Abstract
MicroRNAs are small, noncoding single-stranded RNAs that have emerged as important posttranscriptional regulators of gene expression, with an essential role in vertebrate development and different biological processes. This review highlights the recent advances in the function of miRNAs and their roles in bone remodeling and bone diseases. MicroRNAs (miRNAs) are a class of small (∼22 nt), noncoding single-stranded RNAs that have emerged as important posttranscriptional regulators of gene expression. They are essential for vertebrate development and play critical roles in different biological processes related to cell differentiation, activity, metabolism, and apoptosis. A rising number of experimental reports now indicate that miRNAs contribute to every step of osteogenesis and bone homeostasis, from embryonic skeletal development to maintenance of adult bone tissue, by regulating the growth, differentiation, and activity of different cell systems inside and outside the skeleton. Importantly, emerging information from animal studies suggests that targeting miRNAs might become an attractive and new therapeutic approach for osteoporosis or other skeletal diseases, even though there are still major concerns related to potential off target effects and the need of efficient delivery methods in vivo. Moreover, besides their recognized effects at the cellular level, evidence is also gathering that miRNAs are excreted and can circulate in the blood or other body fluids with potential paracrine or endocrine functions. Thus, they could represent suitable candidates for becoming sensitive disease biomarkers in different pathologic conditions, including skeletal disorders. Despite these promising perspectives more work remains to be done until miRNAs can serve as robust therapeutic targets or established diagnostic tools for precision medicine in skeletal disorders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The pathogenesis of many bone disorders is, at least in part, genetically determined. Indeed, complex regulatory mechanisms and transcriptional activities are necessary to support the expression of genes that regulate the activity of bone cells in response to different stimuli (e.g., hormones, growth factors, cytokines). Apart from monogenic bone diseases, it is now well established that in complex, multifactorial disorders with a recognized hereditary component, such as osteoporosis, the associated genetic variants have a limited impact on gene expression and explain only a small fraction of the disease risk [1]. Moreover, a larger proportion of the variants associated to many human traits or diseases, as indicated by the recent genome-wide association studies, fall in loci which do not encode proteins, suggesting that additional mechanisms other than gene-gene and gene-environment interactions might be involved.
Importantly, the technological progresses in genetics occurred over the past decade have changed dramatically our perspective on eukaryotic gene expression regulation. In fact, several heritable, “epigenetic” changes in gene expression have been characterized for most genes that do not imply a modification in nucleotide sequence. These involve DNA methylation and histone modifications that modulate gene transcription and noncoding RNAs (ncRNAs) that act at the posttranscriptional level. Moreover, while only 1–2% of the human genome is transcribed into proteins, several studies documented pervasive transcription across 70–90% of the human genome, suggesting that ncRNAs represent most of the human transcriptome [2, 3]. It has been recently established that aside from around 21,000 protein coding genes and over 1,100,000 genomic repetitive elements (e.g., Alu repeat elements), the human transcriptome includes more than 11,000 pseudogenes, about 9000 small RNAs, and around 10,000–32,000 long ncRNAs (lncRNAs) [4, 5]. The small ncRNA class includes both housekeeping and regulatory RNAs, such as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), PIWI-interacting RNAs (piRNAs), and small interfering RNAs (siRNAs). More recently, circular RNAs (circRNAs) have emerged as new potential gene regulators [3]. They are known to sequester miRNAs and thus modulate cellular functions and, possibly, disease mechanisms.
The importance of this noncoding transcriptome in determining the greater complexity of higher eukaryotes and disease pathogenesis has become increasingly clear in recent years. While the function of lncRNAs and circRNAs remains poorly understood, an increasing number of reports have highlighted the key role of miRNAs as important regulators of various biological processes. Here, we provide an overview of the function of miRNAs and their roles in bone remodeling and bone diseases.
Biogenesis and function of microRNAs
miRNAs are a class of endogenous, evolutionary conserved, short RNAs (generally 20–24 nucleotides long) that have essential roles in regulating gene expression, with potential major implications in several human diseases. Indeed, up to 60% of protein-coding genes are regulated by miRNAs at the posttranscriptional level by blocking messenger RNA (mRNA) translation or by inducing mRNA degradation [6].
It has been estimated that miRNAs constitute around 1–5% of the human genome [7]. Most of the miRNA genes are transcribed as independent transcriptional units, having their own promoter and other regulatory elements. However, one fourth of the miRNA genes is located in intronic regions of several coding genes [8] and hence are cotranscribed and coregulated from a common promoter. At the cellular level, mature miRNAs are generated by the sequential cleavage of longer poly-adenylated transcripts (pri-miRNAs) that are transcribed from the intragenic or intergenic DNA regions by RNA polymerases II and, less frequently, III [9]. The initial cleavage step occurs in the nucleus where pri-miRNAs are processed by the ribonuclease II called Drosha or the double-stranded DNA-binding protein DGCR8 (Di George syndrome critical gene 8) giving rise to pre-miRNAs (about 70 nucleotides long). Then, pre-miRNAs are exported to the cytoplasm by the export receptor complex exportin5-RAN/GTP and processed by the ribonuclease III endonuclease named Dicer and the coregulator Ago2 (an Argonaut protein) to form small double-stranded miRNAs. Both these steps of miRNAs biogenesis are crucial for life, since either Drosha or Dicer deletion results in early embryonic lethality [10, 11], and conditional deletion of these enzymes in specific cell types affects cellular function and the development of many tissues, including bone [12–15]. Finally, the duplex miRNAs are converted into mature single-stranded miRNAs and integrated into the RNA-induced silencing complex (RISC), which acts on the complementary 3′-UTRs of target mRNAs by either promoting their degradation or inhibiting their translation [16]. The two strands of each miRNA duplex are respectively called miRNA-5p and miRNA-3p (formerly defined as miRNA and star*-miRNA). It was firstly assumed that in most species, the 5p strand acts as the guide-strand and is incorporated into RISC, while the star 3p strand is degraded. However, both arms of the precursor have the potential to produce functional mature miRNAs and the dominant product may change in relation to the species, tissues, or developmental stages [17].
Since the first report in 1993, the miRNA database has grown exponentially, with an actual size of 35,828 mature miRNAs from 223 species (miRBase release 21 June 2014). The human genome is estimated to encode more than 1800 miRNAs, and nearly two thirds of human protein-coding genes show miRNA-binding affinity that is generally conserved across most mammalian species [6]. While the first described miRNAs were named by the gene where they are located (e.g., lin-4 and let-7 of Caenorhabditis elegans), a numerical nomenclature has been now adopted for all miRNA sequences. Moreover, similar to the proteins, miRNAs deriving from the identical ancestor in the phylogenetic tree can be grouped into a family (e.g., miRNA-29, a, b, c) based on sequence similarity in the seed regions (8 nucleotides, with positions 2 to 7 being 99% conserved). Furthermore, miRNA genes are frequently expressed individually, but many exist in clusters of two to seven genes. Generally, a miRNA cluster refers to a set of miRNAs, which are close to each other in the genome, have the same promoter and/or are transcribed as a single primary transcript. Experimental results suggest that miRNA clusters may be expressed cotranscriptionally, which indicates that they are under control of common regulatory sequences [18]. Thus, not only the genomic location, but if a group of miRNAs has a similar pattern of expression, they can be considered to be in the same cluster.
From the functional point of view, most miRNAs are thought to act primarily as mRNA destabilization sequences or as translational repressors by pairing with specific partially complementary 3′-UTR regulatory elements on mRNAs, although target sites in the coding region and 5′-UTR can also be functional [19]. However, there is also evidence indicating that, at least in some circumstances, miRNAs can also enhance translation [20] or that positive transcriptional regulation can be produced by certain miRNAs targeting sites in promoter regions [21]. Importantly, a single gene can be targeted by a cluster of miRNAs and each single miRNA can regulate different protein coding genes.
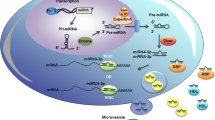
The interest in the field of miRNAs has further increased by the discovery that they do not only exert their action on the intracellular level but they also exist extracellularly and can circulate in the blood flow as protein-bound miRNAs, free circulating RNAs (e.g., released by dead cells), or within secreted microvescicles [22, 23], thus claiming their potential as novel diagnostic and prognostic markers of many diseases. Indeed, the pivotal role of miRNAs in regulating gene expression and disease mechanisms is now reflected by various achievements in biomedical research and the impact they are starting to have on patient management. Remarkable examples include either the development of diagnostic and prognostic miRNA expression signatures for particular human pathologies or the prospect of miRNA-based therapeutics for viral diseases and cancer, which have entered clinical trials (e.g., hsa-miR-34a-5p replacement therapy in liver cancer patients or antisense agents directed against hsa-miR-122-5p for treating hepatitis C infection) [24, 25].
MicroRNAs and bone cells
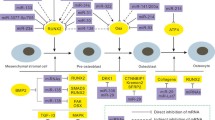
Many reports now indicate that miRNAs contribute to every step of osteogenesis and bone homeostasis, from embryonic skeletal development to maintenance of adult bone tissue, by regulating the growth, differentiation, and activity of different cell systems inside and outside the skeleton. While most of the identified miRNAs appears to specifically regulate cells from the osteoblast or the osteoclast lineage, some miRNAs have been involved in the regulation of bone resorption as well as bone formation (a list of major miRNAs affecting both osteoblast or osteoclast function is given in Table 1). However, despite the rising number of experimental reports about this issue, our understanding on the exact mechanisms through which miRNAs regulate the interplay between the different cell types in the bone remodeling unit under physiologic conditions or in bone diseases remains limited.
Functions of microRNAs on osteoclast formation and activity
Osteoclasts are bone-specific multinucleated cells, which are responsible for bone resorption. They derive from hematopoietic stem cells and monocyte-macrophage precursors, with receptor activator of nuclear factor κB ligand (RANKL) and macrophage colony-stimulating factor (M-CSF) being the most important factors for osteoclast differentiation. Notably, the complete loss of miRNA activity in osteoclast precursors results in a block of mature osteoclast formation. Indeed, specific deletion of crucial enzymes involved in miRNA biogenesis in mononuclear osteoclasts precursors and in mature multinucleated osteoclasts resulted in similar skeletal phenotype characterized by increased bone mass due to a reduction in the number and activity of osteoclasts [12, 14, 15]. Osteoclast formation from hematopoietic stem cells was also affected. The same results were obtained when siRNAs were used to silence components of the Drosha or RISC enzyme complexes in bone marrow precursor cells [26].
A recent mice study demonstrated that from the early to the late differentiation stages of osteoclastogenesis, 49 miRNA are upregulated and 44 were downregulated [27]. Computational analyses predicted mTOR, PI3 kinase/AKT, cell-matrix interactions, actin cytoskeleton organization, focal adhesion, and axon guidance pathways to be the main targets of the seven miRNA clusters deriving from these 93 miRNAs. Other studies indicated that specific miRNAs are operative mostly at the commitment stage of osteoclastogenesis, acting on the differentiation and recruitment of cells derived from the hematopoietic stem line [reviewed in 28, 29].
An updated list of relevant miRNAs in osteoclast formation and/or activity and their potential targets is reported in Table 2. Among the different implicated miRNAs, miRNA-21 has been the most extensively investigated [23, 24]. This miRNA is highly expressed in osteoclast precursors and its expression levels are strongly upregulated during osteoclastogenesis [15]. Transcription factors c-Fos and PU.I, which are crucial modulators of osteoclast formation, trigger miRNA-21 transcription by acting on a specific promoter. At the same time, miRNA-21 is able to downregulate programmed cell death 4 (PDCD4) protein levels that exert an inhibitory effect on c-Fos. Therefore, a positive feedback loop involves c-FOS/miRNA-21/PDCD4 and promotes RANKL-induced osteoclastogenesis. Moreover, miRNA-21 has been also implicated in mediating the inhibitory effects on osteoclastogenesis and in promoting osteoclast apoptosis induced by estrogen [30]. In fact, estrogen suppresses miRNA-21 biogenesis and thus increases the protein levels of another miRNA-21 target, Fas ligand (FasL) that induces osteoclast apoptosis [30]. Different studies indicated that the miRNA-29 family (miRNA-29a-3p, miRNA-29b-3p, miRNA-29c-3p) is a key mediator of osteoclast differentiation. During osteoclastogenesis from either bone marrow monocytes (BMMs) or macrophage cell line RAW264.7, all miRNA-29 family members are increased, while their knockdown inhibits the commitment and migration of osteoclast precursors without interfering with mature osteoclast functions [31]. It has been suggested that the stimulatory effect of miRNA-29 family members on osteoclast formation is mainly mediated by the posttranscriptional suppression of the target protein nuclear factor I-A (NFIA), which is a negative regulator of M-CSF receptor [32]. Indeed, it has been also predicted that members of miRNA-29 family may regulate the expression of more than 6000 genes [28]. Other miRNA-29 target proteins within the macrophage-osteoclast lineages include the calcitonin receptor and mRNAs critical for cytoskeletal organization (e.g., cell division control protein 42 and G protein-coupled receptor 85) [29]. However, conflicting reports were also published since pre-miRNA-29a treatment in rats significantly reduced glucocorticoid-induced bone loss while the suppression of miRNA-29b increased bone resorption in vitro [28, 29]. Albeit these latter evidences indicate a more complex role of some miRNA-29 family members on osteoclast activity and bone remodeling, a parallel effect of miRNA-29 family members on Wnt signaling and osteoblast activity was also described [28, 29], suggesting that the positive effects on bone density and strength observed in the animal model could be, at least in part, mediated by the osteoblasts. Another relevant miRNA in osteoclast biology is miRNA-31, that is widely expressed by a variety of tissues with more than 200 potential targets, most of them involved in cell mobility, polarity and cytoskeletal dynamics [28]. Importantly, miRNA-31 is highly upregulated during RANKL-induced osteoclastogenesis (by up to 18-fold in murine bone marrow cells) and its inhibition by specific antagomirs suppresses terminal osteoclast formation and bone resorption. This effect could be related to the targeting of RhoA that plays a key role in acting ring formation in osteoclasts. Moreover, other studies have evidenced a parallel inhibitory effect of miRNA-31 on osteogenesis and bone formation (see “Functions of microRNAs on osteogenesis and bone formation”). In a miRNA expression profile analysis during osteoclastogenesis from human peripheral blood mononuclear cells (PBMCs), a major effect of miRNA-148a on osteoclast formation was evidenced, likely mediated by a negative regulation of V-maf musculo aponeurotic fibrosarcoma oncogene homolog B (MAFB) [33], which acts on NFATc1, c-Fos, and other regulatory factors of osteoclast differentiation. Consistent with this observation, a suppression of bone resorption together with an increase in bone mass were observed in the ovariectomized (OVX) mice model following the injection with miRNA-148a antagomiRs [33].
Other positive regulators of osteoclastogenesis include miRNA-183, through inhibiting heme oxygenase 1 (HO-1) expression, miRNA-214, that is supposed to target the phosphatase and tensin homolog (PTEN)/Pl3k/Akt pathway, and miR-9718, that is preferentially expressed in bone and leads to posttranslational suppression of protein inhibitor of activated STAT3 (PIAS3), a known inhibitor of NFATc1 and osteoclastogenesis [reviewed in 29]. The important pro-osteoclastogenic role of miRNA-214 has been also recently highlighted in vivo, in the osteoclast-specific miR-214 transgenic mice, that shows increased osteoclast activity and reduced bone density [34].
In contrast, miRNA-7-5p, miRNA-26, miRNA-34, miRNA-124, miRNA-125a, miRNA-146a, miRNA-155, miRNA 218-5p, and miRNA-503 demonstrated an inhibitory function on osteoclast formation, at least in experimental conditions [reviewed in 29]. While miRNA-124 and miRNA-218-5p may act as intrinsic negative regulators of NFATc1, a master stimulator of osteoclastogenesis, miRNA-7-5p and miRNA-26a are supposed to act upstream NFATc1, through the inhibition of DC-STAMP, which is also involved in the fusion of osteoclast precursors into mature osteoclasts, either directly (miRNA-7-5p) or indirectly (miRNA-26a), through the suppression of the DC-STAMP inhibitor connective tissue growth factor/CCN family 2 (CTGF/CCN2). A study by Guo et al. demonstrated a significant downregulation of miRNA-125a during M-CSF and RANKL-induced osteoclastogenesis of PBMCs, while miRNA-125a overexpression inhibited osteoclast formation [35]. Similarly, transfection of a miRNA-125a antagomiR into PBMCs promoted osteoclast differentiation. Tumor necrosis factor receptor-associated factor 6 (TRAF6), a transduction factor for RANKL/RANK/NFATc1 signal, was confirmed to be a target of miRNA-125a. Of interest, in the same in vitro experiments, NFATc1 was able to reduce miRNA-125a transcription, thus suggesting the presence of a TRAF6/NFATc1/miRNA-125a regulatory feedback loop within the osteoclast. Furthermore, it is known that interferon β (IFN-β) may be induced during osteoclast differentiation via a c-Fos-dependent mechanism downstream of the RANKL-RANK signal transduction cascade, and that this increase in IFN-β, in turn, inhibits osteoclastogenesis [36]. An experimental in vitro study in BMMs found that miRNA-155 is an IFN-β-induced miRNA, mediating the suppressive effect of IFN-β on osteoclast differentiation by targeting suppressor of cytokine signaling 1 (SOCS1) and microphthalmia transcription factor (MITF), two regulators of osteoclastogenesis [37]. The role of miRNA-146a on osteoclastogenesis has been investigated either in vitro and in vivo by Nakasa et al. who demonstrated that transfection of double-stranded miRNA-146a in PBMCs derived from healthy individuals suppressed their MCS-F and RANKL-induced differentiation in osteoclasts, while intravenous injection of miRNA-146a prevented bone erosion in a mice model of collagen-induced arthritis [38].
Among other relevant miRNAs for osteoclast biology, miRNA-34a is highly conserved across species and its expression is downregulated during osteoclast differentiation from either BMMs or PBMCs [39]. Transgenic mice specifically expressing miRNA-34a in osteoclasts showed reduced bone resorption and higher bone density, while the opposite phenotype was observed in miRNA-34a knockout model [39]. Moreover, OVX-induced osteoporosis was effectively attenuated by miRNA-34a nanoparticle treatment [39], further evidencing that this miRNA could become an important target for the suppression of osteoclast activity and bone loss. From the molecular point of view, transforming growth factor-β-induced factor 2 (Tgif2) was identified as an essential and direct miRNA-34a target due to its pro-osteoclastogenic effect. Consistent with this hypothesis, Tgif2 deletion reduced bone resorption and abolished miRNA-34a regulation. On the other side, miRNA-503 was demonstrated to be a relevant miRNA on osteoclast biology acting through a direct inhibition of RANK [40]. In fact, in the OVX mice model silencing of miRNA-503 using a specific antagomiR increased RANK protein expression, promoted bone resorption, and decreased bone mass, whereas overexpression of miRNA-503 inhibited bone resorption and prevented bone loss. Of interest, following OVX, there is a decline in miRNA-503 levels while estrogen replacement increases its expression.
Despite miRNA-223 has been investigated in different experimental observations, there are still conflicting evidences showing either stimulatory or inhibitory effects on osteoclast differentiation [reviewed in 28, 29]. This miRNA is almost exclusively expressed in the hematopoietic system, including the myeloid cell lineages and mononuclear osteoclast precursors [41]. Some in vitro evidences suggested that miRNA-223 is increased during osteoclastogenesis and is regulated by the transcription factor PU.I that is also expressed in osteoclast precursors as a response to M-CSF stimulation. In those studies, miRNA-223 was reported to inhibit nuclear factor I-A (NFI-A) expression, thus enhancing osteoclast formation [14]. Notably, a positive feedback loop between PU.I, miRNA-223, NFI-A, and MCS-F receptor was also described [42]. When an antisense was used to suppress miRNA-223 levels, either osteoclast differentiation or osteoclast bone resorbing activity were decreased [14]. In contrast with the above observations, other reports indicated that, in case of miRNA-223 overexpression, osteoclastogenesis is inhibited in either PBMCs or RAW264.7 cells [28, 29]. This effect could be mediated by the downregulation of IKKα, a key factor in noncanonical NFkB pathway. Thus, it is likely that appropriate miRNA-223 expression levels should be kept during osteoclastogenesis and that either a suppression or an excessive increase in this miRNA may have negative effects on osteoclast formation.
Notably, in a more recent and sophisticated study, a microarray analysis was performed to detect the expression profiles of all ncRNAs at different stages during osteoclastogenesis of RAW264.7 cells, revealing a complex interaction among ncRNAs in osteoclast formation and activity [43]. For pre-osteoclasts 1643 lncRNAs, 147 circRNAs and 119 miRNAs were upregulated and 2705 lncRNAs, 109 circRNAs, and 941 miRNAs were downregulated; for mature osteoclasts 1896 lncRNAs, 78 circRNAs and 38 miRNAs were upregulated and 2706 lncRNAs, 135 circRNAs, and 24 miRNAs were downregulated, while for activated osteoclasts 2716 lncRNAs, 78 circRNAs and 38 miRNAs were upregulated and 3124 lncRNAs, 45 circRNAs, and 975 miRNAs were downregulated. However, most of the associated miRNAs were novel, since they were not associated with osteoclast formation and/or activity in the previous studies described above.
Functions of microRNAs on osteogenesis and bone formation
Osteoblasts are differentiated cells derived from the mesenchymal stem cell (MSC) line, which is also the precursor line common to adipocytes, chondrocytes, and myocytes. Indeed, osteoblastogenesis is a multistep process. After osteogenic induction, MSCs differentiate to proliferative pre-osteoblasts before becoming mature osteoblasts. In the terminal step of differentiation, a small fraction of osteoblasts further differentiate into osteocytes, whereas the remaining undergoes apoptosis. Essentially, two families of growth factors act as the major regulators of osteoblast differentiation from MSCs, the Wnt family and the bone morphogenetic proteins (BMPs). Among the several downstream effectors of these signaling pathways, the essential transcription factors for osteoblastogenesis include Runx2, Osterix (Osx), and different classes of homeodomain proteins. To date, the effects of miRNAs on osteoblast differentiation and bone formation have been more extensively investigated than on osteoclasts [reviewed in 44–46].
Experimental studies in animal models indicated that alterations in miRNA processing in either chondrocytes or osteoblasts have a negative effect on bone [13, 44]. For example, the conditional deletion of Dicer in mesenchymal osteoprogenitors cells (from either the osteoblastic or chondrocyte lineages) resulted in marked skeletal deformities and lethality during late gestation in mice models. When Dicer was ablated in mature osteoblasts, the mice were viable but exhibited a reduction in osteoblast number together with delayed bone development and mineralization [13]. Surprisingly, a progressive increase in cortical bone was observed in these mice after 1 month of age, mainly due to increased collagen deposition in the extracellular matrix. A similar reduction in osteoblast number was observed in case of conditional deletion of Dicer in committed pre-osteoblasts, leading to decreased mineralization, without major abnormalities in trabecular or cortical bone volume [45]. However, the latter model also evidenced that deletion of Dicer in mouse osteoprogenitors, but not in mature osteoblasts, disrupts the integrity of the hematopoietic stem cell niche.
An increasing number of miRNAs have been identified that exert a major impact on the regulation of osteoblast differentiation from MSCs and bone formation by targeting specific osteogenic factors or negative regulators of osteogenesis. Indeed, miRNA expression patterns appear to differ in undifferentiated MSC progenitors and the respective fully differentiated cells (e.g., osteoblasts vs. adipocytes vs. chondrocytes) suggesting that miRNAs are crucial for the commitment of MSCs into specific lineages.
miRNAs with an inhibitory role on osteogenesis and osteoblast function
Differential miRNA expression analyses revealed that miRNA-335 is highly expressed in undifferentiated human MSCs, while its levels decreases during MSCs differentiation in osteoblasts or other cell lineages [46]. Overexpression of miR-335 in human MSCs also inhibited their proliferation and migration, together with their osteogenic and adipogenic potential. However, other reports evidenced variable expression of miRNA-335-5p during osteoblast differentiation, indicating an increase in this miRNA at earlier time points following induction of differentiation and then a progressive decrease, particularly in late-stage osteoblasts and osteocytes [47]. By a different approach, pivotal studies in either mice or human bone marrow MSCs (BM-MSCs) compared the expression of representative miRNAs in the derived osteoblasts and chondroblasts [48, 49]. While selective upregulation of miRNA-96 and miRNA-199b was evident during osteogenesis, miRNA-199a and miRNA-124a were strongly upregulated during chondrogenesis. Likewise, different miRNAs regulate the differentiation of mesenchymal progenitor cells and bone marrow stromal cells (BMSCs) towards adipogenesis while decreasing osteogenesis. Of interest, most of these miRNAs exert a modulatory effect on components of the canonical Wnt signaling pathway, which appears to act as a major molecular switch between adipogenesis and osteogenesis of MSCs. A specific high-throughput miRNA assay in cellular models of activation and repression of Wnt signaling evidenced 18 and 29 miRNAs that might promote or repress adipogenesis, respectively [50]. Among these miRNAs, miRNA-210 was shown to block at transcription level Tcf712, a transcription factor triggering the downstream responsive genes of Wnt signaling. More recently, miRNA-188 was identified as a key regulator of the age-related switch between osteogenesis and adipogenesis of BM-MSCs [51]. Its levels were markedly higher in BM-MSCs from aged compared with young mice and humans. With aging, animal lacking miRNA-188 showed a decreased fat accumulation in bone marrow as well as reduced rates of bone loss; conversely, transgenic overexpression of miRNA-188 determined greater age-associated bone loss and fat accumulation in mice. Consistent with these observations, treatment of aging mice with antagomiR-188 via a BM-MSC-specific aptamer increased bone formation, thus providing a potential strategy for future treatment options for senile osteoporosis. From the molecular point of view, this miRNA posttranscriptionally inhibits hystone deacetylase 9 (HDAC9) and RPTOR-independent companion of MTOR complex 2 (Rictor) expression and thus upregulates PPARγ expression promoting adipogenic differentiation of BMMSCs. Likewise, miRNA-206a, known as key factor for muscle differentiation, is progressively downregulated during osteogenesis and its overexpression in different cell lines or animal models was associated with reduced osteoblast differentiation [43]. These negative effects of miRNA-206 on osteoblast development and bone formation have been related to the suppression of the gap junction protein connexin-43 (Cx43) that is crucial for the functional network among osteoblasts, osteocytes, and osteoprogenitors cells [52].
Many other miRNAs were shown to act primarily as inhibitors of osteoblast differentiation at either early-lineage commitment of MSCs or later differentiation stages of committed pre-osteoblast cells by targeting Runx2 or additional relevant osteogenic factors. An initial miRNA profiling during BMP2 induced differentiation of mouse C2C12 mesenchymal cells demonstrated that 22 of 25 miRNAs, which significantly changed in response to BMP2 were downregulated [53]. Among these miRNAs two key transducers of BMP2 osteogenic signal were identified, miRNA-133, which directly targets Runx2 and miRNA-135, which targets Smad5. Other miRNAs have been implicated to the inhibition of osteoblastogenesis by targeting different components of the BMP signaling pathway or directly Runx2. Known inhibitors of BMPs include miRNA-370, which decreases the expression of BMP2, miRNA-26a, miRNA-199a-3p that reduces osteoblast differentiation targeting Smad1 [reviewed in 54], miRNA-27a repressing BMP2, BMPR1A, and Smad9 expression [55], and miRNA-100 targeting BMPR2 [56]. Members of the miRNA-497∼195 cluster and particularly miRNA-195–5p have been also associated to the inhibition of osteoblastogenesis during postnatal bone development and the differentiation of primary calvaria osteoblasts, through the downregulation of multiple BMP-responsive genes [57], while the inhibitory effect of miRNA-30 family on BMP-induced osteogenesis has been related to a contemporary inhibition of Smad1 and Runx2 [58]. Additional in vitro studies showed that several miRNAs are implicated in the suppression of Runx2 during chondrogenesis and osteogenesis. In particular, a panel of 11 Runx2-targeting miRNAs (miRNA-23a, miRNA-30c, miRNA-34c, miRNA-133a, miRNA-135a, miRNA-137, miRNA-204, miRNA-205, miRNA-217, miRNA-218, and miRNA-338) was shown to be expressed in a lineage-related pattern in MSCs [59]. During both osteogenic and chondrogenic differentiation, these miRNAs, in general, are inversely expressed relative to Runx2, and in experimental conditions, each of them was able to attenuate Runx2 protein accumulation. Importantly, during the initial steps of endochondral bone formation, these miRNAs are generally highly expressed in pre-chondrocytes, in order to promote chondrogenesis; subsequently, when cells differentiate into hypertrophic chondrocytes within the growth plate, they become downregulated. As a result, Runx2 increases in order to promote bone formation. In the differentiation process of osteoblasts, the same miRNAs are downregulated until the late stages, when their increase is required to inhibit Runx2 and permit the final stage of osteoblast maturation. Additional studies confirmed the direct effect of miRNA-23a and miRNA-204 or its homolog miRNA-211 on the suppression of Runx2 levels [reviewed in 54]. Thus, tight regulation of Runx2 protein by miRNAs appears critical for osteoblastogenesis and normal bone formation. At the same time, Runx2 negatively regulates expression of the miRNA cluster 23a∼27a∼24-2, establishing a feed-forward mechanism necessary for osteoblast differentiation and activity, while an upregulation of miRNA-23 is relevant for the maintenance of the osteocyte phenotype [60]. A subsequent study demonstrated that Runx2 can also upregulate miRNA-1192, which in turn enhances Runx2-induced osteogenic differentiation, possibly through the downregulation of heparin-binding EGF-like growth factor [61]. The latter is also a target of an additional pro-osteblastogenic miRNA, miRNA-96 [62].
Further reports demonstrated that miRNA-93, miRNA-143, miRNA-145, and miRNA-637 all act downstream of Runx-2 and suppress osteoblast differentiation and bone mineralization by targeting Osx [reviewed in 54]. Notably, a study by Chen et al. evidenced that, during the process of Osx-controlled osteogenesis, Osx has the ability to coordinately modulate Runx2, sclerostin, alkaline phosphatase and the transcription factor Dlx5 at levels appropriate for optimal osteoblast differentiation and function, at least in part, through regulation of specific miRNAs, including the downregulation of miRNA-133a and miRNA-204/211 that are known repressors of Runx2 [63].
More recently, miRNA-103a has been identified as the first mechanosensitive miRNA that regulates osteoblast differentiation by directly targeting Runx2 [64]. In fact, miRNA-103a and its host gene PANK3 were both downregulated during cyclic mechanical stretch-induced osteoblast differentiation, whereas Runx2 protein expression was upregulated. Further, the perturbation of this miRNA also had a significant effect on osteoblast activity and matrix mineralization. Consistent with these in vitro data, an inhibitory role of miRNA-103a in regulating bone formation in hind limb unloading mice was reported and pretreatment with antagomiR-103a partly rescued bone loss caused by mechanical unloading. Taken together, all these studies demonstrate the existence of a tight and reciprocal interplay between the master osteogenic transcription factors like Runx2 or Osx and several miRNAs, during osteoblast differentiation. Conversely, a suppression of osteoblastogenesis together with an increased apoptosis of cells of the osteoblastic lineages have been related to miRNA-182 and miRNA-705, which represses FoxO1, a known antioxidant playing a crucial role in redox balance and osteogenesis [65, 66]. By the modulation of the ERK-dependent pathway, that also plays a key role in the transcriptional control of bone formation, miRNA-138 was involved in the negative regulation of human MSC osteogenic differentiation [67]. Indeed, in vivo ectopic bone formation was enhanced by 60% when miRNA-138 was antagonized in the mice model. Target prediction analysis and experimental validation by luciferase reporter assay confirmed focal adhesion kinase (FAK), an activator of ERKs, as a bona fide target of miRNA-138.
Interestingly, together with a suppressive effect on osteoclastogenesis, as described by Krzeszinski et al. [39], it has been reported that overexpression of miRNA-34a may also exert an inhibitory effect on early commitment and late osteoblast differentiation of human MSCs in vitro and in a preclinical in vivo model of heterotopic bone formation, by targeting Jagged1 (JAG1), a ligand for Notch 1 [68]. Similarly, miRNA-31 not only exerts a stimulatory effect on osteoclast maturation but also negatively affects osteogenesis, as demonstrated by different experimental reports. In particular, the overexpression of miRNA-31 repressed whereas its downregulation enhanced the osteogenesis of human MSCs [69]. These inhibitory effects of miRNA-31 on osteoblast formation have been related to the inhibition of the osteogenic factors Osx, Frizzled-3, and special AT-rich sequence-binding protein 2 (SATB2) [69–71]. Moreover, recent reports suggested that miRNA-31 is highly expressed by the senescent endothelial cells or the human MSC-derived adipocytes and can be released by these cells through extracellular vesicles leading to the suppression of osteogenesis [71, 72].
Thus, several sets of miRNAs may suppress osteogenesis and bone formation by different mechanisms and by targeting most of the known signaling pathways involved in osteoblast biology, at least in experimental conditions (Table 3).
miRNAs with a stimulatory role on osteogenesis and osteoblast function
On the other side, a lower number of miRNAs has been associated to a positive regulation of osteogenesis and osteoblast differentiation (Table 4). Among them, miRNA-20a and members of the miRNA-29 family have been shown to be crucial for osteoblast differentiation. In fact, miRNA-20a promotes the osteogenesis of MSCs by activating BMP/Runx2 signaling through silencing peroxisome proliferator-activated receptors γ (PPARγ), Bambi and Crim1 [reviewed in 73]. Further analysis demonstrated that this miRNA enhances the expression levels of several osteogenic factors, such as BMPs, Runx2, and Osx, but also typical osteoblastic markers like osteocalcin and osteopontin. The osteogenic effect of miRNA-29a seems to be mediated by targeting and suppressing inhibitors of the Wnt-signaling pathway such as Dickkopf-1 (DKK1), Kremen 2, and secreted frizzled related protein 2 [74]. Additional reports evidenced that this miRNA, together with miRNA-29c, is able to suppress osteonectin production [75]. Moreover, at least two miRNA profiling studies identified miRNA-29 and miRNA-26 families to be upregulated through stages of osteoblast differentiation by targeting many collagens and extracellular matrix proteins [76, 77]. In particular, miRNA-29b was shown to be a key regulator of development of the osteoblast phenotype by directly downregulating known inhibitors of osteoblast differentiation, such as HDAC4, TGFβ3, ACVR2A, CTNNBIP1, and DUSP2 proteins [76]. Notwithstanding some discrepancies among these studies, one key finding is that, together with their documented pro-osteoclastogenic effects, the miRNA-29 family members also retain a positive pro-osteoblastogenic action.
Other miRNAs have been demonstrated to stimulate the osteoblast by repressing HDACs, such as miRNA-140 (which targets HDAC4) and miRNA-2861 (which targets HADA5) [reviewed in 54]. Both HDAC4 and HDAC5 are well-known enhancer of Runx2 degradation, thus explaining the positive effects of both miRNAs on osteoblast development. Notably, in vivo silencing of miRNA-2861 using a specific antagomir inhibited bone formation in mice and an inactivating homozygous mutation in pre-miRNA-2861 was shown to cause primary osteoporosis in two related adolescents [78].
Importantly, while downregulation of osteogenesis by miRNAs appears to occur by several mechanisms and at different steps of osteoblast differentiation, most of the miRNAs acting as positive regulators of osteoblastogenesis described to date appear to modulate components of the Wnt signaling pathway or downstream effectors such as Runx2. In fact, similar pro-osteoblastogenic effects on the Wnt pathway to those described above for miRNA-29 have been associated to miRNA-335-5p (which suppresses DKK1) [47] and miRNA-218 (which acts through the inhibition of either DKK1 or sclerostin) [79], leading to a suppression of Wnt inhibitors. By acting downstream of the LRp5/LRP6 receptor complex, other positive modulators of the Wnt signaling within the osteoblastic cell line include Let-7f and miRNA-27, which increase β-catenin stability and its accumulation within the osteoblast [reviwed in 54]. At the same time, a study by Tamura et al. showed that miRNA-34b-5p and miRNA-34c are upregulated by the activation of canonical Wnt signaling in multipotent premyoblast C2C12 cells and thereby contribute to osteoblast differentiation and activity as downstream effectors of Wnt proteins [80]. Indeed, in a previous study, miRNA-34c was also induced by BMP2 during osteoblast differentiation of the same C2C12 cell line [81]. This miRNA was shown to target multiple components of the Notch signaling pathway, such as Notch1, Notch2, and Jag1 in a direct manner, but also to influence osteoclast differentiation in a noncell autonomous fashion. As a result, mice with osteoblast-specific gain of miRNA-34c showed an age-dependent osteoporosis due to defective mineralization and proliferation of osteoblasts and increased osteoclastogenesis [81]. This phenotype is consistent with the effects of loss of Notch in osteoblasts, previously described in mice models [82].
More recently, the miRNA-17~92a cluster (miRNAs 17, 18a, 19a, 20a, 19b, and 92a) at 13q31-q32 has been confirmed to play a relevant role in the regulation of bone formation by multiple in vitro and in vivo evidences [83–85]. This cluster is essential for vertebrate development and germline deletions of MIR17HG have been detected in patients with Feingold syndrome, an autosomal dominant disease characterized by microcephaly, short stature, and digital abnormalities. Most miRNAs within this cluster are expressed at high level in bone tissue and osteoblasts and positively affect osteoblast differentiation from different MSC lines [83, 84]. While its complete loss results in smaller embryos and immediate postnatal death, miRNA-17~92 haplo-insufficiency caused low bone mass due to impaired osteoblast activity in mice [84]. Moreover, conditional disruption of miRNA17~92 cluster in collagen type I-producing osteoblasts resulted in reduced longitudinal growth, decreased bone size, reduced periosteal bone formation, and impaired bone anabolic response to exercise [85]. Expression levels of Runx2 and periostin, known targets for some of the miRNAs of this cluster, were also significantly reduced in the periosteal tissue of conditional knockout mice compared with the wild-type mice.
Other miRNAs with documented positive effects on osteoblasts include miRNA-15b, which suppresses Smurf1 (a promoter of Runx2 degradation by the proteasome) [86], miRNA-181 via repression of TGF-β signaling molecules [87], miRNA-194 through modulating STAT1-mediated Runx2 nuclear translocation [88], miRNA-210, that inhibits the TGF-beta/activin signaling pathway targeting activin A receptor type 1B (AcvR1b) [89], and miRNA-23, acting as a potential inhibitor of TNF-induced osteoblast apoptosis by repressing the expression of Fas, which is involved in the extrinsic pathway of cell apoptosis [90]. In contrast, the miRNA-126/PDGFR-α system was recently shown to regulate the migratory behavior of human osteoblasts, without exerting major effects on cell survival and differentiation [91].
Interestingly, together with the known pro-osteoclastogenic role (as described above), miRNA-21 has been also confirmed to promote the osteoblast differentiation of MSCs by repressing the negative regulator Spry1 [92]. Levels of this miRNA have been described to decrease in response of inflammatory cytokines such as tumor necrosis factor α (TNF-α).
Modulation of miRNAs affecting osteogenesis by hormones and bone active agents
Some experimental evidences suggested that bone active hormones such as estrogen and vitamin D might influence miRNA expression and function in osteoblast lineage cells, with potential therapeutic implications. While estrogen has been shown to upregulate members of the miRNA-17-92a cluster and reduce osteoblast apoptosis [83], some miRNAs were differentially regulated in primary cultures of human osteoblasts following treatment with 1,25 dihydroxy-vitamin D [93]. These include miR-637, targeting the type 4 collagen alpha 1 (COL4A1) and miR-1228, which suppresses bone matrix protein 2-inducible protein kinase (BMP2K) by inhibition of protein translation. Finally, a single preliminary report described an altered expression of different miRNAs relevant to osteoblast differentiation and activity (e.g., miRNA-26a and miRNA-133a) following ibandronate treatment of osteoblast-like, periodontal ligament stem cells [94].
MicroRNAs and bone disorders
As the evidence for the important role of miRNAs in the regulation of bone and mineral homeostasis increased, the investigation of the potential associations between circulating or tissue-specific miRNAs and skeletal disorders has become a new field of investigation. In particular, circulating miRNAs are considered highly stable molecules even in harsh conditions such as boiling, acid or alkaline pH, and freeze/thaw cycles so that their expression levels in serum are reproducible and consistent among individuals [23]. Thus, they could represent suitable candidates for becoming sensitive biomarkers in different pathologic conditions.
miRNA expression profiles in osteoporosis
Despite many studies have analyzed the role of miRNAs in other common disorders (e.g., cancer and cardiovascular diseases), the data in osteoporosis are limited and in most part inconclusive. Indeed, most of the available information has derived from studies performed in different samples (e.g., serum, circulating monocytes or human BM-MSCs, and bone tissue specimens) and in different conditions (i.e., low BMD or fracture and selection of control samples with or without osteoarthritis) or ethnic groups (Table 5). A consistent variation in the number of screened miRNAs and in the employed technology platforms also exists, making difficult to perform a comparison across these studies [108, 109]. Moreover, the majority of circulating miRNAs is not tissue specific and in part derives from blood cells, so that the biological interpretation of their variation in osteoporosis is challenging. However, notwithstanding the limitations described above and the consistent differences in most of the associations reported to date, taken all together, these studies suggest that perturbations in either circulating or skeletal miRNA levels are present in osteoporosis and might be linked to altered bone metabolism and fracture risk. Of interest, some of the circulating miRNAs associated with osteoporosis and fractures (e.g., miRNA-21 and miRNA-27) have been also associated with the regulation of muscle mass and sarcopenia in elderly individuals [110].
Circulating miRNA signatures in osteoporosis
The first evidence of a peculiar miRNA signature in patients with discordant BMD status comes from a TaqMan miRNA array analysis in monocytes derived from peripheral blood of postmenopausal women with high or low BMD, as assessed by DXA analysis [95]. Among the 365 tested miRNAs, miRNA-133a was significantly highly expressed in the low BMD group. Of interest, this miRNA is encoded by two different genes within two loci previously associated with osteoporosis, 18q11.2 and 20q13 [111, 112]. Since a similar analysis in isolated B-cells derived from the same subjects did not evidence any difference in miRNA-133a expression, it was speculated that this miRNA might be a monocyte-specific marker for osteoporosis. Indeed, circulating monocytes are relevant for bone metabolism since they can differentiate into osteoclasts, and bioinformatic analysis identified three potential osteoclast-related target genes of miRNA-133a (CXCL11, CXCR3, and SLC39A1). Moreover, further validation analysis of four marginally upregulated miRNAs from the same cohort evidenced miRNA-422a as an additional BMD-associated miRNA, which may potentially target four genes involved in the inhibition of osteoclastogenesis, CBL, CD226, PAG1, and TOB2 [97].
In a different study, Chen et al. performed microRNA profiling (721 miRNAs) in freshly isolated CD14+ PBMCs in a sample of postmenopausal women and identified miRNA-503 as the most significantly downregulated miRNA in patients with osteoporosis [40]. Among the other relevant associations, and consistent with the previous study, miRNA-133a was upregulated in osteoporotic women. In order to better investigate the skeletal effects of miRNA-503, either cellular or animal studies were also performed. Of interest, RANK was confirmed to be a target of this miRNA and miRNA-503 overexpression or silencing inhibited or stimulated RANKL-induced osteoclastogenesis in the CD14+ PBMCs, respectively. Moreover, overexpression of miRNA-503 with agomiR inhibited bone resorption and prevented bone loss in the OVX mice model.
By using different approaches, two studies investigated variation in miRNA profiles in BM-MSCs of patients with osteoporosis, as compared to age-matched controls. In the first study, a comparative microarray analysis (covering 1040 miRNAs) was performed between BM-MSCs obtained from premenopausal women and postmenopausal women with osteoporosis demonstrating a decrease in miRNA-21 levels after menopause, which was also consistent with experimental observations in the OVX mice model [92]. Additional experimental analyses revealed that a suppression of miRNA-21 might contribute to the impairment of bone formation by elevated TNF-α in estrogen-deficiency-induced osteoporosis. The positive effect of miRNA-21 on bone formation might occur via the repression of Spry1, a negative regulator of the FGF and ERK-MAPK signaling pathways, which are established as being involved in promoting osteogenesis of MSCs. In a subsequent study, a candidate miRNA approach was employed, with the selection of miRNA-125b, because of its role as crucial transcriptional regulator of genes that are involved in cell proliferation or differentiation processes of various cell lineages [98]. A significant upregulation of this miRNA was observed in BM-MSCs derived from patients with senile osteoporosis than in controls. Consistent with this observation, a miRNA-125b mimic was able to suppress the osteogenic differentiation of these cells, mainly by a downregulation of Runx2.
More recent studies mainly investigated miRNA expression profiles in sera of patients with or without osteoporosis [88–92]. Of interest, concordant associations were observed concerning some miRNAs (e.g., miRNA-21, miRNA-125b, and miRNA-133a) even though most of the reported associations have not been replicated among these studies or the previous evidences in monocytes and BM-MSCs. In an interesting study by Seeliger et al., either serum or bone tissue miRNA profiling was performed [99]. In the first analysis, a panel of 83 miRNAs was screened between two pooled samples from ten osteoportic hip fracture patients and ten controls. Then, replication analysis of informative miRNAs was performed by RT-PCR in a larger sample of sera (30 osteoporotic and 30 nonosteoporotic patients), as well as in bone tissue samples from 20 osteoporotic and 20 nonosteoporotic subjects. Overall, after validation, nine miRNAs (miRNA-21, miRNA-23a, miRNA-24, miRNA-93, miRNA-100, miRNA-122a, miRNA-124a, miRNA-125b, and miRNA-148a) were found to be significantly upregulated in the serum of patients with osteoporosis. Of these miRNAs, five were also upregulated in bone tissue samples of osteoporotic patients (miRNA-21, miRNA-23a, miRNA-24, miRNA-100, and miRNA-125b). Conversely, miRNA-25 was only upregulated in osteoporotic bone.
Two additional studies specifically investigated miRNA expression profile in serum of patients with osteoporotic fractures as compared to controls, leading to completely different outcomes [102, 103]. In the first study performed in seven Caucasian patients suffering from recent osteoporotic fractures and seven controls, a screening of 175 miRNAs led to the identification of differentially expressed miRNAs, of which three were upregulated (miRNA-10a-5p, miRNA-10b-5p, and miRNA-22-3p) and three downregulated (miRNA-133b, miRNA-328-3p, and let-7g-5p) in fractured patients [103]. Subsequent validation in a larger sample (n = 23) confirmed a significant effect for miRNA-22-3p, miRNA-328-3p, and let-7g-5p. While both miRNA-22-3p and the let-7 family have been previously associated with osteogenesis of MSCs in vitro, miRNA-328-3p was shown to target the expression of CD44 in macrophages [113]. The second study analyzed a panel of 179 most expressed miRNAs in human serum in two RNA pools from eight Spanish women with osteoporotic hip fracture with respect to five women with osteoarthritis also undergoing surgery for hip prosthesis implantation [102]. Further validation in a cohort of 15 fracture patients and 12 osteoarthritic subjects showed that three miRNAs (miRNA-21-5p, miRNA-122-5p, and miRNA-125-5b) were valuable upregulated biomarkers for osteoporotic fractures. All the three associations were consistent with the results previously reported by Seeliger et al. Moreover, expression levels of miRNA-21-5p were positively and highly correlated with CTX, a marker of bone resorption. By a different approach, Li et al. investigated three candidate miRNAs (miRNA-21, miRNA-133a, and miRNA-146a) in plasma samples from 120 Chinese postmenopausal women divided into osteoporotic, osteopenic, and normal according to total hip BMD levels [100]. An upregulation of miRNA-133a was observed, which is somewhat consistent with the previous results by Wang et al. [95] in circulating monocytes. However, a downregulation of miRNA-21 was also demonstrated in osteoporotic women, on the opposite of what described in the Spanish cohort of patients with osteoporotic fractures [102]. Apart the obvious difference in ethnicity between the two studies, a likely explanation of these conflicting results might be related to the complex role of miRNA-21 on bone biology, acting either as a promoter of osteogenesis or as an inducer of osteoclastogenesis, so that marked variation on its expression profile could be expected following a fracture. In a more recent study comparing the miRNA expression profiles (microarray platform covering 851 human miRNAs) of a sample of 81 postmenopausal osteoporotic women with 74 healthy premenopausal women, miRNA-27a was one of the most strongly downregulated miRNAs in the serum of osteoporotic patients, and thus was selected for experimental analysis [105]. This miRNA was upregulated during osteoblastic differentiation of either human or mice MSCs, while a downregulation was observed in case of adipocyte induction. Moreover, silencing of miRNA-27a decreased bone formation parameters in mice without significant effects on osteoclasts and bone resorption. Bioinformatic analysis followed by luciferase assay demonstrated that myocyte enhancer factor 2c (Mef2c) was the direct target of miRNA-27a. These results were consistent with previous experimental observations [54, 114], suggesting that a reduction in miRNA-27a might be involved in the aged-related decrease in osteogenic differentiation of MSCs, thus contributing to the pathogenesis of senile osteoporosis. On the other side, recent analyses also demonstrated that miRNA-31 levels increases with age in serum as well as in microvescicles derived from human senescent endothelial cells and are significantly higher in plasma from osteoporotic patients than in age-matched controls [71]. These results are consist with the in vitro evidences indicating a pro-osteoclastogenic and anti-osteoblastogenic role of miRNA-31 [28, 69, 70], thus suggesting that the age-related increase in this miRNA might also be relevant for senile osteoporosis.
Finally, two more recent and complex studies investigated whether miRNAs or combinations of miRNAs can discriminate best fracture status in different conditions of bone fragility. In the first of these reports, Heilmeier et al. performed a miRNA expression analysis in two different cohorts of fractured women with postmenopausal osteoporosis or diabetic bone disease [106]. While postmenopausal osteoporosis is generally associated with bone loss and enhanced bone turnover, diabetes is characterized by increased bone fragility in the presence of normal BMD levels and depressed bone turnover [115]. Overall, 20 diabetic women with and 20 without fragility fractures were enrolled in the diabetic group, while a similar sample of 20 postmenopausal women with and 20 without osteoporotic fractures was tested in the nondiabetic group. All the four groups had similar BMD levels at the femoral neck, mostly within the osteopenic range. Remarkably, 48 out of 375 tested miRNAs were identified to be differentially expressed between type 2 diabetic women with or without fractures and 23 miRNAs between nondiabetic women with and without fractures. Of these, 18 miRNAs showed the same patterns of regulation in either diabetic or nondiabetic patients with fractures. Further analyses with multivariate classification models led to the identification of potential candidate miRNA signatures that could best differentiate fracture status in patients with or without diabetes. Among the most overrepresented miRNAs, one (miRNA-382-3p, which was downregulated) was common between diabetic and nondiabetic fractured patients while three (miRNA-96-5p, miRNA-181-5p, and miRNA-550a-5p, all upregulated) were specific among the diabetic signatures and two (miRNA-188-3p and miRNA-942, both downregulated) among the osteoporotic signatures. While either miRNA-96 or miRNA-188 was previously related with osteogenesis (as described above), the other miRNAs were not previously associated with osteoporosis nor with specific alterations in bone cell homeostasis, even though most of them have been involved in oxidative stress response and mitochondrial dysfunction. Preliminary in vitro analyses in human-adipose-tissue-derived MSCs suggested an inhibitory effect of miRNA-550a-5p or a stimulatory effect of miRNA-382-3p on osteogenesis, respectively [106]. In a subsequent analysis, Kocijan et al. assessed circulating miRNA signatures in male and female subjects with idiopathic or postmenopausal osteoporotic fractures [107]. Based on the results from previous published studies, 187 miRNAs were selected for analysis. Importantly, to avoid the potential influence of fracture healing on miRNA profiles, all samples were collected after at least 6 months from the occurrence of the last fracture. Overall, a common set of 3 (miRNA-152-3p, miRNA-335-5p, and miRNA-320a) and 16 (let-7b-5p, miRNA-7-5p, miRNA-16-5p, miRNA-19a-3p, miRNA-19b-3p, miRNA-29b-3p, miRNA-30e-5p, miRNA-93-5p, miRNA-140-5p, miRNA-215-5p, miRNA-186-5p, miRNA-324-3p, miRNA-365a-3p, miRNA-378a-5p, miRNA-532-5p, and miRNA-550a-3p) miRNAs were, respectively, upregulated and downregulated in fracture groups of men, premenopausal and postmenopausal women than in the respective age- and sex-matched controls. Among these miRNAs, eight (miRNA-152-3p, miRNA-335-5p, miRNA-19a-3p, miRNA-19b-3p, miRNA-30e-5p, miRNA-140-5p, miRNA-324-3p, and miRNA-550a-3p) were confirmed to be excellent discriminators of fractures regardless of age and gender, with a higher predictive power than BMD or bone turnover markers. While miR324-3p, miRNA19a-3p, and miRNA-19b-3p have not yet been associated with bone remodeling, most of the other miRNAs were previously related with osteogenesis. The results from this important study provided for the first time specific evidence for the robustness of a diagnostic signature for osteoporosis based on microRNAs.
Bone-specific miRNA signatures of patients with osteoporotic fractures
In an initial analysis of bone specimens from 40 aged patients with fractures, Wang et al. evidenced a positive correlation between expression levels of miRNA-214, but not the other 33 examined miRNAs, with the bone formation markers osteocalcin and alkaline phosphatase [96]. In keeping with this observation, in vitro osteoblast activity and matrix mineralization were promoted by antagomiR-214 and suppressed by agomiR-214, and further analysis suggested that activating transcription factor 4 (ATF4, encoding for a relevant transcription factor required for osteoblast function) could be a functional target of miRNA-214 and may mediate its effects in bone formation. Moreover, osteoblast-specific manipulation of miRNA-214 levels by miRNA-214 antagomiR treatment in miRNA-214 transgenic, OVX, or hind limb-unloaded mice confirmed the inhibitory role of miRNA-214 in regulating bone formation.
Following this report and that of Seeliger et al. [99], other evidences demonstrated peculiar miRNA signatures within the bone tissue specimens of patients undergoing hip replacement for osteoporotic fractures, even though with different results [101, 104]. Given the practical and ethical difficulties to obtain bone samples from nonosteoporotic subjects, in both studies, nonfractured patients with hip osteoarthritis were selected as controls. In the first study, tissue levels of 760 miRNAs were analyzed in bone specimens of eight hip fracture patients and eight osteoarthritis controls [101]. Five miRNAs with statistically significant differences in expression in the discovery stage (miRNA-187, miRNA-193a-3p, miRNA-214, miRNA-518f, and miRNA-636) were selected for the replication stage (sample of 19 hip fracture patients and 19 controls) and two of them, miRNA-187 and miRNA-518f, were confirmed to be significantly downregulated or upregulated in patients with osteoporotic fractures, respectively. In a second study, fresh trabecular bone samples from 12 postmenopausal women undergoing hip replacement due to either osteoporotic fracture (n = 6) or osteoarthritis in the absence of osteoporosis (n = 6) were tested with a miRNA array targeting all human, mouse, or rat miRNAs registered in the miRBASE 18.0 [104]. In addition, a complementary array was made from human primary osteoblasts obtained from postmenopausal women after knee replacement due to osteoarthritis. Overall, 790 and 315 different miRNAs were detected in fresh bone samples and in primary osteoblasts, respectively, 293 of which were common to both groups. The eight miRNAs with the lowest p values (miRNA-675-5p, miRNA-30c-1-3p, miRNA-483-5p, miRNA-542-5p, miRNA-142-3p, miRNA-223-3p, miRNA-32-3p, and miRNA-320a) were then assayed in a validation cohort, and two (miRNA-320a and miRNA-483-5p) were confirmed to be upregulated in the osteoporotic samples. Both these miRNAs were expressed in primary osteoblasts suggesting a possible role in the regulation of bone formation; miRNA-320a has already known to target β-catenin [116], while miRNA-483-5p appeared to downregulate IGF2 levels in osteoblast cultures.
miRNA variants as genetic determinants of osteoporosis
There are increasing reports of genetic variants that can affect or interfere with miRNAs function in different conditions, including bone disorders [117, 118]. In 2009, Li et al. provided the first and unique evidence that a mutation in a miRNA precursor is associated with early onset osteoporosis [78]. Following experimental analyses that lead to the identification of miRNA-2861 as a specific miRNA of the osteoblast lineage that promotes osteogenesis, they screened miRNA-2861 levels in bone from patients with osteoporosis and fractures and found undetectable levels of this miRNA in a 15-year-old boy and in his 17-year-old sister. Both individuals had a history of repeated fragility fractures. Mutational screening identified a common homozygous C-G mutation in the stem of pre-miRNA-2861, blocking the expression of mature miRNA-2861 in vitro. Levels of HDAC5, a target of miRNA-2861, were elevated and Runx2 protein expression was decreased in both mutated patients in comparison with controls. Further mutational screening in 357 normal children, 396 healthy adults, and 369 adult patients with osteoporosis did not evidence any other mutation in pre-miRNA-2861.
More commonly, either single-nucleotide polymorphisms (SNPs) or alternative poly-adenylation can affect miRNA binding of a given transcript from different individuals and tissues, thus emerging as major factors that potentially contribute to variations in miRNA-mRNA interplay. To date, a single study analyzed the skeletal effects of a SNP in pri-miRNA-34b/c (rs4938723) [119], which had been previously shown to significantly affect promoter transcriptional efficiency, leading to aberrant expression of miRNA-34b/c [120]. In a sample of 310 Chinese osteoporotic patients and 371 controls, the presence of the CC and CT/CC pri-miRNA-34b/c genotypes were associated with a significantly reduced risk of osteoporosis compared with the TT genotype. Indeed, as recently reviewed by Dole et al. [118], functional SNPs have been already identified for many miRNAs associated with bone cell activity and/or osteoporosis (e.g., miRNA-27a, miRNA-124, miRNA-125a, miRNA-125b, miRNA-146, miRNA-186a) and were demonstrated to affect the respective miRNA levels with potential implications on different disorders. However, their role on bone biology remains to be investigated.
Likewise, SNPs present at or near miRNA binding sites in protein coding genes could also affect miRNA function, creating or eliminating mRNA binding sites and thus potentially leading to differential protein expression of target genes. In a first analysis by Lei et al., 568 known SNPs within 3′-UTRs of target mRNAs were screened in relation to osteoporosis in a sample of 997 white Caucasian individuals [121]. After replication analyses in a larger cohort of 1728 subjects, three SNPs (rs6854081, rs1048201, and rs7683093) in the fibroblast growth factor 2 (FGF-2) gene were significantly associated with femoral neck BMD. These SNPs reside within nine predicted miRNA target sites (miRNA-25, miRNA-32, miRNA-92, miRNA-92b, miRNA-146a, miRNA-146b, miRNA-363, miRNA-367, and miRNA-545). Subsequent gene expression analyses in monocytes or B-cells from selected individuals consistently demonstrated depressed expression of the FGF2 gene in subjects with high BMD compared with subjects with low BMD. Since most of the 3′-UTRs SNPs of target mRNAs regions are not covered by the available commercial SNPs arrays, it is likely that other relevant associations remain to be discovered.
In another study, Dole et al. showed that a SNP in the 3′-UTR of the osteonectin gene (rs1054204, 1599C/G), previously associated with male idiopathic osteoporosis, creates a target site for miRNA-433 [122]. The presence of the less common 1599G allele related with osteoporotic risk, repressed osteonectin posttranslational regulation by creating the new target site. Osteonectin is known to suppress adipogenic differentiation of MSCs and osteonectin null mice develop low turnover osteoporosis. In keeping with these observations, knock-in mice with the protective allele 1599C displayed higher osteopontin levels in bone, associated with higher bone formation rate, higher trabecular bone volume and greater increases in cortical bone volume in response to teriparatide, compared with mice carrying the 1599G allele [110].
Given the multifactorial and polygenic nature of osteoporosis it is likely that in the future an extended analysis of genetic variations associated with miRNAs and their predicted binding sites could improve our knowledge about the genetics of this disorder, as well as the accuracy of fracture prediction tools.
miRNA expression profiles in other bone disorders
To date, the role of miRNAs in disorders of bone metabolism other than osteoporosis has been poorly investigated. A preliminary screening of more than 100 bone-related miRNAs was performed in serum of 22 patients with osteogenesis imperfecta (OI) compared with 10 healthy controls, with the identification of 11 differently expressed miRNAs [123]. Of them, three (miRNA-26a, miRNA-30e, and miRNA-21) were upregulated and eight (miRNA-34c, miRNA-29a, miRNA-29b, miRNA-489, miRNA-133a, miRNA-145, miRNA-210, and miRNA-1297) downregulated in OI cases. As is evident, several of the OI-associated miRNAs were also described in serum from patients with osteoporosis. A more recent analysis investigated whether expression levels of miRNA-29b may affect the phenotype of OI patients with collagen type 1 mutations [124]. The selection of this miRNA was due to its documented role on bone metabolism and particularly for its COL1A1-dependent regulation of collagen protein accumulation during mineralization, as observed in vitro [76]. However, either COL1A1 or miRNA-29b expression was severely reduced in both type I and type III OI patients. Thus, it was speculated that reduced COL1A1 levels observed in OI are not sufficient for the induction of miRNA-29b.
To identify potential microRNA-target pairs associated with osteopetrosis, Ou et al. applied a combined approach including deep sequencing, quantitative proteomics, and bioinformatics analyses in PBMCs from six affected patients with mutations in CLCN7 and nine age- and sex-matched healthy donors [125]. Overall, 123 differently expressed microRNAs, 173 differently expressed proteins, and 117 computationally predicted miRNA-target pairs with reciprocally expressed level in PBMCs were found in the two sample groups. Among the miRNAs with significantly increased expression level, miRNA-23a was the most abundant, while miRNA-29b-3p was among the most significantly downregulated miRNA in osteopetrosis cohort. Both miRNAs have been previously involved in repressing or stimulating osteoblast differentiation, respectively [54, 73, 126]. Moreover, one of the predicted “miRNA-target pairs,” miRNA-320a and ADP-ribosylation factor 1 (Arf1), was chosen to be further tested in vitro. Notably, Arfs are a family of ubiquitously expressed Ras-like GTPases that have key roles in vesicular transport processes and osteoclast function [127]. The use of miRNA-320a mimics in cell cultures suppressed Arf1 expression, demonstrating that Arf1 is a target of miRNA-320a. Thus, it was speculated that in CLCN7-related osteopetrosis, downregulation of miRNA-320a might occur as a response to a defective chloride channel in order to increase Arf1 levels and improve osteoclast functions. Based on these results, it could be possible that variable expression levels of miRNA-320a or other miRNAs among patients with osteopetrosis might account for differences in their clinical phenotype.
Furthermore, at least three reports evidenced an altered miRNA expression pattern in parathyroid tumors or parathyroid hyperplasia with respect to normal parathyroid tissue [128–130]. Despite some overlaps in the differentially expressed miRNAs between the different parathyroid diseases as compared with normal parathyroid samples, some miRNAs were unique to parathyroid carcinoma or parathyroid adenoma, and a limited number to parathyroid hyperplasia. Generally, up to 60% of miRNAs were downregulated in parathyroid tumors, while in parathyroid hyperplasia, most miRNAs were upregulated [129]. Importantly, in most instances, miRNA profiling showed distinct differentially expressed miRNAs by tumor type, which might result as helpful adjunct to distinguish parathyroid adenoma from carcinoma. In an extended analysis, 91 miRNAs were differentially expressed between adenoma and carcinoma, and the most informative in this respect were miRNA-26b, miRNA-30b, and miRNA-126* [129]. In the two other reports, different associations were provided, with three relevant miRNAs (miRNA-296, miRNA-503, and miRNA-222) or two miRNA clusters in chromosome 19 being the most differentially expressed in parathyroid cancer [128, 130].
In order to further characterize the potential implication of miRNAs on the parathyroid gland, a parathyroid-specific Dicer1 knockout mouse, where parathyroid miRNA maturation is blocked, was recently generated [131]. Despite these mice showed normal calcium and PTH levels under physiologic conditions, they did not respond to acute hypocalcemia by increasing their PTH levels. Moreover, they also had a blunted response to chronic hypocalcemia with a fourfold diminished increase in PTH and absent parathyroid cell proliferation compared to control mice. Similarly, a blunted increase in either PTH or FGF-23 levels was observed in these mice than in controls after induction of uremia. Overall, these results suggest that the response of the parathyroid to both acute or chronic hypocalcemia and uremia is dependent upon parathyroid Dicer and thus intact miRNA function, further indicating a potential role of miRNAs in parathyroid diseases and, possibly, in other disorders of calcium and phosphate metabolism.
The role of miRNAs in other common disorders of bone and mineral metabolism such as Paget’s disease of bone or osteomalacia remains to be investigated. Indeed, a recent abstract presentation evidenced major changes in the expression profile of several miRNAs in either PBMCs or serum from patients with Paget’s disease of bone, as compared to osteoporotic patients or healthy controls, particularly in the presence of mutation in SQSTM1 gene [132].
Taken together, all the above evidences suggest a relevant involvement of miRNAs in many disorders of bone metabolism. While their role as specific disease biomarkers could be of limited relevance for most of these conditions, it is likely that an extended characterization of miRNA profiles among different skeletal disorders might reveal useful for the identification of new therapeutic targets as well as for a better understanding of the variable phenotype characteristics frequently observed even in patients with the same genetic defect.
Importantly, a more consistent deregulation of miRNAs profiles has been widely described in skeletal neoplasia such as osteosarcoma or bone metastases, and in myeloma bone disease, with the identification of several relevant miRNAs. This issue has been revised in many previous articles [133, 134] and is beyond the scope of this review.
Conclusions and future directions
miRNAs have a huge potential either for the diagnosis or treatment of several disorders. In fact, they are increasingly recognized as important regulatory modulators of a large number of biological functions and their expression profile often differs under diverse pathologic conditions. However, the broad and important functions of these regulators for skeletal biology are only now becoming apparent.
Despite additional information is certainly required to have a more clear picture about the in vivo effects of miRNAs on skeletal biology, cumulative evidence from experimental studies has highlighted their crucial role in the commitment of osteogenesis from MSCs precursors as well as in the modulation of either osteoblast or osteoclast formation and activity, thus providing new perspectives on the regulation of skeletal homeostasis. In this respect, compelling future steps will necessary include the improving of our knowledge on how the numerous miRNAs integrate each other within the skeleton to regulate bone homeostasis.
Importantly, available information from some studies in mice with the use of miRNA mimics or antagomiRs also indicates that targeting miRNAs might become an attractive and new therapeutic approach for osteoporosis or other skeletal diseases as well as for tissue engineering application in bone regeneration and repair. However, there are a number of hurdles that greatly hamper the therapeutic use of miRNAs at this stage, mainly concerning the way of efficient miRNA delivery to bone in vivo and the off target effects, since each single miRNA can regulate many target genes within common relevant pathways to extraskeletal tissues. Indeed, some of the miRNAs with potential skeletal benefits (e.g., miRNA-29a) have been also demonstrated to have a crucial role in tumorigenesis acting as protooncogenes, making unlikely their use for therapeutic purposes in bone diseases.
At the same time, it will be essential to improve our knowledge about the potential application of circulating miRNAs (e.g., in serum, PBMCs, or exosomes) as biomarkers for osteoporosis or other skeletal disorders through large-scale prospective studies, with the inclusion of fractures as endpoints, and taking into account that lifestyle, physical activity, diet, and in some instances, circadian rhythm may all influence the expression profile of circulating miRNAs [109].
References
Roberts SB, Wootton E, De Ferrari L, Albagha OM, Salter DM (2015) Epigenetics of osteoarticular diseases: recent developments. Rheumatol Int 35:1293–1305. doi:10.1007/s00296-015-3260-y
Jacquier A (2009) The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10:833–844. doi:10.1038/nrg2683
Tay Y, Rinn J, Pandolfi PP (2014) The multilayered complexity of ceRNA crosstalk and competition. Nature 505:344–352. doi:10.1038/nature12986
Nagano T, Fraser P (2011) No-nonsense functions for long noncoding RNAs. Cell 145:178–181. doi:10.1016/j.cell.2011.03.014
Häsler J, Strub K (2006) Alu elements as regulators of gene expression. Nucleic Acids Res 34:5491–5497. doi:10.1093/nar/gkl706
Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105. doi:10.1101/gr.082701.108
Macfarlane LA, Murphy PR (2010) MicroRNA: biogenesis, function and role in cancer. Curr Genomics 11:537–561. doi:10.2174/138920210793175895
Li SC, Tang P, Lin WC (2007) Intronic microRNA: discovery and biological implications. DNA Cell Biol 26:195–207. doi:10.1089/dna.2006.0558
Winter J, Jung S, Keller S, Gregory RI, Diederichs S (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11:228–234. doi:10.1038/ncb0309-228
Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ (2003) Dicer is essential for mouse development. Nat Genet 35:215–217. doi:10.1038/ng1253
Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN (2004) The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev 18:3016–3027. doi:10.1101/gad.1262504
Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T, Kato S, Miyasaka N, Ezura Y, Noda M (2010) Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem 109:866–875. doi:10.1002/jcb.22228
Gaur T, Hussain S, Mudhasani R, Parulkar I, Colby JL, Frederick D, Kream BE, van Wijnen AJ, Stein JL, Stein GS, Jones SN, Lian JB (2010) Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol 340:10–21. doi:10.1016/j.ydbio.2010.01.008
Sugatani T, Hruska KA (2009) Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem 284:4667–4678. doi:10.1074/jbc.M805777200
Sugatani T, Vacher J, Hruska KA (2011) A microRNA expression signature of osteoclastogenesis. Blood 117:3648–3657. doi:10.1182/blood-2010-10-311415
Lai EC (2002) Micro RNAs are complementary to 3 UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30:363–364. doi:10.1038/ng865
Marco A, Macpherson JI, Ronshaugen M, Griffiths-Jones S (2012) MicroRNAs from the same precursor have different targeting properties. Silence 3:8. doi:10.1186/1758-907X-3-8
Baskerville S, Bartel DP (2005) Microarray profiling of microRNAs reveals frequent co-expression with neighboring miRNAs and host genes. RNA 11:241–247. doi:10.1261/rna.7240905
Lytle JR, Yario TA, Steitz JA (2007) Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5 UTR as in the 3 UTR. Proc Natl Acad Sci U S A 104:9667–9672. doi:10.1073/pnas.0703820104
Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318:1931–1934. doi:10.1126/science.1149460
Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A 105:1608–1613. doi:10.1073/pnas.0707594105
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi:10.1038/ncb1596
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105:10513–10518. doi:10.1073/pnas.0804549105
Bader AG (2012) miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet 3:120. doi:10.3389/fgene.2012.00120
Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR (2013) Treatment of HCV infection by targeting microRNA. N Engl J Med 368:1685–1694. doi:10.1056/NEJMoa1209026
Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y (2012) MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol 8:212–227. doi:10.1038/nrendo.2011.234
Franceschetti T, Dole NS, Kessler CB, Lee SK, Delany AM (2014) Pathway analysis of microRNA expression profile during murine osteoclastogenesis. PLoS One 9:e107262. doi:10.1371/journal.pone.0107262
Tang P, Xiong Q, Ge W, Zhang L (2014) The role of microRNAs in osteoclasts and osteoporosis. RNA Biol 11:1355–1363. doi:10.1080/15476286.2014.996462
Ji X, Chen X, Yu X (2016) MicroRNAs in osteoclastogenesis and function: potential therapeutic targets for osteoporosis. Int J Mol Sci1 7:349. doi:10.3390/ijms17030349
Sugatani T, Hruska KA (2013) Down-regulation of miR-21 biogenesis by estrogen action contributes to osteoclastic apoptosis. J Cell Biochem 114:1217–1222. doi:10.1002/jcb.24471
Franceschetti T, Kessler CB, Lee SK, Delany AM (2013) miR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J Biol Chem 288:33347–33360. doi:10.1074/jbc.M113.484568
Wang FS, Chuang PC, Lin CL, Chen MW, Ke HJ, Chang YH, Chen YS, Wu SL, Ko JY (2013) MicroRNA-29a protects against glucocorticoid-induced bone loss and fragility in rats by orchestrating bone acquisition and resorption. Arthritis Rheum 65:1530–1540. doi:10.1002/art.37948
Cheng P, Chen C, He HB, Hu R, Zhou HD, Xie H, Zhu W, Dai RC, Wu XP, Liao EY, Luo XZ (2013) miR-148a regulates osteoclastogenesis by targeting v-maf musculo aponeurotic fibrosarcoma oncogenes homolog b. J Bone Miner Res 28:1180–1190. doi:10.1002/jbmr.1845
Zhao C, Sun W, Zhang P, Ling S, Li Y, Zhao D, Peng J, Wang A, Li Q, Song J, Wang C, Xu X, Xu Z, Zhong G, Han B, Chang YZ, Li Y (2015) miR-214 promotes osteoclastogenesis by targeting pten/pi3k/akt pathway. RNA Biol 12:343–353. doi:10.1080/15476286.2015.1017205
Guo LJ, Liao L, Yang L, Li Y, Jiang TJ (2014) miR-125a TNF receptor-associated factor 6 to inhibit osteoclastogenesis. Exp Cell Res 321:142–152. doi:10.1016/j.yexcr.2013.12.001
Takayanagi H, Kim S, Matsuo K, Suzuki H, Suzuki T, Sato K, Yocochi T, Oda H, Nakamura K, Ida N, Wagner EF, Taniguchi T (2002) RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature 416:744–749. doi:10.1038/416744a
Zhang J, Zhao H, Chen J, Xia B, Jin Y, Wei W, Shen J, Huang Y (2012) Interferon-β-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett 586:3255–3262. doi:10.1016/j.febslet.2012.06.047
Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M (2011) The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum 63:1582–1590. doi:10.1002/art.30321
Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, Sood AK, Mendell JT, Wan Y (2014) miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and tgif2. Nature 512:431–435. doi:10.1038/nature13375
Chen C, Cheng P, Xie H, Zhou HD, Wu XP, Liao EY, Luo XH (2014) MiR-503 regulates osteoclastogenesis via targeting RANK. J Bone Miner Res 29:338–347. doi:10.1002/jbmr.2032
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414. doi:10.1016/j.cell.2007.04.040
Fazi F, Rosa A, Fatica A, Gelmetti V, de Marchis ML, Nervi C, Bozzoni I (2005) A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and c/EBPα regulates human granulopoiesis. Cell 123:819–831. doi:10.1016/j.cell.2005.09.023
Dou C, Cao Z, Yang B, Ding N, Hou T, Luo F, Kang F, Li J, Yang X, Jiang H, Xiang J, Quan H, Xu J, Dong S (2016) Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci Rep 6:21499. doi:10.1038/srep21499
Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M, Kronenberg HM (2008) Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A 105:1949–1954. doi:10.1073/pnas.0707900105
Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, Ebert BL, Al-Shahrour F, Hasserjian RP, Scadden EO, Aung Z, Matza M, Merkenschlager M, Lin C, Rommens JM, Scadden DT (2010) Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature 464:852–857. doi:10.1038/nature08851
Tomé M, López-Romero P, Albo C, Sepúlveda JC, Fernández Gutiérrez B, Dopazo A, Bernad A, González MA (2011) miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ 18:985–995. doi:10.1038/cdd.2010.167
Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J, Chen J (2011) Effects of miR-335-5p in modulating osteogenic differentiation by specifically down regulating Wnt antagonist DKK1. J Bone Miner Res 26:1953–1963. doi:10.1002/jbmr.377
Suomi S, Taipaleenmäki H, Seppänen A, Ripatti T, Väänänen K, Hentunen T, Säämänen AM, Laitala-Leinonen T (2008) MicroRNAs regulate osteogenesis and chondrogenesis of mouse bone marrow stromal cells. Gene Regul Syst Bio 2:177–191
Laine SK, Alm JJ, Virtanen SP, Aro HT, Laitala-Leinonen TK (2012) MicroRNAs miR-96, miR-124, and miR-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J Cell Biochem 113:2687–2695. doi:10.1002/jcb.24144
Qin L, Chen Y, Niu Y, Chen W, Wang Q, Xiao S, Li A, Xie Y, Li J, Zhao X, He Z, Mo D (2010) A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics 11:320. doi:10.1186/1471-2164-11-320
Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY, Xia ZY, Zhou HD, Cao X, Xie H, Liao EY, Luo XH (2015) MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest 125:1509–1522. doi:10.172/JCI77716
Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, Iwasaki M, Sunamura S, Takeuchi Y, Fukumoto S, Saito K, Nakamura T, Siomi H, Ito H, Arai Y, Shinomiya K, Takeda S (2009) A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci U S A 106:20794–20799. doi:10.1073/pnas.0909311106
Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS (2008) A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A 105:13906–13911. doi:10.1073/pnas.0804438105
Zhao X, Xu D, Li Y, Zhang J, Liu T, Ji Y, Wang J, Zhou G, Xie X (2014) MicroRNAs regulate bone metabolism. J Bone Miner Metab 32:221–231. doi:10.1007/s00774-013-0537-7
Gong Y, Xu F, Zhang L, Qian Y, Chen J, Huang H, Yu Y (2014) MicroRNA expression signature for Satb2-induced osteogenic differentiation in bone marrow stromal cells. Mol Cell Biochem 387:227–239. doi:10.1007/s11010-013-1888-z
Zeng Y, Qu X, Li H, Huang S, Wang S, Xu Q, Lin R, Han Q, Li J, Zhao RC (2012) MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Lett 586:2375–2381. doi:10.1016/j.febslet.2012.05.049
Grünhagen J, Bhushan R, Degenkolbe E, Jäger M, Knaus P, Mundlos S, Robinson PN, Ott CE (2015) MiR-497-195 cluster microRNAs regulate osteoblast differentiation by targeting BMP signaling. J Bone Miner Res 30:796–808. doi:10.1002/jbmr.2412
Wu T, Zhou H, Hong Y, Li J, Jiang X, Huang H (2012) miR-30 family members negatively regulate osteoblast differentiation. J Biol Chem 287:7503–7511. doi:10.1074/jbc.M111.292722
Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS (2011) A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A 108:9863–9868. doi:10.1073/pnas.1018493108
Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB (2010) A network connecting Runx2, SATB2, and the miR-23a*27a*24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A 107:19879–19884. doi:10.1073/pnas.1007698107
Yu S, Geng Q, Ma J, Sun F, Yu Y, Pan Q, Hong A (2013) Heparin-binding EGF-like growth factor and miR-1192 exert opposite effect on Runx2-induced osteogenic differentiation. Cell Death Dis 4:e868. doi:10.1038/cddis.2013.363
Yang M, Pan Y, Zhou Y (2014) miR-96 promotes osteogenic differentiation by suppressing HBEGF-EGFR signaling in osteoblastic cells. FEBS Lett 588:4761–4768. doi:10.1016/j.febslet.2014.11.008
Chen Q, Liu W, Sinha KM, Yasuda H, de Crombrugghe B (2013) Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor Osterix. PLoS One8:e58104. doi:10.1371/journal.pone.0058104
Zuo B, Zhu J, Li J, Wang C, Zhao X, Cai G, Li Z, Peng J, Wang P, Shen C, Huang Y, Xu J, Zhang X, Chen X (2015) microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J Bone Miner Res 30:330–345. doi:10.1002/jbmr.2352
Kim KM, Park SJ, Jung SH, Kim EJ, Jogeswar G, Ajita J, Rhee Y, Kim CH, Lim SK (2012) miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J Bone Miner Res 27:1669–1679. doi:10.1002/jbmr.1604
Liao L, Su X, Yang X, Hu C, Li B, Lv Y, Shuai Y, Jing H, Deng Z, Jin Y (2016) TNF-α inhibits FoxO1 by up-regulating miR-705 to aggravate oxidative damage in bone marrow-derived mesenchymal stem cells during osteoporosis. Stem Cells 34:1054–1067. doi:10.1002/stem.2274
Eskildsen T, Taipaleenmäki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M (2011) MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A 108:6139–6144. doi:10.1073/pnas.1016758108
Chen L, Holmstrøm K, Qiu W, Ditzel N, Shi K, Hokland L, Kassem M (2014) MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells 32:902–912. doi:10.1002/stem.1615
Xie Q, Wang Z, Bi X, Zhou H, Wang Y, Gu P, Fan X (2014) Effects of miR-31 on the osteogenesis of human mesenchymal stem cells. Biochem Biophys Res Commun 446:98–104. doi:10.1016/j.bbrc.2014.02.058
Baglìo SR, Devescovi V, Granchi D, Baldini N (2013) MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene 527:321–331. doi:10.1016/j.gene.2013.06.021
Weilner S, Schraml E, Wieser M, Messner P, Schneider K, Wassermann K, Micutkova L, Fortschegger K, Maier AB, Westendorp R, Resch H, Wolbank S, Redl H, Jansen-Dürr P, Pietschmann P, Grillari-Voglauer R, Grillari J (2016) Secreted microvesicular miR-31 inhibits osteogenic differentiation of mesenchymal stem cells. Aging Cell 15:744–754. doi:10.1111/acel.12484
Martin PJ, Haren N, Ghali O, Clabaut A, Chauveau C, Hardouin P, Broux O (2015) Adipogenic RNAs are transferred in osteoblasts via bone marrow adipocytes-derived extracellular vesicles (EVs). BMC Cell Biol 16:10. doi:10.1186/s12860-015-0057-5
Sun M, Zhou X, Chen L, Huang S, Leung V, Wu N, Pan H, Zhen W, Lu W, Peng S (2016) The regulatory roles of microRNAs in bone remodeling and perspectives as biomarkers in osteoporosis. Biomed Res Int 2016:1652417. doi:10.1155/2016/1652417
Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM (2010) miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem 285:25221–25231. doi:10.1074/jbc.M110.116137
Kapinas K, Kessler CB, Delany AM (2009) MiR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J Cell Biochem 108:216–224. doi:10.1002/jcb.22243
Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB (2009) Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem 284:15676–15684. doi:10.1074/jbc.M809787200
Trompeter HI, Dreesen J, Hermann E, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, Wernet P (2013) MicroRNAs miR-26a, miR-26b, and miR-29b accelerate osteogenic differentiation of unrestricted somatic stem cells from human cord blood. BMC Genomics 14:111. doi:10.1186/1471-2164-14-111
Li H, Xie H, Liu W, Hu R, Huang B, Tan YF, Xu K, Sheng ZF, Zhou HD, Wu XP, Luo XH (2009) A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest 119:3666–3677. doi:10.1172/JCI39832
Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang W, Jafferji M, Gordon JA, Li Z, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB (2012) miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem 287:42084–42092. doi:10.1074/jbc.M112.377515
Tamura M, Uyama M, Sugiyama Y, Sato M (2013) Canonical Wnt signaling activates miR-34 expression during osteoblastic differentiation. Mol Med Rep 8:1807–1811. doi:10.3892/mmr.2013.1713
Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH (2012) miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet 21:2991–3000. doi:10.1093/hmg/dds129
Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, Boyce BF, Lee B (2008) Dimorphic effects of Notch signaling in bone homeostasis. Nat Med 14:299–305. doi:10.1038/nm1712
Guo L, Xu J, Qi J, Zhang L, Wang J, Liang J, Qian N, Zhou H, Wei L, Deng L (2013) MicroRNA-17-92a up-regulation by estrogen leads to Bim targeting and inhibition of osteoblast apoptosis. J Cell Sci 126:978–988. doi:10.1242/jcs.117515
Zhou M, Ma J, Chen S, Chen X, Yu X (2014) MicroRNA-17-92 cluster regulates osteoblast proliferation and differentiation. Endocrine 45:302–310. doi:10.1007/s12020-013-9986-y
Mohan S, Wergedal JE, Das S, Kesavan C (2015) Conditional disruption of miR17-92 cluster in collagen type I-producing osteoblasts results in reduced periosteal bone formation and bone anabolic response to exercise. Physiol Genomics 47:33–43. doi:10.1152/physiolgenomics.00107.2014
Vimalraj S, Partridge NC, Selvamurugan N (2014) A positive role of microRNA-15b on regulation of osteoblast differentiation. J Cell Physiol 229:1236–1244. doi:10.1002/jcp.24557
Bhushan R, Grünhagen J, Becker J, Robinson PN, Ott CE, Knaus P (2013) miR-181a promotes osteoblastic differentiation through repression of TGF-β signaling molecules. Int J Biochem Cell Biol 45:696–705. doi:10.1016/j.biocel.2012.12.008
Li J, He X, Wei W, Zhou X (2015) MicroRNA-194 promotes osteoblast differentiation via down regulating STAT1. Biochem Biophys Res Commun 460:482–488. doi:10.1016/j.bbrc.2015.03.059
Mizuno Y, Tokuzawa Y, Ninomiya Y, Yagi K, Yatsuka-Kanesaki Y, Suda T, Fukuda T, Katagiri T, Kondoh Y, Amemiya T, Tashiro H, Okazaki Y (2009) miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett 583:2263–2268. doi:10.1016/j.febslet.2009.06.006
Dong J, Cui X, Jiang Z, Sun J (2013) MicroRNA-23a modulates tumor necrosis factor-alpha-induced osteoblasts apoptosis by directly targeting Fas. J Cell Biochem 114:2738–2745. doi:10.1002/jcb.24622
Schmidt Y, Simunovic F, Strassburg S, Pfeifer D, Stark GB, Finkenzeller G (2015) miR-126 regulates platelet-derived growth factor receptor-α expression and migration of primary human osteoblasts. Biol Chem 396:61–70. doi:10.1515/hsz-2014-0168
Yang N, Wang G, Hu C, Shi Y, Liao L, Shi S, Cai Y, Cheng S, Wang X, Liu Y, Tang L, Ding Y, Jin Y (2013) Tumor necrosis factor α suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis. J Bone Miner Res 28:559–573. doi:10.1002/jbmr.1798
Lisse TS, Chun RF, Rieger S, Adams JS, Hewison M (2013) Vitamin D activation of functionally distinct regulatory miRNAs in primary human osteoblasts. J Bone Miner Res 28:1478–1488. doi:10.1002/jbmr.1882
Zhou Q, Zhao ZN, Cheng JT, Zhang B, Xu J, Huang F, Zhao RN, Chen YJ (2011) Ibandronate promotes osteogenic differentiation of periodontal ligament stem cells by regulating the expression of microRNAs. Biochem Biophys Res Commun 404:127–132. doi:10.1016/j.bbrc.2010.11.079
Wang Y, Li L, Moore BT, Peng XH, Fang X, Lappe JM, Recker RR, Xiao P (2012) MiR-133a in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. PLoS One 7:e34641. doi:10.1371/journal.pone.0034641
Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G, Cao H, Wu H, Song J, Pan X, Sun Q, Ling S, Li Y, Zhu M, Zhang P, Peng S, Xie X, Tang T, Hong A, Bian Z, Bai Y, Lu A, Li Y, He F, Zhang G, Li Y (2013) miR-214 targets ATF4 to inhibit bone formation. Nat Med 19:93–100. doi:10.1038/nm.3026
Cao Z, Moore BT, Wang Y, Peng XH, Lappe JM, Recker RR, Xiao P (2014) MiR-422a as a potential cellular microRNA biomarker for postmenopausal osteoporosis. PLoS One 9:e97098. doi:10.1371/journal.pone.0097098
Chen S, Yang L, Jie Q, Lin YS, Meng GL, Fan JZ, Zhang JK, Fan J, Luo ZJ, Liu J (2014) MicroRNA-125b suppresses the proliferation and osteogenic differentiation of human bone marrow derived mesenchymal stem cells. Mol Med Rep 9:1820–1826. doi:10.3892/mmr.2014.2024
Seeliger C, Karpinski K, Haug AT, Vester H, Schmitt A, Bauer JS, van Griensven M (2014) Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J Bone Miner Res 29:1718–1728. doi:10.1002/jbmr.2175
Li H, Wang Z, Fu Q, Zhang J (2014) Plasma miRNA levels correlate with sensitivity to bone mineral density in postmenopausal osteoporosis patients. Biomarkers 19:553–556. doi:10.3109/1354750X.2014.935957
Garmilla-Ezquerra P, Sañudo C, Delgado-Calle J, Pérez-Nuñez MI, Sumillera M, Riancho JA (2015) Analysis of the bone microRNome in osteoporotic fractures. Calcif Tissue Int 96:30–37. doi:10.1007/s00223-014-9935-7
Panach L, Mifsut D, Tarín JJ, Cano A, García-Pérez MÁ (2015) Serum circulating microRNAs as biomarkers of osteoporotic fracture. Calcif Tissue Int 97:495–505. doi:10.1007/s00223-015-0036-z
Weilner S, Skalicky S, Salzer B, Keider V, Wagner M, Hildner F, Gabriel C, Dovjak P, Pietschmann P, Grillari-Voglauer R, Grillari J, Hackl M (2015) Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone 79:43–51. doi:10.1016/j.bone.2015.05.027
De-Ugarte L, Yoskovitz G, Balcells S, Güerri-Fernández R, Martinez-Diaz S, Mellibovsky L, Urreizti R, Nogués X, Grinberg D, García-Giralt N, Díez-Pérez A (2015) MiRNA profiling of whole trabecular bone: identification of osteoporosis-related changes in MiRNAs in human hip bones. BMC Med Genet 8:75. doi:10.1186/s12920-015-0149-2
You L, Pan L, Chen L, Gu W, Chen J (2016) MiR-27a is essential for the shift from osteogenic differentiation to adipogenic differentiation of mesenchymal stem cells in postmenopausal osteoporosis. Cell Physiol Biochem 39:253–265. doi:10.1159/000445621
Heilmeier U, Hackl M, Skalicky S, Weilner S, Schroeder F, Vierlinger K, Patsch J, Baum T, Oberbauer E, Lobach I, Burghardt A, Schwartz A, Grillari J, Link T (2016) Serum microRNAs are indicative of skeletal fractures in postmenopausal women with and without type 2 diabetes and influence osteogenic and adipogenic differentiation of adipose-tissue derived mesenchymal stem cells in vitro. J Bone Miner Res. doi:10.1002/jbmr.2897
Kocijan R, Muschitz C, Geiger E, Skalicky S, Baierl A, Dormann R, Plachel F, Feichtinger X, Heimel P, Fahrleitner-Pammer A, Grillari J, Redl H, Resch H, Hackl M (2016) Circulating microRNA signatures in patients with idiopathic and postmenopausal osteoporosis and fragility fractures. J Clin Endocrinol Metab 101:4125–4134. doi:10.1210/jc.2016-2365
Zampetaki A, Mayr M (2012) Analytical challenges and technical limitations in assessing circulating miRNAs. Thromb Haemost 108:592–598. doi:10.1160/TH12-02-0097
Hackl M, Heilmeier U, Weilner S, Grillari J (2016) Circulating microRNAs as novel biomarkers for bone diseases—complex signatures for multifactorial diseases? Mol Cell Endocrinol 432:83–95. doi:10.1016/j.mce.2015.10.015
Brown DM, Goljanek-Whysall K (2015) microRNAs: modulators of the underlying pathophysiology of sarcopenia? Ageing Res Rev 24:263–273. doi:10.1016/j.arr.2015.08.007
Hsu YH, Zillikens MC, Wilson SG, Farber CR, Demissie S, Soranzo N, Bianchi EN, Grundberg E, Liang L, Richards JB, Estrada K, Zhou Y, van Nas A, Moffatt MF, Zhai G, Hofman A, van Meurs JB, Pols HA, Price RI, Nilsson O, Pastinen T, Cupples LA, Lusis AJ, Schadt EE, Ferrari S, Uitterlinden AG, Rivadeneira F, Spector TD, Karakis D, Kiel DP (2010) An integration of genome-wide association study and gene expression profiling toprioritize the discovery of novel susceptibility loci for osteoporosis-related traits. PLoS Genet 6:e1000977. doi:10.137/journal.pgen.1000977
Ralston SH, Galwey N, MacKay I, Albagha OM, Cardon L, Compston JE, Cooper C, Duncan E, Keen R, LAngdahl B, McLellan A, O’Riordan J, Pols HA, Reid DM, Uitterlinden AG, Wass J, Bennett ST (2005) Loci for regulation of bone mineral density in men and women identified by genomewide linkage scan: the FAMOS study. Hum Mol Genet 14:943–951. doi:10.1093/hmg/ddi088
Ishimoto T, Sugihara H, Watanabe M, Sawayama H, Iwatsuki M, Baba Y, Okabe H, Hidaka K, Yokoyama N, Miyake K, Yoshikawa M, Nagano O, Komohara Y, Takeya M, Saya H, Baba H (2014) Macrophage-derived reactive oxygen species suppress miR-328 targeting CD44 in cancer cells and promote redox adaptation. Carcinogenesis 35:1003–1011. doi:10.1093/carcin/bgt402
Wang T, Xu Z (2010) miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem Biophys Res Commun 402:186–189. doi:10.1016/j.bbrc.2010.08.031
Merlotti D, Gennari L, Dotta F, Lauro D, Nuti R (2010) Mechanisms of impaired bone strength in type 1 and 2 diabetes. Nutr Metab Cardiovasc Dis 20:683–690. doi:10.1016/j.numecd.2010.07.008
Sun JY, Huang Y, Li JP, Zhang X, Wang L, Meng YL, Yan B, Bian YQ, Zhao J, Wang WZ, Yang AG, Zhang R (2012) MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting beta-catenin. Biochem Biophys Res Commun 420:787–792. doi:10.1016/j.bbrc.2012.03.075
Afonso-Grunz F, Müller S (2015) Principles of miRNA-mRNA interactions: beyond sequence complementarity. Cell Mol Life Sci 72:3127–3141. doi:10.1007/s00018-015-1922-2
Dole NS, Delany AM (2016) MicroRNA variants as genetic determinants of bone mass. Bone 84:57–68. doi:10.1016/j.bone.2015.12.016
Jia F, Sun R, Li J, Li Q, Chen G, Fu W (2016) Interactions of pri-miRNA-34b/c and TP53 polymorphisms on the risk of osteoporosis. Genet Test Mol Biomarkers 20:398–401. doi:10.1089/gtmb.2015.0282
Zhang S, Qian J, Cao Q, Li P, Wang M, Wang J, Ju X, Meng X, Lu Q, Shao P, Zhang Z, Qin C, Yin C (2014) A potentially functional polymorphism in the promoter region of miR-34b/c is associated with renal cell cancer risk in a Chinese population. Mutagenesis 29:149–154. doi:10.1093/mutage/geu001
Lei SF, Papasian CJ, Deng HW (2011) Polymorphisms in predicted miRNA binding sites and osteoporosis. J Bone Miner Res 26:72–78. doi:10.1002/jbmr.186
Dole NS, Kapinas K, Kessler CB, Yee S, Adams DJ, Pereira RC, Delany AM (2015) A single nucleotide polymorphism in osteonectin 3 untranslated region regulates bone volume and is targeted by miR-433. J Bone Miner Res 30:723–732. doi:10.1002/jbmr.2378
Wang Z, Lu Y, Zhang X, Ren X, Wang Y, Li Z, Xu C, Han J (2012) Serum microRNA is a promising biomarker for osteogenesis imperfecta. Intractable Rare Dis Res 1:81–85. doi:10.5582/irdr.2012.v1.2.81
Kaneto CM, Lima PS, Zanette DL, Prata KL, Pina Neto JM, de Paula FJ, Silva WA Jr (2014) COL1A1 and miR-29b show lower expression levels during osteoblast differentiation of bone marrow stromal cells from Osteogenesis Imperfecta patients. BMC Med Genet 15:45. doi:10.1186/1471-2350-15-45
Ou M, Zhang X, Dai Y, Gao J, Zhu M, Yang X, Li Y, Yang T, Ding M (2014) Identification of potential microRNA-target pairs associated with osteopetrosis by deep sequencing, iTRAQ proteomics and bioinformatics. Eur J Hum Genet 22:625–632. doi:10.1038/ejhg.2013.221
Taipaleenmäki H, Bjerre Hokland L, Chen L, Kauppinen S, Kassem M (2012) Mechanisms in endocrinology: micro-RNAs: targets for enhancing osteoblast differentiation and bone formation. Eur J Endocrinol 166:359–371. doi:10.1530/EJE-11-0646
Heckel T, Czupalla C, Expirto Santo AI, Anitei M, Arantzazu Sanchez-Fernadez M, Mosch K, Krause E, Hoflack B (2009) Src-dependent repression of ARF6 is required to maintain podosome-rich sealing zones in bone-digesting osteoclasts. Proc Natl Acad Sci 106:1451–1456. doi:10.1073/pnas.0804464106
Corbetta S, Vaira V, Guarnieri V, Scillitani A, Eller-Vainicher C, Ferrero S, Vicentini L, Chiodini I, Bisceglia M, Beck-Peccoz P, Bosari S, Spada A (2010) Differential expression of microRNAs in human parathyroid carcinomas compared with normal parathyroid tissue. Endocr Relat Cancer 17:135–146. doi:10.1677/ERC-09-0134
Rahbari R, Holloway AK, He M, Khanafshar E, Clark OH, Kebebew E (2011) Identification of differentially expressed microRNA in parathyroid tumors. Ann Surg Oncol 18:1158–1165. doi:10.1245/s10434-010-1359-7
Vaira V, Elli F, Forno I, Guarnieri V, Verdelli C, Ferrero S, Scillitani A, Vicentini L, Cetani F, Mantovani G, Spada A, Bosari S, Corbetta S (2012) The microRNA cluster C19MC is deregulated in parathyroid tumours. J Mol Endocrinol 49:115–124. doi:10.1530/JME-11-0189
Shilo V, Ben-Dov IZ, Nechama M, Silver J, Naveh-Many T (2015) Parathyroid-specific deletion of dicer-dependent microRNAs abrogates the response of the parathyroid to acute and chronic hypocalcemia and uremia. FASEB J 29:3964–3976. doi:10.1096/fj.15-274191
Bianciardi S, Merlotti D, Sebastiani G, Valentini M, Gonnelli S, Caffarelli C, Evangelista IA, Cenci S, Nuti R, Dotta F, Gennari L (2016) Micro-RNA expression profiling in Paget’s disease of bone. Bone Abstracts 5:P452. doi:10.1530/boneabs.5.P452
Croset M, Kan C, Clézardin P (2015) Tumour-derived miRNAs and bone metastasis. Bonekey Rep 4:688. doi:10.1038/bonekey.2015.56
Nugent M (2015) microRNA and bone cancer. Adv Exp Med Biol 889:201–230. doi:10.1007/978-3-319-23730-5_11
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None.
Rights and permissions
About this article
Cite this article
Gennari, L., Bianciardi, S. & Merlotti, D. MicroRNAs in bone diseases. Osteoporos Int 28, 1191–1213 (2017). https://doi.org/10.1007/s00198-016-3847-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00198-016-3847-5