Abstract
MAM-2201 is a synthetic cannabinoid that is increasingly found in recreational drug abusers and cases of severe intoxication. Thus, characterization of the metabolic pathways of MAM-2201 is necessary to predict individual pharmacokinetics and toxicity differences, and to avoid toxic drug-drug interactions. Collectively, 19 phase 1 metabolites of MAM-2201 were identified using liquid chromatography–Orbitrap mass spectrometry following human liver microsomal incubations in the presence of NADPH: 7 hydroxy-MAM-2201 (M1–M7), 4 dihydroxy-MAM-2201 (M8–M11), dihydrodiol-MAM-2201 (M12), N-(5-hydroxypentyl)-MAM-2201 (M13), hydroxy-M13 (M14), N-dealkyl-MAM-2201 (M15), 2 hydroxy-M15 (M16, M17), MAM-2201 N-pentanoic acid (M18), and hydroxy-M18 (M19). On the basis of intrinsic clearance values in human liver microsomes, hydroxy-MAM-2201 (M1), N-(5-hydroxypentyl)-MAM-2201 (M13), and hydroxy-M13 (M14) were the major metabolites. Based on an enzyme kinetics study using human cDNA-expressed cytochrome P450 (CYP) enzymes and an immunoinhibition study using selective CYP antibodies in human liver microsomes, CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A4 enzymes were responsible for MAM-2201 metabolism. The CYP3A4 enzyme played a prominent role in MAM-2201 metabolism, and CYP1A2, CYP2B6, CYP2C8, and CYP2C9 enzymes played major roles in the formation of some metabolites. MAM-2201 is extensively metabolized by multiple CYP enzymes, indicating that MAM-2201 and its metabolites should be used as markers of MAM-2201 abuse and toxicity.

In vitro metabolic pathways of MAM-2201 were characterized in human liver microsomes and recombinant CYPs using LC-HRMS analysis. Total 19 phase I metabolites were identified with predominant contribution of CYP3A4
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Synthetic cannabinoids, a group of substances with generally similar chemical structures binding to the cannabinoid receptor type 1 (CB1) or type 2 (CB2), were first identified in “herbal mixtures” in 2008 [1–3]. Structure–activity relationships for synthetic cannabinoids have been established [4–8], and JWH compounds (JWH-018, JWH-122, and JWH-073) have been modified as follows: introduction of a fluorine atom (AM-2201, MAM-2201, and EAM-2201) [9] and substitution of the naphthyl group for a cyclopropyl group (UR-144 and XLR-11) [10, 11], adamantyl group (APICA and 5F-APICA) [10], or quinolinyl group (QUPIC and QUCHIC) [12, 13]. Synthetic cannabinoids have been found in seized herbal materials, and their continual emergence on recreational and illicit drug markets has become a global issue [14–19].
MAM-2201, [1-(5-fluoropentyl)-1H-indol-3-yl](4-methyl-1-naphthalenyl)-methanone, is a third-generation synthetic cannabinoid detected as an active ingredient in illegal herbal incense blends [10, 16, 18, 20, 21]. MAM-2201 exerts potent pharmacological actions on brain function and causes psychoactive and intoxicating effects [22–24]. MAM-2201 has been detected in recreational users and intoxication cases, when plasma concentrations ranged from <0.1 to 49 ng/mL, indicating its extensive metabolism [19–21, 25–27]. N-(5-hydroxypentyl)-MAM-2201, N-(4-hydroxyfluoropentyl)-MAM-2201, MAM-2201 pentanoic acid, and dealkyl-MAM-2201 have been identified as the metabolites of MAM-2201 in urine and hair samples from MAM-2201 abusers [27–32].
Incubation of EAM-2201, a structural analogue of MAM-2201, with human liver microsomes formed 39 phase 1 metabolites [33]; however, in vitro metabolism of MAM-2201 in human liver microsomes resulted in only three metabolites, i.e., N-(5-hydroxypentyl)-MAM-2201, N-(4-hydroxyfluoropentyl)-MAM-2201, and MAM-2201 pentanoic acid [27]. Therefore, it is necessary to identify additional in vitro metabolites of MAM-2201 and separate the metabolites with the same molecular ions ([M + H]+). To predict individual differences in synthetic cannabinoid toxicity and to avoid toxic drug-drug interactions, the drug-metabolizing enzymes of the derivatives AM-2201 and EAM-2201 were characterized using major human cytochrome P450 (CYP) enzymes [33, 34]. Conversion of AM-2201 to 4-hydroxyfluoropentyl-AM-2201, AM-2201 pentanoic acid, and 5-hydroxypentyl-AM-2201 was catalyzed by CYP1A2, CYP2C9, and CYP2C19, respectively [33]. Twenty-eight metabolites were formed from EAM-2201 by CYP1A2, CYP2B6, CYP2C8/9/19, CYP2D6, and CYP3A4 [34]; however, there are no reports of the CYP enzymes responsible for MAM-2201 metabolism.
In the present study, 19 MAM-2201 metabolites were identified for the first time after human liver microsome incubation of MAM-2201 in the presence of NADPH using liquid chromatography–Orbitrap mass spectrometry (LC-Orbitrap MS) and an analytical method for the simultaneous determination of 19 MAM-2201 metabolites in human liver microsome mixtures was described using LC-tandem mass spectrometric (LC-MS/MS). To predict individual differences in MAM-2201 pharmacokinetics and toxicity, the CYP enzymes involved in MAM-2201 metabolism were also characterized using enzyme kinetics with human cDNA-expressed CYP enzymes and immunoinhibition assays with human CYP-selective antibodies in human liver microsomes.
Materials and methods
Reagents
MAM-2201 and its metabolites such as N-(4-hydroxyfluoropentyl)-MAM-2201, N-(5-hydroxypentyl)-MAM-2201, and MAM-2201 pentanoic acid were purchased from Cayman Chemical Company (Ann Arbor, MI, USA). Reduced nicotinamide adenine dinucleotide phosphate (NADPH) was obtained from Sigma-Aldrich Co. (St. Louis, MO, USA). UltraPool human liver microsomes (150 donors), human cDNA-expressed CYP enzymes (CYPs 1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, and 3A4), and human-specific antibodies for the immunoinhibition of human CYPs (anti-CYP1A2, anti-CYP2B6, anti-CYP2C8, anti-CYP2C19, anti-CYP2D6, and anti-CYP3A4) were obtained from Corning Life Sciences (Woburn, MA, USA). Homoegonol was obtained from Toronto Research Chemicals (Toronto, Ontario, Canada). Methanol and water (LC-MS grade) were from Fisher Scientific (Fair Lawn, NJ, USA). Other chemicals used were of the highest quality available.
Identification of MAM-2201 metabolites in human liver microsomes
Incubation mixtures containing 240 μL of 50 mM potassium phosphate buffer (pH 7.4), 12 μL of 250 mM magnesium chloride, 30 μL of human liver microsomes (3 mg/mL total protein), 15 μL of 1 mM NADPH, and 3 μL of 2 mM MAM-2201 (20 μM final) were prepared in a total incubation volume of 300 μL. Control incubations were conducted under the same conditions with MAM-2201 in the absence of NADPH. The reaction mixtures were incubated at 37 °C for 20 min in a shaking water bath, and the reaction was terminated by adding 300 μL of ice-cold methanol. The reaction mixture was then centrifuged (10,000×g, 4 min, 4 °C), and 500 μL of supernatant was evaporated under an N2 gas stream. The residue was dissolved in 100 μL of 40% methanol, and a 5 μL aliquot was injected into the LC-Orbitrap MS system.
Metabolism of MAM-2201 by human cDNA-expressed CYP enzymes
Screening of the major human CYP enzymes responsible for the metabolism of MAM-2201 was performed with reaction mixtures containing 10 μL of nine different human cDNA-expressed CYP enzymes (CYPs 1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, and 3A4; 4 pmol), 1 μL of 250 μM MAM-2201 (2.5 μM final), 4 μL of 250 mM magnesium chloride, and 80 μL of 50 mM potassium phosphate buffer (pH 7.4). The reaction was initiated by adding 5 μL of 1 mM NADPH, and the mixtures (final volume, 100 μL) were incubated in triplicate for 20 min at 37 °C in a shaking water bath. The reaction was stopped by adding 100 μL of ice-cold methanol containing internal standard (homoegonol, 70 ng/mL). The mixtures were centrifuged (10,000×g, 4 min, 4 °C), and 150 μL of supernatant was evaporated under an N2 gas stream. The residue was dissolved in 50 μL of 40% methanol, and an aliquot (5 μL) was injected into the LC-MS/MS system.
For the enzyme kinetic experiments, 1 μL of MAM-2201 at various concentrations (final concentrations of 0.125 to 25 μM; final acetonitrile concentration not exceeding 1.0%, v/v) was incubated with 10 μL of human cDNA-expressed CYP enzymes (CYPs 1A2, 2B6, 2C8, 2C9, 2C19, 2D6, and 3A4; 2 pmol), 5 μL of 1 mM NADPH, 4 μL of 250 mM magnesium chloride, and 80 μL of 50 mM potassium phosphate buffer (pH 7.4) for 15 min at 37 °C in a shaking water bath. After addition of 100 μL of ice-cold methanol containing homoegonol (70 ng/mL), the mixture was centrifuged (10,000×g, 4 min, 4 °C) and 150 μL of supernatant was evaporated under an N2 gas stream. The residue was dissolved in 50 μL of 40% methanol, and an aliquot (5 μL) was injected into the LC-MS/MS system.
Immunoinhibition of MAM-2201 metabolism with CYP antibodies in human liver microsomes
Immunoinhibition experiments were performed by incubating ultrapool human liver microsomes with various amounts of human CYP-selective antibodies including anti-CYP1A2, anti-CYP2B6, anti-CYP2C8, anti-CYP2C19, anti-CYP2D6, and anti-CYP3A4 for 20 min on ice, and then, the reaction was initiated by the addition of 50 mM potassium phosphate buffer (pH 7.4), MAM-2201 (final concentration of 2.5 μM), 250 mM magnesium chloride, and 1 mM NADPH. Control incubations were performed using liver microsomes and 25 mM Tris buffer instead of a CYP-selective antibody.
LC-MS analysis
A Q-Exactive Orbitrap mass spectrometer equipped with an Accela UPLC system (Thermo Fisher Scientific Inc., Waltham, MA, USA) was used for the separation and identification of MAM-2201 and its metabolites. MAM-2201 and its metabolites were separated on a Halo C18 column (2.7 μm, 2.1 mm i.d. × 100 mm; Advanced Materials Technology, Wilmington, DE, USA) using a gradient elution of 0.1% formic acid in 5% methanol (mobile phase A) and 0.1% formic acid in 95% methanol (mobile phase B) at a flow rate of 0.3 mL/min: 40% mobile phase B for 0.5 min, 40 to 60% mobile phase B for 14.5 min, 60 to 95% mobile phase B for 9 min, 95% mobile phase B for 3 min, 95 to 40% mobile phase B for 0.1 min, and 40% mobile phase B for 2.9 min. The column and autosampler were maintained at 40 °C and 4 °C, respectively.
The mass spectra for MAM-2201 and its metabolites were obtained with an electrospray ionization source (ESI) in positive mode. The ESI source settings for MAM-2201 and its metabolites were optimized as follows: sheath gas flow rate, 35 (arbitrary units); auxiliary gas flow rate, 10 (arbitrary units); spray voltage, 4 kV; and auxiliary gas heater temperature, 300 °C. Data were acquired using the Xcalibur software (Thermo Fisher Scientific Inc.). Full MS scan data were obtained from m/z 100 to 600 at a resolution of 70,000, and data-dependent MS/MS spectra were acquired at a resolution of 35,000 with a normalized collision energy of 40 eV. The proposed compound structures were determined using the Mass Frontier software (ver. 6.0; HighChem Ltd., Slovakia) with product ions of MAM-2201 and its metabolites.
The quantification of each metabolite was performed using an Agilent 6460 triple quadrupole mass spectrometer coupled with Agilent 1290 Infinity LC (Agilent Technologies, Santa Clara, CA, USA). Chromatographic separation was performed as described above, and the ESI source for MAM-2201 and its metabolites was operated in the positive mode setting, as follows: gas temperature, 350 °C; gas flow, 10 L/min; nebulizer gas pressure, 35 psi; sheath gas temperature, 350 °C; sheath gas flow, 11 L/min; capillary voltage, 3500 V; and nozzle voltage, 0 V. Selected reaction monitoring (SRM) of the analytes was performed using N2 gas as the collision gas set to 32, and SRM mode was applied using the mass transition of each protonated molecular ion to the most abundant product ion (the first diagnostic product ions in Table 1). The Mass Hunter software (Agilent Technologies) was used for the LC-MS/MS system control and data processing.
Calibration standards for MAM-2201 and the three metabolites, N-(4-hydroxyfluoropentyl)-MAM-2201, N-(5-hydroxypentyl)-MAM-2201, and MAM-2201 pentanoic acid, were prepared at seven different concentrations in a blank microsomal incubation mixture. The calibration curves for MAM-2201 and three metabolites were linear over 0.005–25 pmol. The coefficients of variation (precision) and accuracy of the quality control samples at 0.005, 0.015, 0.15, and 15 pmol of MAM-2201 and three metabolites in blank microsomal incubation mixture were ≤11.5 and 97.5 to 101.6%, respectively. Authentic standards of 16 metabolites (except M2, M13, and M18) were not available; thus, the quantification of these 16 metabolites was performed using the standard curve of N-(4-hydroxypentyl)-MAM-2201. Consequently, a limitation exists in the accurate interpretation of the enzyme kinetic data for 16 metabolites because their concentration was calculated from the calibration curve for N-(4-hydroxypentyl)-MAM-2201.
Data analysis
Results are presented as the average of three determinations obtained from human liver microsomes and human cDNA-expressed CYP enzymes. The apparent kinetic parameters (K m and V max) were determined using the Michaelis–Menten equation [V = V max⋅S/(K m + S)], the Hill equation [V = V max⋅Sn/(K m n + Sn)], or the substrate-inhibition equation [V = V max/(1 + K m/S) + S/K i)] using the Enzyme Kinetics program (ver. 1.3; Systat Software Inc.). In the equations above, V is the velocity of the reaction at substrate concentration [S], V max is the maximum velocity, K m is the substrate concentration at which the reaction velocity is 50% of V max, and n is the Hill coefficient. The intrinsic clearance (Cl int) of in vitro incubation was calculated as V max/K m .
The relative contributions of CYP isoforms to the formation of metabolites (M1-M19) from MAM-2201 in human liver microsomes were determined using the relative activity factor (RAF) [35]. The RAF incorporates the hepatic abundance of each CYP isoform and the differences in activity per unit enzyme between human cDNA-expressed CYPs and human liver microsomal CYPs [35]. The RAF for each CYP isoform was defined as V max,HLM /V max,CYP (pmol CYP/mg protein), where V max,HLM is the maximum velocity of the CYP probe of interest in human liver microsomes (pmol/min/mg protein) and V max,CYP is the maximum velocity of the CYP probe of interest in human cDNA-expressed CYP enzymes (pmol/min/nmol CYP). The velocity for MAM-2201 metabolites catalyzed by multiple CYPs can be described as a linear combination of velocity functions for each CYP isoform (Velocity CYPi ) weighted for the RAFs: \( {Velocity}_{HLM}=\sum_{i=1}^n{Velocity}_{CYPi}\times {RAF}_{CYPi} \). The relative contribution of each CYP isoform to MAM-2201 metabolism activity in human liver microsomes is as follows: contribution (%) = (Velocity CYPi /Velocity HLM ) × 100.
Results and discussion
Identification of MAM-2201 metabolites in human liver microsomes
Nineteen metabolites (M1-M19) were identified by LC-Orbitrap MS analysis after incubation of MAM-2201 with human liver microsomes in the presence of NADPH (Fig. 1). The exact masses of [M + H]+ ion and diagnostic product ions and the retention times of MAM-2201 and its possible metabolites (M1–M19) are listed in Table 1. Diagnostic product ions are listed in order of relative abundance.
Because many metabolites of MAM-2201 have the same [M + H]+ ions including M1–M7 at m/z 390.1864, M8–M11 at m/z 406.1813, and M17–M18 at m/z 302.1176 (Table 1), chromatographic separation of the metabolites was necessary to unambiguously identify the structure of the metabolites. MAM-2201 and its 19 metabolites were well separated on a Halo C18 column using a gradient elution of methanol and 0.1% formic acid (Fig. 1, Table 1).
The MS/MS spectrum of MAM-2201 showed four characteristic product ions at m/z 169.0647 [(4-methylnaphthalen-1-yl)(oxo)methylium ion], m/z 141.0698 (4-methylnaphthalen-1-ylium ion), m/z 232.1129 (loss of 4-methyl-1-naphthalenyl moiety from [M + H]+ ion of MAM-2201), and m/z 144.0443 (loss of 5-fluoropentyl moiety from m/z 232.1129) (Fig. 2), serving as diagnostic product ions for the identification of MAM-2201 metabolites.
M1–M7 showed the [M + H]+ ion at m/z 390.1864, 16 amu more than the [M + H]+ ion of MAM-2201, indicating hydroxylation of MAM-2201 (Fig. 1). On the basis of MS/MS spectra, seven hydroxy-MAM-2201 (M1–M7) were classified into three categories due to hydroxylation positions at the methylnaphthalene, indole, and pentyl moieties. M1, M5, and M6 showed characteristic product ions at m/z 185.0595 [hydroxy-(4-methylnaphthalen-1-yl)(oxo)methylium ion], m/z 157.0646 (hydroxy-4-methylnaphthalen-1-ylium ion), m/z 232.1129, and m/z 144.0443, suggesting that hydroxylation in M1, M5, and M6 occurred at the methylnaphthalene moiety, but the exact hydroxylation positions were not identified (Fig. 2). M2 and M3 showed product ions at m/z 248.1078 (loss of 4-methyl-1-naphthalenyl moiety from [M + H]+ ions of M2 and M3), m/z 144.0443 (loss of hydroxy-5-fluoropentyl moiety from m/z 248.1078), m/z 169.0647, and m/z 141.0698, indicating that hydroxylation occurred in the 5-fluoropentyl group of MAM-2201, but the exact hydroxylation position of M3 was not identified (Fig. 2). M2 was identified as N-(4-hydroxyfluoropentyl)-MAM-2201 by comparison with the retention time and the product scan spectrum of the corresponding authentic standard. M4 and M7 produced characteristic product ions at m/z 248.1078 (loss of 4-methyl-1-naphthalenyl moiety from [M + H]+ ions of M4 and M7), m/z 160.0392 (loss of 5-fluoropentyl moiety from m/z 248.1078), m/z 169.0647, and m/z 141.0698, indicating that hydroxylation of M4 and M7 occurred at the indole moiety, but the accurate hydroxylation positions were not identified (Fig. 2).
Four MAM-2001 metabolites (M8–M11) showed the [M + H]+ ion at m/z 406.1813, 32 amu higher than MAM-2201, indicating that M8–M11 may be dihydroxy-MAM-2201 (Fig. 1). M8 showed product ions at m/z 185.0596, m/z 248.1078, and m/z 160.0391, indicating that M8 was dihydroxylated at both the methylnaphthalene and indole moieties (Fig. 2). M9 and M10 showed product ions at m/z 185.0596, m/z 157.0647, m/z 248.1078, and m/z 144.0443 (Fig. 2), suggesting that M9 and M10 were dihydroxylated at both the methylnaphthalene and 5-fluoropentyl moieties. M9 was identified as the major metabolite after incubation of N-(4-hydroxyfluoropentyl)-MAM-2201 (M2) with human liver microsomes in the presence of NADPH, so M9 was tentatively identified as hydroxy-M2, but the exact hydroxylation site at the methylnaphthalene moiety could not be identified (Fig. 2). M11 produced product ions at m/z 232.1129, m/z 201.0544 [dihydroxy-(4-methylnaphthalen-1-yl)(oxo)methylium ion], m/z 183.0439 (a loss of H2O from m/z 201.0544), m/z 155.0490 (a loss of CO from m/z 183.0439), and m/z 144.0444, indicating dihydroxylation of MAM-2201 at the methylnaphthalene moiety (Fig. 2).
M12 showed the [M + H]+ ion at m/z 408.1975, which is 34 amu higher than MAM-2201 (Fig. 1), and generated product ions at m/z 390.1860 (loss of H2O from [M + H]+ ions of M12), m/z 232.1129, m/z 203.0700, m/z 185.0596 (a loss of H2O from m/z 203.0700), m/z 157.0647 (a loss of CO from m/z 185.0596), and m/z 144.0442, suggesting the formation of a dihydrodiol metabolite (Fig. 3). These results indicate that M12 is dihydrodiol-MAM-2201 via epoxide formation at the naphthalene ring and subsequent hydrolysis of the epoxide moiety. The hydroxyl positions of M12 were not accurately identified.
Product scan spectra of dihydrodiol-MAM-2201 (M12), N-(5-hydroxypentyl)-MAM-2201 (M13), hydroxy-M13 (M14), N-dealkyl-MAM-2201 (M15), hydroxy-M15 (M16 and M17), MAM-2201 pentanoic acid (M18), and hydroxy-M18 (M19) formed after human liver microsome incubation of MAM-2201 in the presence of NADPH for 20 min at 37 °C using an LC-Q-Exactive mass spectrometer in a normalized collision energy at 40 eV
M13 showed the [M + H]+ ion at m/z 372.1958, which was 2 amu lower than the MAM-2201 [M + H]+ ion (Fig. 1). M13 produced product ions at m/z 169.0647, m/z 141.0697, m/z 144.0442, and m/z 230.1172 (loss of m/z 141.0697 from [M + H]+ ion of M13), indicating that M13 was formed from MAM-2201 via oxidative defluorination (Fig. 3). M13 was identified as N-(5-hydroxypentyl)-MAM-2201 by comparison with the retention time and the product scan spectrum of the corresponding authentic standard. Human liver microsome incubation of M13 in the presence of NADPH resulted in the formation of M14, M15, M18, and M19.
M14 showed the [M + H]+ ion at m/z 388.1907, which was 16 amu higher than the [M + H]+ ion of M13 (Fig. 1), and produced product ions at m/z 185.0595, m/z 230.1173, m/z 157.0646, and m/z 144.0443, indicating additional hydroxylation of M13 at the methylnaphthalene moiety (Fig. 3). M14 was also identified as the major metabolite after human liver microsome incubation with M13 and NADPH, supporting that M14 was tentatively identified as hydroxy-M13.
M15 showed the [M + H]+ ion at m/z 286.1226, which was 88 amu lower than the [M + H]+ ion of MAM-2201, and the product ions at m/z 144.0443, m/z 169.0647, m/z 141.0697, and m/z 116.0496 (Fig. 1, Table 1), indicating that M15 was N-dealkyl-MAM-2201 due to the loss of the 5-fluoropentyl group (Fig. 3).
M16 and M17 showed the [M + H]+ ion at m/z 302.1176, which was 16 amu higher than the [M + H]+ ion of M15, indicating hydroxylation of M15 (Fig. 1, Table 1). M16 showed product ions at m/z 185.0596, m/z 157.0647, m/z 144.0443, and m/z 116.0497, indicating hydroxylation of the methylnaphthalene moiety in M15 (Fig. 3). M17 showed product ions at m/z 169.0647, m/z 141.0698, and m/z 160.0392, indicating hydroxylation of the indole moiety in M15 (Fig. 3).
M18 showed the [M + H]+ ion at m/z 386.1751 and product ions at m/z 144.0443, m/z 141.0697, m/z 169.0647, and m/z 244.0965 (Figs. 1 and 3), indicating that M18 was formed via the biotransformation of a fluoropentyl group to pentanoic acid. M18 was identified as MAM-2201 pentanoic acid by comparison with the retention time and the product scan spectrum of the corresponding authentic standard.
M19 showed the [M + H]+ ion at m/z 402.1700, which was 16 amu higher than [M + H]+ ion of M18, and product ions at m/z 144.0442, m/z 185.0595, and m/z 244.0965 (Fig. 1, Table 1), indicating hydroxylation of M18 at the methylnaphthalene moiety (Fig. 3). M19 was also identified as the major metabolite after human liver microsome incubation of MAM-2201 pentanoic acid (M18) in the presence of NADPH, supporting that M19 was tentatively identified as hydroxy-M18, but the exact hydroxylation site at methylnaphthalene moiety could not be determined.
Based on these results, the proposed possible metabolic pathways of MAM-2201 in human liver microsomes are shown in Fig. 4, as follows: monohydroxylation of the methylnaphthalene moiety to M1, M5, and M6; monohydroxylation of the indole moiety to M4 and M7; monohydroxylation of the pentyl moiety to M2 and M3; dihydroxylation of the methylnaphthalene moiety to M11; dihydroxylation of the methylnaphthalene and indole moieties to M8; dihydroxylation of the methylnaphthalene and pentyl moieties to M9 and M10; oxidative defluorination to M13; dihydrodiol formation to M12; N-dealkylation to M15; carboxylation to M18; and hydroxylation of M2, M13, M18, and M15 at the methylnaphthalene moiety to M9, M14, M19, and M16, respectively.
Characterization of CYP enzymes responsible for MAM-2201 metabolism
Reaction phenotyping of the CYP enzymes responsible for MAM-2201 metabolism was performed by metabolism study using human cDNA-expressed CYP enzymes and immunoinhibition study in human liver microsomes. Metabolite screening of MAM-2201 with nine human cDNA-expressed CYP enzymes revealed that CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A4 enzymes were responsible for MAM-2201 metabolism (Table 2). CYP2A6 and CYP2E1 enzymes were not involved in MAM-2201 metabolism.
The formation rates of 19 metabolites (M1-M19) from MAM-2201 versus the MAM-2201 concentration in the presence of NADPH in human liver microsomes and human cDNA-expressed CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A4 enzymes demonstrated a better fit to single enzyme kinetics, Hill, or the substrate-inhibition equation per each metabolite. Enzyme kinetic parameters, such as K m, V max, n, K i, and Cl int values, for the formation of the 19 metabolites from MAM-2201 in human liver microsomes and cDNA-expressed CYPs are summarized in Table 3.
Cl int values for the formation of hydroxy-MAM-2201 (M1), N-(5-hydroxypentyl)-MAM-2201 (M13), and hydroxy-M13 (M14) (19,731, 18,059, and 13,125 nL/min/mg protein, respectively) were higher than Cl int values of the other 16 metabolites (32–1881 nL/min/mg protein) in human liver microsomes. These results indicate that hydroxylation to M1, oxidative defluorination to M13, and hydroxylation of M13 to M14 were the major metabolic pathways of MAM-2201 in human liver microsomes. N-(5-hydroxypentyl)-MAM-2201 (M13) was identified as a major metabolite in human liver microsomes and human urine [28], but hydroxy-MAM-2201 (M1) and hydroxy-M13 (M14) were for the first time identified as major in vitro metabolites of MAM-2201 in this study. From these results, hydroxy-MAM-2201 (M1) and hydroxy-M13 (M14) can be used with N-(5-hydroxypentyl)-MAM-2201 (M13), N-(4-hydroxyfluoropentyl)-MAM-2201 (M2), and MAM-2201 pentanoic acid (M18) as abuse biomarkers of MAM-2201.
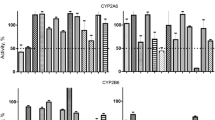
CYP3A4 enzyme was involved in the formation of 19 metabolites from MAM-2201 and played the major role in the formation of hydroxy-MAM2201 (M3-M7), dihydroxy-MAM-2201 (M8-M11), dihydrodiol-MAM-2201 (M12), hydroxy-M13 (M14), dealkyl-MAM-2201 (M15), hydroxy-M15 (M16), MAM-2201 pentanoic acid (M18), and hydroxy-M18 (M19) (Table 3). CYP1A2 enzyme played the major role in the formation of hydroxy-MAM-2201 (M3, M4, M5, M7), dihydroxy-MAM-2201 (M8), dihydrodiol-MAM-2201 (M12), hydroxy-M13 (M14), dealkyl-MAM-2201 (M15), and hydroxy-M15 (M16, M17). CYP2B6 enzyme played the major role in the formation of N-(4-hydroxyfluoropentyl)-MAM-2201 (M2) and hydroxy-MAM-2201 such as M4 and M6 (Table 3). CYP2C8 enzyme played the major role in the formation of N-(4-hydroxyfluoropentyl)-MAM-2201 (M2), hydroxy-MAM-2201 (M3), N-(5-hydroxypentyl)-MAM-2201 (M13), and MAM-2201 pentanoic acid (M18). CYP2C9 enzyme played a prominent role in the formation of hydroxy-MAM-2201 (M1). CYP2C19 and CYP2D6 enzymes showed a little contribution to the formation of the metabolites from MAM-2201 (Table 3).
To further investigate the CYP enzymes responsible for MAM-2201 metabolism, immunoinhibition studies were performed via pretreatment of human liver microsomes with antibodies to CYP1A2, CYP2B6, CYP2C8, CYP2C19, CYP2D6, and CYP3A4 (Fig. 5).
Effects of human-specific CYP antibodies on the metabolism of MAM-2201 to 12 major metabolites. Ultrapool human liver microsomes (0.2 mg protein/mL) were preincubated with anti-CYP1A2 (white circle), anti-CYP2B6 (black circle), anti-CYP2C8 (white triangle), anti-CYP2C19 (black triangle), anti-CYP2D6 (white square), and anti-CYP3A4 (black square). Data shown are the average of duplicate determinations
The relative contributions of CYP2C8, CYP2C9, and CYP3A4 enzymes to the formation of N-(5-hydroxypentyl)-MAM-2201 (M13), a major metabolite of MAM-2201, were 88.1, 5.4, and 2.4%, respectively (Table 3). The CYP2C8 antibody potently inhibited the formation of N-(5-hydroxypentyl)-MAM-2201 (M13) in human liver microsomes by up to 73% (Fig. 5). These results indicate that CYP2C8 plays a more prominent role in the oxidative defluorination of MAM-2201 to N-(5-hydroxypentyl)-MAM-2201 (M13) than CYP2C9 or CYP3A4 in human liver microsomes.
The relative contributions of CYP2C9, CYP3A4, and CYP1A2 to the formation of hydroxy-MAM-2201 (M1), one of the major metabolites, from MAM-2201 were 87.0, 7.3, and 3.8%, respectively (Table 3). The CYP3A4 antibody inhibited the formation of hydroxy-MAM-2201 (M1) in human liver microsomes by up to 30% (Fig. 5). These results indicate that CYP2C9 and CYP3A4 play major roles in the hydroxylation of MAM-2201 to hydroxy-MAM-2201 (M1).
Formation of hydroxy-M13 (M14), one of the major metabolites from MAM-2201, was mediated by CYP3A4 and CYP1A2 on the basis of the relative contributions of CYP3A4 (63.3%) and CYP1A2 (31.8%) (Table 3). M14 was identified as a major metabolite after incubation of M13 with human liver microsomes. The formation of hydroxy-M13 (M14) from MAM-2201 in human liver microsomes was inhibited by CYP3A4 antibody by up to 30%, but was inhibited negligibly by CYP1A2 antibody (Fig. 5). Based on these results, CYP3A4 plays a prominent role in the hydroxylation of M13 to hydroxy-M13 (M14).
Formation of N-(4-hydroxyfluoropentyl)-MAM-2201 (M2) from MAM-2201 was mediated by CYP2B6 and CYP2C8 on the basis of the relative contributions of CYP2B6 (34.4%) and CYP2C8 (55.8%) (Table 3) and the inhibition of M2 formation by CYP2B6 and CYP2C8 antibodies, up to 45 and 60%, respectively (Fig. 5).
Formation of MAM-2201 pentanoic acid (M18) from MAM-2201 was mediated by CYP2C8 and CYP3A4 on the basis of the relative contributions of CYP2C8 (86.7%) and CYP3A4 (11.3%) (Table 3) and the inhibition of M18 formation by CYP2C8 and CYP3A4 antibodies in human liver microsomes, up to 80 and 35%, respectively.
The relative contributions of CYP3A4 to the formation of dihydroxy-MAM-2201 metabolites such as M8, M9, M10, and M11 from MAM-2201 were 58.5, 96.0, 97.1, and 100.0%, respectively (Table 3), and the formation of M8–M11 from MAM-2201 was potently inhibited by CYP3A4 antibody in human liver microsomes. These results indicate that CYP3A4 is responsible for the formation of dihydroxy-MAM-2201 metabolites (M8–M11) from MAM-2201.
The relative contributions of CYP3A4 to the hydroxylation of MAM-2201 to hydroxy-MAM-2201 metabolites such as M3, M5, M6, and M7 were 44.8, 57.9, 90.2, and 36.7%, respectively (Table 3), and the CYP3A4 antibody potently inhibited the formation of M3, M5, M6, and M7 by 65 to 90% in human liver microsomes. CYP3A4 plays a more prominent role in the hydroxylation of MAM-2201 to M3, M5, M6, and M7 than other CYP enzymes. However, CYP1A2 played a major role in the formation of hydroxy-MAM-2201 (M4) from MAM-2201 with a contribution from CYP3A4 and CYP2B6 on the basis of the relative contribution of each CYP enzyme and the immunoinhibition study.
CYP1A2 and CYP3A4 were the major enzymes involved in the formation of dihydrodiol-MAM-2201 (M12) from MAM-2201 on the basis of the relative contributions of CYP1A2 (45.1%) and CYP3A4 (35.4%) (Table 3) and potent inhibition by CYP3A4 and CYP1A2 antibodies (Fig. 5).
The major enzymes responsible for the formation of dealkyl-MAM-2201 (M15) from MAM-2201 were CYP1A2 and CYP3A4 based on the high contributions of CYP1A2 (66.8%) and CYP3A4 (25.1%) (Table 3) and immunoinhibition by CYP3A4 and CYP1A2 antibodies, respectively (Fig. 5).
The major enzymes involved in the formation of the 19 MAM-2201 metabolites were CYP3A4, CYP1A2, CYP2B6, CYP2C8, and CYP2C9. These polymorphic enzymes were responsible for significant inter-individual differences in MAM-2201 pharmacokinetics [36–40]. CYP3A4 enzyme, which is abundantly expressed in the gastrointestinal tract and liver, was the prominent enzyme responsible for the formation of 19 metabolites from MAM-2201. Therefore, MAM-2201 is extensively metabolized by first-pass metabolism in humans, supporting the very low concentrations of MAM-2201 found in urine and plasma samples of MAM-2201 abusers [20, 21, 25, 27, 28, 31].
Conclusions
On the basis of the exact mass of [M + H]+ ion and the diagnostic product ions, 19 metabolites of MAM-2201 from human liver microsomes were identified, including 7 hydroxy-MAM-2201 (M1–M7), 3 dihydroxy-MAM-2201 (M8–M11), dihydrodiol-MAM-2201 (M12), N-(5-hydroxypentyl)-MAM-2201 (M13), hydroxy-M13 (M14), N-dealkyl-MAM-2201 (M15), 2 hydroxy-M15 (M16 and M17), MAM-2201 N-pentanoic acid (M18), and hydroxy-M18 (M19) (Table 1, Fig. 4). Multiple CYP enzymes such as CYP3A4, CYP1A2, CYP2B6, CYP2C8/9/19, and CYP2D6 were involved in MAM-2201 metabolism. CYP3A4 played a more prominent role, with moderate contributions of CYP1A2, CYP2B6, CYP2C8, and CYP2C9 in MAM-2201 metabolism. The metabolic pathways of MAM-2201 may be useful in the development of analytical methods for monitoring MAM-2201 abuse in biological samples such as urine and plasma. Such pathways will also help to predict the individual differences in MAM-2201 pharmacokinetics and toxicity.
References
Auwarter V, Dresen S, Weinmann W, Muller M, Putz M, Ferreiros N. ‘Spice’ and other herbal blends: harmless incense or cannabinoid designer drugs? J Mass Spectrom. 2009;44:832–7.
Uchiyama N, Kikura-Hanajiri R, Goda Y. Identification of a novel cannabimimetic phenylacetylindole, cannabipiperidiethanone, as a designer drug in a herbal product and its affinity for cannabinoid CB(1) and CB(2) receptors. Chem Pharm Bull. 2011;59:1203–5.
Uchiyama N, Kikura-Hanajiri R, Kawahara N, Haishima Y, Goda Y. Identification of a cannabinoid analog as a new type of designer drug in a herbal product. Chem Pharm Bull. 2009;57:439–41.
Compton DR, Rice KC, De Costa BR, Razdan RK, Melvin LS, Johnson MR, et al. Cannabinoid structure-activity relationships: correlation of receptor binding and in vivo activities. J Pharmacol Exp Ther. 1993;265:218–26.
Melvin LS, Milne GM, Johnson MR, Subramaniam B, Wilken GH, Howlett AC. Structure-activity relationships for cannabinoid receptor-binding and analgesic activity: studies of bicyclic cannabinoid analogs. Mol Pharmacol. 1993;44:1008–15.
Eissenstat MA, Bell MR, D’Ambra TE, Alexander EJ, Daum SJ, Ackerman JH, et al. Aminoalkylindoles: structure-activity relationships of novel cannabinoid mimetics. J Med Chem. 1995;38:3094–105.
Wiley JL, Compton DR, Dai D, Lainton JA, Phillips M, Huffman JW, et al. Structure-activity relationships of indole- and pyrrole-derived cannabinoids. J Pharmacol Exp Ther. 1998;285:995–1004.
Page D, Balaux E, Boisvert L, Liu Z, Milburn C, Tremblay M, et al. Novel benzimidazole derivatives as selective CB2 agonists. Bioorg Med Chem Lett. 2008;18:3695–700.
Nakajima J, Takahashi M, Nonaka R, Seto T, Suzuki J, Yoshida M, et al. Identification and quantitation of a benzoylindole (2-methoxyphenyl)(1-pentyl-1H-indol-3-yl)methanone and a naphthoylindole 1-(5-fluoropentyl-1H-indol-3-yl)-(naphthalene-1-yl)methanone (AM-2201) found in illegal products obtained via the Internet and their cannabimimetic effects evaluated by in vitro [35S]GTPγS binding assays. Forensic Toxicol. 2011;29:132–41.
Uchiyama N, Kawamura M, Kikura-Hanajiri R, Goda Y. URB-754: a new class of designer drug and 12 synthetic cannabinoids detected in illegal products. Forensic Sci Int. 2013;227:21–32.
Wohlfarth A, Pang S, Zhu M, Gandhi AS, Scheidweiler KB, Liu HF, et al. First metabolic profile of XLR-11, a novel synthetic cannabinoid, obtained by using human hepatocytes and high-resolution mass spectrometry. Clin Chem. 2013;59:1638–48.
Uchiyama N, Matsuda S, Kawamura M, Kikura-Hanajiri R, Goda Y. Two new-type cannabimimetic quinolinyl carboxylates, QUPIC and QUCHIC, two new cannabimimetic carboxamide derivatives, ADB-FUBINACA and ADBICA, and five synthetic cannabinoids detected with a thiophene derivative α-PVT and an opioid receptor agonist AH-7921 identified in illegal products. Forensic Toxicol. 2013;31:223–40.
Shevyrin V, Melkozerov V, Nevero A, Eltsov O, Shafran Y. Analytical characterization of some synthetic cannabinoids, derivatives of indole-3-carboxylic acid. Forensic Sci Int. 2013;232:1–10.
Chung H, Choi H, Heo S, Kim E, Lee J. Synthetic cannabinoids abused in South Korea: drug identifications by the National Forensic Service from 2009 to June 2013. Forensic Toxicol. 2013;32:82–8.
Helander A, Backberg M, Hulten P, Al-Saffar Y, Beck O. Detection of new psychoactive substance use among emergency room patients: results from the Swedish STRIDA project. Forensic Sci Int. 2014;243:23–9.
Seely KA, Lapoint J, Moran JH, Fattore L. Spice drugs are more than harmless herbal blends: a review of the pharmacology and toxicology of synthetic cannabinoids. Prog Neuro-Psychopharmacol Biol Psychiatry. 2012;39:234–43.
Andreeva-Gateva PA, Nankova VH, Angelova VT, Gatev TN. Synthetic cannabimimetics in Bulgaria 2010–2013. Drug Alcohol Depend. 2015;157:200–4.
Langer N, Lindigkeit R, Schiebel HM, Ernst L, Beuerle T. Identification and quantification of synthetic cannabinoids in ‘spice-like’ herbal mixtures: a snapshot of the German situation in the autumn of 2012. Drug Test Anal. 2014;6:59–71.
Musshoff F, Madea B, Kernbach-Wighton G, Bicker W, Kneisel S, Hutter M, et al. Driving under the influence of synthetic cannabinoids (“Spice”): a case series. Int J Legal Med. 2014;128:59–64.
Lonati D, Buscaglia E, Papa P, Valli A, Coccini T, Giampreti A, et al. MAM-2201 (analytically confirmed) intoxication after “Synthacaine” consumption. Ann Emerg Med. 2014;64:629–32.
Derungs A, Schwaninger AE, Mansella G, Bingisser R, Kraemer T, Liechti ME. Symptoms, toxicities, and analytical results for a patient after smoking herbs containing the novel synthetic cannabinoid MAM-2201. Forensic Toxicol. 2012;31:164–71.
Irie T, Kikura-Hanajiri R, Usami M, Uchiyama N, Goda Y, Sekino Y. MAM-2201, a synthetic cannabinoid drug of abuse, suppresses the synaptic input to cerebellar Purkinje cells via activation of presynaptic CB1 receptors. Neuropharmacology. 2015;95:479–91.
Zaitsu K, Hayashi Y, Suzuki K, Nakayama H, Hattori N, Takahara R, et al. Metabolome disruption of the rat cerebrum induced by the acute toxic effects of the synthetic cannabinoid MAM-2201. Life Sci. 2015;137:49–55.
Tomiyama K, Funada M. Cytotoxicity of synthetic cannabinoids on primary neuronal cells of the forebrain: the involvement of cannabinoid CB1 receptors and apoptotic cell death. Toxicol Appl Pharmacol. 2014;274:17–23.
Saito T, Namera A, Miura N, Ohta S, Miyazaki S, Osawa M, et al. A fatal case of MAM-2201 poisoning. Forensic Toxicol. 2013;31:333–7.
Kronstrand R, Roman M, Andersson M, Eklund A. Toxicological findings of synthetic cannabinoids in recreational users. J Anal Toxicol. 2013;37:534–41.
Kim J, Park Y, Park M, Kim E, Yang W, Baeck S, et al. Simultaneous determination of five naphthoylindole-based synthetic cannabinoids and metabolites and their deposition in human and rat hair. J Pharm Biomed Anal. 2015;102:162–75.
Jang M, Shin I, Yang W, Chang H, Yoo HH, Lee J, et al. Determination of major metabolites of MAM-2201 and JWH-122 in in vitro and in vivo studies to distinguish their intake. Forensic Sci Int. 2014;244:85–91.
Kronstrand R, Brinkhagen L, Birath-Karlsson C, Roman M, Josefsson M. LC-QTOF-MS as a superior strategy to immunoassay for the comprehensive analysis of synthetic cannabinoids in urine. Anal Bioanal Chem. 2014;406:3599–609.
Scheidweiler KB, Jarvis MJ, Huestis MA. Nontargeted SWATH acquisition for identifying 47 synthetic cannabinoid metabolites in human urine by liquid chromatography-high-resolution tandem mass spectrometry. Anal Bioanal Chem. 2015;407:883–97.
Zaitsu K, Nakayama H, Yamanaka M, Hisatsune K, Taki K, Asano T, et al. High-resolution mass spectrometric determination of the synthetic cannabinoids MAM-2201, AM-2201, AM-2232, and their metabolites in postmortem plasma and urine by LC/Q-TOFMS. Int J Legal Med. 2015;129:1233–45.
Berg T, Kaur L, Risnes A, Havig SM, Karinen R. Determination of a selection of synthetic cannabinoids and metabolites in urine by UHPSFC-MS/MS and by UHPLC-MS/MS. Drug Test Anal. 2015. doi:10.1002/dta.1844.
Chimalakonda KC, Seely KA, Bratton SM, Brents LK, Moran CL, Endres GW, et al. Cytochrome P450-mediated oxidative metabolism of abused synthetic cannabinoids found in K2/Spice: identification of novel cannabinoid receptor ligands. Drug Metab Dispos. 2012;40:2174–84.
Kim JH, Kim HS, Kong TY, Lee JY, Kim JY, In MK, et al. In vitro metabolism of a novel synthetic cannabinoid, EAM-2201, in human liver microsomes and human recombinant cytochrome P450s. J Pharm Biomed Anal. 2016;119:50–8.
Crespi CL. Xenobiotic-metabolizing human cells as tools for pharmacological and toxicological research. In: Bernard T, Urs AM, editors. Advances in drug research, vol. 26. London: Academic; 1995. p. 179–235.
Werk AN, Cascorbi I. Functional gene variants of CYP3A4. Clin Pharmacol Ther. 2014;96:340–8.
Chaudhry SR, Muhammad S, Eidens M, Klemm M, Khan D, Efferth T, et al. Pharmacogenetic prediction of individual variability in drug response based on CYP2D6, CYP2C9 and CYP2C19 genetic polymorphisms. Curr Drug Metab. 2014;15:711–8.
Holstein A, Beil W, Kovacs P. CYP2C metabolism of oral antidiabetic drugs-impact on pharmacokinetics, drug interactions and pharmacogenetic aspects. Expert Opin Drug Metab Toxicol. 2012;8:1549–63.
Zanger UM, Klein K. Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front Genet. 2013;4:24.
McGraw J, Waller D. Cytochrome P450 variations in different ethnic populations. Expert Opin Drug Metab Toxicol. 2012;8:371–82.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2015M3A9E1028325 and NRF-2014R1A2A2A01002582).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The studies have been approved by the appropriate ethic committee and have been performed in accordance with the ethical standards.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Tae Yeon Kong and Ju-Hyun Kim contributed equally to this work.
Rights and permissions
About this article
Cite this article
Kong, T.Y., Kim, JH., Choi, W.G. et al. Metabolic characterization of (1-(5-fluoropentyl)-1H-indol-3-yl)(4-methyl-1-naphthalenyl)-methanone (MAM-2201) using human liver microsomes and cDNA-overexpressed cytochrome P450 enzymes. Anal Bioanal Chem 409, 1667–1680 (2017). https://doi.org/10.1007/s00216-016-0113-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-0113-9