Abstract
The proline-glutamic acid and proline-proline-glutamic acid (PE/PPE) family of proteins is widespread in pathogenic mycobacteria and plays different roles in mycobacterial physiology. While several PE/PPE family proteins have been studied, the exact function of most PE/PPE proteins in the physiology of Mycobacterium tuberculosis (Mtb) remains unknown. PE_PGRS47 belongs to the PE/PPE family of proteins reported to help Mtb evade protective host immune responses. In this study, we demonstrate a novel role of PE_PGRS47. Heterologous expression of the pe_pgrs47 gene in a non-pathogenic Mycobacterium smegmatis, intrinsically deficient of PE_PGRS protein, exhibits modulated colony morphology and cell wall lipid profile leading to a marked susceptibility to multiple antibiotics and environmental stressors. Using ethidium bromide/Nile red uptake assays, Mycobacterium smegmatis expressing PE_PGRS47 showed higher cell wall permeability than the control strain. Overall, these data suggested that PE_PGRS47 is cell surface exposed and influences cell wall integrity and the formation of mycobacterial colonies, ultimately potentiating the efficacy of lethal stresses against mycobacteria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tuberculosis (TB), attributed to Mycobacterium tuberculosis (Mtb) infection, remains a significant threat to public health worldwide. Although intense chemotherapy has been used as a TB treatment for decades, this terrible epidemic still claims the lives of ~ 1.4 million, and there were ~ 10.6 million new cases in 2021 (WHO 2022). One reason for the success of Mtb is its remarkable capability against diverse stresses within macrophages, enabling the bacillus to persist for long periods (Dong et al. 2012). Understanding the pathogen's survival strategies is critical for developing better TB vaccines and drugs.
Sequence analysis of the complete genomic DNA of Mtb led to the identification of a unique multigenic family of proline-glutamic acid and proline-proline-glutamic acid proteins (PE/PPE); this family consists of the PE, PPE, and PE_PGRS subfamilies and accounts for about ten percent of the coding capacity of the Mtb genome (Cole et al. 1998). The proteins of the PE_PGRS subfamily consist of a highly conserved PE domain and a variable PGRS domain. PE indicates an N-terminal Pro-Glu sequence, and PGRS refers to a polymorphic GC-rich repetitive sequence (Deng et al. 2017). The subfamily of PE_PGRS proteins, mainly restricted to pathogenic mycobacteria (Kohli et al. 2012), has been implicated in Mtb pathogenesis for several decades (Meena 2015). The expression pattern, primary origin, and pathogenic role of a few PE_PGRS subfamily proteins have been well studied and reviewed (Fishbein et al. 2015; Kohli et al. 2012; Li et al. 2019; Meena 2015; Sampson 2011; Tian and Jian-Ping 2010). However, the exact role of most PE_PGRS members in the physiology and pathogenesis of Mtb remains unknown.
PE_PGRS proteins were initially thought to serve similar functions because of their high sequence homology. In contrast, studies on individual PE_PGRS proteins indicated that some PE_PGRS proteins could carry out several unrelated tasks. For example, Deng et al. reported that PE_PGRS41 disturbed the innate immune response by inhibiting macrophage autophagy and apoptosis (Deng et al. 2017). On the other hand, PE_PGRS41 severely impaired bacterial resistance to various environmental stresses and antibiotics by affecting cell wall integrity (Deng et al. 2017). PE_PGRS33 protein, localized on the bacillus cell envelope, participates in cell adhesion and influences mycobacterial cell structures (Delogu et al. 2004; Neyrolles et al. 2011; Ramakrishnan et al. 2000). PE_PGRS33 also enhances the persistence of recombinant Mycobacterium smegmatis (M. smegmatis) within host cells and might be responsible for the induction of host cell necrosis and apoptosis by interacting with Toll-like receptor 2 (Basu et al. 2006; Dheenadhayalan et al. 2006). Furthermore, PE_PGRS33 has been reported to be a secreted protein that targets host mitochondria and causes apoptosis in mouse splenocytes or macrophages (Dheenadhayalan et al. 2006; Vallecillo and Espitia 2009). Previous reports have demonstrated that many other members of the PE/PPE family, such as PE11 (Deng et al. 2015; Rastogi et al. 2017; Singh et al. 2016), PPE44 (Bonanni et al. 2005; Yu et al. 2017), PPE38 (Dong et al. 2012; Wang et al. 2013), and PE_PGRS30 (Chatrath et al. 2011, 2016, 2014; Iantomasi et al. 2012), could also perform several unrelated tasks. These studies reveal the need to explore the exact roles of individual PE_PGRS proteins.
PE_PGRS47 (Rv2741) was predicted to be involved in the Mtb-host interaction in our recent study (Li et al. 2015b). Subsequently, Saini et al. reported that PE_PGRS47 is a crucial virulence factor in manipulating innate and adaptive immune responses in infected macrophages (Saini et al. 2016). In the present study, we explored the function of PE_PGRS47 by overexpressing it in the widely used model strain M. smegmatis. For this purpose, the pe_pgrs47 gene was cloned in a mycobacterial expression plasmid and electroporated into M. smegmatis. We found that PE_PGRS47 is a cell surface-exposed protein and influences cell wall integrity and the formation of mycobacterial colonies, ultimately potentiating the efficacy of lethal stresses against mycobacteria. The present study illustrates for the first time the role of PE_PGRS47 protein on the growth of mycobacteria under stress conditions.
Materials and methods
M. smegmatis culture and transformation
M. smegmatis mc2155 bacteria were grown in Middlebrook 7H9 medium (BD Difco, USA) supplemented with 0.2% (v/v) glycerol, 0.2% (w/v) glucose, and 0.05% (v/v) Tween 80 (Sangon Biotech, China). The culture was allowed to grow to prepare competent cells until the mid-log phase. The culture was centrifuged, washed four times with 10% glycerol, and resuspended in 1/100th of the culture volume in ice-cold 10% glycerol; 400 μl aliquots of the cells were stored at -80 °C until further use. Before the transformation, the cells were thawed on ice, and 1 μg plasmid was added. The mixture of cells and plasmids was incubated on ice for 10 min, transferred to a prechilled cuvette with a 1 mm gap width, and transformed at the setting of 2.5 kV and 1000 Ω (Li et al. 2008). One milliliter of the culture medium was added immediately, and the cells were incubated at 37 °C for 4 h. The transformants were selected on 7H10 agar plates containing 20 μg/ml kanamycin (Sangon Biotech, China).
Construction of recombinant M. smegmatis expressing PE_PGRS47
The full-length pe_pgrs47 gene was PCR amplified from Mtb H37Rv genomic DNA using specific primers (forward primer, 5'-GGGGAATTCATGTCATTTGTGATCGCG-3' and reverse primer, 5'-TTAAAGCTTTCACAGATCCTCTTCAGAGATGAGTTTCTGCTCGCTAGGCAGCAATCCGT-3') by PrimeSTAR HS DNA Polymerase with GC Buffer (TAKARA, Japan). PCR cycle conditions were: initial denaturation 95 °C 3 min, followed by 30 cycles of 98 °C 10 s, annealing 55 °C 5 s, extension 72 °C 100 s; and final extension 72 °C 5 min. The right-size PCR product was digested with EcoRI and HindIII and inserted into the mycobacterial expression vector pMV261 (Alibaud et al. 2011) to generate the recombinant plasmid pMV261-PE_PGRS47 (Supplementary data 1). The recombinant plasmid was confirmed by restriction digestion and DNA sequencing (BGI, China). M. smegmatis underwent electroporation with pMV261-PE_PGRS47 to generate recombinant M. smegmatis (MS-PE_PGRS47), and the transformants were selected on a 7H10 agar plate containing 20 μg/mL kanamycin. M. smegmatis transformed with the pMV261 vector (MS-pMV) was used as a control group.
Expression of PE_PGRS47 in M. smegmatis
The MS-pMV and MS-PE_PGRS47 strains were grown in 7H9 medium supplemented with 0.2% glucose, 0.2% glycerol, and 0.05% Tween 80 until the absorbance at 600 nm reached 0.6–0.8. Bacterial cell pellets were harvested and washed thrice with phosphate-buffered saline with Tween 80 (PBS-T). The resulting pellets were then resuspended in lysis buffer (50 mM Tris–Cl (pH 8.0), 300 mM NaCl, 1 mM PMSF, and 1 mM DTT) and vortexed. Bacterial cells were lysed using an ultrasonic sonicator, and the lysed cells were centrifuged to collect the supernatant. Samples were subjected to SDS-PAGE, and the Myc-tagged PE_PGRS47 protein was detected by the mouse anti-Myc antibody (TIANGEN, China).
Sub-cellular localization of PE_PGRS47 protein
Sub-cellular fractionation was performed as previously described (Li et al. 2022, 2014). In brief, recombinant M. smegmatis strains were grown in 7H9 medium at 37 °C to an OD600 of 0.6–0.8. Bacteria were harvested by centrifugation. The whole cells were sonicated and centrifuged at 3,000 g for 30 min to obtain the whole cell lysates (WCL) from the supernatant. WCL was centrifuged at 27,000 g for 30 min. The pellet from this centrifugation step was considered the cell wall fraction (CW), and the supernatant was supposed to be the cell membrane and cytosolic (CM + Cy) fractions. All centrifugation steps were performed at 4 °C.
Equal amounts of protein from fractionated samples were subjected to Western blot analysis. The Myc-tagged PE_PGRS47 protein was detected by an anti-Myc antibody (1:1000, catalog number: AF0033, Beyotime, China). Native M. smegmatis GroEL, which contains a string of endogenous histidines (Rengarajan et al. 2008), was detected by an anti-His primary antibody (1:1000, catalog number: AB102-01, TIANGEN, China). GroEL served as a cytosol marker protein of M. smegmatis. (Bashiri et al. 2012).
Minimal inhibitory concentration (MIC) of antibiotics
The MIC was determined using the serial two-fold dilution method as described previously (Dong et al. 2020; Palomino et al. 2002; Zeng et al. 2016). The highest antibiotic concentration was prepared, and 200 μl was added to the first row of a 96-well microtiter plate. This sample was serially diluted by half by mixing with an equal volume of bacterial culture in the subsequent rows until the second to the last row. The last row was the control without antibiotics. The M. smegmatis strains were grown in replicates in 7H9 medium to an OD600 of 0.8, and 1% of the original bacteria was inoculated with 100 μl of the prepared culture with or without antibiotics. Following 3 days of incubation at 37 °C, 30 μL of 0.01% resazurin solution was added to each well, then the 96-well plates were cultured 12 h. The color change of each well was visually inspected. The minimum antibiotic concentration at which resazurin remains blue, its concentration was defined as the MIC of an anti-tuberculosis drug.
Survival curves
Mid-exponential phase cultures of MS-pMV and MS-PE_PGRS47 were diluted in 7H9 medium, grown at 37 °C, and treated with antibiotics or sodium dodecyl sulfate (SDS) at the various concentrations as indicated. After the indicated time, the cell survival was estimated by a colony formation assay on MB 7H10 agar containing kanamycin. The percentage of recovered colony-forming units (CFU) was determined relative to the untreated control sample when the antibiotics were added. All experiments were repeated at least three times.
In vitro growth
For in vitro growth curves, cultures were inoculated in triplicate with a starting absorbance (OD600) of ~ 0.02. The OD600 was measured at various time points over a 70–80 h growth period.
Disk diffusion assay
The disk diffusion method was used to qualitatively measure the differences in SDS sensitivity between MS-pMV and MS-PE_PGRS47 (Li et al. 2016a). Mid-exponential phase cultures were used to prepare the cell lawns as previously described (Bauer et al. 1966). An indicated concentration of SDS was spotted on 5.5 mm-diameter Whatman filter disks placed on the bacterial lawn. After 3–4 days of incubation, the diameter of the zone of complete inhibition was determined from a photograph. All experiments were repeated at least three times.
Colony morphology
To examine the colony morphology, we plated the MS-PE_PGRS47 and MS-pMV strains on MB 7H10 plates with or without 0.05% (v/v) Tween 80 and incubated for 5–6 days at 37 °C. A Nikon Digital camera (Japan) was used to take photomicrographs of the colonies.
Ethidium bromide and Nile red uptake assays
Mid-exponential phase cultures of the MS-PE_PGRS47 and MS-pMV strains were centrifuged and washed with PBS containing 0.05% Tween 80. Bacterial cell concentration was adjusted to an absorbance of ~ 0.4 at 600 nm. The bacterial cells were incubated in PBS with 2 μg/mL ethidium bromide and 2 μM Nile red in a 96-well black fluoroplate. The accumulation of these dyes was measured with an excitation of 545 nm and emission of 600 nm for ethidium bromide and an excitation of 540 nm and emission of 630 nm for Nile Red.
Fatty acid analysis
M. smegmatis strains (MS-pMV and MS-PE_PGRS47) were grown at 37 °C in 500 ml Middlebrook 7H9 broth supplemented with 0.5% glycerol, 0.05% Tween 80 and 0.2% glucose until an absorbance of 0.8 at 600 nm was reached. Fatty acids were extracted according to the standard procedure (Lewis et al. 2000). The fatty acid concentrations were determined by GC–MS (Thermo Fisher Trace GC 1310-ISQ LT single quadrupole EI MS, A1-1310 autosampler) with a Thermo TG-5MS capillary column. The detailed analytical procedure has been described previously (Deng et al. 2015). In short, the column temperature program was as follows: initial temperature 80 °C, held for 1 min; 10 °C/min to 200 °C, 5 °C/min to 250 °C and 2 °C/min at 270 °C, respectively. High-purity helium served as the carrier gas in constant-flow mode at a 1.2 mL/min column flow rate. 70 eV was set for the electron impact energy. The fatty acid peaks were identified by comparing the retention times with those of a mixture of standard fatty acid methyl esters.
Statistical analysis
Data were analyzed using Student’s two-tailed t-test. Statistical significance was defined as a P value of 0.05. Error bars represent the standard deviation (SD). GraphPad Prism 5.02 software (GraphPad software) was used for statistical analysis.
Results and discussion
The PE_PGRS47 protein expresses on the cell envelope of recombinant MS-PE_PGRS47 strain
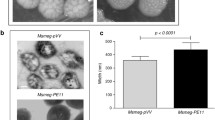
The open reading frame of Mtb pe_pgrs47 is about 1.6 kbp (Fig. 1A) and encodes a protein of approximately 44 kDa. To find out how PE_PGRS47 influences mycobacterial physiology, we expressed the pe_pgrs47 gene in the non-pathogenic M. smegmatis strain (MS-PE_PGRS47), which is widely used as a surrogate bacterium to study Mtb proteins (Li et al. 2015a, 2016b; Singh et al. 2016; Wang et al. 2015). The MS-PE_PGRS47 strain was designed to express a Myc-tagged PE_PGRS47 fusion protein, but the MS-pMV strain harbored the empty vector alone. Immunoblot assay revealed that a protein band (approximately 50 kDa) representing the Myc- PE_PGRS47 protein was detectable in the cell lysates of the MS-PE_PGRS47 strain but not the MS-pMV strain (Fig. 1B). The data demonstrated that the PE_PGRS47 protein of Mtb was expressed in the recombinant MS-PE_PGRS47 strain. We further confirmed that the PE_PGRS47 protein was localized to the cell wall fraction of the MS-PE_PGRS47 strain (Fig. 1C). To evaluate the effect of PE_PGRS47 on the growth of recombinant M. smegmatis, we then compared the growth kinetics of MS-pMV and MS-PE_PGRS47, as overexpression of a heterologous protein is known to cause a metabolic burden on the host cell, sometimes hindering the bacterial growth (Bentley et al. 1990; Daim et al. 2011). As shown in Fig. 1D, these two recombinant bacteria present similar growth kinetics, indicating that the expression of the PE_PGRS47 protein did not influence the growth of recombinant M. smegmatis.
The PE_PGRS47 protein expresses on the cell envelope of recombinant M. smegmatis. A PCR amplification of the pe_pgrs47 gene from M. tuberculosis H37Rv genome. B Western blot with mouse anti-Myc antibody demonstrated the expression of Myc-tagged PE_PGRS47 protein in recombinant M. smegmatis. C Cell fractionation experiments were performed to determine the sub-cellular localization of PE_PGRS47. WCL represents whole cell lysates, CM + Cy represents cytoplasmic membrane and cytoplasm, CW represents cell wall. Cytosolic GroEL was detected as a cytoplasmic control. D The growth of MS-pMV and MS-PE_PGRS47 at 37 °C in Middlebrook 7H9 liquid medium was monitored by determining OD600 at 6 h or 12 h intervals. Experiments were performed three times, and similar results were obtained. Error bars indicate the standard deviation
PE_PGRS47 reduced the survival of MS-PE_PGRS47 following exposure to antibiotics
The Mtb cell wall is an effective permeable barrier to antibiotics (Antony T. Vincent et al. 2018). PE_PGRS47 was a cell surface-exposed protein. To study the sensitivity of MS-PE_PGRS47 against antibiotics, MS-pMV and MS-PE_PGRS47 were treated with various anti-TB drugs, and the MICs of each antibiotic were determined using the broth dilution method (Liu and Nikaido 1999; Ren and Liu 2006). We showed that the MS-PE_PGRS47 and MS-pMV strains had comparable sensitivity to ciprofloxacin (CIP), ofloxacin (OFL), chloramphenicol (CHL), norfloxacin (NOR) and isoniazid (INH). Nevertheless, MS-PE_PGRS47 was hypersensitive to streptomycin (STR), vancomycin (VAN), gentamycin (GEN), erythromycin (ERY), and rifampin (RIP) (Table 1). The MIC of VAN, GEN, and RIP were 2, 2, and 4 µg/ml, respectively, for MS-pMV and 1, 1, and 2 µg/ml, respectively, for MS-PE_PGRS47. Furthermore, MS-PE_PGRS47 was highly susceptible to STR and ERY. MICs of the MS-PE_PGRS47 strains were 6- to eightfold lower than those of the MS-pMV strain for these two antibiotics. The data thus suggest a novel role of PE_PGRS47 protein in antibiotics susceptibility.
To determine whether PE_PGRS47 decreases mycobacterial survival during lethal antibiotic stress, we treated MS-pMV and MS-PE_PGRS47 strains with various concentrations of antibiotics (Fig. 2). The data confirmed that the PE_PGRS47 protein contributes to the lethal activity of GEN, RIP, STR, and ERY. The sensitivity of MS-PE_PGRS47 to INH remained unchanged. VAN, an antibiotic that blocks cell wall synthesis, was tested to determine whether PE_PGRS47 affects the cell wall or the membrane stress in mycobacteria. VAN was reported to bind to the terminal D-ala-D-ala of the pentapeptide chain on the peptidoglycan precursor molecules outside the cell membrane, affecting the cell wall synthesis machinery in mycobacteria (Li et al. 2016a; Zeng et al. 2016). Killing curve experiments revealed that MS-PE_PGRS47 is more susceptible to VAN than MS-pMV, indicating that PE_PGRS47 most likely affects the cell wall integrity.
PE_PGRS47 expression reduced bacterial survival following exposure to antibiotics. The control strain (MS-pMV) and recombinant strain (MS-PE_PGRS47) were diluted in MB 7H9 medium and then treated with streptomycin (0, 0.1, 0.8, and 2.0 μg/ml) (A), vancomycin (0, 5.0, 10.0, and 20.0 μg/ml) (B), gentamycin (0, 2.0, 10.0, and 40.0 μg/ml) (C), erythromycin (0, 4.0, 8.0, and 32.0 μg/ml) (D), isoniazid (0, 4.0, 8.0, and 32.0 μg/ml) (E), and rifampicin (0, 20.0, 50.0, and 80.0 μg/ml) (F) for 6 h, respectively. Then, a ten-fold dilution of the bacteria was spotted on MB 7H10 supplemented with kanamycin, and the MS-pMV and MS-PE_PGRS47 bacteria were counted after 3–4 days of cultivation. Data are means ± SD of technical triplicate from one representative of three or more independent experiments (*p < 0.05, **p < 0.01)
PE_PGRS47 alters colony morphology and increases sensitivity to environmental stresses of recombinant MS-PE_PGRS47 strain
Many PE/PPE family proteins are localized to the cell envelope (Cascioferro et al. 2007; Delogu et al. 2004; Deng et al. 2014; Dona et al. 2013; Neyrolles et al. 2011), suggesting a link between these PE/PPE family proteins and the cell wall structure. We showed above that the MS-PE_PGRS47 strain has a significant defect in cell wall integrity when treated with the cell wall-acting antibiotic VAN. Based on this result, we hypothesized that the PE_PGRS47 protein might affect the cell wall permeability because it harms the integrity of the cell wall.
Colony morphology is a complex phenotype affected by cell–cell interaction (Shi et al. 2011). The bacterial cell wall is responsible for the particular colony morphology and contributes to antibiotic susceptibility and virulence (Kuze and Uchihira 1984; Singh et al. 2016). To determine whether the PE_PGRS47 protein affects the colony morphology, we plated MS-pMV and MS-PE_PGRS47 strains on MB 7H10 agar plates supplemented with or without Tween 80 which is a detergent to prevent clumping of mycobacteria. We observed no difference in colony morphology between these two strains with Tween 80 (Fig. 3A). However, MS-PE_PGRS47 cells were wider and drier than MS-pMV cells without Tween 80 (Fig. 3A).
PE_PGRS47 alters colony morphology and increases sensitivity to environmental stresses of recombinant M. smegmatis. A MS-pMV and MS-PE_PGRS47 were grown at 37 °C on MB 7H10 agar supplemented with or without 0.05% (v/v) Tween 80 (T80). Pictures were taken on day 5. B A mid-exponential phase culture was prepared as described in methods, and 10 μl of SDS, H2O2, and lysosome at the indicated concentrations were spotted on the Whatman disks. Pictures were taken on day 3. C The diameter zone of complete inhibition in B was measured. D The growth of MS-pMV and MS-PE_PGRS47 after treatment with different pH gradients was monitored by determining OD600 at 6 h or 12 h intervals. The mid-log-phase cultures of MS-pMV and MS-PE_PGRS47 were either treated with medium alone or subjected to SDS for 8 h E or H2O2 for 12 h G at the indicated concentrations. The mid-log-phase cultures of MS-pMV and MS-PE_PGRS47 were incubated in MB 7H9 supplemented with 0.075% (w/v) SDS F or 10 mM H2O2 H for the indicated time. Then, the recombinant strains were plated onto 7H10 plates by serially ten-fold dilution, and the bacterial CFUs were counted after 3–4 days of cultivation. Data are means ± SD of technical triplicate from one representative of three or more independent experiments (*p < 0.05, **p < 0.01)
Colony morphology is a subjective measurement (Garces et al. 2010), so we sought more objective methods to evaluate the cell wall architecture in MS-pMV and MS-PE_PGRS47 strains. We assessed the ability of MS-PE_PGRS47 to resist stressors that mimic the intracellular environment. We found that PE_PGRS47 does not influence the resistance to lysozyme and acidic conditions (Fig. 3B-D). However, data shown in Fig. 3B, C reveal that MS-PE_PGRS47 is more susceptible to SDS and H2O2 treatment as compared to the control MS-pMV. The data in Fig. 3E–H further indicates that MS-PE_PGRS47 offered lower resistance against SDS detergent and H2O2 than MS-pMV. In transmission electron microscopy analysis, we observed a thinner envelope of MS-PE_PGRS47 cells compared to MS-pMV (Fig. S2), which is also indicative of alteration in the cell wall composition of M. smegmatis expressing PE_PGRS47. These results indicate that the expression of PE_PGRS47 alters the morphology and cell wall architecture of the M. smegmatis strain.
Our results are consistent with studies of other PE/PPE family members showing that different proteins involved in cell wall biosynthesis have distinct effects on susceptibility and permeability analysis. The susceptibility phenotype might be easily concluded from the gene function in some cases. For example, PE11, which increases the number of glycolipids in the cell wall of bacteria, causes resistance to cell wall stress (Singh et al. 2016). In many cases, however, the link between the PE_PGRS proteins and the resistance to various cell wall stressors is not apparent (Chatrath et al. 2014; Deng et al. 2017; Yu et al. 2017), reflecting our limited understanding of the cell wall assembly in mycobacteria. Recently, Strong et al. (2020) also reported that PE_PGRS47 is present on the cell envelope of mycobacteria when expressed in M. smegmatis (Strong et al. 2020). Interestingly, Strong et al. (2020) observed no increased sensitivity of M. smegmatis expressing PE_PGRS47 to various antibiotics and no alteration of the lipid profile in contrast to our study. This may be due to the different types and positions of the tag used while expressing PE_PGRS47 in M. smegmatis. Different methods used in the two studies may also have contributed to the inconsistency. For example, the sensitivity of GC–MS is higher than that of thin-layer chromatography. In any case, however, further genetic assays should be performed to determine the specific cell wall defect caused by the loss of PE_PGRS47 activity.
PE_PGRS47 expression in MS-PE_PGRS47 strain results in increased cell wall permeability
The increased sensitivity observed in the MS-PE_PGRS47 strain to antibiotics and stresses suggested possible changes in the cell wall permeability. To prove this, we used dye influx/accumulation assays to detect cell wall permeability. Nile red (NR) and ethidium bromide (EB) were used as reference dyes for hydrophobic and hydrophilic compounds that could permeate the mycobacterial cell membrane and cell wall freely (Chuang et al. 2015; Rodrigues et al. 2012). The results revealed that NR and EB accumulated more rapidly in MS-PE_PGRS47 cells than in control MS-pMV cells (Fig. 4A, B), indicating increased cell wall permeability or inhibition of efflux pump of MS-PE_PGRS47 strain. To further determine the specific reason, efflux pumps specific inhibitor carbonyl cyanide m-chlorophenyl hydrazone (CCCP) was added to cultivated strains. However, the addition of CCCP led to a similar increase in NR and EB accumulation in MS-pMV and MS-PE_PGRS47 strain (Fig. 4C, D). The data showed that PE_PGRS47 increased the permeability of the cell envelope without affecting the efflux pump.
PE_PGRS47 expression in M. smegmatis increases cell wall permeability. A Mid-log-phase cultures of MS-pMV and MS-PE_PGRS47 were incubated in PBS with 25 mM glucose and 2 μg/mL EB for indicated time. B Mid-log-phase cultures of MS-pMV and MS-PE_PGRS47 were incubated in PBS containing 25 mM glucose and 2 μM NR. The assay of accumulation of EB and NR over time in M. smegmatis was conducted at 37 °C. C Mid-log-phase cultures of MS-pMV and MS-PE_PGRS47 were incubated in PBS with 25 mM glucose and 2 μg/mL EB and co-incubated with 0.625 μg/mL CCCP. D Mid-log-phase cultures of MS-pMV and MS-PE_PGRS47 were incubated in PBS containing 25 mM glucose and 2 μM NR and co-incubated with 0.625 μg/mL CCCP. E Quantification of total fatty acid methyl esters extracted from the MS-pMV and MS-PE_PGRS47 estimated by GC/MS analysis. Data are means ± SD of technical triplicate from one representative out of three or more independent experiments (*p < 0.05, **p < 0.01, ***p < 0.001)
The increased NR and EB influx might be due to an altered fatty acids profile leading to a higher accumulation rate in MS-PE_PGRS47 than MS-pMV. Therefore, GC–MS assay was used to analyze the fatty acids content of MS-pMV and MS-PE_PGRS47 at the same growth stage. Thirty-two fatty acid methyl esters were identified in both strains from C8 to C24 (Fig. S1). The data showed that MS-PE_PGRS47 had decreased quantities of total fatty acids in the cell envelope (Fig. 4E). The data suggest that the overexpression of PE_PGRS47 significantly reduced amounts of total fatty acids and increased the cell wall permeability, which further confirmed that the cell wall localization of PE_PGRS47 altered the cell wall structure of the recombinant MS-PE_PGRS47 strain.
Conclusions
This study found that M. smegmatis expressing PE_PGRS47 is more sensitive to SDS and H2O2 that mimic the intracellular environment. However, this phenotype is not correlated with increased survival within host cells, as PE_PGRS47 enhanced the intracellular survival of mycobacteria in macrophages (Strong et al. 2021) and mice models (Saini et al. 2016). Given the involvement of PE_PGRS47 in fatty acid metabolism, it is reasonable to speculate that this protein may be a dual-functional protein. On the one hand, PE_PGRS47 suppresses the host immune response during the active phase of tuberculosis. On the other hand, this protein is involved in lipid metabolism during the latent phase, thereby providing energy for the dormant Mtb, as is the case for the LipY, which is a lipase that hydrolyzes triacylglycerol stored in lipid inclusion bodies (Saxena et al. 2013). Nevertheless, the detailed role of the PE_PGRS47 in physiology and pathogenicity should be further investigated by the deletion of the gene in Mtb.
In sum, our findings revealed a novel role of the PE_PGRS47 protein. The role of PE_PGRS47 in altering the colony morphology and the cell wall permeability of mycobacteria was confirmed, as evidenced by the increased susceptibility of recombinant MS-PE_PGRS47 strain to antibiotics and antimicrobial agents, including SDS and H2O2. Heterologous expression of PE_PGRS47 significantly decreased quantities of total fatty acids, which further confirmed that the cell wall localization of PE_PGRS47 altered the cell wall structure of the recombinant MS-PE_PGRS47 strain. This study suggested that PE_PGRS47 is a cell surface-exposed protein that influences cell wall permeability and the formation of mycobacterial colonies, ultimately potentiating the efficacy of lethal stresses against mycobacteria.
Data availability
The datasets used or analyzed during the current study are available from the corresponding authors upon reasonable request.
References
Alibaud L, Rombouts Y, Trivelli X et al (2011) A Mycobacterium marinum TesA mutant defective for major cell wall-associated lipids is highly attenuated in Dictyostelium discoideum and zebrafish embryos. Mol Microbiol 80(4):919–934. https://doi.org/10.1111/j.1365-2958.2011.07618.x
Bashiri G, Perkowski EF, Turner AP, Feltcher ME, Braunstein M, Baker EN (2012) Tat-dependent translocation of an F(420)-binding protein of Mycobacterium tuberculosis. PLoS ONE 7(10):e45003. https://doi.org/10.1371/journal.pone.0045003PONE-D-12-04036
Basu S, Pathak SK, Banerjee A et al (2006) Execution of macrophage apoptosis by PE_PGRS33 of Mycobacterium tuberculosis is mediated by toll-like receptor 2-dependent release of tumor necrosis factor-α. J Biol Chem 282(2):1039–1050. https://doi.org/10.1074/jbc.M604379200
Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Tech Bull Regist Med Technol 36(3):49–52
Bentley WE, Mirjalili N, Andersen DC, Davis RH, Kompala DS (1990) Plasmid-encoded protein: the principal factor in the “metabolic burden” associated with recombinant bacteria. Biotechnol Bioeng 35(7):668–681. https://doi.org/10.1002/bit.260350704
Bonanni D, Rindi L, Lari N, Garzelli C (2005) Immunogenicity of mycobacterial PPE44 (Rv2770c) in Mycobacterium bovis BCG-infected mice. J Med Microbiol 54(Pt 5):443–448. https://doi.org/10.1099/jmm.0.45960-0
Cascioferro A, Delogu G, Colone M et al (2007) PE is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. Mol Microbiol 66(6):1536–1547. https://doi.org/10.1111/j.1365-2958.2007.06023.x
Chatrath S, Gupta VK, Dixit A, Garg LC (2011) The Rv1651c-encoded PE_PGRS30 protein expressed in Mycobacterium smegmatis exhibits polar localization and modulates its growth profile. FEMS Microbiol Lett 322(2):194–199. https://doi.org/10.1111/j.1574-6968.2011.02354.x
Chatrath S, Gupta VK, Garg LC (2014) The PGRS domain is responsible for translocation of PE_PGRS30 to cell poles while the PE and the C-terminal domains localize it to the cell wall. FEBS Lett 588(6):990–994. https://doi.org/10.1016/j.febslet.2014.01.059S0014-5793(14)00102-1
Chatrath S, Gupta VK, Dixit A, Garg LC (2016) PE_PGRS30 of Mycobacterium tuberculosis mediates suppression of proinflammatory immune response in macrophages through its PGRS and PE domains. Microbes Infect 18(9):536–542. https://doi.org/10.1016/j.micinf.2016.04.004
Chuang Y-M, Bandyopadhyay N, Rifat D, Rubin H, Bader JS, Karakousis PC (2015) Deficiency of the novel exopolyphosphatase Rv1026/PPX2 leads to metabolic downshift and altered cell wall permeability in Mycobacterium tuberculosis. Mbio 6(2):e02428-e2514. https://doi.org/10.1128/mBio.02428-14
Cole ST, Brosch R, Parkhill J et al (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393(6685):537–544. https://doi.org/10.1038/31159
Daim S, Kawamura I, Tsuchiya K et al (2011) Expression of the Mycobacterium tuberculosis PPE37 protein in Mycobacterium smegmatis induces low tumour necrosis factor alpha and interleukin 6 production in murine macrophages. J Med Microbiol 60(5):582–591. https://doi.org/10.1099/jmm.0.026047-0
Delogu G, Pusceddu C, Bua A, Fadda G, Brennan MJ, Zanetti S (2004) Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol Microbiol 52(3):725–733. https://doi.org/10.1111/j.1365-2958.2004.04007.x
Deng W, Li W, Zeng J et al (2014) Mycobacterium tuberculosis PPE family protein Rv1808 manipulates cytokines profile via co-activation of MAPK and NF-kappaB signaling pathways. Cell Physiol Biochem 33(2):273–288. https://doi.org/10.1159/000356668
Deng W, Zeng J, Xiang X, Li P, Xie J (2015) PE11 (Rv1169c) selectively alters fatty acid components of Mycobacterium smegmatis and host cell interleukin-6 level accompanied with cell death. Front Microbiol 6:613. https://doi.org/10.3389/fmicb.2015.00613
Deng W, Long Q, Zeng J et al (2017) Mycobacterium tuberculosis PE_PGRS41 enhances the intracellular survival of M. smegmatis within macrophages via blocking innate immunity and inhibition of host defense. Sci Rep 7:46716. https://doi.org/10.1038/srep46716
Dheenadhayalan V, Delogu G, Brennan MJ (2006) Expression of the PE_PGRS 33 protein in Mycobacterium smegmatis triggers necrosis in macrophages and enhanced mycobacterial survival. Microbes Infect 8(1):262–272. https://doi.org/10.1016/j.micinf.2005.06.021
Dona V, Ventura M, Sali M et al (2013) The PPE domain of PPE17 is responsible for its surface localization and can be used to express heterologous proteins on the mycobacterial surface. PLoS ONE 8(3):e57517. https://doi.org/10.1371/journal.pone.0057517PONE-D-12-26841
Dong D, Wang D, Li M et al (2012) PPE38 modulates the innate immune response and is required for Mycobacterium marinum virulence. Infect Immun 80(1):43–54. https://doi.org/10.1128/IAI.05249-11
Dong W, Wang R, Li P et al (2020) Orphan response regulator Rv3143 increases antibiotic sensitivity by regulating cell wall permeability in Mycobacterium smegmatis. Arch Biochem Biophys 692:108522. https://doi.org/10.1016/j.abb.2020.108522
Fishbein S, van Wyk N, Warren RM, Sampson SL (2015) Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol 96(5):901–916. https://doi.org/10.1111/mmi.12981
Garces A, Atmakuri K, Chase MR et al (2010) EspA acts as a critical mediator of ESX1-dependent virulence in Mycobacterium tuberculosis by affecting bacterial cell wall integrity. PLoS Pathog 6(6):e1000957. https://doi.org/10.1371/journal.ppat.1000957
Iantomasi R, Sali M, Cascioferro A et al (2012) PE_PGRS30 is required for the full virulence of Mycobacterium tuberculosis. Cell Microbiol 14(3):356–367. https://doi.org/10.1111/j.1462-5822.2011.01721.x
Kohli S, Singh Y, Sharma K, Mittal A, Ehtesham NZ, Hasnain SE (2012) Comparative genomic and proteomic analyses of PE/PPE multigene family of Mycobacterium tuberculosis H37Rv and H37Ra reveal novel and interesting differences with implications in virulence. Nucleic Acids Res 40(15):7113–7122. https://doi.org/10.1093/nar/gks465
Kuze F, Uchihira F (1984) Various colony-formers of Mycobacterium avium-intracellulare. Eur J Respir Dis 65(6):402–410
Lewis T, Nichols PD, McMeekin TA (2000) Evaluation of extraction methods for recovery of fatty acids from lipid-producing microheterotrophs. J Microbiol Methods 43(2):107–116. https://doi.org/10.1016/S0167-7012(00)00217-7
Li JM, Li N, Zhu DY, Wan LG, He YL, Yang C (2008) Isocitrate lyase from Mycobacterium tuberculosis promotes survival of Mycobacterium smegmatis within macrophage by suppressing cell apoptosis. Chin Med J (engl) 121(12):1114–1119
Li W, Zhao Q, Deng W, Chen T, Liu M, Xie J (2014) Mycobacterium tuberculosis Rv3402c enhances mycobacterial survival within macrophages and modulates the host pro-inflammatory cytokines production via NF-Kappa B/ERK/p38 signaling. PLoS ONE 9(4):e94418. https://doi.org/10.1371/journal.pone.0094418PONE-D-13-36639
Li J, Chai QY, Zhang Y et al (2015a) Mycobacterium tuberculosis Mce3E suppresses host innate immune responses by targeting ERK1/2 signaling. J Immunol 194(8):3756–3767. https://doi.org/10.4049/jimmunol.1402679jimmunol.1402679
Li W, Fan X, Long Q, Xie L, Xie J (2015b) Mycobacterium tuberculosis effectors involved in host-pathogen interaction revealed by a multiple scales integrative pipeline. Infect Genet Evol 32:1–11. https://doi.org/10.1016/j.meegid.2015.02.014
Li Q, Zhou M, Fan X, Yan J, Li W, Xie J (2016a) Mycobacteriophage SWU1 gp39 can potentiate multiple antibiotics against Mycobacterium via altering the cell wall permeability. Sci Rep 6:28701. https://doi.org/10.1038/srep28701srep28701
Li W, Liu M, Xie J (2016b) Rv3369 induces cytokine interleukin-1beta production and enhances Mycobacterium smegmatis intracellular survival. J Interferon Cytokine Res 36(2):140–147. https://doi.org/10.1089/jir.2015.0090
Li W, Deng W, Xie J (2019) Expression and regulatory networks of Mycobacterium tuberculosis PE/PPE family antigens. J Cell Physiol 234(6):7742–7751. https://doi.org/10.1002/jcp.27608
Li W, Deng W, Zhang N, Peng H, Xu Y (2022) Mycobacterium tuberculosis Rv2387 facilitates mycobacterial survival by silencing TLR2/p38/JNK signaling. Pathogens 11(9):981. https://doi.org/10.3390/pathogens11090981
Liu J, Nikaido H (1999) A mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids accumulates meromycolates. Proc Natl Acad Sci U S A 96(7):4011–4016. https://doi.org/10.1073/pnas.96.7.4011
Meena LS (2015) An overview to understand the role of PE_PGRS family proteins in Mycobacterium tuberculosis H37 Rv and their potential as new drug targets. Biotechnol Appl Biochem 62(2):145–153. https://doi.org/10.1002/bab.1266
Neyrolles O, Cascioferro A, Daleke MH et al (2011) Functional dissection of the PE domain responsible for translocation of PE_PGRS33 across the mycobacterial cell wall. PLoS ONE 6(11):e27713. https://doi.org/10.1371/journal.pone.0027713
Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F (2002) Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46(8):2720–2722. https://doi.org/10.1128/AAC.46.8.2720-2722.2002
Ramakrishnan L, Federspiel NA, Falkow S (2000) Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288(5470):1436–1439. https://doi.org/10.1126/science.288.5470.1436
Rastogi S, Singh AK, Pant G, Mitra K, Sashidhara KV, Krishnan MY (2017) Down-regulation of PE11, a cell wall associated esterase, enhances the biofilm growth of Mycobacterium tuberculosis and reduces cell wall virulence lipid levels. Microbiology 163(1):52–61. https://doi.org/10.1099/mic.0.000417
Ren H, Liu J (2006) AsnB is involved in natural resistance of Mycobacterium smegmatis to multiple drugs. Antimicrob Agents Chemother 50(1):250–255. https://doi.org/10.1128/AAC.50.1.250-255.2006
Rengarajan J, Murphy E, Park A et al (2008) Mycobacterium tuberculosis Rv2224c modulates innate immune responses. Proc Natl Acad Sci U S A 105(1):264–269. https://doi.org/10.1073/pnas.0710601105
Rodrigues L, Machado D, Couto I, Amaral L, Viveiros M (2012) Contribution of efflux activity to isoniazid resistance in the Mycobacterium tuberculosis complex. Infect Genet Evol 12(4):695–700. https://doi.org/10.1016/j.meegid.2011.08.009
Saini NK, Baena A, Ng TW et al (2016) Suppression of autophagy and antigen presentation by Mycobacterium tuberculosis PE_PGRS47. Nat Microbiol 1(9):16133. https://doi.org/10.1038/nmicrobiol.2016.133nmicrobiol2016133
Sampson SL (2011) Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin Dev Immunol 2011:497203. https://doi.org/10.1155/2011/497203
Saxena AK, Roy KK, Singh S et al (2013) Identification and characterisation of small-molecule inhibitors of Rv3097c-encoded lipase (LipY) of Mycobacterium tuberculosis that selectively inhibit growth of bacilli in hypoxia. Int J Antimicrob Agents 42(1):27–35. https://doi.org/10.1016/j.ijantimicag.2013.03.007
Shi T, Fu T, Xie J (2011) Polyphosphate deficiency affects the sliding motility and biofilm formation of Mycobacterium smegmatis. Curr Microbiol 63(5):470–476. https://doi.org/10.1007/s00284-011-0004-4
Singh P, Rao RN, Reddy JR et al (2016) PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci Rep 6:21624. https://doi.org/10.1038/srep21624srep21624
Strong EJ, Jurcic Smith KL, Saini NK, Ng TW, Porcelli SA, Lee S (2020) Identification of autophagy-inhibiting factors of Mycobacterium tuberculosis by high-throughput loss-of-function screening. Infect Immun 88(12):e00269–20. https://doi.org/10.1128/IAI.00269-20
Strong EJ, Ng TW, Porcelli SA, Lee S, Stallings CL (2021) Mycobacterium tuberculosis PE_PGRS20 and PE_PGRS47 Proteins Inhibit Autophagy by Interaction with Rab1A. mSphere 6(4):e00549–21. https://doi.org/10.1128/mSphere.00549-21
Tian C, Jian-Ping X (2010) Roles of PE_PGRS family in Mycobacterium tuberculosis pathogenesis and novel measures against tuberculosis. Microb Pathogenesis 49(6):311–314. https://doi.org/10.1016/j.micpath.2010.07.004S0882-4010(10)00128-2
Vallecillo AJ, Espitia C (2009) Expression of Mycobacterium tuberculosis pe_pgrs33 is repressed during stationary phase and stress conditions, and its transcription is mediated by sigma factor A. Microb Pathog 46(3):119–127. https://doi.org/10.1016/j.micpath.2008.11.003S0882-4010(08)00151-4
Vincent AT, Nyongesa S, Morneau I, Reed MB, Tocheva EI, Veyrier FJ (2018) The mycobacterial cell envelope: a relict from the past or the result of recent evolution? Front Microbiol 9(2341):1–9. https://doi.org/10.3389/fmicb.2018.02341
Wang H, Dong D, Tang S, Chen X, Gao Q (2013) PPE38 of Mycobacterium marinum triggers the cross-talk of multiple pathways involved in the host response, as revealed by subcellular quantitative proteomics. J Proteome Res 12(5):2055–2066. https://doi.org/10.1021/pr301017e
Wang J, Li B-X, Ge P-P et al (2015) Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat Immunol 16:237–245. https://doi.org/10.1038/ni.3096
WHO (2022) Global tuberculosis report 2022. Geneva: World Health Organization, 2022. https://www.who.int/publications/i/item/9789240061729
Yu Z, Zhang C, Zhou M et al (2017) Mycobacterium tuberculosis PPE44 (Rv2770c) is involved in response to multiple stresses and promotes the macrophage expression of IL-12 p40 and IL-6 via the p38, ERK, and NF-kappaB signaling axis. Int Immunopharmacol 50:319–329. https://doi.org/10.1016/j.intimp.2017.06.028
Zeng J, Deng W, Yang W et al (2016) Mycobacterium tuberculosis Rv1152 is a novel GntR family transcriptional regulator involved in intrinsic vancomycin resistance and is a potential vancomycin adjuvant target. Sci Rep 6:28002. https://doi.org/10.1038/srep28002
Funding
This study was funded by the National Natural Science Foundation (81601740), the Applied Basic Research Program of Science & Technology Department of Sichuan Province (2018JY0108), the Doctoral Scientific Research Foundation of Neijing Normal University (15B10), and the grant of Neijiang Normal University (17CZ01).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. WL, ZY, and NZ performed the experiments. WL, ZZ, and XX analyzed the data. All the authors contributed to the writing of this manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Yusuf Akhter.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, W., Yan, Z., Zhang, N. et al. Novel role of PE_PGRS47 in the alteration of mycobacterial cell wall integrity and drug resistance. Arch Microbiol 205, 174 (2023). https://doi.org/10.1007/s00203-023-03515-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-023-03515-x