for flurazepam are shown in Fig. 3. As shown in Fig. 3, no peak is observed in the blank urine. Additionally, the chronogram of diazepam-d5 (IS) is shown in Fig. S1.
According to our previous reports [33, 41, 42], intra- and inter-day truenesses (i.e., relative errors) and precisions (i.e., relative standard error) for each drug were validated using the QC samples. The detail of validation procedures is shown in supplementary materials. Both of truenesses and precisions were 28% or less, demonstrating the satisfactory reliability, and repeatability of the method. As mentioned in our previous reports [30], PESI shows high reproducibility of the matrix effect like flow injection analysis, resulting that there is no need to select each IS compound for each targeted drug. Thus, we selected diazepam-d5 as the IS compound for quantifying the targeted drugs in this method because of its availability, and the validation results for the target drugs were satisfactory as described above. Additionally, the sensitivities for IS did not change, as determined through a validation study (i.e., 3.7% relative errors for IS through the analyses of 300 urine samples), suggesting the ruggedness of the method. Since Hiraoka et al. demonstrated that PESI requires a considerably small amount of samples like nano-ESI [37], the ionization source tends not to be stained, resulting in such ruggedness of the method.
Application of the RaDPi-U to postmortem urine samples
We applied the RaDPi-U to three postmortem urine samples (samples 1–3) to evaluate its applicability. As shown in Table 5, the RaDPi-U successfully detects drugs in all samples within 3 min per sample, proving that the RaDPi-U achieves a rapid-fire drug screening test for human urine. The representative chronogram for sample 1 is shown in Fig. 4, and the other chronograms for samples 2 and 3 are shown in Fig. S2 and S3, respectively.
To visually compare the results, positive (detected) and negative (not detected) results are shown in Fig. 4; for instance, the chronograms for risperidone (143 ng/mL), dihydrocodeine (101 ng/mL), and etizolam (not detected) are shown. Immediately after the analysis was finished, the qualitative results for the detected drugs with their quantitative values were automatically reported via the built-in software, enabling us to perform rapid drug screening within 3 min.
Limitations of this study and future perspectives
Although this study demonstrated that RaDPi-U has high feasibility for drug screening tests in urine, only three samples were available in this study for bioethical reasons. In particular, we could not make a receiver operating characteristic curve to evaluate, and thus, further study is required to apply the RaDPi-U to more real forensic samples, particularly drug-related crime. Additionally, only 40 forensically important drugs were targeted in this study; as the preliminary study, we focused on unchanged form of drugs though the number of targeted drugs, especially their metabolites, will be continuously increased in future studies. This method can be extended to forensic blood samples, and the development of the next method for blood samples, named RaDPi-B, is underway.
Conclusions
We constructed the rapid-fire drug screening method for urine samples using PESI-MS/MS, named RaDPi-U. The workflow of the RaDPi-U was just three steps without tedious sample pretreatment, and the analytical results for the 40 forensically important drugs can be acquired within 3 min. The validation results for the target compounds were satisfactory, demonstrating the good quantitativity, and sensitivity of the method. To evaluate the practicality of this method, we applied RaDPi-U to three postmortem urine samples, resulting in the successful detection of drugs in each sample. Quantitative results were automatically reported via the built-in software, and chronograms were visually understandable. Since the RaDPi-U is a highly user-friendly technique, it will be useful as the rapid-fire drug screening method not only in the forensic fields but also in clinical toxicology. Further, we have implemented an analysis pipeline for multivariate analysis and the data visualization of the multivariate data acquired with PESI-MS/MS [45]; thus, the data obtained with the present method can be linked to similar rapid data analysis in the future.
References
Schütz H, Paine A, Erdmann F, Weiler G, Verhoff MA. Immunoassays for drug screening in urine. Forensic Sci Med Pathol. 2006;2(2):75–83. https://doi.org/10.1385/FSMP:2:2:75.
DeRienz RT, Holler JM, Manos ME, Jemionek J, Past MR. Evaluation of four immunoassay screening kits for the detection of benzodiazepines in urine*. J Anal Toxicol. 2008;32(6):433–7. https://doi.org/10.1093/jat/32.6.433.
Papoutsis II, Athanaselis SA, Nikolaou PD, Pistos CM, Spiliopoulou CA, Maravelias CP. Development and validation of an EI–GC–MS method for the determination of benzodiazepine drugs and their metabolites in blood: applications in clinical and forensic toxicology. J Pharm Biomed Anal. 2010;52(4):609–14. https://doi.org/10.1016/j.jpba.2010.01.027.
Meyer MR, Peters FT, Maurer HH. Automated mass spectral deconvolution and identification system for GC-MS screening for drugs, poisons, and metabolites in urine. Clin Chem. 2010;56(4):575–84. https://doi.org/10.1373/clinchem.2009.135517.
Feng J, Wang L, Dai I, Harmon T, Bernert JT. Simultaneous determination of multiple drugs of abuse and relevant metabolites in urine by LC-MS-MS*. J Anal Toxicol. 2007;31(7):359–68. https://doi.org/10.1093/jat/31.7.359.
Vindenes V, Lund HME, Andresen W, Gjerde H, Ikdahl SE, Christophersen AS, et al. Detection of drugs of abuse in simultaneously collected oral fluid, urine and blood from Norwegian drug drivers. Forensic Sci Int. 2012;219(1):165–71. https://doi.org/10.1016/j.forsciint.2012.01.001.
Guale F, Shahreza S, Walterscheid JP, Chen H-H, Arndt C, Kelly AT, et al. Validation of LC–TOF-MS screening for drugs, metabolites, and collateral compounds in forensic toxicology specimens. J Anal Toxicol. 2012;37(1):17–24. https://doi.org/10.1093/jat/bks084.
Leporati M, Capra P, Brizio P, Ciccotelli V, Abete MC, Vincenti M. Fit-for-purpose in veterinary drug residue analysis: development and validation of an LC-MS/MS method for the screening of thirty illicit drugs in bovine urine. J Sep Sci. 2012;35(3):400–9. https://doi.org/10.1002/jssc.201100691.
Montenarh D, Hopf M, Maurer HH, Schmidt P, Ewald AH. Development and validation of a multi-analyte LC-MS/MS approach for quantification of neuroleptics in whole blood, plasma, and serum. Drug Test Anal. 2016;8(10):1080–9. https://doi.org/10.1002/dta.1923.
Punt AM, Stienstra NA, van Kleef MEA, Lafeber M, Spiering W, Blankestijn PJ, et al. Screening of cardiovascular agents in plasma with LC-MS/MS: a valuable tool for objective drug adherence assessment. J Chromatogr B Analyt Technol Biomed Life Sci. 2019;1121:103–10. https://doi.org/10.1016/j.jchromb.2019.05.013.
Giorgetti A, Barone R, Pelletti G, Garagnani M, Pascali J, Haschimi B, et al. Development and validation of a rapid LC-MS/MS method for the detection of 182 novel psychoactive substances in whole blood. Drug Test Anal. 2022;14(2):202–23. https://doi.org/10.1002/dta.3170.
Sofalvi S, Lavins ES, Kaspar CK, Michel HM, Mitchell-Mata CL, Huestis MA, et al. Development and validation of an LC–MS-MS method for the detection of 40 benzodiazepines and three Z-Drugs in blood and urine by solid-phase extraction. J Anal Toxicol. 2020;44(7):708–17. https://doi.org/10.1093/jat/bkaa072.
Kronstrand R, Nystrom I, Strandberg J, Druid H. Screening for drugs of abuse in hair with ion spray LC-MS-MS. Forensic Sci Int. 2004;145(2–3):183–90. https://doi.org/10.1016/j.forsciint.2004.04.034.
Regester LE, Chmiel JD, Holler JM, Vorce SP, Levine B, Bosy TZ. Determination of designer drug cross-reactivity on five commercial immunoassay screening kits. J Anal Toxicol. 2014;39(2):144–51. https://doi.org/10.1093/jat/bku133.
Zaitsu K. Chapter 1 - Introduction to ambient ionization mass spectrometry. In: Zaitsu K, editor. Ambient ionization mass spectrometry in life sciences. Elsevier; 2019. p. 1–32.
Huang M-Z, Cheng S-C, Cho Y-T, Shiea J. Ambient ionization mass spectrometry: a tutorial. Anal Chim Acta. 2011;702(1):1–15. https://doi.org/10.1016/j.aca.2011.06.017.
Feider CL, Krieger A, DeHoog RJ, Eberlin LS. Ambient ionization mass spectrometry: recent developments and applications. Anal Chem. 2019;91(7):4266–90. https://doi.org/10.1021/acs.analchem.9b00807.
Vasiljevic T, Gómez-Ríos GA, Pawliszyn J. Single-use Poly(etheretherketone) solid-phase microextraction-transmission mode devices for rapid screening and quantitation of drugs of abuse in oral fluid and urine via direct analysis in real-time tandem mass spectrometry. Anal Chem. 2018;90(1):952–60. https://doi.org/10.1021/acs.analchem.7b04005.
Jett R, Skaggs C, Manicke NE. Drug screening method development for paper spray coupled to a triple quadrupole mass spectrometer. Anal Methods. 2017;9(34):5037–43. https://doi.org/10.1039/C7AY01009E.
Rydberg M, Dowling S, Manicke NE. Automated and high-throughput urine drug screening using paper spray mass spectrometry. J Anal Toxicol. 2022;00:1–7. https://doi.org/10.1093/jat/bkac053.
Brown H, Oktem B, Windom A, Doroshenko V, Evans-Nguyen K. Direct Analysis in Real Time (DART) and a portable mass spectrometer for rapid identification of common and designer drugs on-site. Forensic Chem. 2016;1:66–73. https://doi.org/10.1016/j.forc.2016.07.002.
Nie H, Li X, Hua Z, Pan W, Bai Y, Fu X. Rapid screening and determination of 11 new psychoactive substances by direct analysis in real time mass spectrometry and liquid chromatography/quadrupole time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2016;30(S1):141–6. https://doi.org/10.1002/rcm.7629.
Kennedy JH, Palaty J, Gill CG, Wiseman JM. Rapid analysis of fentanyls and other novel psychoactive substances in substance use disorder patient urine using paper spray mass spectrometry. Rapid Commun Mass Spectrom. 2018;32(15):1280–6. https://doi.org/10.1002/rcm.8164.
McKenna J, Jett R, Shanks K, Manicke NE. Toxicological drug screening using paper spray high-resolution tandem mass spectrometry (HR-MS/MS). J Anal Toxicol. 2018;42(5):300–10. https://doi.org/10.1093/jat/bky001.
Bianchi F, Agazzi S, Riboni N, Erdal N, Hakkarainen M, Ilag LL, et al. Novel sample-substrates for the determination of new psychoactive substances in oral fluid by desorption electrospray ionization-high resolution mass spectrometry. Talanta. 2019;202:136–44. https://doi.org/10.1016/j.talanta.2019.04.057.
Zhang Y, Zhang W, Xin G, Liu L, Duan X, Liu C. Rapid screening of nine illicit drugs in human blood and urine by direct analysis in real-time mass spectrometry. J Forensic Sci Med. 2019;5(3):136–40.
Morato NM, Pirro V, Fedick PW, Cooks RG. Quantitative swab touch spray mass spectrometry for oral fluid drug testing. Anal Chem. 2019;91(11):7450–7. https://doi.org/10.1021/acs.analchem.9b01637.
Wiseman JM, Evans CA, Bowen CL, Kennedy JH. Direct analysis of dried blood spots utilizing desorption electrospray ionization (DESI) mass spectrometry. Analyst. 2010;135(4):720–5. https://doi.org/10.1039/b922329k
Shamraeva MA, Bormotov DS, Shamarina EV, Bocharov KV, Peregudova OV, Pekov SI, et al. Spherical sampler probes enhance the robustness of ambient ionization mass spectrometry for rapid drugs screening. Molecules. 2022;27(3). https://www.ncbi.nlm.nih.gov/pubmed/35164211
Zaitsu K, Hayashi Y, Murata T, Ohara T, Nakagiri K, Kusano M, et al. Intact endogenous metabolite analysis of mice liver by probe electrospray ionization/triple quadrupole tandem mass spectrometry and its preliminary application to in vivo real-time analysis. Anal Chem. 2016;88(7):3556–61. https://doi.org/10.1021/acs.analchem.5b04046.
Hayashi Y, Zaitsu K, Murata T, Ohara T, Moreau S, Kusano M, et al. Intact metabolite profiling of mouse brain by probe electrospray ionization/triple quadrupole tandem mass spectrometry (PESI/MS/MS) and its potential use for local distribution analysis of the brain. Anal Chim Acta. 2017;983:160–5. https://doi.org/10.1016/j.aca.2017.06.047.
Zaitsu K, Hayashi Y, Murata T, Yokota K, Ohara T, Kusano M, et al. In vivo real-time monitoring system using probe electrospray ionization/tandem mass spectrometry for metabolites in mouse brain. Anal Chem. 2018;90(7):4695–701. https://doi.org/10.1021/acs.analchem.7b05291.
Hisatsune K, Murata T, Ogata K, Hida M, Ishii A, Tsuchihashi H, et al. RECiQ: a rapid and easy method for determining cyanide intoxication by cyanide and 2-Aminothiazoline-4-carboxylic acid quantification in the human blood using probe electrospray ionization tandem mass spectrometry. ACS Omega. 2020;5(36):23351–7. https://doi.org/10.1021/acsomega.0c03229.
Zaitsu K, Eguchi S, Ohara T, Kondo K, Ishii A, Tsuchihashi H, et al. PiTMaP: a new analytical platform for high-throughput direct metabolome analysis by probe electrospray ionization/tandem mass spectrometry using an R software-based data pipeline. Anal Chem. 2020;92(12):8514–22. https://doi.org/10.1021/acs.analchem.0c01271.
Kawakami D, Tsuchiya M, Murata T, Iguchi A, Zaitsu K. Rapid quantification of extracellular neurotransmitters in mouse brain by PESI/MS/MS and longitudinal data analysis using the R and Stan-based Bayesian state-space model. Talanta. 2021;234: 122620. https://doi.org/10.1016/j.talanta.2021.122620.
Ishibashi M, Zaitsu K, Yoshikawa I, Otagaki S, Matsumoto S, Oikawa A, et al. High-throughput analysis of anthocyanins in horticultural crops using probe electrospray ionization tandem mass spectrometry (PESI/MS/MS). Hortic Res. 2023;10(4):uhad039. https://doi.org/10.1093/hr/uhad039.
Hiraoka K, Nishidate K, Mori K, Asakawa D, Suzuki S. Development of probe electrospray using a solid needle. Rapid Commun Mass Spectrom. 2007;21(18):3139–44. https://doi.org/10.1002/rcm.3201.
Usui K, Kobayashi H, Fujita Y, Kubota E, Hanazawa T, Yoshizawa T, et al. An ultra-rapid drug screening method for acetaminophen in blood serum based on probe electrospray ionization-tandem mass spectrometry. J Food Drug Anal. 2019;27(3):786–92. https://doi.org/10.1016/j.jfda.2019.02.001.
Usui K, Minami E, Fujita Y, Kubota E, Kobayashi H, Hanazawa T, et al. Application of probe electrospray ionization-tandem mass spectrometry to ultra-rapid determination of glufosinate and glyphosate in human serum. J Pharm Biomed Anal. 2019;174:175–81. https://doi.org/10.1016/j.jpba.2019.05.040.
Griffeuille P, Dulaurent S, Moreau S, Saint-Marcoux F. Driving under the influence of drugs: PESI for the measurement of illicit drugs in saliva. Column. 2021;17(05):13–7.
Taniguchi M, Minatani T, Miyazaki H, Tsuchihashi H, Zaitsu K. A highly sensitive quantification method for 12 plant toxins in human serum using liquid chromatography tandem mass spectrometry with a quick solid-phase extraction technique. J Pharm Biomed Anal. 2021;192: 113676. https://doi.org/10.1016/j.jpba.2020.113676.
Taniguchi M, Takamura N, Watanabe T, Ishimaru R, Chinaka S, Miki A, et al. Easily operable quantification method of 21 plant-derived alkaloids in human serum by automatic sample preparation and liquid chromatography-tandem mass spectrometry. Chromatographia. 2022;85(12):1051–63. https://doi.org/10.1007/s10337-022-04212-5.
Gómez-Ríos GA, Pawliszyn J. Development of coated blade spray ionization mass spectrometry for the quantitation of target analytes present in complex matrices. Angew Chem Int Ed. 2014;53(52):14503–7. https://doi.org/10.1002/anie.201407057.
Thirukumaran M, Singh V, Arao Y, Fujito Y, Nishimura M, Ogura T, et al. Solid-phase microextraction- probe electrospray ionization devices for screening and quantitating drugs of abuse in small amounts of biofluids. Talanta. 2021;231:122317. https://doi.org/10.1016/j.talanta.2021.122317
Zaitsu K, Eguchi S, Iguchi A. Bioinformatics and data science for mass spectrometry data analysis. Med Mass Spectrom. 2023;7(1):17–25.
Acknowledgements
We are very grateful to Dr. Y. Hayashi, Ms. M. Shibata, and Ms. K. Furui for their assistance with data acquisition and sample preparation, and Dr. Issey Takahashi for making a great graphical abstract.
Author information
Authors and Affiliations
Contributions
Kazuaki Hisatsune: methodology, validation, formal analysis, investigation, data curation, writing — original draft, visualization. Tasuku Murata: methodology, validation, formal analysis, investigation, data curation. Masaru Taniguchi: investigation, writing — review and editing, visualization. Tomomi Asano: writing — review and editing, visualization. Koretsugu Ogata: methodology, validation, formal analysis, investigation, data curation. Akira Iguchi: data curation, writing — review and editing, visualization. Kei Zaitsu: conceptualization, methodology, writing — original draft, supervision, project administration, funding acquisition.
Corresponding authors
Ethics declarations
Ethics approval
This work was approved by the Ethics Committee of Nagoya University Graduate School of Medicine (approval number: 2017-0175).
Source of biological material
Postmortem urine samples were obtained from three anonymous drug abusers by autopsy with the approval of the Ethics Committee of Nagoya University Graduate School of Medicine (approval number: 2017-0175).
Competing interests
The authors declare are no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hisatsune, K., Murata, T., Taniguchi, M. et al. Development of a rapid-fire drug screening method by probe electrospray ionization tandem mass spectrometry for human urine (RaDPi-U). Anal Bioanal Chem 416, 2503–2513 (2024). https://doi.org/10.1007/s00216-024-05215-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-024-05215-x