Abstract
Wheat blast, caused by Pyricularia oryzae pathotype Triticum, is one of the most notorious diseases of wheat. In this study, a total of twenty-four monoconidial isolates representing four major wheat blast affected districts, namely Chuadanga, Meherpur, Kustia and Jhenaidah of Bangladesh were analyzed. Eight RAPD and four ISSR primers being used for genetic diversity assay produced a total of 94 bands of which 85% were polymorphic. UPGMA dendrogram based on combined dataset (RAPD and ISSR) separated all the isolates into two main clusters having similarity ranged from 64 to 93%. Principal coordinate analysis showed congruent result with cluster analysis. However, clustering of the isolates according to their origin and plant part sampled was not apparent in the dendrogram. The genetic diversity indices unveiled that genetic diversity in P. oryzae populations is low. Average Nei's gene diversity (h) and Shannon’s Information Index (I) calculated for isolates from each district were found 0.16 and 0.24, respectively. The population structure analysis of the isolates revealed the presence of two sub-populations with admixture of alleles. Analysis of molecular variance indicated that significantly higher level of variation (96%) in the population was present within districts while a relatively low proportion (4%) of the variation was detected among districts. Knowledge generated in this study will give a pace in the development of appropriate wheat blast management strategies to control this disease in Bangladesh.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Being the biggest commercial crop in the world, wheat has a key contribution to the global food security. It provides 20% of food calories and protein of majority of the population in developing countries (Saharan et al. 2016). Like the other part of the world, demand of wheat has been increasing rapidly in South Asian countries day by day. In Bangladesh, wheat is the second most important grain crop after rice from consumption and economic point of view (Hossain and Silva 2013).
Wheat blast or ‘brusone’, an explosive disease of wheat, is caused by ascomycetous fungus Pyricularia oryzae Cavara (Teleomorh: Magnaporthe oryzae B. C. Couch) pathotype Triticum which can cause up to 100% yield loss under favorable climatic conditions (Cruz and Valent 2017). Presence of wheat blast was first reported in Paraná state of Brazil in 1985 (Igarashi et al.1986) and then it has spread to the adjacent countries with time (Barea and Toledo 1996; Viedma 2005; Alberione et al. 2008; Cabrera and Gutiérrez 2007). Previously, this disease was restricted to some South American countries such as Brazil, Bolivia, Paraguay, Argentina, and Uruguay except to one report in the experimental field in USA in 2011 (Callaway 2016). Emergence of deadly wheat blast was confirmed in Bangladesh in 2016, which was its first epidemic outside South America (Callaway 2016; Islam et al. 2016; Malaker et al. 2016). The first occurrence of wheat blast damaged 15,000 hectares of wheat fields that was approximately 15% of Bangladesh’s total wheat area (Islam et al. 2016). Since then, wheat blast has become a potential threat to wheat production for Bangladesh and other South Asian countries due to the possibility of its spread to the neighboring countries. Peng et al. (2011) warned that attention should be given to prevent pathogen dispersal to protect wheat production which came true when blast was reported from Bangladesh in 2016. Seed-borne inoculum might facilitate the long-distance dispersal of P. oryzae and allow it to invade different agroecosystems (Gomes et al. 2017; Goulart et al. 1995). However, P. oryzae conidia dispersed by air seem to play a major role in wheat blast epidemiology at the field scale (Urashima et al. 1993). Favorable climatic condition coupled with susceptible cultivars may cause severe damage to wheat production. In a recent study, applying the climate analog analysis, Mottaleb et al. (2018) warned that out of 40.9 million ha of total wheat-producing land in India, Bangladesh, and Pakistan, more than 17% of the area (6.9 million ha) is vulnerable to wheat blast.
Polymerase chain reaction (PCR)-based molecular markers are useful tools for estimating genetic variation within populations of phytopathogens (Vakalounakis and Fragkiadakis 1999; Kolmer and Liu, 2000). Random amplified polymorphic DNA (RAPD), inter simple sequence repeats (ISSRs), amplified fragment length polymorphism (AFLP), restriction fragment length polymorphism (RFLP) and simple sequence repeat (SSRs/microsatellites) are the most commonly used molecular markers to study the genetic variation in the genome of Pyricularia spp. populations (Sharma et al. 2002; Séré et al. 2007; Ashtiani et al. 2016). RAPD and ISSR fingerprinting methods are inexpensive, less time consuming to AFLP, RFLP and SSRs, highly polymorphic and they do not require any prior sequence information (Godwin et al. 1997; Gillings and Holley 1997). Moreover, application of RAPD markers coupled with other reproducible markers such as ISSR provides more reliable assessments of genetic diversity in pathogen populations (Schlotterer 2004; Ashtiani et al. 2016).
Previous studies on wheat blast pathogen found considerable morphological variations among the isolates (Al Noman et al. 2021) and also revealed that the isolates with high virulence were prevalent in Bangladesh P. oryzae population (Noman and Shamsi 2021). Evolutionary potential and the ability to adopt resistant cultivars and fungicide treatments have a relationship with the genetic diversity in plant pathogen population. Although wheat cultivars IPR 85, CD 113 and BR 18 from Brazil and cultivar Paragua CIAT and Parapeti CIAT from Bolivia showed considerable resistance against blast, cultivar Milan derived from CIMMYT (International Maize and Wheat Improvement Center) line showed the highest level of resistance (Kohli et al. 2011; Islam et al. 2019). Wheat cultivation using resistant varieties might be an effective strategy against wheat blast. But disease resistance can be affected by high genetic diversity of the pathogen. Therefore, a fundamental understanding on the extent of genetic variation in plant pathogen populations is essential in disease epidemiology and developing breeding strategies to develop cultivars with durable resistance. However, to the best of our knowledge, no study has been reported on the genetic diversity of wheat blast pathogen from Bangladesh after its emergence in 2016. Again, report on genetic diversity of wheat-infecting P. oryzae populations around the globe is also scanty. Considering the above facts, the present investigation had been undertaken to gain insight into the genetic diversity and genetic structure of P. oryzae isolates from major wheat blast affected districts of Bangladesh through RAPD and ISSR.
Materials and methods
Collection of wheat blast samples
Infected leaves and panicles showing characteristics blast symptoms were collected from the four highly wheat blast affected districts of Bangladesh namely Chuadanga, Meherpur, Kustia and Jhenaidah during the tenure of 2017–18 wheat season (Supplementary Fig. S1). Samples were collected in brown paper bags from the fields and stored at 4 °C in the laboratory for further analyses.
Isolation and maintenance of monoconidial cultures
A total of twenty-four monoconidial isolates were isolated from the field samples following single conidium isolation technique as described by Al Noman and Shamsi (2021). Monoconidial cultures were obtained by transferring single germinating conidium from 3% water agar to potato dextrose agar (PDA) plate using fine glass needle and then incubating at 25 °C with a 12-h photoperiod for 14 days. Details of the isolates are articulated in Table 1. Spores of different isolates were micro-photographed at 40X under compound microscope facilitated with digital camera (Nikon optiphot-2 trinocular microscope, Japan) using ImageFocus Alpha software. Photos of colony and conidia of the isolates are shown in Supplementary Figs. S5–S8. All the isolates were maintained in PDA slants at 4 °C. For long-term storage, isolates were grown on sterile filter paper disks and kept at – 80 °C.
Genomic DNA extraction
DNA was extracted from 1 g of fungal mycelium collected from 10-day-old monoconidial cultures of each isolate following method as described by Amer et al. (2011). DNA quality was checked by running on 1% agarose gel and the quantity was measured using NanoDrop (Thermo Scientific) spectrophotometer. Extracted DNA was stored at – 20 °C for further analyses.
ITS rDNA amplification and sequencing
The internal transcribe spacer (ITS) regions of sixteen P. oryzae isolates were amplified using ITS1 as forward and ITS4 as reverse primer (White et al. 1990). The PCR was performed in a 25 μL reaction mixture containing 1.0 μL forward and reverse primer, 12.5 μL master mix (Clever Scientific Ltd., Warwickshire, UK), 2.0 μL template DNA and 8.5 μL nuclease free water. The PCR consists of 30 cycles involving initial denaturation step at 94˚C for 5 min, followed by 94, 54 and 72 °C each for 30 s, with a final extension step of 5 min at 72 °C and ended with 4 °C. The PCR products were electrophoresed in 1% agarose gel. Amplified PCR products were sequenced through automated sequencer (SeqStudio Genetic Analyzer, Thermo Fisher Scientific, USA) in the Centre for Advanced Research in Sciences (CARS), University of Dhaka, Dhaka, 1000. The obtained sequences were compared with the sequences already available in the National Center for Biotechnology Information (NCBI) using BLASTn tool (http://www.ncbi.nlm.nih.gov/BLAST) and these sequences were deposited in the GenBank database.
RAPD and ISSR amplification
Seven RAPD primers OPA-03, OPA-10, OPA-13, OPB-10, OPB-13, OPF-04 and OPM-20 (Macrogen, Inc., Seoul, South Korea) and four ISSR primers including ISSR5, ISSR7, UBC881 and UBC890 (Eurogentech, Germany) (Table 2) that produced consistent and reproducible profiles and showed high polymorphism in the previous studies (Sharma et al. 2002; Chadha and Gopalakrishna 2007; Vanaraj et al. 2013; Srivastava et al. 2014; Ashtiani et al. 2016) for Pyricularia spp. were chosen to determine DNA polymorphism among P. oryzae isolates in this study. PCR amplifications were performed in a 25 µL reaction volume containing 12.5 µL of PCR master mix (Clever Scientific Ltd., Warwickshire, UK), 1.0 µL of RAPD or ISSR primer, 9.5 µL of nuclease free water and 2.0 µL of genomic DNA at 15 ng/μL. PCR amplification using RAPD primers was conducted with the following program: 45 cycles of 94 °C for 1 min (denaturation), 35 °C for 1.5 min (annealing) and 72 °C for 2 min (extension) with the initial denaturing of 94 °C for 4 min and final extension of 72 °C for 10 min. Thermal cycling profiles for ISSR were:45 cycles of 94 °C for 1 min (denaturation), 55 °C for 1.5 min (annealing) and 72 °C for 2 min (extension) with the initial denaturing of 94 °C for 5 min and final extension of 72 °C for 10 min.
Gel electrophoresis and staining
The amplified PCR products of RAPD and ISSR were separated electrophoretically on 1% agarose gel, stained with ethidium bromide and visualized under UV light. The sizes of the amplicons were estimated by comparing with 1 kb DNA ladder (Clever Scientific Ltd., Warwickshire, UK).
Data analyses
Bands generated in the RAPD and ISSR analyses were expressed in binary characters and scored manually as 1 for presence and 0 for absence of the band. Polymorphic information content (PIC) was calculated as described by Smith et al. (1997). Marker index (MI) was calculated as the product of PIC and effective multiplex ratio (EMR), where EMR is the product of the fraction of polymorphic loci and the number of polymorphic loci (Milbourne et al. 1997; Prevost and Wilkinson 1999). Observed number of alleles (Na), effective number of alleles (Ne), Nei's gene diversity (h) and Shannon’s information index (I) were calculated using POPGENE version 1.31 (Yeh et al. 1999). Dendrograms were constructed by NTSYSpc version 2.1 (Exeter Software, New York, USA) using the SAHN clustering program with the unweighted pair group method using arithmetic averages (UPGMA) method. Principal coordinate analysis (PCoA) was performed using DARwin version 6.0.21 (Perrier and Jacquemoud-Collet 2006) software. Population structure and individual clustering (K) of the isolates were inferred using the Bayesian clustering method by means of STRUCTURE version 2.3.4 (Pritchard et al. 2000). The number of hypothetical sub-populations (K) was set to vary between 1 and 10. ΔK method was applied to best estimate K and was computed using Structure Harvester ver. 0.56.3 (Earl 2012). GenAlEx version 6.5 (Peakall and Smouse 2012) was utilized for the analysis of molecular variance (AMOVA) with 9999 permutations.
Results
Morphological and molecular identification
All the isolates produced characteristic pyriform-shaped asexual conidia (Supplementary Figs. S7 and S8). Those conidia were three celled with each having single nucleus per cell and hyaline to pale gray in color. Identification based on morphological characters was performed by comparing with available literature (Cruz and Valent 2017; Urashima 2010; Islam et al. 2016). Molecular identification was done by matching the obtained ITS sequences with the already available sequences of P. oryzae in NCBI database. Newly generated sequences of this study were submitted to NCBI GenBank and the obtained accession numbers are given in Table 1.
RAPD analysis
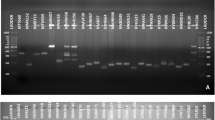
Eight RAPD primers generated a total of 69 bands ranging from 250 to 3200 bp with an average of 8.63 bands per primer (Table 2; Fig. 1 and Supplementary Figs. S2 and S3). All the primers generated polymorphic bands among the isolates. Average PIC value, as a measure of informativeness of genetic markers, was found 0.86. The highest MI value (12.32) was shown by the primer OPB-10 and lowest (3.92) was observed for primer OPB-13. Effective number of alleles (Ne) ranged between 1.22 and 1.63. Primer OPM-20 showed the highest number of effective alleles per locus (1.63) and the lowest alleles were observed in OPB-18 with the value of 1.22. The mean value of the Nei’s gene diversity(h) was 0.24, with the highest value for primer OPB-10 (0.35) and the lowest for OPB-18 primer (0.16).
ISSR analysis
Four ISSR primers used in this study could produce a total of 25 fragments varying from 250 to 4000 bp with an average of 6.25 bands per primer (Table 2; Fig. 1 and Supplimentary Fig. S4). Primer ISSR5 generated maximum number of bands (10 bands) whereas primers UBC 890, ISSR7 and UBC 881 produced 6, 5 and 4 bands, respectively. In total, 11 Polymorphic bands were obtained with an average of 2.27 polymorphic fragments per primer. Average percentage of polymorphic bands was 36.67% with the highest value for the primer ISSR5 (70%). ISSR5 also displayed highest PIC and MI value (0.64 and 3.14, respectively). However, primer ISSR7 revealed maximum effective number of alleles (Ne) and Nei's gene diversity (h).
A total of 94 bands were generated by RAPD and ISSR markers cumulatively of which 80 fragments (85%) were polymorphic. The combined average PIC and MI values were found 0.59 and 4.26, respectively. Average values of effective number of alleles (Ne) and Nei's gene diversity (h) for both the RAPD and ISSR primers were 1.32 and 0.2, respectively.
Cluster analysis
UPGMA dendrogram based on cumulative data of both RAPD and ISSR markers was generated to analyze the genetic relationship among the isolates. Cluster analysis showed distinction among the isolates by dividing them into two major clusters namely cluster A and cluster B (Fig. 2). The range of similarity among the isolates was 64–93% indicating a high degree of diversity among them. Cluster A is further divided into two sub-clusters viz., sub-cluster A1 and sub-cluster A2. Sub-cluster A1 consisted of sixteen isolates from four studied districts and sub-cluster A2 contained three isolates from Kustia, Jhenaidah and Meherpur each. Cluster B comprised of five isolates all from Meherpur district. On the basis of RAPD and ISSR analyses, it was evident that due to the genetic variation among the isolates, a clustering according to the origin of the isolates and plant organs sampled was not apparent in the dendrogram.
Principal coordinate analysis (PCoA)
PCoA analysis was performed to evaluate the dissimilarity of the isolates and to verify the clustering pattern from the dendrogram. Both RAPD and ISSR fingerprinting data of the isolates were used to prepare a scatter plot obtained from principal coordinate analysis. Here, PCoA result was consistent with the cluster analysis (Figs. 2, 3), i.e., isolates were grouped in the scatter plot as in the UPGMA dendrogram. Major clusters and sub-clusters (A1a, AIb, A2 and B) in the dendrogram are shown in PCoA to demonstrate the consistency of results between these two methods.
Population genetic diversity
The genetic diversity indices calculated for P. oryzae isolates from four different districts are represented in Table 3. The percentage of polymorphic bands fell in the range of 27.7–74.47%, with an average of 43.88%. The observed number of alleles (Na), effective number of alleles (Ne) and Nei's gene diversity (h) ranged from 1.28 to 1.74, 1.2 to 1.42 and 0.11 to 0.25, respectively, for the four different populations (Meherpur, Chuadanga, Kustia and Jhenaidah). Isolates from Meherpur district showed highest Na (1.74), Ne (1.42) and h (0.25) values while these values were lowest for isolates from Kustia (Na = 1.74, Ne = 1.2 and h = 0.11). Based on the Shannon’s information Index (I), the highest diversity was revealed in P. oryzae population from Meherpur (I = 0.38), whereas it was lowest for Kustia (I = 0.17) population. Average Nei's gene diversity (h) and Shannon’s information Index (I) were found 0.16 and 0.24, respectively, for studied P. oryzae populations.
Analysis of molecular variance (AMOVA) conducted using both RAPD and ISSR data indicated that most of the variation (96%) in the population was attributable to the differences among isolates within districts, while a relatively low proportion (4%) of the variation was detected for among districts (Table 4).
Structure analysis
Cumulative RAPD and ISSR data set was also used to infer population structure and number of sub-populations among P. oryzae isolates. Structure analysis on the basis of maximum likelihood and delta K values revealed existence of two sub-populations with genetic admixture and are represented by two different colors (Fig. 4).
Discussion
Multilocus RAPD and ISSR fingerprinting methods have been widely used for the determination of genetic variation among P. oryzae isolates in rice (Sharma et al. 2002; Chadha and Gopalakrishna 2007; Ashtiani et al. 2016) as well as other plant pathogenic fungi (Bayraktar et al. 2008; Baysal et al. 2010; Nasehi et al. 2014; Singh et al. 2019). Cumulative data of RAPD and ISSR markers were utilized to analyze genetic variation among wheat infecting P. oryzae isolates from the studied districts of Bangladesh. Such use of combined marker fingerprints to assess genetic variability in fungal population has been previously demonstrated in Fusarium oxysporum f. sp. ciceris (Bayraktar et al. 2008), Alternaria alternata (Kandan et al. 2014), Fusarium solani (Arif et al. 2015), Bipolaris sorghicola (Kandan et al. 2016) and Fusarium oxysporum f. sp. carthami (Singh et al. 2019). In this study, RAPD and ISSR markers combinedly produced a total of 94 bands of which 85% were polymorphic. The amplification of large number of polymorphic bands suggested that the primer sets used in this study could be of significance for the measurement of genetic diversity in P. oryzae population. UPGMA dendrogram based on both RAPD and ISSR data revealed considerable genetic variation among the isolates. Principal coordinate analysis (PCoA) showed congruent result with cluster analysis. The topology of the dendrogram suggested that isolates from same district were distributed in different clusters and sub-clusters, depicting presence of no clear correlation between clustering of the isolates in the dendrogram and their area of origins. Similar type of result had been documented in previous studies on Pyricularia spp. isolates from different countries (Consolo et al. 2008; Chadha & Gopalakrishna 2005; Ngueko et al. 2004; El-Wahsh et al. 2016). Again, Maciel et al. (2014) found no subdivision among the wheat-infecting P. oryzae populations in Brazil while elucidating population structure of the wheat blast pathogen using 11 microsatellite loci. Such lack of correlation in our study implies that genetic variation among the isolates is independent of their area of origin. Possible explanation for such finding might be the geographical closeness of the study areas or P. oryzae population need more time to evolve region specific races.
The population structure analysis showed admixture of alleles and resulted in two sub-populations. Analysis of molecular variance (AMOVA) indicated that higher level of variation (96%) in the population was present within districts while a relatively low proportion (4%) of the variation was detected among districts. Structure analysis based on cumulative RAPD and ISSR data provided no specific demarcation of genetic grouping in different populations and little amount of variation (4%) among districts was observed in AMOVA. These results highlight lower level of genetic divergence of P. oryzae isolates among regions sampled and are also consistent with the gene flow across the districts investigated. Here, human-mediated movement of infested seeds or the dispersal of the pathogen via airborne inoculum may facilitate gene flow among the study areas.
Average Nei's gene diversity (h) and Shannon’s information Index (I) calculated for isolates from each district were found 0.16 and 0.24, respectively, indicating that the genetic diversity in the studied P. oryzae populations is low. This can be explained by the fact that environmental factors may not play a vital role in generating high genetic variation, as the studied districts were geographically close and may share almost same climatic conditions. However, the source of genetic diversity among the isolates is unclear. Mutations, sexual recombination, parasexual recombination, action of transposable elements, selection or hyphal anastomosis can be the possible source of genetic diversity in pathogen population (Zeigler et al. 1997; Park et al. 2003). Furthermore, Maciel et al. (2014) found evidence for mixed reproductive system characterized by both sexual and asexual reproduction in wheat infecting P. oryzae population in Brazil. However, no evidence of sexual recombination of P. oryzae causing wheat blast has been reported yet from Bangladesh. This also explains low genetic diversity found among P. oryzae populations in this study.
In conclusion, this study indicates the presence of low genetic diversity among P. oryzae populations in Bangladesh and reveals that the larger part of variability in the population was attributable to the differences among isolates within districts. As wheat blast pathogen is on move, rapid development of efficient and appropriate disease management strategies including development of blast resistant variety becomes a crying need. Genetic diversity in pathogen population may affect disease resistance and also limit the efficacy of fungicide treatments. Hence, knowledge on genetic variation and genetic structure of plant pathogen populations is one of the prerequisites to provide proper strategies to control and manage plant diseases (McDonald and Linde 2002). Findings of this study will be helpful to understand the genetic nature of Bangladesh P. oryzae isolates and give a pace in the development of appropriate wheat blast management strategies in Bangladesh.
References
Al Noman MA, Shamsi S (2021) An improved and effective protocol for monoconidial isolation of Pyricularia oryzae. J Plant Pathol 103:317–320
Al Noman MA, Hosen S, Shamsi S (2021) Morphological and molecular characterization of Pyricularia oryzae isolates causing wheat blast in Bangladesh. Indian Phytopathol 74:123–131
Alberione E, Bainotti C, Cettour I, Salines J (2008) Evaluation of diseases in summer planting wheat in the Argentine-Campana NEA 2007/2008. 7th National Congress of Wheat. Santa Rosa, La Pampa, Argentina
Amer OE, Mahmoud MA, El-Samawaty ARMA, Sayed SRM (2011) Non-liquid nitrogen based method for isolation of DNA from filamentous fungi. Afr J Biotechnol 10(65):14337–14341
Arif M, Zaidi NW, Haq QM, Singh YP, Taj G, Kar CS, Singh US (2015) Morphological and comparative genomic analyses of pathogenic and non-pathogenic Fusarium solani isolated from Dalbergia sissoo. Mol Biol Rep 42:1107–1122
Ashtiani FA, Kadir J, Nasehi A, Rahaghi SRH, Vadamalai G, Rambe SK (2016) Characterisation of Magnaporthe oryzae isolates from rice in peninsular Malaysia. Czech J Genet Plant Breed 52:145–156
Barea G, Toledo J (1996) Identificación y zonificación de piricularia o bruzone (Pyricularia oryzae) en el cultivo del trigo en el dpto. de Santa Cruz. In: CIAT. Informe Técnico. Proyecto de Investigación Trigo, Santa Cruz, pp 76–86
Bayraktar H, Dolar FS, Maden S (2008) Use of RAPD and ISSR markers in detection of genetic variation and population structure among Fusarium oxysporum f. sp. ciceris isolates on chickpea in Turkey. J Phytopathol 156:146–154
Baysal Ö, Siragusa M, Gümrükcü E, Zengin S, Carimi F, Sajeva M, Da Silva JA (2010) Molecular characterization of Fusarium oxysporum f. sp. melongenae by ISSR and RAPD markers on eggplant. Biochem Gen 48:524–537
Cabrera MG, Gutiérrez S (2007) Primer registro de Pyricularia grisea en cultivos de trigo del NE de Argentina. Jornada de Actualización en Enfermedades de Trigo. IFSC Press, Lavallol, Buenos Aires
Callaway E (2016) Devastating wheat fungus appears in Asia for first time. Nature 532:421–422
Chadha S, Gopalakrishna T (2005) Genetic diversity of Indian isolates of rice blast pathogen (Magnaporthe grisea) using molecular markers. Curr Sci 88:1466–1469
Chadha S, Gopalakrishna T (2007) Comparative assessment of REMAP and ISSR marker assays for genetic polymorphism studies in Magnaporthe grisea. Curr Sci 93:688–692
Consolo VF, Cordo CA, Salerno GL (2008) DNA fingerprint and pathotype diversity of Pyricularia oryzae populations from Argentina. Australas Plant Pathol 37:357
Cruz CD, Valent B (2017) Wheat blast disease: danger on the move. Trop Plant Pathol 42:210–222
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Gillings M, Holley M (1997) Amplification of anonymous DNA fragments using pairs of long primers generates reproducible DNA fingerprints that are sensitive to genetic variation. Electrophoresis 18:1512–1518
Godwin ID, Aitken AB, Smith LW (1997) Application of inter simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis 18:1524–1528
Gomes DP, Rocha VS, Pereira OL, Souza MAD (2017) Damage of wheat blast on the productivity and quality of seeds as a function of the initial inoculum in the field. J Seed Sci 39:66–74
Goulart ACP, Paiva FA, Andrade PJM (1995) Relationship between incidence of blast in wheat seeds and the presence of Pyricularia grisea in the harvested seeds. Fitopatol Bras 20:184–189
Hossain A, Silva JAT (2013) Wheat production in Bangladesh: its future in the light of global warming. AoB Plants 5:42
Igarashi S, Utiamada CM, Igarashi LC, Kazuma AH, Lopes RS (1986) Pyricularia emtrigo. 1. Ocorrência de Pyricularia sp. no estado do Paraná. Fitopatol Bras 11:351–352
Islam MT, Croll D, Gladieux P, SoanesDM PA, Bhattacharjee P, Hossain MS, Gupta DR, Rahman MM, Mahboob MG, Cook N, Salam MU, Surovy MZ, Sancho VB, Maciel JLN, Nhani A, Castroagudin VL, Reges JTD, Ceresini PC, Ravel S, Kellner R, Fournier E, Tharreau D, Lebrun MH, Mcdonald BA, Stitt T, Swan D, Talbot NJ, Saunders DGO, Win J, Kamoun S (2016) Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol 14:11
Islam M, Arif M, Hassan L, Robin A (2019) Wheat blast disease: Bangladesh and global perspectives of blast resistance. J Bangl Agric Univ 17(2):122–132
Kandan A, Akhtar J, Singh B, Dev U, Goley R, Chand D, Roy A, Rajkumar S, Agarwal PC (2014) Genetic diversity analysis of Alternaria alternata isolates infecting different crops using URP and ISSR markers. Indian J Plant Prot 42:229–236
Kandan A, Akhtar J, Singh B, Pal D, Chand D, Rajkumar S, Agarwal P (2016) Genetic diversity analysis of fungal pathogen Bipolaris sorghicola infecting Sorghum bicolor in India. J Environ Biol 37:1323–1330
Kohli MM, Mehta YR, Guzman E, De Viedma L, Cubilla LE (2011) Pyricularia blast—a threat to wheat cultivation. Czech J Genet Plant Breed 47:130–134
Kolmer JA, Liu JQ (2000) Virulence and molecular polymorphisms in international collections of the wheat leaf rust fungus Puccinia triticina. Phytopathology 90:427–436
Maciel JLN, Ceresini PC, Castroagudin VL, Zala M, Kema GH, McDonald BA (2014) Population structure and pathotype diversity of the wheat blast pathogen Magnaporthe oryzae 25 years after its emergence in Brazil. Phytopathology 104(1):95–107
Malaker PK, Barma NCD, Tiwari TP, Collis WJ, Duveiller E, Singh PK, Joshi AK, Singh RP, Braun HJ, Peterson GL, Pedley KF, Farman ML, Valent B (2016) First report of wheat blast caused by Magnaporthe oryzae pathotype triticum in Bangladesh. Plant Dis 100:2330
McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379
Milbourne D, Meyer R, Bradshaw JE, Baird E, Bonar N, Provan J, Powell W, Waugh R (1997) Comparison of PCR based marker systems for the analysis of genetic relationships in cultivated potato. Mol Breed 3:127–136
Mottaleb KA, Singh PK, Sonder K, Kruseman G, Tiwari TP, Barma NCD, Malaker PK, Braun H, Erenstein O (2018) Threat of wheat blast to South Asia’s food security: an ex-ante analysis. PLoS ONE 13:e0197555. https://doi.org/10.1371/journal.pone.0197555
Nasehi A, Kadir J, Nasr-Esfahani M, Abed-Ashtiani F, Mui-Yun W, Rambe SKH, Golkhandan E (2014) Analysis of genetic and virulence variability of Stemphylium lycopersici associated with leaf spot of vegetable crops. Eur J Plant Pathol 140:261–273
Ngueko RB, Ying S, Hong-kai W, Fu-cheng L, Tong X (2004) Genetic diversity analysis of Magnaporthe grisea from some blast nurseries of Hunan province using random amplified polymorphic DNA. J Zhejiang Univ 30:355–362
Noman MAA, Shamsi S (2021) Virulence analysis of Pyricularia oryzae isolates causing wheat blast in Bangladesh. Dhaka Univ J Biol Sci 30(2):151–157
Park SY, Milgroom MG, Han SS, Kang S, Lee YH (2003) Diversity of pathotypes and DNA fingerprint haplotypes in populations of Magnaporthe grisea in Korea over two decades. Phytopathology 93:1378–1385
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Peng JL, Zhou YL, He ZH (2011) Global warning against the spread of wheat blast. J Triticeae Crops 31:989–993
Perrier X, Jacquemoud-Collet JP (2006) DARwin software: dissimilarity analysis and representation for windows. http://darwin.Cirad.Fr/Darwin. Accessed 11 Mar 2017
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Saharan MS, Bhardwaj SC, Chatrath R, Sharma P, Choudhary AK, Gupta RK (2016) Wheat blast disease—an overview. J Wheat Res 8:1–5
Schlotterer C (2004) The evolution of molecular markers – just a matter of fashion? Nature 5:63–69
Séré Y, Onasanya A, Afolabi A, Mignouna HD, Akator K (2007) Genetic diversity of the blast fungus, Magnaporthe grisea (Hebert) Barr, in Burkina Faso. Afr J Biotechnol 6:2568–2577
Sharma TR, Chauhan RS, Singh BM, Paul R, Sagar V, Rathour R (2002) RAPD and pathotype analyses of Magnaporthe grisea populations from the north-western Himalayan region of India. J Phytopathol 150:649–656
Singh N, Anand G, Kapoor R (2019) Virulence and genetic diversity among Fusarium oxysporum f. sp. carthami isolates of India using multilocus RAPD and ISSR markers. Trop Plant Pathol 44:409–422
Smith JS, Chin EC, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theor Appl Genet 95:163–173
Srivastava D, Shamim MD, Kumar D, Pandey P, Khan NA, Singh SN (2014) Morphological and molecular characterization of Pyricularia oryzae causing blast disease in rice (Oryza sativa) from north India. Int J Sci Res Publ 4(7):2250–3153
Urashima AS (2010) Blast. In: Bockus WW, Bowden RL, Hunger RM, Morrill WL, Murray TD, Smiley RW (eds) Compendium of wheat diseases and pests. American Phytopathological Society, Saint Paul, pp 22–23
Urashima AS, Igarashi S, Kato H (1993) Host range, mating type, and fertility of Pyricularia grisea from wheat in Brazil. Plant Dis 12:11–16
Vakalounakis DJ, Fragkiadakis GA (1999) Genetic diversity of Fusarium oxysporum isolates from cucumber: differentiation by vegetative compatibility, pathogenicity and RAPD fingerprinting. Phytopathology 89:161–168
Vanaraj P, Kandasamy S, Ambalavanan S, Ramalingam R, Sabariyappan R (2013) Variability in Pyricularia oryzae from different rice growing regions of Tamil Nadu, India. Afr J Microbiol 7(26):3379–3388
Viedma LQ (2005) Wheat blast occurrence in Paraguay. Phytopathology 95:S152
White TJ, Burns T, Lee S, Taylor J (1990) Application and direct sequencing of fungal ribosomal RNA genes for phylogenetics: a guide to methods and amplifications. Academic Press, San Diego
Yeh FC, Yang RC, Boyle TB, Ye ZH, Mao JX (1999) POPGENE 1.31, user-friendly shareware for population genetic analysis. In: Molecular biology and biotechnology center. University of Alberta, Edmonton
Zeigler RS, Scott RP, Leung H, Bordeos AA, Kumar J (1997) Evidence of parasexual exchange of DNA in rice blast fungus challenges its clonality. Phytopatology 87:248–287
Acknowledgements
The first author (Al Noman MA) expresses his deepest gratitude to the Ministry of Science and Technology, Govt. of the People’s Republic of Bangladesh for providing financial assistance to this research through NST fellowship program. The authors are thankful to Dr. Md. Zabed Hossain, Professor, Dept. of Botany, University of Dhaka for providing NTSYSpc software.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human participants and/or animals
The present research did not involve any experimentation on human or animals.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Noman, M.A.A., Hosen, S. & Shamsi, S. Elucidating genetic diversity and population structure of Pyricularia oryzae isolates causing wheat blast in Bangladesh. Arch Microbiol 204, 134 (2022). https://doi.org/10.1007/s00203-021-02752-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-021-02752-2