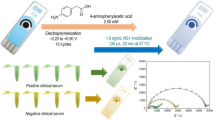
Abstract
A straightforward in situ detection method for dengue infection was demonstrated through the molecular imprinting of a dengue nonstructural protein 1 (NS1) epitope into an electropolymerized molecularly imprinted polyterthiophene (E-MIP) film sensor. The key enabling step in the sensor fabrication is based on an epitope imprinting strategy, in which short peptide sequences derived from the original target molecules were employed as the main template for detection and analysis. The formation of the E-MIP sensor films was facilitated using cyclic voltammetry (CV) and monitored in situ by electrochemical quartz crystal microbalance (EC-QCM). Surface properties were analyzed using different techniques including atomic force microscopy (AFM), X-ray photoelectron spectroscopy (XPS), and polarization modulation-infrared reflection-adsorption (PM-IRRAS). The standard calibration curve (R = 0.9830) was generated for the detection of the epitope, Ac-VHTWTEQYKFQ-NH2, with a linear range of 0.2 to 30 μg/mL and detection limit of 0.073 μg/mL. A separate calibration curve (R = 0.9786) was obtained using spiked buffered solutions of dengue NS1 protein, which resulted in a linear range of 0.2 to 10 μg/mL and a detection limit of 0.056 μg/mL. The fabricated E-MIP sensor exhibited long-term stability, high sensitivity, and good selectivity towards the targeted molecules. These results indicated that the formation of the exact and stable cavity imprints in terms of size, shape, and functionalities was successful. In our future work, we aim to use our E-MIP sensors for NS1 detection in real-life samples such as serum and blood.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dengue infection is a vector-borne systemic disease with a burden level almost equivalent to that of tuberculosis [1]. It is prevalent in 100 subtropical and tropical countries where the temperature is optimum for the proliferation of the virus [2]. There are approximately 390 million cases of dengue infection occurring yearly with a case fatality rate equivalent to 5% [3, 4]. The cause of dengue infection is the virus from the Flaviviridae family carried by mosquito vectors principally by the species Aedes aegypti and Aedes albopictus [5]. This virus has four serotypes, namely, DENV1, DENV2, DENV3, and DENV4, all of which are genetically distinct from one another, but are closely related in terms of genome structure. This distinction is the reason why the natural lifetime immunity from one serotype does not guarantee immunity from the other serotypes [4, 6]. Dengue fever, also known as breakdown fever, is the most common manifestation of dengue infection. Severe fatal manifestations of dengue infection include dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) [7]. Presently, no clinically certified antiviral medicine against dengue infection is available. Supportive and symptomatic treatments are the only procedures available to prevent further complications triggered by dengue infection [8], where careful monitoring of the blood platelet count and hematocrit baseline is extremely vital [9].
To decrease the chance of dengue complications, it is critical to detect the infection as early as possible. Currently, the available diagnostic procedures for the detection of dengue infection include virus isolation [1, 7, 10], serological tests [1, 10], and nucleic acid amplification tests (NAAT). From these methods, only virus isolation and NAAT allow detection at the onset of the dengue infection. Serological tests such as IgM antibody-capture enzyme-linked immunosorbent assay (MAC-ELISA) require the presence of the dengue antibodies, which are not existent until the third day of the ailment [11]. Other than these, serological tests have cross-reactivities with other flaviviruses causing false positives [1]. Virus isolation is the most applicable for early detection. The disadvantage of virus isolation, however, is that the process is time-consuming and false positives are very likely if the blood samples are not taken from the onset or near the onset of the illness [1, 7, 10]. NAAT are also common in dengue detection with two major procedures for analysis—the reverse transcriptase polymerase chain reaction (RT-PCR) and real-time polymerase chain reaction (real-time PCR). Similar to virus isolation, these methods can do early detection, but these procedures take longer time to be processed and the facilities for NAAT are not always readily available [12, 13].
Recent studies presented another early diagnostic method involving the dengue virus nonstructural protein 1 (NS1), a glycoprotein secreted by flaviviruses and circulates in the blood serum in high concentrations. Since NS1 appears in the blood at the onset of the illness, it is the most appropriate biomarker for the early detection of dengue [14,15,16,17]. Detection of NS1 can be done in many ways such as electrochemical detection [18, 19], fluorescence [20], and surface plasmon waveguide spectroscopy [21]. Most of these methods require the entire NS1 protein for analysis and thus require an extremely efficient antibody immobilization for sensing. If the immobilization of the sensing element or the antibodies is not efficient, the selectivity and specificity of the sensor decrease. Another problem with using antibodies as the sensing element is their stability, including the lifetime and the applicability of the fabricated sensors [22]. These molecules require proper temperature and storage conditions to keep their stability and, in turn, their performance. Adding the issue on the cost of the sensors also intensifies the impracticality of using the whole NS1 protein for dengue detection. To answer these challenges, this paper presents a method of detecting dengue virus that provides better sensor stability, good sensitivity and selectivity, and low-cost sensors. The detection strategy is based on the concepts of molecularly imprinted polymers and epitope imprinting.
Molecular imprinting is a robust technique capable of creating molecule imprints on a given polymer matrix [23,24,25,26,27,28,29,30,31,32]. These imprints are complementary in size, shape, and functionality as the target analyte mimicking the action of antibodies. The specificity and binding interactivity of molecularly imprinted polymers (MIP) are highly comparable to those of antibodies making them suitable alternatives for biosensing [33]. The major advantage of MIP materials over antibodies in sensor fabrication is their ability to form physically and chemically stable sensing elements, whose binding sites are highly specific to the target molecule [34, 35]. This physicochemical stability increases the range of solvents at which sensing can be done [36]. Tuning the interactions and properties is also possible and easier because of the vast number of monomers available for the fabrication of MIP materials [22, 35]. Imprinted polymers are also easier to prepare and are relatively less expensive, making them valuable in the sensor industry.
Preparation of MIPs involves the polymerization of a functional monomer in the presence of the target template and crosslinking agent. During polymerization, two important events occur concurrently. The first is the orientation of the monomers towards their interacting template molecules, whose stability is dictated by the strength of the interaction between the monomer and template. After the pre-organization process, the monomers polymerize resulting in the immobilization of the orientation of the functional groups of the polymer with respect to the template molecules. Upon completion of the polymerization process, the template molecules are removed using appropriate solvents leaving the orientations formed during the polymerization untouched [22, 37].
There are two significant challenges in imprinting large molecules such as NS1. The first challenge is the efficiency of specific binding. Due to the presence of many functional sites, the specific binding between the molecules involved is very weak and unstable. The second challenge is the limited mobility of the large template molecule in and out of the highly crosslinked polymer matrix [38, 39]. To solve this issue, the technique of epitope imprinting can be employed. Epitope imprinting is a strategy in molecular imprinting commonly used for protein imprinting, which does not require the entire protein for templating. The strategy is based on the principle of antibody-antigen interaction, wherein an antibody detects only a certain portion of the antigen called an epitope [22, 40]. Epitopes are small portions of the entire protein that are usually exposed, making them easy targets for detection. This strategy was first introduced by Rachkov et al. [38] in which only part of the original target analyte is used to create the cavities of the polymer matrix. One thing that must be considered in choosing an epitope for imprinting is the epitope length. This is very crucial to attain a unique recognition since peptide epitopes with very long amino acid sequences are too flexible structures. Flexible structures can usually damage imprinting and rebinding process, so they are commonly avoided in epitope imprinting. The optimum length of epitope for imprinting is between 7 and 12 amino acids [41]. This strategy offers several advantages over other imprinting protocols: (1) epitope imprinting is highly favorable for molecules whose molecular orientation changes with changing solution conditions; and (2) it decreases the binding affinity energy required for interaction as compared to the binding affinity energy needed if the whole structure is used [36, 42].
Experimental section
Materials
The monomer used for imprinting, (2,2’:5’,2″-terthiophene)-3’-acetic acid [G03TCOOH], was synthesized following the procedure of Yassar et al. [43]. The synthesis procedure is described in detail in the Supporting information (SI), while the reaction scheme and the corresponding NMR spectra are shown in Figures S1 and S2. All reagents needed for the synthesis were obtained from Sigma-Aldrich. The epitope template [Ac-VHTWTEQYKFQ-NH2] was prepared following the standard procedure of solid-phase peptide synthesis elaborated by Mäde et al. [37]. The synthesis procedure is described in SI, while the HPLC profiles and mass spectrum are shown in Figures S3 and S4. For the epitope synthesis, the Fmoc-terminated amino acids were purchased from GL Biochem (Shanghai, China). The rest of the epitope synthesis reagents were purchased from Sigma-Aldrich. Terthiophene synthesis reagents were all used as received without further purification. For the preparation of the polyterthiophene films, the supporting electrolyte, tetrabutylammoniumhexafluorophosphate (TBAH), and epitope solvent, phosphate buffer saline, were purchased from Fisher Scientific and Sigma-Aldrich, respectively. The imprinting solvent, acetonitrile, was also obtained from Fisher Scientific.
The substrate used for the analysis is a standard AT-cut 5-MHz gold-coated quartz crystal microbalance (QCM) chips (25.4 mm, 25 °C) with a chromium adhesion layer obtained from INFICON Inc. (East Syracuse, NY). Prior to use, all QCM crystals were cleaned using piranha solution (i.e., 1:3 v/v H2O2/H2SO4) for 60 s and thoroughly rinsed with Milli-Q water. The crystals were dried under N2 gas and treated via O2 plasma cleaning using March Plasmod GCM 200 plasma cleaner at 10 W for 30 s.
Preparation of VHTWTEQYKFQ-imprinted MIP sensor films
Preparation of molecularly imprinted polymer films was completed in a standard three-electrode electrochemical cell shown in Fig. 1: Ag/AgCl electrode as the reference, platinum wire as the counter, and the gold-coated QCM chips as the working electrode. The pre-polymer mixture used for MIP preparation contains 5.00 mM monomer in acetonitrile and 0.14 mM template in 20.0 mM phosphate buffer (pH 7.4), both mixed with 0.10 M TBAH. The non-imprinted polymer (NIP) counterpart was prepared in a similar manner, but in the absence of a template. All solutions were allowed to stand overnight prior to analysis to ensure that the molecules in the solution have adequate time for orientation and pre-organization. The polymers were directly deposited onto the gold-coated QCM chips via cyclic voltammetry using AutolabGeneral Purpose Electrochemical System PGSTAT302 unit (MetrohmAutolab). The electrochemical polymerization and deposition were completed by sweeping the potential between 0 and 1.1 V at a scan rate of 50 mV/s for 10 cycles. All QCM ships were washed thoroughly with acetonitrile and dried under nitrogen after analysis. Lastly, the templates were removed via potential washing at 0.7 V for 10 min [44].
Monitoring of MIP and NIP electropolymerization
The progress of the deposition of MIP and NIP materials onto the substrate surface was completed using electrochemical-quartz crystal microbalance (EC-QCM) technique. The experiment was completed using a Maxtex RQCM-quartz crystal microbalance research system (INFICON, Inc.) with a built-in phase lock oscillator, a measurement resolution of < 0.4 ng/cm2, and frequency range of 3.8 to 6 MHz. The potentiostat used in tandem with the QCM equipment is the Amel 2049 potentiostat together with Powerlab System (Milano, Italy). The experiment uses a three-electrode system with Ag/AgCl as the reference, Pt as the counter, and AT-cut, 25.4-mm-diameter Au-coated QCM crystals (INFICON, Inc.) as the working electrode.
Characterization of MIP and NIP sensor films
Different surface chemistry techniques were used to characterize the MIP- and NIP-coated films. The first two techniques—redox probing and electrochemical impedance spectroscopy (EIS) —were carried out using the same equipment used in electropolymerization. Redox probing was performed by scanning between − 0.2 and 0.6 V at 50 mV/s. EIS was performed at an AC amplitude of 10 mV. All electrochemical analyses were performed using 20.0 mM potassium ferricyanide and K3Fe(CN)6 (Sigma Aldrich) in 0.1 M KCl (Sigma Aldrich), following the three-electrode electrochemical cell setup in Fig. 1.
Surface morphology and thickness were analyzed using atomic force microscopy (AFM) and ellipsometry, respectively. AFM images were obtained from PicoScan 2500 AFM (Agilent Technologies) equipped with a piezo scanner set to scan films at 1–1.5 lines/s. Tapping mode was applied during the collection of AFM data using high-resolution NSG30 silicon AFM probes from NT-MDT Spectrum Instruments with a resonant frequency of 290 to 410 kHz. Processing of AFM micrographs was completed using Gwyddion Software. Ellipsometric measurements were carried out using an Optrel Multiskop ellipsometer (Germany) running at a fixed wavelength of 632.8 nm with an angle of 60° with respect to the surface normal. The values of Δ and ψ obtained from the experiments were then fitted using package software of the equipment (Elli, Optrel) with an assumption that the refractive index of the polymer matrix is 1.5.
Further surface characterization was completed using polarization modulation-infrared reflection-adsorption (PM-IRRAS) spectroscopy and X-ray photoelectron spectroscopy (XPS). Experiments on PM-IRRAS were completed using Cary 600 Fourier transform infrared (FTIR) series (Agilent Technologies). The PM-IRRAS accessory incorporates a helium–neon (He–Ne) laser operating in the visible region at 832.8 nm. Also, the detector of PM-IRRAS requires continuous cryogenic cooling. The XPS spectra of the films were collected using the Physical Electronics Model 5700 XPS instrument that uses a monochromatic Al KR X-ray source (1486.6 eV) operating at 350 W. All data from XPS were processed and analyzed using the Multipak software.
Sensor performance analysis
Series of solutions from 0.0 to 30.0 μg/mL were prepared for the rebinding analysis of Ac-VHTWTEQYKFQ-NH2 to the prepared sensor films. Both MIP and NIP films were subjected to rebinding analysis to check the possible non-specific binding of the analyte to the polymer matrix. Monitoring of the response of both films was done in situ using QCM200 (Stanford Research Systems, Inc.).
Similarly, the selectivity analysis was conducted in situ using the following compounds: (1) angiotensin II human (Sigma), (2) glycyl glycine (Sigma), (3) bovine serum albumin (Sigma), and (4) fibrinogen (Merck Millipore). The concentration of all solutions for selectivity was 10.0 μg/mL.
Results and discussion
Preparation of epitope-imprinted polymer sensor films via electropolymerization
The preparation of epitope-imprinted sensor films is illustrated in Fig. 2, highlighting the use of a terthiophene-based monomer (G03TCOOH). The physical appearance of the fabricated sensor films is shown in Figure S5. Terthiophene compounds are known for its low oxidation potential, making it highly suitable for electrochemical functionalization and modification. Terthiophenes oxidize at 1.1 V [45], while its base unit, thiophene, oxidizes at 1.6 V [46], making the former more suitable for Au-coated working substrates whose oxidation potential is 1.3 V.
For G03TCOOH, the electropolymerization was conducted by sweeping the potential from 0 to 1.1 V—a process known as anodic electropolymerization. This method allows the direct grafting of the conducting polymer onto the surface of the working substrate in the absence of catalyst and initiators, and allows film thickness control via deposition charge [47, 48]. The oxidation half reaction of the process presented an oxidation peak at approximately 0.90 V as shown in Fig. 3A, which represents the one-electron transfer process forming terthiophene cation radicals. The formation of this cation radical is more favorable when the electropolymerization scan rate is low and the concentration of the terthiophene monomer is in the order of 10–3 M. This half reaction is highly irreversible pushing the reaction towards the formation of more radical cations, which in turn initiates the series of coupling reaction forming polythiophene/polyterthiophenes. The termination of the process is represented by the reduction half reaction. Two reduction peaks were observed representing two different allowable termination processes forming the polymer.
The oxidation onsets between the non-imprinted polymers (NIP) and MIP were compared to distinguish the polymerization of the two polymer films. In the presence of the template, the required amount of current needed to drive the polymerization forward is higher, compared to the system with no templates (Fig. 3A, B). The difference in the shape of the voltammograms also suggested the disparity in the composition of the polymer films. A free monomer scan was also conducted after the process to confirm the stability of the polymer film surface.
The actual material deposition was monitored using EC-QCM. The main feature of EC-QCM analysis is the staircase behavior common to many electropolymerization processes. In the plot of ΔF (frequency) vs time presented in Fig. 3D, each stair denotes a single cycle in the process. The immediate decrease in ΔF corresponds to the oxidation half scan (from 0 V to 1.1 V), followed by a slight increase in ΔF, signaling the reduction half scan (from 1.1 to 0 V) of the polymerization. This oscillating behavior indicates the continuous capture and release of the supporting electrolyte anion, PF6─, during polymerization [42]. The EC-QCM data also provides another distinguishing feature between NIP and MIP films, which is the difference between the ΔF shifts after polymerization. MIP EC-QCM data in Fig. 3C showed a total deposited mass of 13.572 μg/cm (ΔF = − 770.91 Hz). This value is higher compared to NIP having a total deposited mass of 10.607 μg/cm (ΔF = − 147.35 Hz). The values obtained provide evidence of the presence of the epitope template in the deposited material.
Electrochemical characterization of MIP and NIP sensor films
One method of checking the material deposition on the Au-gold surface is by monitoring the electrochemical behavior of the surface before and after the process. The standard used for this analysis is K3Fe(CN)6 due to its well-established voltammogram. The peak intensities of the K3Fe(CN)6 redox reaction are highly affected by the amount of conducting surface area exposed for analysis. In the absence of deposited materials, the signature peaks of K3Fe(CN)6 can be observed clearly. As the amount of deposited material increases, the peak current describing the redox process of K3Fe(CN)6 decreases. When the substrate is completely covered with the deposited materials, the redox peak currents disappear. This phenomenon was observed during the electropolymerization of both MIP and NIP films (Fig. 4A, B). Upon template removal, cavities were formed, creating a porous polymer surface. These cavities allow transfer of electrons for the redox of K3Fe(CN)6. This observable phenomenon supports the claim on the removal of template by potential washing. Figure S6 shows the rebinding analysis after template removal.
Another process that can be used for electrochemical monitoring is EIS [49]. The Nyquist plots (imaginary impedance component Z” against the real impedance component Z’ at each excitation frequency) show the different responses of the surface of the sensor film. Prior to deposition, the spectrum of bare Au is observed as a straight plot with a very small semicircle in the high-frequency range as shown in Fig. 4C and D. Upon deposition, the impedance of the system increases as shown by the increase in the semicircle diameter of the plot. This change in impedance can be attributed to the surface being covered by the polymer materials, causing a blockage for electron transfer. Upon template removal, it was observed that the impedance decreased. This result is expected since theoretically there should be changes in the porosity of the surface due to the process of potential washing. The Rct values for bare Au, after NIP deposition, and after washing are ~ 150, ~ 500, and ~ 400 Ω, respectively. Figure 4D displays the Nyquist plot for the MIP sensor film, showing the increase in the system’s impedance after the MIP deposition. However, the increase was large such that the plot did not project a complete semicircle. This result suggests that the surface impedance of the system was drastically changed because of the deposition of the MIP with the template. Upon template removal, the impedance decreased, suggesting that the originally filled substrate housed cavities that allowed the passage of electrons. This hypothesis supports the claim that the templates were removed from the polymer matrix. The Rct values for bare Au, after MIP deposition, and after washing are ~ 100, ~ 900, and ~ 600 Ω, respectively.
Surface characterization of MIP and NIP sensor films
The surface roughness of the sensor films was analyzed by AFM using the tapping mode. In general, the MIP sensor film exhibited a rougher surface due to the presence of imprinted cavities throughout its surface, unlike NIP sensor film, which lacks these imprints. This expected trend was clearly observed in the AFM images obtained from the sensor films (after epitope removal) as shown in Fig. 5A and B. The surface roughness for the NIP film was observed to be ~ 5.73 nm. Consequently, MIP film had a surface roughness of ~ 6.13 nm. The values coincide with the expected trend between the MIP and NIP sensor films. The surface stability after polymerization and after potential washing was analyzed by CV and the results are shown in Figure S7. It can be observed that potential washing does not cause damage on the surface of the films.
The film thickness was also measured using ellipsometry (Table 1) and the values suggest that templates were present on the film surface, thereby increasing the film thickness, compared to the NIP counterpart. Such result is expected since the process entails a type of surface imprinting, meaning most of the templates are protruding from the surface. This phenomenon causes the thickness to be higher. However, after template removal, the thicknesses of the MIP and NIP became closer to each other, with the MIP film thickness slightly lower by 2 nm. Also, this result is expected as the preparation method for the two films was similar, except for the presence of the template in the MIP preparation. The slightly smaller value for the MIP is due to the presence of its imprinted cavities, unlike in NIP.
Elemental analysis for the films was also conducted using XPS (Fig. 5D). The point of comparison is the presence of the nitrogen peak in the wide spectrum for the template. Other elements representing the monomer and template were also detected. The sulfur peak confirms the presence of the terthiophene monomer on the surface of the QCM chip. The expected decrease in intensity after template removal was observed, which also supports the removal of the majority of the template from the sensor surface.
Further analysis with PM-IRRAS, an FTIR technique highly advantageous for monolayers and thin films, was conducted. Figure 5C presents the PM-IRRAS spectrum of MIP sensor film in comparison with the spectrum of MIP with template film. Three major signals are important for analysis here. Two of these signals, which suggest the presence of the polyterthiophene material on the surface of the film, were attributed to the C = O and C = C stretch between 1685–1665 cm−1 and 1540–1520 cm−1, respectively. The last signal, attributed to the N–H bending, supports the presence of the nitrogen detected by XPS. This N–H peak was observed in the spectrum of unwashed MIP sensor film at 1485–1475 cm−1 [50]. Its disappearance in the spectrum of the washed MIP suggests the removal of the peptide templates from the surface of the film, further supporting the information obtained from XPS.
Sensor performance
The rebinding capacity of the imprinted films for the detection of the target was measured using QCM. The QCM curves for the rebinding of Ac-VHTWTEQYKFQ-NH2 on NIP and MIP sensor films are shown in Figure S8. Both films were subjected to adsorption studies to examine the extent of non-specific binding interactions of the analyte to the NIP film and these interactions were measured in terms of changes in the frequency of the quartz crystal (ΔF) against time (t). Figure 6A presents the time-dependent QCM response of the two films in the presence of 10.0 μg/mL VHTWTEQYKFQ. It can be observed that there is a significant difference between the binding activities of the two films. This result supports the presence of imprinted cavities within the polymer matrix. Without these analyte-specific cavities, only non-specific binding should be observed, which is true for the case of the NIP film. Furthermore, this plot proves that the non-specific binding of VHTWTEQYKFQ to the polymer matrix does not significantly affect the QCM response with the imprinted sensor film. The response of the films was further investigated by measuring their QCM responses in the presence of increasing concentration of the analyte. Figure 6B presents the changes in the frequency of the quartz crystal for both films as the concentration of the injected epitope template was increased. The response of the NIP film clearly suggests that the film has only non-specific binding with respect to the analyte. Even at large concentrations, only a small portion of the injected sample was bound in the polymer matrix. MIP sensor films, on the other hand, showed good response in the presence of the analyte. This difference in ΔF was distinctly presented in Fig. 6B together with the imprinting factors (IF) for each concentration. IF defines the rebinding behavior difference between the NIP and MIP sensor films. As IF values increase, the binding interactions occurring in the absence of the imprinted cavities become insignificant or negligible.
Sensor response towards the presence of the epitope analyte and dengue NS1 protein. A QCM response of the analyte with MIP and NIP sensor film with 10.0 μg/mL analyte, B QCM response and imprinting factors (i.e., numbers below the bars) comparison with increasing concentration of the analyte and the calibration curve for the rebinding activity of C VHTWTEQYKFQ and D dengue NS1 protein in phosphate buffer (pH 7.4, 20 mM) with the fabricated MIP sensor film
The linearity of the MIP response with the epitope template is presented in Fig. 6B and C. The calculated calibration sensitivity of the system was ─2.27 Hz per unit concentration (μg/mL) with a linear correlation factor (R2) of 0.9830. The limits of detection and quantification were calculated using the following equations:
Values 72.6 and 242.0 ng/mL for the limits of detection and quantification, respectively, were obtained.
The selectivity of the sensor was tested using different peptide sequences and small proteins: angiotensin II human, bovine serum albumin, fibrinogen, and glycyl glycine. Figure 7 presents the response of the compounds tested together with the response of the original template molecule. Also, the response of the protein target, NS1, was included in the plot. Compared to the original template molecule and NS1, the response of the other compounds was relatively smaller. The signals for the test compounds, however, show that these compounds have non-specific binding interaction to the sensor film. Nonetheless, the response is weak and insignificant as compared to the target molecules.
Another important analysis for the performance testing of MIP sensor films is the analysis with solutions spiked with NS1 antigen. The range of concentration used was between 0.2 and 10.0 μg/mL of NS1 in phosphate buffer solution. It can be observed in Fig. 6D that the fabricated MIP sensor film had good response to the incoming NS1 antigen molecules. This result suggests that the epitope-imprinted cavities were able to bind the target protein. The limit of detection for the analysis is 56.1 ng/mL, while the limit of quantification calculated was 187.15 ng/mL. The values were significantly lower than those of the reported values of NS1 in the blood serum, suggesting the potential of the proposed system and method of NS1 antigen diagnosis.
The detection limit achieved for this QCM-based MIP sensor film was reasonable and highly applicable for the NS1 detection in the blood serum. The value in the literature for the elevated level of NS1 in serum within the first 72 h is 600 ng/mL [51]. Another study reported medium level values of 104 ng/mL for dengue fever and 156 ng/mL for dengue hemorrhagic fever [19]. The detection limit obtained was significantly lower as compared to the reported values, making this QCM detection method practical and operational for dengue infection detection. Our initial assessment of the sensing capability of our MIPs should lead to a future work that focuses on the detection of dengue infection in real blood samples. The analysis with more complex media often requires extensive sample purification and analyte concentration. The goal of our current work is to set the method of MIP use, which we believe is unique as it employs the epitope approach in a most effective manner.
Conclusion
A facile in situ QCM detection method for dengue infection that is highly specific and selective to its target analyte has been demonstrated using E-MIP of polyterthiophene and NS1 epitope. Unlike other detection methods, the sensor film developed in this study does not incorporate antibodies in the sensing element of the detection procedure. Instead, a more stable, cost-effective, and simple sensing element was established making the fabricated sensor films for dengue more applicable for industrial and commercial purposes. The developed method incorporated the strategy of epitope imprinting, which allows the creation of NS1-specific cavities without actually imprinting the original protein. Although real blood serums were not used in this study, we aim to further use our developed E-MIP sensors for NS1 detection in such real-world samples for a more practical application in a future work. Detection using blood serum may result in possible interference with other serum proteins, albumin, lyzozymes, etc., which will require further long and tedious sample separation, purification, and recalibration of all the measurements. Most real-time and real blood studies require pre-testing purification or serial dilution to result in a meaningful analyte study. In addition, we will not be able to reliably account for changes in the RI and differential experiments with changes in the viscosity of blood, which would require a different model and setup with the current QCM. While serum analysis is important, many studies reported in the literature did not explore real blood serum and found their data more than appropriate for examining the capability of their sensing material for dengue detection. Likewise, our initial assessment of the sensing capability of our E-MIPs should lead to a future work that focuses on the detection of dengue infection in real blood samples. The goal of our current work is to set the method of MIP use, which we believe is unique as it employs the epitope approach in a most effective manner rather than focusing on sample purification. Furthermore, our method of preparing sensor films via electrochemistry shows a promising start for developing other sensor films for other illnesses such as malaria, tuberculosis, and Japanese encephalitis, all of which are viral illnesses plaguing the world population. Further analysis for this system can be conducted such as analysis of NS1-spiked blood samples and clinical testing for practical laboratory usage. Also, another future prospect of this developed method is to incorporate it in small handy sensor devices, basically developing a point-of-care diagnostic system for dengue infection.
Change history
08 December 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00216-021-03818-2
References
Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martínez E. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–16.
Boutayeb A. The double burden of communicable and non-communicable diseases in developing countries. Trans R Soc Trop Med Hyg. 2006;100:191–9.
Alera MT, Srikiatkhachorn A, Velasco JM, Tac-An IA, Lago CB, Clapham HE, Fernandez S, Levy JW, Thaisomboonsuk B, Klungthong C. Incidence of dengue virus infection in adults and children in a prospective longitudinal cohort in the Philippines. PLoS Negl Trop Dis. 2016;10:e0004337.
Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O. The global distribution and burden of dengue. Nature. 2013;496:504–7.
Cheng MS, Ho JS, Tan CH, Wong JPS, Ng LC, Toh C-S. Development of an electrochemical membrane-based nanobiosensor for ultrasensitive detection of dengue virus. Anal Chim Acta. 2012;725:74–80.
Osorio JE, Wallace D, Stinchcomb DT. A recombinant, chimeric tetravalent dengue vaccine candidate based on a dengue virus serotype 2 backbone. Expert Rev Vaccines. 2016;15:497–508.
Oliveira MD, Nogueira ML, Correia MT, Coelho LC, Andrade CA. Detection of dengue virus serotypes on the surface of gold electrode based on Cratylia mollis lectin affinity. Sens Actuators B Chem. 2011;155:789–95.
Wiwanitkit V. Dengue fever: diagnosis and treatment. Expert Rev Anti Infect Ther. 2010;8:841–5.
Rajapakse S, Rodrigo C, Rajapakse A. Treatment of dengue fever. Infect Drug Resist. 2012;5:103.
Roche RR, Alvarez M, Guzmán MG, Morier L, Kourí G. Comparison of rapid centrifugation assay with conventional tissue culture method for isolation of dengue 2 virus in C6/36-HT cells. J Clin Microbiol. 2000;38:3508–10.
Fernandez R, Vazquez S. Serological diagnosis of dengue by an ELISA inhibition method (EIM). Mem Inst Oswaldo Cruz. 1990;85:347–51.
Johnson BW, Russell BJ, Lanciotti RS. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J Clin Microbiol. 2005;43:4977–83.
Conceição TM, Da Poian AT, Sorgine MH. A real-time PCR procedure for detection of dengue virus serotypes 1, 2, and 3, and their quantitation in clinical and laboratory samples. J Virol Methods. 2010;163:1–9.
Sergio I, Flores-Aguilar H, González-Mateos S, López-Martinez I, Alpuche-Aranda C, Ludert JE, del Angel RM. Determination of viremia and concentration of circulating nonstructural protein 1 in patients infected with dengue virus in Mexico. Am J Trop Med Hyg. 2013;88:446–54.
Zaytseva NV, Montagna RA, Lee EM, Baeumner AJ. Multi-analyte single-membrane biosensor for the serotype-specific detection of dengue virus. Anal Bioanal Chem. 2004;380:46–53.
Navakul K, Sangma C, Yenchitsomanus P, Chunta S, Lieberzeit PA. Enhancing sensitivity of QCM for dengue type 1 virus detection using graphene-based polymer composites. Anal Bioanal Chem. 2021 1–8
Hatta PM, Supawat C, Patsamon R, Kriengsak L, Werasak S. Dengue NS1 detection in pediatric serum using microfluidic paper-based analytical devices. Anal Bioanal Chem. 2020;412:2915–25.
Silva M, Dias A, Cordeiro M, Marques E Jr, Goulart M, Dutra R. A thiophene-modified screen printed electrode for detection of dengue virus NS1 protein. Talanta. 2014;128:505–10.
Silva MM, Dias AC, Silva BV, Gomes-Filho SL, Kubota LT, Goulart MO, Dutra RF. Electrochemical detection of dengue virus NS1 protein with a poly (allylamine)/carbon nanotube layered immunoelectrode. J Chem Technol Biotechnol. 2015;90:194–200.
Neeraja M, Lakshmi V, Lavanya V, Priyanka E, Parida M, Dash P, Sharma S, Rao PL, Reddy G. Rapid detection and differentiation of dengue virus serotypes by NS1 specific reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay in patients presenting to a tertiary care hospital in Hyderabad, India. J Virol Methods. 2015;211:22–31.
Wong WR, Sekaran SD, Adikan FRM, Berini P. Detection of dengue NS1 antigen using long-range surface plasmon waveguides. Biosens Bioelectron. 2016;78:132–9.
Schirhagl R. Bioapplications for molecularly imprinted polymers. Anal Chem. 2014;86:250–61.
Ahmad OS, Bedwell TS, Esen C, Garcia-Cruz A, Piletsky SA. Molecularly imprinted polymers in electrochemical and optical sensors. Trends Biotechnol. 2019;37:294–309.
BelBruno JJ. Molecularly imprinted polymers. Chem Rev. 2018;119:94–119.
Benachio I, Lobato A, Gonçalves LM. Employing molecularly imprinted polymers in the development of electroanalytical methodologies for antibiotic determination. J Mol Recognit. 2021;34:e2878.
Dulay JAL, Caldona EB, Camacho AR, Espenilla MDT, Gopez CD, Oribello EAA. Phytoremediation of cadmium contaminated water by Hydrilla (Hydrilla verticillata). SLU Res J. 2010;41:23–33.
Bezdekova J, Zemankova K, Hutarova J, Kociova S, Smerkova K, Adam V, Vaculovicova M. Magnetic molecularly imprinted polymers used for selective isolation and detection of Staphylococcus aureus. Food Chem. 2020;321:126673.
Bhogal S, Mohiuddin I, Kaur K, Lee J, Brown RJ, Malik AK, Kim K-H. Dual-template magnetic molecularly imprinted polymer-based sorbent for simultaneous and selective detection of phenolic endocrine disrupting compounds in foodstuffs. Environ Pollut. 2021;275:116613.
Gonçalves LM. Electropolymerized molecularly imprinted polymers (e-MIPs), perceptions based in recent literature for soon-to-be world-class scientists. Curr Opin Electrochem. 2020.
Gui R, Jin H, Guo H, Wang Z. Recent advances and future prospects in molecularly imprinted polymers-based electrochemical biosensors. Biosens Bioelectron. 2018;100:56–70.
Vergara AV, Pernites RB, Tiu BDB, de Leon ACC, Mangadlao JD, Binag CA, Advincula RC. Capacitive detection of morphine via cathodically electropolymerized, molecularly imprinted poly (p-aminostyrene) films. Macromol Chem Phys. 2016;217:1810–22.
Caldona EB, Sibaen JW, Tactay CB, Mendiola SLD, Abance CB, Añes MP, Serrano FDD, De Guzman MMS. Preparation of spray-coated surfaces from green-formulated superhydrophobic coatings. SN Appl Sci. 2019;1:1657.
Tiu BDB, Krupadam RJ, Advincula RC. Pyrene-imprinted polythiophene sensors for detection of polycyclic aromatic hydrocarbons. Sens Actuators B Chem. 2016;228:693–701.
Lu C-H, Zhang Y, Tang S-F, Fang Z-B, Yang H-H, Chen X, Chen G-N. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens Bioelectron. 2012;31:439–44.
Yang X, Dong X, Zhang K, Yang F, Guo Z. A molecularly imprinted polymer as an antibody mimic with affinity for lysine acetylated peptides. J Mater Chem B. 2016;4:920–8.
Kunath S, Panagiotopoulou M, Maximilien J, Marchyk N, Sänger J, Haupt K. Cell and tissue imaging with molecularly imprinted polymers as plastic antibody mimics. Adv Healthc Mater. 2015;4:1322–6.
Mäde V, Els-Heindl S, Beck-Sickinger AG. Automated solid-phase peptide synthesis to obtain therapeutic peptides. Beilstein J Org Chem. 2014;10:1197–212.
Rachkov A, Minoura N. Towards molecularly imprinted polymers selective to peptides and proteins. The epitope approach. Biochim Biophys Acta BBA-Protein Struct Mol Enzymol. 2001;1544:255–66.
Wei S, Jakusch M, Mizaikoff B. Capturing molecules with templated materials—analysis and rational design of molecularly imprinted polymers. Anal Chim Acta. 2006;578:50–8.
Yang L, Wei W, Xia J, Tao H, Yang P. Capacitive biosensor for glutathione detection based on electropolymerized molecularly imprinted polymer and kinetic investigation of the recognition process. Electroanal Int J Devoted Fundam Pract Asp Electroanal. 2005;17:969–77.
Bossi AM, Sharma PS, Montana L, Zoccatelli G, Laub O, Levi R. Fingerprint-imprinted polymer: rational selection of peptide epitope templates for the determination of proteins by molecularly imprinted polymers. Anal Chem. 2012;84:4036–41.
Tiu BDB, Pernites RB, Tiu SB, Advincula RC. Detection of aspartame via microsphere-patterned and molecularly imprinted polymer arrays. Colloids Surf Physicochem Eng Asp. 2016;495:149–58.
Yassar A, Moustrou C, Youssoufi HK, Samat A, Guglielmetti R, Garnier F. Synthesis and characterization of poly (thiophenes) functionalized by photochromic spironaphthoxazine groups. Macromolecules. 1995;28:4548–53.
Advincula R, Pernites RB. Sensors and separation based on molecular recognition via electropolymerization and colloidal layer templates. 2020.
Pernites RB, Ponnapati RR, Advincula RC. Surface plasmon resonance (SPR) detection of theophylline via electropolymerized molecularly imprinted polythiophenes. Macromolecules. 2010;43:9724–35.
Wei Y, Jang G-W, Chan C (1991) Polymerization of thiophene and its derivatives
Roncali J. Conjugated poly (thiophenes): synthesis, functionalization, and applications. Chem Rev. 1992;92:711–38.
de Leon ACC, Pernites RB, Advincula RC. Superhydrophobic colloidally textured polythiophene film as superior anticorrosion coating. ACS Appl Mater Interfaces. 2012;4:3169–76.
Caldona EB, Smith DW Jr, Wipf DO. Surface electroanalytical approaches to organic polymeric coatings. Polym Int. 2021;70:927–37.
Caldona EB, Zhang M, Liang G, Hollis TK, Webster CE, Smith DW Jr, Wipf DO. Corrosion inhibition of mild steel in acidic medium by simple azole-based aromatic compounds. J Electroanal Chem. 2021;880:114858.
Libraty DH, Young PR, Pickering D, Endy TP, Kalayanarooj S, Green S, Vaughn DW, Nisalak A, Ennis FA, Rothman AL. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J Infect Dis. 2002;186:1165–8.
Acknowledgements
Work (or part of this work) was conducted by ORNL’s Center for Nanophase Materials Sciences by RCA, which is a US Department of Energy Office of Science User Facility. The authors would like to express their gratitude to the following departments: Department of Macromolecular Science and Engineering—Case Western Reserve University, Institute of Chemistry—University of the Philippines (Diliman Campus), Park AFM Systems, and Biolin Scientific.
Funding
This research is financially supported by the following agencies: Philippines-California Advanced Research Institutes – Commission on Higher Education (PCARI-CHED). Department of Energy (DOE), and National Science Foundation (NSF 1608457 and CMMI NM 1333651).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: Unfortunately, in this article the caption to Figure 6 was inadvertently converted.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Buensuceso, C.E., Tiu, B.D.B., Lee, L.P. et al. Electropolymerized-molecularly imprinted polymers (E-MIPS) as sensing elements for the detection of dengue infection. Anal Bioanal Chem 414, 1347–1357 (2022). https://doi.org/10.1007/s00216-021-03757-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03757-y