Abstract
C-reactive protein (CRP), a non-specific acute-phase indicator of inflammation, has been widely recognized for its value in clinical diagnostic applications. With the advancement of testing technologies, there have been many reports on fast, simple, and reliable methods for CRP testing. Among these, the aptamer-based biosensors are the focus and hotspot of research for achieving high-sensitivity analysis of CRP. This review summarizes the progress of in vitro aptamer screening for CRP and the recent advances in aptamer-based CRP sensor applications, thus developing insight for the new CRP aptasensor design strategy.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
C-reactive protein (CRP) is one of the acute-phase proteins synthesized by liver cells, which is not affected by individual differences, body conditions and drugs [1]. In general, the CRP level in the peripheral blood of healthy subjects does not exceed 10 mg/L; however, when the body experiences infection or inflammation, CRP will quickly activate the complement system and participate in the body’s non-specific immunity, then rapidly return to normal level upon clinical intervention [1, 2]. Therefore, the CRP level is considered to be one of the most sensitive pathological state and degree indicators. It is generally believed that when the CRP level is less than 50 mg/L, the body may be infected by a virus or inflammatory injury; when the CRP level exceeds 100 mg/L, the body may be considered to be in a severe bacterial infection state [3]. Because CRP can specifically reflect the state or degree of infection and inflammation, it is often used in pathological diagnosis, the treatment of various diseases, and prognostic monitoring [2]. For example, as an indicator of inflammation, CRP can be used for the diagnosis of cancer (except leukemia) [4], sepsis [5], metabolic syndrome [6], and other inflammation-related diseases. It can also be used to monitor the risk of postoperative infections [7]. In addition, CRP is a common diagnostic indicator of lower respiratory tract infection (LRI), and it is used to identify the infection source of the LRI to reduce the abuse of antibiotics [8]. An elevated high-sensitivity C-reactive protein (hs-CRP) level is directly associated with atherosclerotic thrombosis [9], coronary atherosclerosis [10], venous thromboembolism (VTE) [11], and atrial fibrillation (AF) [12]. Therefore, CRP is listed as an important assessment indicator for the risk of cardiovascular diseases (CVD) [13, 14].
In view of the value of CRP as a pathological diagnostic indicator in clinical application, the detection of CRP has always been a research hotspot in the field of laboratory medicine and bioanalytical chemistry. Immunoassays based on the principle of antigen-antibody binding, such as immunoturbidimetry (ITM), lateral flow assay (LFA), and enzyme-linked immunosorbent assay (ELISA), are currently the mainstream technologies for clinical detection of CRP [15,16,17]. However, antibodies have certain limitations, such as tedious and complex in vivo preparation processes, high preparation costs, large intra-batch variability, and susceptibility to denaturation [18,19,20]. Therefore, in recent years, scientists have invested immense resources in the research of CRP analysis, which is a non-antibody recognition method. Among these studies, aptamer-based analysis has gradually become the alternative approach to the mainstream techniques. An aptamer is a single-stranded nucleic acid (DNA or RNA) obtained by in vitro screening that forms a three-dimensional active conformation through the folding of its secondary and tertiary structures, then binds to the target substance with high affinity [21,22,23]. Compared to antibodies, an aptamer has the advantages of simple synthesis and modification, high affinity, strong specificity, low cost, low molecular weight, good stability, sustainable repeated denaturation, and renaturation. Therefore, it is currently considered to be a superior alternative to antibodies [24].
A biosensor is an analytical device that integrates target-specific identification elements (such as aptamer, antibody, and cell) and physicochemical transducers (such as field effect transistors and piezoelectric crystals) [25]. It has many advantages such as rapid detection, high selectivity, and the simultaneous detection of point-of-care test (POCT) and multiple biomarkers, serving as an ideal detection platform for CRP POCT [26]. Recently, aptamer-based biosensors have been widely used in the analysis of pesticide residues [27], environmental heavy metals [28], and clinical biomarkers [29]. This review will discuss the progress of in vitro aptamer screening in recent years and its application in CRP biosensor research to provide new research ideas for the development of better CRP biosensors.
Advances in CRP in vitro aptamer screening
Generally, aptamers are screened in vitro through a defined iterative procedure, which is called SELEX. The convention SELEX operation steps shown in Fig. 1, however, are time and labor consuming. The originally reported CRP aptamer was obtained by this method after ten rounds of “incubation-screening-isolation-amplification” cycle [30]. With the development of materials science and analysis technology, some new SELEX libraries (such as primer-free SELEX library, SOMAmer) and SELEX variant (such as M-SELEX, GO-SELEX, and MARAS) have been reported. The affinity (expressed by a dissociation constant, referred to as Kd value) of CRP aptamers obtained from these new SELEX libraries or SELEX variants can reach the level of nanomoles per liter or even picomoles per liter, which is superior to antibody affinities reported (0.44 μmol/L and 0.55 μmol/L, respectively) [31]. The application of these new SELEX libraries or SELEX variants can greatly reduce the loss of time and improve screening efficiency. This chapter briefly introduces the SELEX library of CRP aptamer SELEX and the optimization strategy of in vitro screening methods and analyzes the aptamer sequence and the relevant information (Table 1).
An illustration of the key steps of a typical SELEX protocol. SELEX is a comprehensive procedure consisting of an iterative procedure, which includes SELEX library design, incubation with target and separation of binding/unbinding sequences, elution of aptamers, amplification of eluted aptamers, and aptamer identification
Optimization of the separation method in vitro
A successful SELEX heavily relies on the efficient separation of library sequences bound and unbound to targets. In the first SELEX report, the separation of free sequences was achieved through a nitrocellulose membrane: after the SELEX library was incubated with the target molecular protein, the mixture was filtered by a nitrocellulose membrane, the bound sequences were fixed on the nitrocellulose membrane surface, and the remaining free sequences were directly passed through the nitrocellulose membrane [32]. However, due to the non-specific binding of free nucleic acids to the large surface area of the nitrocellulose membrane, the enrichment and screening of high-target affinity sequences are seriously hindered. Generally, it takes 10–20 rounds of repeated artificial screening cycles to obtain the target aptamer, which is time consuming and laborious.
To simplify the operation procedure and improve separation efficiency, researchers have developed a more efficient CRP aptamer SELEX scheme by introducing new materials such as microfluidic chips, magnetic beads, and graphene.
M-SELEX program
M-SELEX is an automated SELEX screening platform, which was proposed and first demonstrated by Lou et al. [33]. It uses microfluidic technology to integrate the unit operations of traditional SELEX on a chip with the size of several centimeters, so as to shorten the SELEX screening cycle and realize the automatic screening of the aptamer.
Based on the M-SELEX principle, a localized magnetic gradient field via ferromagnetic patterns integrated into the microchannel to separate the nucleic acid sequences binding/unbound to the target coated magnetic beads [34] was developed a microfluidic chip which can be used for aptamer screening. As a demonstration, CRP was used as an epitope to bind to magnetic beads for aptamer screening, finally successfully screening sequence B (Table 1). Huang et al. used the SPR technique to characterize the affinity of sequence B, and found that the dissociation constant (Kd) between sequence B and CRP was 3.51 nmol, which was better than the CRP-specific antibody [34]. In addition, the immunoassay confirmed that the aptamer could produce a corresponding chemiluminescence signal in response to the change of CRP concentration, and the detection limit for CRP detection was experimentally found to be 0.0125 mg/L. The linear range (R2 = 0.97) was identified in concentrations ranging from 0.0125 to 10 mg/L. The coefficient of variation for these measurements is about 5.7%, which is very suitable for the development of an aptasensor.
Similarly, on the basis of the M-SELEX project, Wu et al. also independently developed a new microfluidic SELEX screening chip by using photolithography and wet chemical etching technology [35]. They used a 40-mer random sequence with 16-mer primers fixed at both ends as a SELEX starting library to screen CRP-specific aptamers, and finally obtained sequence C (Table 1). The dissociation constant of sequence C with CRP was 22.1 nmol/L, which proved that sequence C had high affinity. In addition, five control proteins including IgG, HSA, hemoglobin, transferrin, and myoglobin were used to confirm that sequence C only bound to CRP and changed its conformation. No other similar reaction was found. It is proved that sequence C can specifically recognize CRP protein in complex serum matrix, and has high specificity and potential to develop an aptasensor [35].
MARAS program
The MARAS program is a rapid program developed by Lai et al., which can selectively separate a suitable aptamer [36, 37]. The principle is to use two sets of Helmholtz coils placed in an orthogonal direction to prepare a controllable external rotating magnetic field, which makes the magnetic particles fixed with CRP rotate in it to generate centrifugal force, hence removing the nucleic acid sequence not bound with CRP. The MARAS program can be divided into two steps, including a forward screening program and a reverse screening program; the former is to enrich the aptamer sequence with high affinity for CRP by using CRP magnetic beads, while the latter is to remove the aptamer sequence that can bind to other serum proteins by removing CRP serum magnetic beads, ensuring that the target aptamer only has a specific binding effect on CRP.
It is worth mentioning that, unlike traditional SELEX and other SELEX variants, the MARAS program can control the affinity between the target aptamer and the target by changing the frequency of the external magnetic field, so the MARAS program can complete the separation step in only a few hours without repetitive periodic screening and separation. Because of the correlation between the external magnetic field frequency and the aptamer, Li et al. established a window MARAS protocol [38]. Its principle is to establish the relationship between the frequency of the external magnetic field and the target affinity, directly determine the required frequency of the external magnetic field in MARAS for aptamers with preset target affinity, and further shorten the aptamer screening time.
GO-SELEX program
In the existing SELEX screening, most of the target proteins will be fixed on the ELISA plate or beads by a covalent bond or an ion bond to improve the screening efficiency. However, some studies have found that the screening programs using CRP immobilization tend to change the active conformation of the proteins or cause CRP depolymerization, thereby reducing the affinity of the screened aptamers [39, 40]. GO-SELEX is a SELEX variant developed by Park et al. to avoid the negative impact of the target-immobilization strategy [41]. Its working principle is as follows: the fixed SELEX library strategy is used to replace the target material fixation strategy, and the non-specific adsorption between graphene oxide and the nucleic acid sequence is used to protect its secondary and tertiary structure from being changed, thus enhancing the affinity of the aptamer. Therefore, the GO-SELEX program can not only avoid the structural changes of the nucleic acid library, but also prevent the aggregation or modification of the target substance [41], thus enhancing the affinity of the screened nucleic acid sequence to the pentamer CRP.
Yang et al. used DNA with fixed primers at both ends and a 40-mer-long random sequence in the middle as the SELEX starting library, and screened the CRP aptamer with the GO-SELEX program [42]. The final sequence F is the product of removing the constant region at both ends of the selected nucleic acid chain (Table 1). In addition, Yang’s team also selected five control proteins (IgG, HSA, Hb, TRF, and Myo) to verify their specificity. The results showed that sequence F only had binding response with CRP, which confirmed that the aptamer had good specificity. Yang also participated in the establishment of the CRP aptamer enrichment level recognition pattern system based on GO-SELEX [43]. The main content of it is to use a computer support vector classification algorithm to classify CRP aptamers screened by GO-SELEX, and establish the corresponding pattern recognition system to predict the enrichment level of CRP candidate aptamers [43].
Optimization of the SELEX library
The SELEX library is the starting point of the SELEX program. The traditional SELEX library is based on four kinds of deoxyribonucleic acids (A, T, C, G) or ribonucleotides (A, U, C, G) in accordance with the random arrangement and combination of the design sequence of the nucleic acid chain, and the integration of them. At present, it has not been reported that aptamer target protein affinity has a preference for nucleic acid sequence length or a certain nucleotide, so the SELEX library length is mostly determined by researchers after considering their own laboratory conditions. However, the traditional SELEX inventory has some shortcomings, such as the influence of primers at both ends and the restriction of information diversity based on the restriction of the number of nucleotide monomers. Therefore, there are some novel SELEX library designs for CRP aptamer screening.
Primer-free SELEX library
A classic SELEX library consists of a constant region at both ends (about 10–40 mer) and a random sequence in the middle (about 20–80 mer, according to the needs of researchers). The constant region contains primer binding sites for PCR amplification. Ideally, the target substance should only interact with the intermediate random sequence. However, during the SELEX procedure, there is an inevitable interaction between the two primers and the random sequence. Previous studies have also shown that the preset constant region will interfere with the SELEX selection tendency, which will seriously affect the screening results of target aptamers. The preset constant region will interfere with the SELEX selection tendency, which would critically influence which aptamers are selected from the random pool. The primer-annealing sequences regeneration technology based on thermal cycles of hybridization-extension proposed by Wen et al. makes it possible to use a primer-free SELEX library [44]. Based on this technology, the PCR amplification process can be completed without the preset constant region, eliminating the negative impact of the preset constant region on the SELEX selection tendency. In addition, the primer-free SELEX library with the same length has higher information diversity than the primed SELEX library, and it is more likely to screen aptamers with high affinity and specificity.
In the process of CRP aptamer screening, the MARAS program developed by Lai et al. is the best primer-free SELEX library–compatible program [36]. For example, Lai et al. constructed a SELEX library with a 20-mer random sequence without a primer region and screened it in the MARAS program [36]. The obtained sequence D and sequence E (Table 1) were verified by immunoassay. It was confirmed that both sequence D and sequence E could specifically recognize CRP and generate signals in response to the change of CRP concentration by immunoassay. Sequence G was screened by Taso et al. using the same Maras program; they also verified the specificity of sequence G with blind serum samples [45]. The result was consistent with monoclonal antibody–based nephelometry analysis, which indicated that sequence G has high specificity toward targets.
Slow off-rate modified aptamer
The slow off-rate modified aptamer (SOMAmer) is a special chemically modified nucleotide developed by Hopfield et al. [46]. Different from common nucleotides, SOMAmer contains uracil deoxyriboside residues, and its fifth position is replaced by a hydrophobic aromatic functional group similar to an amino acid side chain, so it has more variable physicochemical properties (such as hydrophobicity, size, and shape) and affinity with the target protein. Therefore, SOMAmer is often used in the aptamer screening of biomolecules which are difficult to be screened by the conventional SELEX library. Minagawa et al. used SOMAmer as the SELEX library and finally screened sequence H (Table 1) [47]. The affinity test of sequence H also showed that the Kd value of the aptamer was 6.2 pmol/L by SOMAmer technology, which was also the sequence H with the strongest affinity with CRP.
Advances in aptamer-based sensing assays for CRP
The biosensor is a multi-disciplinary integrated technology that plays an important role in scientific research, industrial production, and even the well-being of the public. It uses a transducer to capture the reaction between the target substance and the identification component, and employs discrete or continuous physicochemical signals to express the degree of reaction, thereby deriving the content of the target substance in the sample [48]. Aptamer, with the superior performance among the available identification components, is highly compatible with biosensors. It can also offer biosensors with diverse sensing strategies and better performance in analysis [25]. Aptamer-based biosensors are also undergoing the fastest development.
To meet the needs of clinical testing, many aptamer-based CRP biosensors have emerged in recent years, such as high-sensitivity SPR aptasensors, field effect transistor (FET) microfluidic aptasensors with automatic sample analysis, label-free electrochemical impedance spectroscopy (EIS) aptasensors, and Blu-ray disc aptasensors suitable for POCT. Their overall development and advancement are summarized in Fig. 2.
This article divides CRP aptasensors into optical aptasensors, electrochemical aptasensors, microfluidic aptasensors, and some other types of aptasensors according to the type of the detected signals. The characteristics of each type of aptasensor are summarized in Table 2.
CRP optical aptasensor
The optical sensor conducts sample analysis by sensing the optical signal changes of a single or multiple light beams in the detection system, with the advantages of compact structure and high sensitivity. The optical aptasensor equipped with the aptamer’s small size effect can also reduce the detection background interference. It is the first type of CRP aptasensor that was developed. According to the difference of optical detection signals, optical aptasensors can be divided into five categories, namely SPR aptasensors, fluorescent aptasensors, optical fiber aptasensors, dark field imaging aptasensors, and colorimetric aptasensors (Fig. 3).
C-reactive protein optical aptasensor: A sensitive surface plasmon resonance aptasensor [51], with permission from Scientific Reports (copyright 2014); B sensitive fluorescent aptasensor [57], with permission from RSC Advances (copyright 2019); C optical fiber aptasensor [60], with permission from Elsevier (copyright 2017); D dark-field imaging aptasensor [62], with permission from RSC Advances (copyright 2019); and E gold nanoparticle colorimetric aptasensor [69], with permission from Elsevier (copyright 2020)
SPR aptasensor
SPR is one of the most effective label-free analyses with which the interaction between the recognition element and the target substance can be measured, and has been widely used in the fields of biomedicine, proteomics, genomics, and bioengineering [49]. The first CRP aptasensor was also developed based on SPR technology. Another kind of CRP SPR aptasensor modified biotin at the end of the aptamer and immobilized it on the surface of a gold membrane modified with streptavidin to analyze the light refraction deviation effect caused by the combination of CRP and the aptamer [50]. The aptasensor has a linear range suitable for clinical application (0–0.1 ppm), high sensitivity (LOD 0.005 ppm), and good reproducibility (CV 11%) in hydroxyethyl piperazine ethane sulfonic acid buffer (HEPES) buffer (pH 6.5, 2 mmol/L Ca2+, 0.005% Tween 20), meeting the precision needs for CRP analysis. With the help of this study, researchers made a strategic improvement to further optimize the detection performance of SPR aptasensors.
In terms of sensitivity improvement, Vance et al. coated the aptamer with quantum dots, which were employed as a light source to amplify the SPR detection signal [51]. The aptasensor has a detection sensitivity of up to 5 × 10−9 mg/L and is currently one of the most sensitive methods in CRP analysis (Fig. 3A). In the context of portable detection, another kind of CRP SPR aptasensor applied hot embossing and doctor blading to reduce the size of the precious metal plane required by SPR technology, making the preparation of SPR aptasensors more convenient [52]. This aptasensor only needs 50 μL CRP solution for quantitative analysis and can also directly read the analysis results with portable instruments such as a mobile phone.
Fluorescence aptasensor
Fluorescence analysis uses fluorescent dyes as tracer material. Within a certain range, the fluorescence intensity in the detection system is correlated with the concentration of the analyte in the sample. The fluorescent aptasensor performs well with small background interference, and has a wide range of linear response detection and high detection sensitivity. It is currently the research focus in the field of bioanalysis [53, 54].
Bernard et al. prepared a fluorescent aptasensor for multi-analyte analysis based on microbeads [55]. After the aptamer combines with the CRP in the sample, it is quickly separated from the buffer system by magnetic separation. After that, the induced antibody binds to it to form an immune complex. The phycoerythrin label on the antibody aggregates to produce fluorescent signals. The aptasensor has a detection range of 0.4–100 mg/L, and the required amount of CRP solution was 50 μL per well. Bernard’s team also performed optimization on process parameters such as time, temperature, buffer systems, etc., involved in the analysis of the aptasensor, and wrote a set of mature instruction manuals, all of which revealed the potential for this aptasensor to be developed into a mature commercial detection kit [56]. However, this method still has its disadvantages, such as low sensitivity. Therefore, Liu’s team [57] developed an ultra-high-sensitivity fluorescent aptasensor based on the combination of the principles of nucleic acid amplification and graphene oxide fluorescence quenching. The CRP in the sample binds to the aptamer and releases the complementary strand, triggering the cutting function of ribonuclease H to shorten the length of fluorescent-labeled RNA fragments, so that it can be separated from graphene oxide adsorption, thereby enhancing the fluorescent signal. The limit of detection (LOD) of the fluorescence aptasensor developed by this strategy is 1 × 10−5 mg/L, which can meet the clinical needs regarding high sensitivity (Fig. 3B).
Optical fiber aptasensor
Optical fiber is a kind of transmission tool for remote optical signal transmission and transportation [58]. Because the optical fiber works by the principle of total reflection of light and is very sensitive to the refractive index of incident light, an alternative that is a cost-effective, compact, and fast aptasensor can be obtained by modifying its surface with the aptamer [58]: when CRP reacts with the aptamer, the refractive index of light will naturally change due to CRP being a biological protein, and this subtle refractive change will occur because the optical fiber is sensitive to identify and transmit to the detector, so it is gradually explored and applied to a new generation of biosensor detection platform [59].
The typical preparation and analysis procedures of optical fiber aptasensors are as follows [60]: a layer of metal or metal oxide film is coated on the surface of the optical fiber transmission end, and the aptamer is then fixed on the film as an identification element. When CRP binds to an aptamer, the light refraction will change, and the refracted light is transmitted to the analyzer through the optical fiber for signal interpretation, thereby completing the sample analysis (Fig. 3C). It is worth mentioning that the optical fiber aptasensor prepared by Zubiate et al. is small, convenient, sensitive, and quick (LOD 0.0625 mg/L; whole detection time 61 s), and can be consecutively and repeatedly used because of the flexible restoration of the aptamer [60]. This strategy can reduce the cost of testing and is very suitable for use in remote areas or rudimentary healthcare facilities. To further enhance the detection sensitivity, Schulze et al. removed the optical fiber outer coating using hydrofluoric acid etching [61]. It enhanced the sensitivity of the refractive index of the sensor’s fiber Bragg grating to the surrounding environment and lowered the CRP detection level to 8.2 × 10−10 mg/L.
Dark field imaging aptasensor
The detection technology based on dark-field imaging can measure the protein content at a single nanoparticle level, developing a testing approach with relatively high precision. The dark-field imaging sensor developed by Zhao et al. uses an indirect strategy to determine the CRP concentration, in which the aptamer binds to CRP and releases the complementary strand to label gold nanoparticle [62]. The sample concentration can be determined by counting the gold nanoparticles in the dark-field imaging under microscopic observation (Fig. 3D). This method eliminates the interference from other proteins, has a high detection accuracy, and can accurately measure CRP serum samples at 3.22 × 10−5 mg/L.
Colorimetric aptasensor
Colorimetric analysis mainly measures the intensity or the change in the chromatic value of the detection liquid for quantitative analysis, which is very suitable for point-of-care tests [63]. The gold nanoparticle has attracted great attention in the colorimetric sensing field because of its special inter-particle optical effect and the catalytic effect similar to chromogenic enzymes [64]. According to different testing strategies, colorimetric aptasensors can be divided into labeled colorimetric aptasensors and label-free colorimetric aptasensors.
In the research and development of the labeled colorimetric aptasensor, our team [65] improved the traditional ELISA method and established a sandwich-like nano-enzyme colorimetric sensor: conjugation of citicoline to BSA to prepare the artificial antibody instead of the first antibody of traditional sandwich ELISA to recognize CRP in samples; the aptamer acts as a second antibody, and nanogold attached to the aptamer was used instead of traditional chromogenic enzyme to catalyze the color reaction between hydrogen peroxide and tetramethylbenzidine. Compared with the traditional ELISA method, this method only needs 50 μL CRP solution for a single detection, with a smaller sample volume and lower inter-batch differences, and can directly measure the CRP level in a diluted blood sample, with the LOD as low as 8 × 10−9 mg/L. However, the formation of a sandwich complex requires the tedious addition of reagents and a lengthy incubation process, which is not conducive to the rapid analysis of clinical CRP samples. Therefore, it is necessary to develop a label-free colorimetric sensor.
With its special inter-particle optical effect, a gold nanoparticle can be directly used as a tracer material to achieve label-free analysis [66], where the aptamer is adsorbed to the gold nanoparticle to form an immunoprobe. After the target substance reacts with the immunoprobe, the gold nanoparticle is released and exposed to the high-concentration saline buffer to gradually cross-link and aggregate. The light absorption peak shifts toward a longer wavelength along with it, and the visual appearance shows a change from the red color of the dispersed state to the blue color of the coagulated state, thereby generating the colorimetric signal [67, 68] (Fig. 3E). Antonio et al. applied this analysis method to the detection of serum CRP in clinical patients [69]. The obtained label-free colorimetric aptasensor was very convenient and fast. A single detection only needs 30 μL sample and 5 min incubation time to complete all the analyses. The LOD of the aptasensor is 1.2 mg/L and the detection range is 0.889–20.7 mg/L, which can be used for high-throughput rapid detection of clinical CVDs.
CRP electrochemical aptasensor
The electrochemical aptasensor fixes the recognition molecule and electrochemical sensing element on the electrode and performs quantitative analysis of CRP by determining the change in electrochemical signal after a sample is added. The electrochemical aptasensor has the advantage of high sensitivity, high specificity, low cost, and easy portability [48, 70]. According to the difference in testing signals, the electrochemical aptasensors can be roughly divided into voltammetric aptasensors, EIS aptasensors, and electrochemiluminescence (ELC) aptasensors (Fig. 4).
C-reactive protein electrochemical aptasensor: A sandwich voltammetric aptasensor [72], with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim (copyright 2009); B electrochemical impedance spectroscopy aptasensor [79]; (a) structure chart and (b) capacitive Nyquist plots of the molecular film showing its high responsiveness to CRP, with permission from the American Chemical Society (copyright 2018); and C electrochemiluminescence aptasensor [84], with permission from Elsevier (copyright 2019)
Voltammetric aptasensor
The voltammetric aptasensor primarily detects the current or voltage signal generated by the oxidative-reductive reaction on the electrode surface. Aptamers have more stable physical and chemical properties than antibodies, and their three-dimensional active structure is not prone to change caused by electric current; hence, it is very suitable for voltammetric detection [71]. Centi et al. established the first CRP electrochemical aptasensor [72]: aptamer-modified magnetic beads were placed on the surface of the electrode and incubated with sample solution, alkaline phosphatase (AP)–labeled antibody solution, and α-naphthyl phosphate successively. If the sample contains CRP, the aptamer will specifically recognize CRP and quantitatively generate an “aptamer-CRP-AP–labeled antibody” sandwich complex on the electrode surface due to the concentration of CRP. AP catalyzes the decomposition of α-naphthyl phosphate and forms current signal, thus realizing the quantitative analysis of CRP (Fig. 4A). The electrochemical aptasensor needs 0.45 mL of CRP solution for each detection, which has good detection precision (CV 8%) and accuracy (highly consistent with the ELISA method). However, the sensitivity is too low (LOD 5.4 × 10−2 mg/L) and cannot reach the high-sensitivity testing requirements in clinical application. To increase the detection sensitivity and achieve quantitative detection, researchers worldwide have adopted a method to replace the tracer material or electrode base material to obtain a wider detection range and lower detection limit.
For example, Wang et al. prepared a tracer material by immobilizing a large amount of Zn2+ on silica microspheres with high specific surface area to improve the sensing process [73]. The response signal of the current can be amplified with this method, with the detection sensitivity of CRP reaching 1.7 × 10−9 mg/L, which meets the requirement of high-sensitivity detection. However, the upper detection limit of this aptasensor is less than 0.125 mg/L, which renders it not directly useable for clinical sample analysis. Another kind of voltammetric aptasensor used a polypyrrole wire mesh with a high specific surface area and high conductivity as the electrode base material to resolve the problem of narrow detection range [74]. It has not only enhanced the detection sensitivity of current (LOD 1.19 × 10−17 mg/L) but also increased the load of recognition molecules, thus obtaining a wide detection range spanning 14 orders of magnitude (1.19 × 10−16–119 mgl/L). Similarly, in order to enhance the detection sensitivity, Wang et al. used NH2-Ni-metal organic frameworks (NH2-Ni-MOFs) as a tracer material and employed its catalytic effect on methylene blue (MB) electron transfer to perform CRP quantitative analysis [75]. The advantage of this method is that NH2-Ni-MOFs can be synthesized through chemical methods, and the particle size of the composite can be controlled. The team also found that NH2-Ni-MOFs with a particle size of 300 nm have the strongest catalytic performance of currents. The voltammetric aptasensor developed based on the above principle has a LOD at 2.9 × 10−11 mg/L, which is currently the most sensitive type of CRP aptasensor [75].
Jarczewska et al. found that most of the voltammetric aptasensors are based on a sandwich complex immunoassay for carrying out analysis [76]. After the aptamer reacts with CRP, it takes a long time to incubate the sandwich complex, which is very time consuming and laborious. Therefore, the team used MB and an aptamer to form a non-sandwich type of voltammetric aptasensor. MB nanoparticles are embedded on the aptamer, and the aptamer binds to CRP to undergo conformational changes, thereby releasing MB nanoparticles with the current catalytic effect and causing current changes in the test. The aptasensor has good specificity and shows a linear response to CRP in the range of 1.9 × 10−8–1.9 × 10−6 mg/L.
EIS aptasensor
The EIS aptasensor detects signals from changes in electrode impedance or capacitance. It is worth noting that, because biomacromolecules have electrical impedance, the EIS aptasensor is generally operated in a label-free mode [77]. Compared with sandwich-type sensors that require the formation of sandwich-like immune complexes for detection, an EIS aptasensor can skip the lengthy incubation process, with a more convenient and faster detection procedure.
Qureshi et al. used the Au-S covalent principle to immobilize the aptamer directly on the gold interdigital (GID) capacitor, and constructed a label-free EIS aptasensor for the first time [77]. The Au-S covalent principle is based on the empty orbitals from Au atoms and lone-pair electrons from sulfur atoms [29]. Thus, the AuNPs can spontaneously form Au-S covalent bioconjugates with thioalcohol by molecular self-assembly, thereby enabling easy and efficient labeling on thiol-containing biomolecules. The LOD of the aptasensor is 1 × 10−4 mg/L with a linear response range of 0.1–0.5 mg/L. To further broaden the detection range, the team then inserted a carbon nanotube layer structure into the original GID electrode–aptamer structure to amplify the EIS signal with its high storage capacity, so that the dynamic CRP detection range of the aptasensor can be expanded to 1.19 × 10−2 mg/L [78]. another CRP EIS aptasensor used redox labeled peptides to replace carbon nanotubes as signal amplifiers to improve the recognition specificity and sensitivity of the detection interface of EIS aptasensor [79]. It utilized its small molecule effect to reduce the LOD of the EIS aptasensor to 1.19 × 10−7 mg/L, therefore compensating for the weakness of low detection accuracy of the EIS aptasensor (Fig. 4B).
ELC aptasensor
The ELC aptasensor mainly measures the luminescence signal generated by the electric current on the electrode surface, combining the strength of the high sensitivity of optical sensors and the easy synthesis of electrochemical sensors [80]. According to the source of ELC signals, it can be divided into label-free ELC aptasensors and labeled ELC aptasensors [81].
The label-free ELC aptasensor is based on the principle that guanine-rich DNA enhances the thioflavin T luminescence signal. Since nucleic acid molecules can be transcribed, replicated, and complemented, various forms of the molecule have been developed. For example, the ELC aptasensor directly screened out guanine-rich CRP aptamer, which is used to build a label-free ELC aptasensor for the one-step detection of biological samples [82]. However, the LOD of this aptasensor is 4.51 × 10−6 mg/L, thus requiring further improvement and optimization. Li et al. improved the ELC sensor using the principles of nucleic acid transcription and replication to further enhance the detection signal [83]. The complementary strand of the guanine-rich aptamer was obtained as a PCR primer. After the CRP reacts with the aptamer, it is released to the padlock strand, T4 ligase, phi29 polymerase, and deoxyribonucleic acid triphosphate system. Rolling circle amplification takes place to increase the amount of guanine-containing DNA, thereby greatly enhancing the ELC signal and realizing the high-sensitivity analysis of CRP in serum samples at a level of 5.94 × 10−10 mg/L.
The labeled ELC aptasensor modifies the ELC signal tracer material on the surface of the identification element to form an immune complex on the surface of the detection electrode, and proportionally generates the ELC detection signal for quantitative analysis. However, under the same linear dimension, the traditionally labeled ELC immune complex is prone to forming a space gap and prolonging the electron transfer path. Another kind of CRP ELC aptasensor employed a two-dimensional porphyrin covalent organic framework as the ELC signal source to shorten the electron transfer path and remove the excessive pore structure, thereby avoiding the loss caused by the recombination of electrons [84]. This aptasensor has a low LOD of 1 × 10−4 mg/L and a wide detection range spanning three orders of magnitude, which meets the detection needs of low-content CRP samples (Fig. 4C).
CRP microfluidic aptasensor
Traditional aptasensor detection requires the manual addition of samples or separation of immune complexes, especially for aptasensors involving sandwich-type structures (such as voltammetric electrochemical aptasensor, labeled fluorescent aptasensor, etc.). After an aptamer recognizes CRP, it is necessary to add a second antibody, the detection substrate, or even enzymes in sequence for incubation, entailing a very tedious and time-consuming process. Microfluidic aptasensor refers to a sensing technology that uses an aptamer as the recognition element to automatically complete the entire process of sample preparation, reaction, separation, and detection on a micrometer-level detection chip integrated with a micro-electromechanical system. With the advantage of automatic detection, a microfluidic aptasensor is currently a hot research topic in vitro analysis and laboratory analysis [85]. According to different detection modes, it can be divided into labeled and label-free microfluidic aptasensors.
Labeled microfluidic aptasensor
Pultar et al. established the first CRP microfluidic aptasensor using a labeled fluorescence analysis combined with a microfluidic chip [86]: the CRP aptamer was incubated on the slide with a dot detector to form separate reaction wells. The microfluidic chip was made by combining the slide and silica gel resin. The sample solution and fluorescein-labeled antibody were added to the slide in turn, and the immune complex of “aptamer-CRP-fluorescein labeled antibody” was formed with the aptamer. The concentration of CRP in the sample can be calculated by directly detecting the fluorescence intensity of the corresponding reaction pore. It is worth mentioning that the aptasensor can determine the number of reaction holes on the slide and the types and concentrations of an aptamer incubated according to the requirements. These reaction wells can even be extended to the pattern of microarray, and the simultaneous detection of different target proteins in multiple parallel control groups or samples can be realized. The team also compared the testing performance of the microfluidic sensor with the CRP-specific antibody and aptamer used as recognition molecules and found that the aptamer-based sensor is compatible with microfluidic technology, and that the combination of the two can obtain a wider detection range (0.01–100 mg/L) and higher recovery rate (70–130%).
Label-free microfluidic aptasensor
FET has the advantage of low signal-to-noise ratio, low power consumption, and easy integration. The FET-based microfluidic chip can perform label-free detection [87]. The most common FET label-free microfluidic chip structure is presented by Yang et al. [88]. A pneumatic micropump, vortex micromixer, pneumatic microinjector, and several microvalves were integrated on the detection chip to quickly and automatically complete the whole process of CRP detection (Fig. 5). Compared with the traditional desktop detection instruments, the FET-based label-free microfluidic CRP aptasensor is not only easy to operate, lightweight, and portable, but can also enable optimization of the detection sensitivity (LOD 0.0125 mg/L, which is about 10 times higher than that of a desktop detector) and shorten detection time (the detection time is only 25 min, about 20% that of a desktop detector). Innovative study has also been carried out on this basis.
C-reactive protein label-free field effect transistor microfluidic aptasensor [88]: A microfluidic chip structure chart and B test process; (a) the magnetic beads labeled with CRP aptamer were incubated with the samples containing CRP in the reaction chamber; (b) after incubation, the magnetic separation method attracted the immune complex to the bottom of the microchannel, and then washed the buffer to remove the interfering substances; (c) the acridine ester–labeledanti-CRP antibody was added and interacted with the immune complex; (d) the magnetic separation method was used to remove the interfering substances The unconjugated acridine ester–labeled anti CRP antibody was removed, and then the color reagent (H2O2) was added for chemiluminescence detection, with permission from Elsevier (copyright 2009)
In terms of clinical POCT applications, Lee et al. [89] considered that the chip developed by Yang et al. [88] required an external solenoid valve as the power source to prevent liquid backflow between the internal chambers and channels of the chip, which did not conform to the requirement of POCT for convenience. Lee’s team [89] prepared a driven micropump that did not require an external power supply. The micropump was based on the floating block structure of PDMS, which is located at the end of the liquid channel. It can generate potential energy by injecting compressed air into the pipeline, thus opening the hydraulic pressure generated by polydimethylsiloxane (PDMS) membrane creep. Lee’s team used this pneumatic micropump to replace the micropump based on solenoid valve control. The improved microfluidic chip does not need to control the external power supply current and internal solenoid valve frequency, which makes the operation more convenient and more suitable for clinical POCT needs.
It is composed of a PDMS base membrane and a floating module on it. The liquid pressure makes the PDMS connecting to the vent valve, and compressed air quickly passes through it to generate driving force. The new microfluidic aptasensor developed thus realizes high-speed transmission by the micropump without external power supply, and it can also prevent pollution from liquid backflow, rendering it more suitable for clinical POCT use.
In view of enhanced stability, the FET microfluidic aptasensors prepared in the above manner fixed the aptamer on the surface of the chip as a recognition element. However, they still use antibodies to immobilize the tracer material for sandwich-type detection, which still has defects such as poor physical and chemical stability of the antibodies. Therefore, Kao et al. innovatively established a double aptamer sandwich aptasensor [90]. Aptamers with different binding sites were utilized to replace the antibody-immobilized tracer material to form a tracer material-aptamer2-CRP aptamer1 immune complex. The aptasensor prepared by this method completely eliminates the defect resulting from unstable antibody properties, and hence has stronger environmental adaptability, with convenient transportation and storage.
With respect to reducing the influence of matrix and background interference, Chen et al. employed AlGaN/GaN material to prepare FET microfluidic chips [91]. The chemical inertness of AlGaN/GaN prevents the diffusion of ions and shields the background interference caused by the high concentration of electrolytes in the matrix, and the biological sample testing is directly performed. However, the aptasensor has a narrow detection range (0.24–1.18 mg/L), so it cannot be used in clinical testing. Therefore, another kind of CRP FET aptasensor constructed an electrical double-layer structure with the AlGaN/GaN FET electrodes, so that the detection solution only reacts with the gate electrode and the separation trench, thereby increasing the concentration of active electrons in the detection electrode, further amplifying the current change signal, and expanding the detection range to 0.34–23.2 mg/L [92].
To expand the clinical practical application, Sinha et al. further enhanced the use of the FET microfluidic aptasensor [93]. They prepared a FET microfluidic chip containing multiple detection chambers, which can detect four biomarkers at the same time to meet the need for multiple analyses.
Other types of CRP aptasensor
An aptamer is stable and compatible with multiple sensing models. Therefore, in addition to the three main aptasensor forms such as electrochemical sensors, optical sensors, and microfluidic sensors, some CRP aptasensors are based on other platforms (isotachophoresis, in vivo detection, Blu-ray disc, and SOMAscan, etc.) have gradually been developed (Fig. 6).
Isotachophoresis (ITP) can simultaneously analyze multiple ionic compounds, with simple sample pretreatment process and fairly compatible operating conditions, so it is widely used in biochemical analysis. Another kind of CRP ITP aptasensor has made an ITP aptasensor that separates the CRP aptamer binding complex by ion spacing technology and analyzes the complex content using ITP, thus shortening the ITP detection cycle to 20 min [94]. This aptasensor has the potential to serve as a simple and fast alternative to immunoassay and become a new platform for clinical POCT.
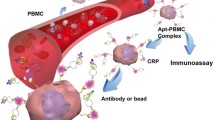
The living cell sensor is a novel CRP aptasensor designed by Hwang et al. [95]. Its principle is to combine an aptamer with the surface of peripheral blood mononuclear cells (PBMCs) to make aptamer/PBMCs conjugate, which can recognize CRP in fluid and form immune complex. Finally, the immune complex was incubated with the fluorescein-labeled CRP antibody to generate a detectable fluorescent signal (Fig. 6A). It is worth mentioning that aptamer modified PBMCs have good physiological activity. Hwang’s team also confirmed that apt PBMCs conjugate can detect CRP content in flowing sheep blood under simulated vascular conditions. Therefore, the living cell sensor is expected to be the first CRP in vivo detection scheme by optimizing CRP aptamer/PBMCs immune complex detection technology. As a kind of non-attaching, non-differentiating, independent immune cells, PBMCs will migrate to the lesions for immune response when the body is injured. Therefore, the living cell sensor is expected to be derived into a continuous monitoring device of clinical pathological state or a monitoring device of wound inflammation state.
As an important portable storage device in daily life, Blu-ray discs can replace traditional chips as the detection substrate. Combined with the high-precision optical signal reading ability of the Blu-ray optical drive, it can be used to develop analytical sensing devices that are easy to popularize at the basic level, shines a light on the new direction for the development of clinical CRP POCT. The Blu-ray disc aptasensor established by Weng et al. [96] is made by fixing the aptamer on the blue light disc (Fig. 6B). The CRP aptamer, myoglobin aptamer, and troponin I aptamer were incubated in non-interference areas of the Blu-ray disc, respectively, so that the aptasensor could obtain the concentration information of three proteins simultaneously when reading the Blu-ray disc, which is very convenient and fast.
SOMAscan is an aptamer-based proteomic detection technology proposed by SomaLogic, Inc. (Colorado, USA). It is also known as an aptamer-based analysis that has been validated in real clinical settings [97, 98]. Kim et al. included CRP in the SOMAscan protein library and prepared an aptasensor based on the SOMAscan platform [99]. It used plasma samples of 52 subjects to test the repeatability and stability of the method. The results showed that the aptasensor had good repeatability (CV < 10%) and stability (ICC or Spearman R ≥ 7.5 within 48 h, ICC or Spearman R ≥ 0.4 after 1 year of storage). The aptasensor confirmed the feasibility of the aptamer analysis method in the clinical application of CRP, especially for plasma samples stored within 1 year, and had the advantages of short detection time, high detection sensitivity, and good reproducibility. It is currently the most feasible sensing strategy for rapid and high-throughput detection of biological samples.
Conclusions and prospects
In recent years, aptamer-based biosensing technology has been developed rapidly in the bioanalytical field and has gradually become an effective method for rapid and accurate clinical identification of the biomarker CRP. Compared to antibodies, the aptamer has the advantage of low steric hindrance, easy modification, stable properties, and low immunogenicity. It can be expanded to a variety of CRP sensing platforms and is considered the best sensor recognition element [18, 25]. Currently, a variety of signal transduction methods have been developed to manufacture aptamer-based CRP biosensors, and it has been proven that they meet the requirements for CRP analysis such as high sensitivity [51], wide linear response range [74], POCT [52, 60], multiplexing [55, 56], etc. They can even be used to conduct in vivo testing [95] and achieve reproducible results in repeated testing [60]. However, there are still certain issues that need to be resolved in order to apply the aptasensor to real clinical testing.
The first problem is the background interference on the aptasensor caused by the matrix during CRP detection. The biological sample matrices have a complex composition, which contains a variety of irrelevant proteins (such as albumin, gamma globulin, etc.) other than CRP. These proteins are easily adsorbed non-specifically on the testing substrate of the aptasensor, resulting in false-positive or false-negative results [50]. Furthermore, aptamers have a highly flexible structure and different three-dimensional folding structures under different buffer conditions. This has both its advantages and disadvantages. The advantages lie in the following: the dynamic flexible structure enables the aptamer to have good physical and chemical properties, which can adapt to long-term storage and transportation at room temperature; the disadvantages lie in the following: the combination of aptamer and CRP must be carried out in a specific buffer solution; for example, the sequence must be involved in Na+ to detect CRP [23]. Considering the uncontrollable accidental error factors in practical operation, the disadvantage of this flexible structure will inevitably affect the repeatability of the aptasensor detection method. Finally, in view of the importance of CRP as an indicator of inflammation and infection in the monitoring of curative effect and prognosis, it is necessary to develop a real-time continuous monitoring system, so that the frequent monitoring of CRP in vivo is similar to the implantable continuous blood glucose monitoring system. The aptamer has low immunogenicity, which makes it possible to detect CRP in vivo. The living cell sensor further confirmed that the aptasensor has the ability to specifically recognize and combine CRP in blood vessels. However, further research is needed to achieve continuous monitoring of CRP in vivo based on the aptasensor.
Currently, two strategies can be adopted to solve these problems. The first is to promote the interaction, integration, and fusion of different disciplines, such as analytical chemistry, clinical medicine, materials science, nanoscience, and physics to design sensors with high-specificity clinical real-time monitoring devices in vivo. The second is to accelerate the development process of an aptamer 3D rapid modeling program and aptamer target molecular structure docking program, promote the research on the binding sites between the CRP and the aptamer, optimize the SELEX screening program, and obtain CRP-specific aptamers with good stability, high affinity, and low buffer impact. This will provide basic guarantee for optimizing the precision and repeatability of the aptamer detection method.
References
Roy N, Ohtani K, Matsuda Y, Mori K, Hwang I, Suzuki Y, et al. Collectin CL-P1 utilizes C-reactive protein for complement activation. Biochim Biophys Acta, Gen Subj. 2016;1860(6):1118–28.
Ansar W, Ghosh S. C-reactive protein and the biology of disease. Immunol Res. 2013;56(1):131–42.
Hong S. National guide to clinical laboratory procedures (fourth edition). 4th ed ed. Beijing: People's Health Publishing House; 2015.
Huang J, Baum Y, Alemozaffar M, Ogan K, Harris W, Kucuk O, et al. C-reactive protein in urologic cancers. Mol Asp Med. 2015;45:28–36.
Prucha M, Bellingan G, Zazula R. Sepsis biomarkers. Clin Chim Acta. 2015;440:97–103.
Mc Causland FR, Claggett B, Burdmann EA, Eckardt KU, Kewalramani R, Levey AS, et al. C-reactive protein and risk of ESRD: results from the trial to reduce cardiovascular events with aranesp therapy (TREAT). Am J Kidney Dis. 2016;68(6):873–81.
Reynolds IS, Boland MR, Reilly F, Deasy A, Majeed MH, Deasy J, et al. C-reactive protein as a predictor of anastomotic leak in the first week after anterior resection for rectal cancer. Color Dis. 2017;19(9):812–8.
Clark TW, Medina MJ, Batham S, Curran MD, Parmar S, Nicholson KG. C-reactive protein level and microbial aetiology in patients hospitalised with acute exacerbation of COPD. Eur Respir J. 2015;45(1):76–86.
Ridker PM. From C-reactive protein to interleukin-6 to interleukin-1 moving upstream to identify novel targets for atheroprotection. Circ Res. 2016;118(1):145–56.
Munkhaugen J, Otterstad JE, Dammen T, Gjertsen E, Moum T, Husebye E, et al. The prevalence and predictors of elevated C-reactive protein after a coronary heart disease event. Eur J Prev Cardiol. 2018;25(9):923–31.
Kunutsor SK, Seidu S, Blom AW, Khunti K, Laukkanen JA. Serum C-reactive protein increases the risk of venous thromboembolism: a prospective study and meta-analysis of published prospective evidence. Eur J Epidemiol. 2017;32(8):657–67.
Kwon CH, Kang JG, Lee HJ, Kim NH, Sung J-W, Cheong E, et al. C-reactive protein and risk of atrial fibrillation in East Asians. Europace. 2017;19(10):1643–9.
Pearson Thomas A, Mensah George A, Alexander RW, Anderson Jeffrey L, Cannon Richard O, Criqui M, et al. Markers of inflammation and cardiovascular disease. Circulation. 2003;107(3):499–511.
Avan A, Sany SBT, Ghayour-Mobarhan M, Rahimi HR, Tajfard M, Ferns G. Serum C-reactive protein in the prediction of cardiovascular diseases: overview of the latest clinical studies and public health practice. J Cell Physiol. 2018;233(11):8508–25.
Chandra P, Suman P, Airon H, Mukherjee M, Kumar P. Prospects and advancements in C-reactive protein detection. World J Meth. 2014;4(1):1–5.
Tang M-Q, Mao X-H, Gong Y-X-Y, Qing L-S, Xie J. Research progress of C-reactive protein analysis. Chinese J Anal Chem. 2020;48(9):1121–30.
Vashist SK, Venkatesh AG, Marion Schneider E, Beaudoin C, Luppa PB, Luong JHT. Bioanalytical advances in assays for C-reactive protein. Biotechnol Adv. 2016;34(3):272–90.
Chen A, Yang S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens Bioelectron. 2015;71:230–42.
Chen C-X, Dai L, Feng H-Y, Wang L, Guo K, Ding L-S, et al. A new strategy for the preparation of antibody against natural glycoside: with astragaloside IV as an example. Fitoterapia. 2020;142:104488.
Kaiser L, Weisser J, Kohl M, Deigner H-P. Small molecule detection with aptamer based lateral flow assays: applying aptamer-C-reactive protein cross-recognition for ampicillin detection. Sci Rep. 2018;8(1):5628.
Dunn MR, Jimenez RM, Chaput JC. Analysis of aptamer discovery and technology. Nat Rev Chem. 2017;1(10):16.
Meng HM, Liu H, Kuai HL, Peng RZ, Mo LT, Zhang XB. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem Soc Rev. 2016;45(9):2583–602.
Zhang Y, Lai BS, Juhas M. Recent advances in aptamer discovery and applications. Molecules. 2019;24(5):941.
Odeh F, Nsairat H, Alshaer W, Ismail MA, Esawi E, Qaqish B, et al. Aptamers chemistry: chemical modifications and conjugation strategies. Molecules. 2019;25(1):3.
Crivianu-Gaita V, Thompson M. Aptamers, antibody scFv, and antibody Fab' fragments: an overview and comparison of three of the most versatile biosensor biorecognition elements. Biosens Bioelectron. 2016;85:32–45.
Pandey CM, Augustine S, Kumar S, Kumar S, Nara S, Srivastava S, et al. Microfluidics based point-of-care diagnostics. Biotechnol J. 2018;13(1):11.
Yan X, Li HX, Su XG. Review of optical sensors for pesticides. Ttac-Trend Anal Chem. 2018;103:1–20.
Farzin L, Shamsipur M, Sheibani S. A review: aptamer-based analytical strategies using the nanomaterials for environmental and human monitoring of toxic heavy metals. Talanta. 2017;174:619–27.
Wang Q-L, Huang W-X, Zhang P-J, Chen L, Lio C-K, Zhou H, et al. Colorimetric determination of the early biomarker hypoxia-inducible factor-1 alpha (HIF-1α) in circulating exosomes by using a gold seed-coated with aptamer-functionalized Au@Au core-shell peroxidase mimic. Microchim Acta. 2019;187(1):61.
Kim S, Ryu J, Yi H, Kim S, Zhang B. Construction of C-reactive protein-binding aptamer as a module of the DNA computing system for diagnosing cardiovascular diseases. Preliminary proceedings of the tenth international meeting on DNA computing (DNA10)2004. p. 334–43.
Meyer MHF, Hartmann M, Keusgen M. SPR-based immunosensor for the CRP detection—a new method to detect a well known protein. Biosens Bioelectron. 2006;21(10):1987–90.
Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Sciences. 1990;249(4968):505–10.
Lou X, Qian J, Xiao Y, Viel L, Gerdon AE, Lagally ET, et al. Micromagnetic selection of aptamers in microfluidic channels. Proc Natl Acad Sci U S A. 2009;106(9):2989–94.
Huang C-J, Lin H-I, Shiesh S-C, Lee G-B. Integrated microfluidic system for rapid screening of CRP aptamers utilizing systematic evolution of ligands by exponential enrichment (SELEX). Biosens Bioelectron. 2010;25(7):1761–6.
Wu B, Jiang R, Wang Q, Huang J, Yang X, Wang K, et al. Detection of C-reactive protein using nanoparticle-enhanced surface plasmon resonance using an aptamer-antibody sandwich assay. Chem Commun. 2016;52(17):3568–71.
Lai JC, Hong CY. A novel protocol for generating high-affinity ssDNA aptamers by using alternating magnetic fields. J Mat Chem B. 2014;2(26):4114–21.
Lai J-C, Hong C-Y. Magnetic-assisted rapid aptamer selection (MARAS) for generating high-affinity DNA aptamer using rotating magnetic fields. ACS Comb Sci. 2014;16(7):321–7.
Li H-H, Wen C-Y, Hong C-Y, Lai J-C. Evaluation of aptamer specificity with or without primers using clinical samples for C-reactive protein by magnetic-assisted rapid aptamer selection. RSC Adv. 2017;7(68):42856–65.
Wang MS, Black JC, Knowles MK, Reed SM. C-reactive protein (CRP) aptamer binds to monomeric but not pentameric form of CRP. Anal Bioanal Chem. 2011;401(4):1309–18.
Wang MS, Reed SM. Direct visualization of electrophoretic mobility shift assays using nanoparticle–aptamer conjugates. Electrophoresis. 2011;33(2):348–51.
Park JW, Tatavarty R, Kim DW, Jung HT, Gu MB. Immobilization-free screening of aptamers assisted by graphene oxide. Chem Commun. 2012;48(15):2071–3.
Yang X-H, Wang Y, Wang K, Wang Q, Wang P, Lin M, et al. DNA aptamer-based surface plasmon resonance sensing of human C-reactive protein. RSC Adv. 2014;4(58):30934–7.
Yu XL, Yu RQ, Yang XH. Pattern recognition of enrichment levels of SELEX-based candidate aptamers for human C-reactive protein. Biomed Eng-Biomed Tech. 2017;62(3):333–8.
Wen JD, Gray DM. Selection of genomic sequences that bind tightly to Ff gene 5 protein: primer-free genomic SELEX. Nucleic Acids Res. 2004;32(22):e182.
Tsao S-M, Lai J-C, Horng H-E, Liu T-C, Hong C-Y. Generation of aptamers from a primer-free randomized ssDNA library using magnetic-assisted rapid aptamer selection. Sci Rep. 2017;7(1):45478.
Hopfield JJ, Yamane T, Yue V, Coutts SM. Direct experimental evidence for kinetic proofreading in amino acylation of tRNAIle. Proc Natl Acad Sci. 1976;73(4):1164.
Minagawa H, Kataoka Y, Fujita H, Kuwahara M, Horii K, Shiratori I, et al. Modified DNA aptamers for C-reactive protein and lactate dehydrogenase-5 with sub-nanomolar affinities. Int J Mol Sci. 2020;21(8):2683.
Felix FS, Angnes L. Electrochemical immunosensors - a powerful tool for analytical applications. Biosens Bioelectron. 2018;102:470–8.
Nguyen HH, Park J, Kang S, Kim M. Surface plasmon resonance: a versatile technique for biosensor applications. Sensors. 2015;15(5):10481–510.
Bini A, Centi S, Tombelli S, Minunni M, Mascini M. Development of an optical RNA-based aptasensor for C-reactive protein. Anal Bioanal Chem. 2008;390(4):1077–86.
Vance SA, Sandros MG. Zeptomole detection of C-reactive protein in serum by a nanoparticle amplified surface plasmon resonance imaging aptasensor. Sci Rep. 2014;4(1):5129.
Walter JG, Eilers A, Alwis LSM, Roth BW, Bremer K. SPR biosensor based on polymer multi-mode optical waveguide and nanoparticle signal enhancement. Sensors. 2020;20(10):11.
Wang RE, Zhang Y, Cai J, Cai W, Gao T. Aptamer-based fluorescent biosensors. Curr Med Chem. 2011;18(27):4175–84.
Wu D, Sedgwick AC, Gunnlaugsson T, Akkaya EU, Yoon J, James TD. Fluorescent chemosensors: the past, present and future. Chem Soc Rev. 2017;46(23):7105–23.
Bernard ED, Nguyen KC, DeRosa MC, Tayabali AF, Aranda-Rodriguez R. Development of a bead-basedaptamer/antibody detection system for C-reactive protein. Anal Biochem. 2015;472:67–74.
Bernard ED, Nguyen KC, DeRosa MC, Tayabali AF, Aranda-Rodriguez R. Incorporating aptamers in the multiple analyte profiling assays (xMAP): detection of C-reactive protein. Methods Mol Biol. 2017;1575:303–22.
Liu Z-Z, Luo D, Ren F-L, Ran F-Y, Chen W, Zhang B-Q, et al. Ultrasensitive fluorescent aptasensor for CRP detection based on the RNase H assisted DNA recycling signal amplification strategy. RSC Adv. 2019;9(21):11960–7.
Kim K, Gu MB, Kang DH, Park JW, Song IH, Jung HS, et al. High-sensitivity detection of oxytetracycline using light scattering agglutination assay with aptasensor. Electrophoresis. 2010;31(18):3115–20.
Wang X-D, Wolfbeis OS. Fiber-optic chemical sensors and biosensors (2013–2015). Anal Chem. 2016;88(1):203–27.
Zubiate P, Zamarreno CR, Sanchez R, Matias IR, Arregui FJ. High sensitive and selective C-reactive protein detection by means of lossy mode resonance based optical fiber devices. Biosens Bioelectron. 2017;93:176–81.
Schulze S, Wehrhold M, Hille C. Femtosecond-pulsed laser written and etched fiber bragg gratings for fiber-optical biosensing. Sensors. 2018;18(9):2844.
Zhao Y-F, Zhao J-R, Jin T, Sun S-Q, Liu W-L, Tan Y. An aptasensor based on the microscopic enumeration of encoding gold nanoparticles for the detection of C-reactive protein. RSC Adv. 2019;9(59):34293–8.
Wang T-T, Lio CK, Huang H, Wang R-Y, Zhou H, Luo P, et al. A feasible image-based colorimetric assay using a smartphone RGB camera for point-of-care monitoring of diabetes. Talanta. 2020;206:120211.
Cliffel DE, Turner BN, Huffman BJ. Nanoparticle-based biologic mimetics. Wiley Interdiscip Rev: Nanomed Nanobiotechnol. 2009;1(1):47–59.
Xie J, Tang M-Q, Chen J, Zhu Y-H, Lei C-B, He H-W, et al. A sandwich ELISA-like detection of C-reactive protein in blood by citicoline-bovine serum albumin conjugate and aptamer-functionalized gold nanoparticles nanozyme. Talanta. 2020;217:121070.
Yuan Z, Hu CC, Chang HT, Lu C. Gold nanoparticles as sensitive optical probes. Analyst. 2016;141(5):1611–26.
Liu B, Liu J. Methods for preparing DNA-functionalized gold nanoparticles, a key reagent of bioanalytical chemistry. Anal Methods. 2017;9(18):2633–43.
Aldewachi H, Chalati T, Woodroofe MN, Bricklebank N, Sharrack B, Gardiner P. Gold nanoparticle-based colorimetric biosensors. Nanoscale. 2018;10(1):18–33.
Antonio M, Ferreira R, Vitorino R, Daniel-da-Silva AL. A simple aptamer-based colorimetric assay for rapid detection of C-reactive protein using gold nanoparticles. Talanta. 2020;214:120868.
Gui RJ, Jin H, Guo HJ, Wang ZH. Recent advances and future prospects in molecularly imprinted polymers-based electrochemical biosensors. Biosens Bioelectron. 2018;100:56–70.
Han K, Liu T, Wang Y-H, Miao P. Electrochemical aptasensors for detection of small molecules, macromolecules, and cells. Rev Anal Chem. 2016;35(4):201–11.
Centi S, Sanmartin LB, Tombelli S, Palchetti I, Mascini M. Detection of C reactive protein (CRP) in serum by an electrochemical aptamer-based sandwich assay. Electroanalysis. 2009;21(11):1309–15.
Wang J, Guo J, Zhang J, Zhang W, Zhang Y. RNA aptamer-based electrochemical aptasensor for C-reactive protein detection using functionalized silica microspheres as immunoprobes. Biosens Bioelectron. 2017;95:100–5.
Lin Z-T, Li Y, Gu J, Wang H, Zhu Z, Hong X, et al. A conductive nanowire-mesh biosensor for ultrasensitive detection of serum C-reactive protein in melanoma. Adv Funct Mater. 2018;28(31).
Wang Z, Dong P, Sun Z-X, Sun C, Bu H-Y, Han J, et al. NH2-Ni-MOF electrocatalysts with tunable size/morphology for ultrasensitive C-reactive protein detection via an aptamer binding induced DNA walker-antibody sandwich assay. J Mater Chem B. 2018;6(16):2426–31.
Jarczewska M, Rebis J, Gorski L, Malinowska E. Development of DNA aptamer-based sensor for electrochemical detection of C-reactive protein. Talanta. 2018;189:45–54.
Qureshi A, Gurbuz Y, Kallempudi S, Niazi JH. Label-free RNA aptamer-based capacitive biosensor for the detection of C-reactive protein. Phys Chem Chem Phys. 2010;12(32):9176–82.
Qureshi A, Roci I, Gurbuz Y, Niazi JH. An aptamer based competition assay for protein detection using CNT activated gold-interdigitated capacitor arrays. Biosens Bioelectron. 2012;34(1):165–70.
Piccoli J, Hein R, El-Sagheer AH, Brown T, Cilli EM, Bueno PR, et al. Redox capacitive assaying of C-reactive protein at a peptide supported aptamer interface. Anal Chem. 2018;90(5):3005–8.
Li X, Li W, Zhang S. Chemiluminescence DNA biosensor based on dual-amplification of thrombin and thiocyanuric acid-gold nanoparticle network. Analyst. 2010;135(2):332–6.
Chen Y, Zhou SW, Li LL, Zhu JJ. Nanomaterials-based sensitive electrochemiluminescence biosensing. Nano Today. 2017;12:98–115.
Wu B, Chen N, Wang Q, Yang X-H, Wang K, Li W-S, et al. A simple label-free aptamer-based method for C-reactive protein detection. Anal Methods. 2016;8(21):4177–80.
Li M-J, Wang H-J, Yuan R, Chai Y-Q. A zirconium-basedmetal–organic framework sensitized by thioflavin-T for sensitive photoelectrochemical detection of C-reactive protein. Chem Commun. 2019;55(72):10772–5.
Zhang X, Chi K-N, Li D-L, Deng Y, Ma Y-C, Xu Q-Q, et al. 2D-porphrinic covalent organic framework-based aptasensor with enhanced photoelectrochemical response for the detection of C-reactive protein. Biosens Bioelectron. 2019;129:64–71.
Li W, Zhang LY, Ge XH, Xu BY, Zhang WX, Qu LL, et al. Microfluidic fabrication of microparticles for biomedical applications. Chem Soc Rev. 2018;47(15):5646–83.
Pultar J, Sauer U, Domnanich P, Preininger C. Aptamer-antibody on-chip sandwich immunoassay for detection of CRP in spiked serum. Biosens Bioelectron. 2009;24(5):1456–61.
Kaisti M. Detection principles of biological and chemical FET sensors. Biosens Bioelectron. 2017;98:437–48.
Yang Y-N, Lin H-I, Wang J-H, Shiesh S-C, Lee G-B. An integrated microfluidic system for C-reactive protein measurement. Biosens Bioelectron. 2009;24(10):3091–6.
Lee W-B, Chen Y-H, Lin H-I, Shiesh S-C, Lee G-B. An integrated microfluidic system for fast, automatic detection of C-reactive protein. Sensors Actuators B Chem. 2011;157(2):710–21.
Kao WC, Chu CH, Chang WH, Wang YL, Lee GB, Ieee. Dual-aptamer assay for C-reactive protein detection by using field-effect transistors on an integrated microfluidic system. 2016 Ieee 11th Annual International Conference on Nano/Micro Engineered and Molecular Systems 2016.
Chen P-C, Chen Y-W, Sarangadharan I, Hsu C-P, Chen C-C, Shiesh S-C, et al. Field-effect transistor-based biosensors and a portable device for personal healthcare. ECS J Solid State Sci Technol. 2017;6(7):Q71–Q6.
Chu C-H, Sarangadharan I, Regmi A, Chen Y-W, Hsu C-P, Chang W-H, et al. Beyond the debye length in high ionic strength solution: direct protein detection with field-effect transistors (FETs) in human serum. Sci Rep. 2017;7(1):5256.
Sinha A, Tai T-Y, Li K-H, Gopinathan P, Chung Y-D, Sarangadharan I, et al. An integrated microfluidic system with field-effect-transistor sensor arrays for detecting multiple cardiovascular biomarkers from clinical samples. Biosens Bioelectron. 2019;129:155–63.
Eid C, Palko JW, Katilius E, Santiago JG. Rapid slow off-rate modified aptamer (SOMAmer)-based detection of C-reactive protein using isotachophoresis and an ionic spacer. Anal Chem. 2015;87(13):6736–43.
Hwang J, Seo Y, Jo Y, Son J, Choi J. Aptamer-conjugated live human immune cell based biosensors for the accurate detection of C-reactive protein. Sci Rep. 2016;6(1):34778.
Weng S, Li X, Niu M, Ge B, Yu H-Z. Blu-ray technology-based quantitative assays for cardiac markers: from disc activation to multiplex detection. Anal Chem. 2016;88(13):6889–96.
Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5(12):17.
De Groote MA, Nahid P, Jarlsberg L, Johnson JL, Weiner M, Muzanyi G, et al. Elucidating novel serum biomarkers associated with pulmonary tuberculosis treatment. PLoS One. 2013;8(4):e61002.
Kim CH, Tworoger SS, Stampfer MJ, Dillon ST, Gu X, Sawyer SJ, et al. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci Rep. 2018;8(1):8382.
Acknowledgements
This work was supported by the Chinese Academy of Sciences (CAS) “Light of West China” Program (2018XBZG_XBQNXZ_A_005), the Faculty Research Grants of Macau University of Science and Technology (FRG-20-004-SKL), and the Innovation Training Program for College Students in Sichuan Province of China (S202013705121).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain any studies with human or animal subjects performed by any of the authors.
Informed consent
Informed consent is not applicable because of the nature of this study.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tang, MQ., Xie, J., Rao, LM. et al. Advances in aptamer-based sensing assays for C-reactive protein. Anal Bioanal Chem 414, 867–884 (2022). https://doi.org/10.1007/s00216-021-03674-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03674-0