Abstract
A general framework for smooth regression of a functional response on one or multiple functional predictors is proposed. Using the mixed model representation of penalized regression expands the scope of function-on-function regression to many realistic scenarios. In particular, the approach can accommodate a densely or sparsely sampled functional response as well as multiple functional predictors that are observed on the same or different domains than the functional response, on a dense or sparse grid, and with or without noise. It also allows for seamless integration of continuous or categorical covariates and provides approximate confidence intervals as a by-product of the mixed model inference. The proposed methods are accompanied by easy to use and robust software implemented in the pffr function of the R package refund. Methodological developments are general, but were inspired by and applied to a diffusion tensor imaging brain tractography dataset.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Research in functional regression, where the responses and/or predictors are curves, has received great attention recently: see, e.g., Ramsay and Silverman (2005, Ch. 12), Ferraty and Vieu (2006, 2009), Aneiros-Pérez and Vieu (2008), Horváth and Kokoszka (2012, Ch. 8). However, the lack of working statistical inferential tools that are flexible enough for many settings encountered in practice and that are fully implemented in software, is a serious methodological and computational gap in the literature. In this paper we propose an estimation and inferential framework to study the association between a functional response and one or multiple functional predictors in a variety of settings. Specifically, we introduce penalized function-on-function regression (PFFR) implemented in the pffr function of the R (2014) package refund (Crainiceanu et al. 2014). The proposed framework is very flexible and accommodates (1) multiple functional predictors observed on the same or different domains than the response, and in various realistic scenarios such as predictors and response observed on dense or sparse grids, as well as (2) linear and nonlinear effects of multiple scalar covariates.
The paper considers linear relationships between the functional response and the functional predictor, so that the effect of the predictor is expressed as the integral of the corresponding covariate weighted by a smooth bivariate coefficient function. Function-on-function linear regression models are well known; see, e.g., Ramsay and Silverman (2005, Ch. 16), and Horváth and Kokoszka (2012, Sect. 8.3).
Most of the work in this area (e.g. Aguilera et al. 1999; Yao et al. 2005b; He et al. 2010; Wu et al. 2010; Horváth and Kokoszka 2012, Sect. 8.3) considers regression with a single functional predictor, or with an additional lagged response functional explanatory process, see Valderrama et al. 2010, and is based on an eigenbasis expansion for both the functional predictor and the corresponding functional coefficient. While these approaches are intuitive, they are also subject to two subtly dangerous problems. First, estimating the number of components used for the expansion of the functional predictors is known to be difficult, and, as noticed in Crainiceanu et al. (2009), the shape and interpretation of the functional parameter can change dramatically when one includes one or two additional principal components. This problem is exacerbated by the fact that eigenfunctions corresponding to smaller eigenvalues tend to be much wigglier in applications. Second, the smoothness of the functional parameter is induced by the choice of basis dimension of the functional predictor. This may lead to strong under-smoothing when the functional parameter is much smoother than the higher order principal components. Furthermore, principal component-based function-on-function regression methods given in the literature currently do not incorporate a large number of functional predictors or effects of additional scalar covariates. While such an extension, at least for linear effects of scalar covariates, might be possible using the hybrid principal component approach of Ramsay and Silverman (2005, Ch. 10), the quality of estimation, optimal scaling of variables and scalability to a large number of functional and/or scalar covariates would certainly need further research. Penalized approaches with basis functions expansions for function-on-function regression (e.g. Ramsay and Silverman 2005, Ch. 14, Ch. 16; Horváth and Kokoszka 2012, p. 130) and their implementations are not currently developed to the fullest generality, focusing on one functional predictor, no scalar covariates (Matsui et al. 2009), or on the concurrent model, for a setting where both variables are observed over the same domain (e.g. Ramsay and Silverman 2005, Ch. 14). Regression models for functional responses with non-linear effect of functional predictors have been considered in Ferraty et al. (2011, 2012). While such modeling approaches are more general in how they account for the effect of the functional predictor, it is not clear how to extend them to account for additional functional or scalar predictors. A current work (Fan et al. 2014) discusses a functional response model with scalar covariates and a single index model for the functional covariates. This is an interesting approach focusing on prediction, though not on inference for the estimated effects. Kadri et al. (2011) consider a nonparametric approach with both functional and scalar covariates based on reproducing kernels, but this short proceedings paper gives no details on implementation, choice of kernel, selection of smoothing parameter, simulations or applications, it does not consider inference in addition to prediction, nor does it provide software. Most of the discussed approaches are furthermore limited to the case of functional responses and functional covariates not being sparsely observed.
We propose a novel solution for the linear function-on-function regression setting. Our approach is inspired by the penalized functional regression (PFR) in Goldsmith et al. (2011), developed for the simpler case of scalar on function regression. It uses basis function expansions of the smooth coefficient function/s, based on pre-determined bases, and applies quadratic roughness penalties to avoid overfitting. The smoothness of the functional coefficients is controlled by smoothing parameters, which are estimated using restricted maximum likelihood (REML) in an associated mixed model. Estimation of the functional coefficients is carried out under the working assumption of independent errors.
To the best of the authors’ knowledge, the developed pffr function is the first publicly available software for fitting function-on-function regression models with two or more functional predictors and additional scalar covariates. The fda package (Ramsay et al. (2014)) in R includes functions linmod and fRegress for function-on-function regression. However, linmod (see Ramsay et al. (2009), Ch. 10.3) cannot handle multiple functional predictors or linear or non-linear effects of scalar covariates. Also, the fRegress function is restricted to concurrent associations, where response and predictors are observed on the same domain. The PACE package (Yao et al. 2005b) in Matlab (2014) does not accommodate multiple functional predictors, nor linear or smooth effects of additional scalar covariates.
The main contributions of this paper thus are the following: (1) It develops a modeling and estimation framework for function-on-function regression flexible enough to accommodate multiple functional covariates, functional responses and/or covariates observed on possibly different non-equidistant or sparse grids, as well as smooth effects of scalar covariates, (2) it describes model-based and bootstrap-based confidence intervals, (3) it provides an open source implementation in the R package refund and describes the implementation in detail, (4) it tests these methods in an application and in extensive simulations, including realistic scenarios, such as covariates or responses observed sparsely and with noise.
The remainder of the article proceeds as follows: We present our PFFR methodology in Sect. 2. Section 3 describes the software implementation of this method in detail. The performance of PFFR in simulations is discussed in Sect. 4. Section 5 presents an application to the motivating diffusion tensor imaging (DTI) tractography dataset. Section 6 provides our discussion.
2 Mixed model representation of function-on-function regression
2.1 Function-on-function regression framework
While the approach is general, the presentation is focused on the case of two functional predictors, which is also relevant in our DTI application in Sect. 5. Let \(Y_i(t_{ij})\) be the functional response for subject \(i\) measured at \(t_{ij} \in \mathcal {T}\), an interval on the real line, \( 1 \le i \le n\), \(1 \le j \le m_{i}\), where \(n\) is the number of curves or subjects and \(m_i\) is the number of observations for curve \(i\). We assume that the observed data for the \(i^{\mathrm{th}}\) subject are \([ \{Y_{i}(t_{ij}) \}_j, \{ X_{i1}(s_{ik})\}_k, \{ X_{i2}(r_{iq}) \}_q, {\varvec{W}}_{i} ]\), where \(\{X_{i1}(s_{ik}): 1\le k \le K_{i}\}\) and \(\{X_{i2}(r_{iq}): 1\le q \le Q_{i}\}\) are functional predictors and \({\varvec{W}}_{i}\) is a \(p \times 1\) vector of scalar covariates. Furthermore, it is assumed that \(\{X_{i1}(s_{ik}) \}\) and \(\{X_{i2}(r_{iq})\}\) are square-integrable, finite sample realizations of some underlying stochastic processes \(\{ X_{1}(s): s \in \mathcal {S} \} \) and \(\{ X_{2}(r): r \in \mathcal {R}\}\) respectively, where \(\mathcal {S}\) and \(\mathcal {R}\) are intervals on the real line. We start with the following model for \(Y_{i}(t_{ij})\):
where the mean function is modeled semiparametrically (Ruppert et al. 2003; Wood 2006) and consists of two components: a linear parametric function \({\varvec{W}}_{i}^{t} {\varvec{\gamma }}\) to account for the covariates \({\varvec{W}}_{i}\), whose effects are assumed linear and constant along the interval \(\mathcal {T}\), where \({\varvec{\gamma }}\) is a p-dimensional parameter, and an overall nonparametric function \(\beta _{0}(t)\). The effect of the functional predictors is captured by the component \(\int _\mathcal {S} \beta _{1}(t_{ij},s) X_{i1}(s) \,ds + \int _\mathcal {R}\beta _{2}(t_{ij},r) X_{i2}(r) \,dr\). The regression parameter functions, \(\beta _1(\cdot ,\cdot )\) and \(\beta _2(\cdot ,\cdot )\), are assumed to be smooth and square integrable over their domains. The errors \(\epsilon _i\) are zero-mean random processes and are uncorrelated with the functional predictors and scalar covariates. The model will be extended in several different ways in the following sections.
2.1.1 Extension to sparsely observed functional responses
Our approach is able to accommodate sparseness of the observed response trajectories. This work is the first, to the best of our knowledge, to consider a sparsely observed functional response within the general setting of multiple functional and scalar predictors. Let \(\{ t_{ij}: j=1,\ldots , m_{i} \}\) be the set of grid points at which the response for curve \(i\) is observed such that \(\cup _{i=1}^{n} [\{ t_{ij} \}_{j=1}^{m_{i}} ]\) is dense in \(\mathcal {T}\). PFFR can easily accommodate such a scenario with only few modifications in the implementation, which we outline in Sect. 3.2. Section 4.2 provides simulation results obtained in this setting.
2.1.2 Extension to corruptly observed functional predictors
When the functional predictors are observed with error or not on the same fine grid, the underlying smooth curves need to be estimated first. If gridpoints are relatively fine, then a popular method is to smooth each trajectory separately using common smoothing approaches (Ramsay and Silverman 2005; Ramsay et al. 2009, Ch. 5). Zhang and Chen (2007) provide justification for local polynomial methods that this approach can estimate the true curves with asymptotically negligible error under some regularity assumptions. If grids of time points are sparse, the smoothing is done by pooling information across curves. We follow an approach similar to Yao et al. (2005a): first consider an undersmooth estimator of the covariance function and then smooth it to remove the measurement error. At the latter step we use penalized splines-based smoothing, as previously employed by Goldsmith et al. (2011). In such cases, the PFFR methodology is recommended to be applied to the reconstructed smooth trajectories, \(\widehat{X}_{i1}(s)\) and \(\widehat{X}_{i2}(r)\), that, in addition, are centered to have zero mean function. This approach is investigated numerically in Sect. 4.3.
2.2 A penalized criterion for function-on-function regression
In this section we introduce a representation of model (1) that allows us to estimate the model using available mixed model software, as detailed in Sect. 2.3. We discuss the case when the predictor curves, \(X_{i1}(\cdot )\) and \(X_{i2}(\cdot )\), are measured densely and without noise. For simplicity of presentation, consider the case when \(K_{i}=K\), \(s_{ik}=s_{k}\), \(Q_{i}=Q\) and \(r_{iq}=r_{q}\) for every \(i, k\) and \(q\). Additionally, assume that the response curves are measured on a common grid, that is \(m_i=m\) and \(t_{ij}=t_j\) for every \(i\) and \(j\).
To start with, we represent the functional intercept using a basis function expansion as \(\beta _0(t) \approx \sum _{l=1}^{\kappa _0} A_{0,l}(t) \beta _{0,l}\), where \(A_{0,l}(\cdot )\) is a known univariate basis and \(\beta _{0,l}\) are the corresponding coefficients. Then, we use numerical integration to approximate the linear “function-on-function” terms; for example, \(\int _{\mathcal {S}} \beta _{1}(t,s) X_{i1}(s) ds\) can be approximated by \(\int _{\mathcal {S}} \beta _{1}(t,s) X_{i1}(s) ds \approx \sum _{k=1}^K \Delta _k\beta _{1}(t,s_{k})X_{i1}(s_{k})\), where \(s_{k}\), \(k=1,\ldots ,K\), forms a grid of points in \(\mathcal {S}\) and \(\Delta _k\) are the lengths of the corresponding intervals. The next step is to expand \(\beta _1(t,s) \approx \sum _{l=1}^{\kappa _1} a_{1,l}(t,s) \beta _{1,l}\), where \(a_{1,l}(\cdot ,\cdot )\) is a bivariate basis and \(\beta _{1,l}\) are the corresponding coefficients. Thus, \(\int _{\mathcal {S}} \beta _{1}(t,s) X_{i1}(s) ds\) can be approximated by
where \(\widetilde{X}_{i1}(s_{k})=\Delta _k X_{i1}(s_{k})\), and the accuracy of this approximation depends on how dense the grid \(\{s_k:k = 1,\ldots ,K\}\) is. Note that \(X_{i1}\) values can be included directly in the function-on-function term and we do not apply a basis expansion reconstruction approach for \(X_{i1}\) if \(X_{i1}\) is observed densely and without noise. Using similar notation, \(\int _{\mathcal {R}} \beta _{2}(t,r) X_{i2}(r) dr\) can be approximated by
Thus, we approximate model (1) by the additive model:
where \(A_{1,l,i}(t)= \sum _{k=1}^{K} a_{1,l}(t,s_{k}) \widetilde{X}_{i1}(s_{k})\) and \(A_{2,l,i}(t)= \sum _{q=1}^{Q} a_{2,l}(t,r_{q}) \widetilde{X}_{i2}(r_{q})\) are known because the predictor functions are observed without noise.
While the presentation is provided in full generality, the various choices involved are crucial when one develops practical software. Our approach to smoothing is to use rich bases that reasonably exceed the maximum complexity of the parameter functions to be estimated and then penalize the roughness of these functions. This translates into choosing a large number of basis functions, \(\kappa _0\), \(\kappa _1\), \(\kappa _2\), and applying the penalties \(\lambda _0P_0({\varvec{\beta }}_0)\), \(\lambda _1P_1({\varvec{\beta }}_1)\), and \(\lambda _2P_2({\varvec{\beta }}_2)\), where \({\varvec{\beta }}_d\) is the vector of all parameters \(\beta _{d,l}\) for \(d=0,1,2\). Thus, if we denote by \(\mu _{i}(t;{\varvec{\gamma }},{\varvec{\beta }}_0,{\varvec{\beta }}_1,{\varvec{\beta }}_2)\) the mean of \(Y_i(t)\), our penalized criterion to be minimized is
This is a penalized least squares or penalized integrated squared error criterion, and is a natural extension of the integrated residual sum of squares criterion discussed in Ramsay and Silverman (2005, Ch. 16.1).
Penalties, \(P_0({\varvec{\beta }}_0)\), \(P_1({\varvec{\beta }}_1)\), and \(P_2({\varvec{\beta }}_2)\) are of known functional form with the amount of shrinkage being controlled by the three scalar smoothing parameters \(\lambda _0\), \(\lambda _1\), and \(\lambda _2\). We employ quadratic penalties, \(P_0({\varvec{\beta }}_0)={\varvec{\beta }}_0^t{\varvec{D}}_0{\varvec{\beta }}_0\), \(P_1({\varvec{\beta }}_1)={\varvec{\beta }}_1^t{\varvec{D}}_1{\varvec{\beta }}_1\), \(P_2({\varvec{\beta }}_2)={\varvec{\beta }}_2^t{\varvec{D}}_2{\varvec{\beta }}_2\), where \({\varvec{D}}_0\), \({\varvec{D}}_1\), \({\varvec{D}}_2\) are known penalty matrices associated with the chosen basis. Examples include integrated square of second derivative penalties (c.f. Wood 2006, Sect. 3.2.2), B-spline bases with a difference penalty (Eilers and Marx 1996), but also thin plate splines, cubic regression splines, cyclic cubic regression splines or Duchon splines (Wood 2006). Among the possible criteria for selection of the smoothing parameters, generalized cross validation (GCV), AIC and REML are most popular. We prefer REML in a particular mixed model due to its favorable comparison to GCV in terms of MSE and stability (Reiss and Ogden 2009; Wood 2011); our preference is also motivated by the theoretical results that restricted maximum likelihood-based selection of the smoothing parameters is more robust than AIC to moderate violations of the independent error assumption, see Krivobokova and Kauermann (2007). Estimation of smoothing parameters and model parameters are described next.
2.3 Mixed model representation of function-on-function regression
We note that the penalized criterion (3) with the quadratic penalty choices described in the previous section becomes
where \(\sigma ^2_\epsilon \) is the variance of the errors \(\epsilon _i(t)\) in model (1). In the literature on semiparametric regression (see e.g. Ruppert et al. 2003), such a penalized log-likelihood has a well-established connection to mixed model estimation, where the \(\beta \) coefficients are treated during estimation as if they were random effects. The intuition behind this approach is that a penalty can be thought of as similar to a prior in a Bayesian setting or a normal distribution for random effects in a mixed model context, in terms of incorporating prior knowledge (here of smoothness), and in fact the quadratic penalty corresponds to the logarithm of a normal distribution density for the coefficients. Mathematically, by denoting \(\sigma ^2_0=\sigma ^2_\epsilon /\lambda _0\), \(\sigma ^2_1=\sigma ^2_\epsilon /\lambda _1\), \(\sigma ^2_2=\sigma ^2_\epsilon /\lambda _2\) and following arguments identical to those in Ruppert et al. (2003, Sect. 4.9) minimizing the penalized criterion (3) is equivalent to obtaining the best linear unbiased predictor in the mixed model
where \({\varvec{D}}_0\), \({\varvec{D}}_1\), and \({\varvec{D}}_2\) are known, and shrinkage of the functional parameters is controlled by \(\sigma ^2_0\), \(\sigma ^2_1\), and \(\sigma ^2_2\), respectively. There is a slight abuse of notation in model (4), as matrices \({\varvec{D}}_0\), \({\varvec{D}}_1\), and \({\varvec{D}}_2\) are typically not invertible. Indeed, in many cases, only a subset of the coefficients are being penalized or the penalty matrix is rank deficient. These are well known problems in penalized regression and the standard solution (Wood 2006, Ch. 6.6) is to either replace the inverse with a particular generalized inverse or to separate the coefficients that are penalized from the ones that are not. For example, for the penalty corresponding to \(\beta _{1}\), the eigendecomposition for matrix \({\varvec{D}}_1\) is used to split the coefficients into two kinds: one kind that is not penalized, i.e. in the null-space of the penalty matrix, which would be estimated as fixed coefficients, and the other kind, say \({\varvec{b}}_1^R\) that is penalized (not in the null-space of the penalty) and that would be treated as random effects during estimation using the distributional assumption \({\varvec{b}}_1^{R}\sim N({\varvec{0}},\sigma ^2_1{\varvec{D}}_{1+}^{-1})\), where \({\varvec{D}}_{1+}\) is a projection of \({\varvec{D}}_1\) into the cone of positive definite matrices; see Sect. 6.6.1, Wood (2006), for a detailed presentation of this partitioning of penalized coefficient vectors into a mixed model representation with fixed and random components for producing smooth estimators for the parameters.
Replacing the penalized approach with the mixed model (4) has many favorable consequences. First, it allows the estimation of the smoothing parameters as certain variance ratios in this mixed model, \(\lambda _k = \sigma _\epsilon ^2 / \sigma _k^2\), \(k=0, 1, 2\), using REML estimation. This has favorable theoretical properties as discussed in the previous subsection and avoids computationally costly cross-validation. Second, models can naturally be extended in a likelihood framework to adapt to different levels of data complexity, such as the addition of smooth effects of scalar covariates. This enables us to expand the scope of function-on-function regression substantially. Third, inferential methods originally developed for mixed models transfer to function-on-function regression. In particular, approximate confidence intervals (Wahba 1983; Nychka 1988; Ruppert et al. 2003) can be obtained as a by-product of the fitting algorithm. This provides a statistically sound solution to an important problem that is currently unaddressed in function-on-function regression. We discuss implementation of this inferential tool when independent identically distributed (i.i.d.) errors in model (1) are assumed. Online Appendix A discusses first steps towards related inference tools for settings with non-i.i.d. errors. Moreover, tests on the shape of the association between responses and covariates are available as well. For suitably chosen bases and penalties, likelihood ratio tests of linearity versus non-linearity, or constancy versus non-constancy, of the coefficient surfaces along the lines described in Crainiceanu and Ruppert (2004), Greven et al. (2008) can be performed with the R package RLRsim (Scheipl et al. 2008).
We propose to fit model (4) using frequentist model software based on REML estimation of the variance components. The robust mgcv package (Wood 2014) in R (R Development Core Team 2014), which is designed for penalized regression and has a built-in capability to construct the penalty matrices that are appropriate for the specified spline bases can be used for model fitting.
3 Function-on-function regression via mixed models software
3.1 Implementation for functional regression with densely observed functional response
We now turn our attention to implementation. In particular, we explain how model (4) can be fit using the mgcv package (Wood 2014) in R. Note that our representation of the function-on-function regression model (2) and (3) is a penalized additive model where the original basis functions are re-weighted via the expressions \(A_{1,l,i}(t)= \sum _{k=1}^{K} a_{1,l}(t,s_{k}) \widetilde{X}_{i1}(s_{k})\) and \(A_{2,l,i}(t)= \sum _{q=1}^{Q} a_{2,l}(t,r_{q}) \widetilde{X}_{i2}(r_{q})\). If \(a_{0,l}(\cdot )\), \(a_{1,l}(\cdot ,\cdot )\), and \(a_{2,l}(\cdot ,\cdot )\) are, for example, thin plate spline bases, then the mgcv fit can simply be expressed as

These very short software lines deserve an in-depth explanation to illustrate the direct connection to our mixed model representation of function-on-function regression. In order to fit the model, the response functions \(Y_i(t)\) are stacked in an \(nm\times 1\) dimensional vector \({\varvec{Y}}=\{Y_1(t_1),\ldots ,Y_1(t_m),Y_2(t_1), \ldots ,Y_n(t_m)\}^t\), where \(n\) is the number of curves (or subjects) and \(m\) is the number of observations per curve. This vector is labeled Y. The \(p\times 1\) dimensional vectors of covariates \({\varvec{W}}_i\) are also stacked using the same rules used for \(Y_i(t)\). More precisely, for every subject \(i\) the vector \({\varvec{W}}_i\) is repeated \(m\) times and rows are row-stacked in an \(m\times p\) dimensional matrix \(\widetilde{{\varvec{W}}}_i\). These subject specific matrices are further row-stacked across subjects to form an \(mn\times p\) dimensional matrix \({\varvec{W}}\). This matrix is labeled W. The next step is to define a grid of points for the smooth function \(\beta _0(t)\) in model (1). The grid corresponds exactly to the stacking of the response functions and is defined as the \(mn\times 1\) dimensional vector \({\varvec{t}}_0=(t_1,\ldots ,t_m,\ldots ,t_1,\ldots ,t_m)^t\) obtained by stacking \(n\) repetitions of the grid vector \((t_1,\ldots ,t_m)^t\). This vector is labeled t_0. The expression s(t_0) thus fits a penalized univariate thin-plate spline at the grid points in the vector \({\varvec{t}}_0\).
So far, data manipulation and labeling have been quite straightforward. However, fitting the functional part of the model requires some degree of software customization. We focus on the first functional component and build three \(nm\times K\) dimensional matrices: (1) \({\varvec{t}}_1\), obtained by column-binding \(K\) copies of the \(nm\times 1\) dimensional vector \({\varvec{t}}_0\); this matrix is labeled t_1; (2) \({\varvec{s}}_{0}\) obtained by row-binding \(nm\) copies of the \(1\times K\) dimensional vector \((s_1,\ldots ,s_K)\); this matrix is labeled s_0; and (3) \(\widetilde{{\varvec{X}}}_1\) the \(nm\times K\) dimensional matrix
where each row \(\{ \widetilde{X}_{i1}(s_{1}) , \ldots , \widetilde{X}_{i1}(s_{K}) \}\) is repeated \(m\) times, for \(i=1, \ldots , n\); this matrix is labeled DX1. With these notations, the expression s(t_1,s_0,by=DX1) is essentially building \(\sum _l A_{1,l,i}(t)\beta _{1,l}(t)\), where \(A_{1,l,i}(t)= \sum _{k=1}^{K} a_{1,l}(t,s_{k}) \widetilde{X}_{i1}(s_{k})\). Without the option by=DX1 the expression s(t_1,s_0) would build the bivariate thin-plate spline basis \(a_{1,l}(t,s_{k})\), whereas adding by=DX1 it averages these bivariate bases along the second dimension using the weights \(\widetilde{X}_{i1}(s_{k})\) derived from the first functional predictor. A similar construction is done for the second functional predictor. Smoothing for all three penalized splines is done via REML, as indicated by the option method=“REML”.
The implementation presented above is simple, but the great flexibility of the gam function allows multiple useful extensions. First, it is obvious that the method and software can be adapted to a larger number of functional predictors. The number of basis functions used for each smooth can be adjusted. For example, requiring \(k_0=10\) basis functions for a univariate thin-plate penalized spline for the \(\beta _0(\cdot )\) function can be obtained by replacing s(t_0) with s(t_0,k=10). Moreover, the implementation can accommodate unevenly sampled grid points both for the functional response and predictors. Indeed, nothing in the modeling or implementation requires equally spaced grids. Changing from (isotropic) bivariate thin-plate splines to (anisotropic) tensor product splines based on two univariate bases can be done by replacing \(\mathtt{s(t_1,s_0,by=DX1)}\) with \(\mathtt{te(t_1,s_0,by=DX1)}\). This changes criterion (3) only slightly, such that \({\varvec{\beta }}_{1}\) then has two additive penalties in \(s\) and \(t\) direction, respectively. We can also easily incorporate linear effects of scalar covariates allowed to vary smoothly along \(t\) (varying coefficients), that is \(z_{i} {\beta _{3}}(t)\), using expressions of the type s(t_0,by=Z), where Z is obtained using a strategy similar to the one for functional predictors. For large datasets the function bam is more computationally efficient than gam.
In practice, it is useful to have a dedicated, user-friendly interface that automatically takes care of the stacking, concatenation and multiplication operations described here, calls the appropriate estimation routines in mgcv, and returns a rich model object that can easily be summarized, visualized, and validated. pffr offers a formula-based interface that accepts functional predictors and responses in standard matrix form, i.e., the \(i^{\mathrm{th}}\) row contains the function evaluations for subject \(i\) on an index vector like \({\varvec{t}}_0\) or \({s_{0}}\). It returns a model object whose fit can be summarized, plotted and compared with other model formulations without any programming effort by the user through the convenient and fully documented functions summary, plot and predict. The model formula syntax used to specify models is very similar to the established formula syntax to lower the barrier to entry for users familiar with the mgcv-package, i.e. to specify model (2), we use

where Ymat, X1mat, X2mat are matrices containing the function evaluations, and where ff(Xmat, yind=t) denotes a linear function-on-function term \(\int X(s)\beta (t,s) ds\). A functional intercept \(\beta _0(t)\) is included by default. The term c(W) corresponds to a constant effect of the covariates in \({\varvec{W}}\), i.e., \({\varvec{W^{t}\gamma }}\). By default, pffr associates scalar covariates with an effect varying smoothly on the domain of the response, i.e., \(\tilde{\,}\mathtt{~W}\) yields an effect \({\varvec{W\gamma }}(t)\). Our implementation also supports non-linear effects of scalar covariates \(z_{i}\) that may or may not be constant over the domain of the response, i.e., \(f(z_{i})\) or \(f(z_{i},t)\), specified as c(s(z)) or s(z), respectively, as well as multivariate non-linear effects of scalar covariates \(z_{1i}, z_{2i}\) that may or may not be constant over the domain of the response, i.e., \(f(z_{1i}, z_{2i})\) or \(f(z_{1i}, z_{2i}, t)\), specified as c(te(z1, z2)) or te(z1, z2), respectively.
Prior to applying our pffr procedure, we recommend to center the functional predictors. For each functional predictor, the mean function is estimated (e.g. Ramsay et al. 2009, Sect. 6.1; Bunea et al. 2011) and subtracted from each curve \(i\). When the functional predictors are centered, the functional intercept has the interpretation of the overall mean function for observations with functional predictor values at the respective mean values.
3.2 Implementation for functional regression with sparsely observed functional response
We have already seen in Sect. 3.1 that the class of models covered by our framework is much broader than model (1); specifically the proposed framework accommodates easily multiple functional predictors, varying coefficient effects, and non-linear effects of one or multiple scalar covariates. Here we present extensions to another realistic setting. PFFR can easily accommodate a function-on-function regression scenario when the functional response is sparsely observed, with only few modifications in the fitting procedure. First, the vector of responses, labeled Y, accommodates subject-responses observed at different time points, and has the form \({\varvec{Y}}=\{Y_1(t_{11}),\ldots ,Y_1(t_{1m_{1}}),Y_2(t_{21}), \ldots ,Y_2(t_{2m_{2}}), \ldots ,Y_n(t_{nm_{n}}))\}^t\). Second, the vector labeled t_0 has the form \({\varvec{t}}_0=(t_{11},\ldots ,t_{1m_{1}},\ldots ,t_{nm_{n}})^t\). Third, the matrix of covariates, labeled W, is obtained by taking \(m_i\) copies of the \(1\times p\) row vector of covariates \({\varvec{W}}_i\) for subject \(i\), column-stacking these into a \(m_i\times p\)-dimensional matrix \(\widetilde{{\varvec{W}}}_i\), and then further column-stacking these matrices across subjects. The functional components labeled DX1 and DX2 are constructed using a similar logic. There is no modification in the definitions of the vectors labeled \({\varvec{t}}_{1}\) and \({\varvec{s}}_{0}\). Our implementation in pffr performs these modified pre-processing steps automatically if a suitable data structure for sparsely observed functional responses is provided. With these adjustments, the fitting procedure can proceed with gam as described in Sect. 3.1. Section 4.2 provides simulation results obtained in this setting.
4 Simulation study
We conduct a simulation study to evaluate the performance of PFFR in realistic scenarios including both densely or sparsely observed functional predictors, and for a functional response that is densely or sparsely sampled.
Due to the lack of other available implemented methods to do regression with multiple functional and scalar covariates, we compare PFFR to a sequence of scalar-on-function regressions; see Sect. 4.4 for a comparison with linmod (Ramsay et al. 2009), for the simpler setting of a single functional covariate and no additional scalar covariates. More precisely, for every fixed \(t\), the function-on-function regression model (1) becomes a scalar on function regression that can be fit using, for example, PFR (Goldsmith et al. 2011), as implemented in the R function pfr from the refund package. PFR provides estimates of the functional parameters \(\beta _{1}(t,s)\) and \(\beta _{2}(t,r)\) for every fixed \(t\). Aggregating these functions over \(t\) leads to surface estimators that are then smoothed using a bivariate penalized spline smoother. This approach is labeled modified-PFR in the remainder of the paper. Without bivariate smoothing the modified-PFR was found not to be as competitive. For the modified-PFR method, the upper and lower bound surfaces of the confidence intervals were obtained analogously by smoothing the upper and lower bounds estimated by PFR for each fixed \(t\).
4.1 Densely sampled functional predictors
The curves \(Y_{i}(t)\) were observed on an equally spaced grid \(t \in \{ j/10: j=1,2,...,60\}\) and the functional predictors were observed on equally spaced grids, but on different domains: \(X_{i1}(s)\) on \(s \in \{ k/10: k=1,2,...,50\}\) and \(X_{i2}(r)\) on \(r \in \{ q/10: q=1,2,..., 70\}\). The bivariate functional parameters \(\beta _{1}(t,s)=\text{ cos }(t \pi /3) \text{ sin }(s \pi /5)\) and \(\beta _{2}(t,r)=\sqrt{tr}/4.2\) have comparable range and are displayed in the top left panels of Fig. 2. The functional intercept is \(\beta _{0}(t)=2 e^{-(t-2.5)^2}\) and the random errors \(\epsilon _i(t_{j})\) were simulated i.i.d. \(N(0,\sigma _{\epsilon }^{2})\).
We generated \(n\) functional responses \(Y_{i}(t)\) from model (1) by approximating the integrals via Riemann sums with a dense grid for each domain \(\mathcal {S}\) and \(\mathcal {R}\). For the first functional predictor we considered the following mean zero process \(V_{i1}(s)=X_{i1}(s)+\delta _{i1}(s)\), where \( X_{i1}(s)= \sum _{k=1}^{E_{s}}\{v_{ik_{1}} \text{ sin } (k\pi s/5) + v_{ik_{2}} \text{ cos } ( k \pi s/5 ) \}\) with \(E_{s}=10\), and where \(v_{ik_{1}}, v_{ik_{2}} \sim \) N(0,\(1/k^{4})\) are independent across subjects \(i\). For the second functional predictor we considered \(V_{i2}(r)=X_{i2}(r)+\delta _{i2}(r)\), where \(X_{i2}(r)=\sum _{k=1}^{ E_{r}} (2 \sqrt{2}/(k \pi )) U_{ik} \text{ sin } (k \pi r/7)\) with \(E_{r}=40\), and where \(U_{ik}\sim \) N(0,1), and \(\delta _{i1}(s), \delta _{i2}(r) \sim \text{ N } (0,\sigma _{X}^{2})\) are mutually independent. More precisely, \(X_{i1}(s)\) and \(X_{i2}(r)\) were the true underlying processes used to generate \(Y_i(t)\) from model (1) and \(V_{i1}(s)\) and \(V_{i2}(r)\) were the actually observed functional predictors. The choice of \(X_{i1}(s)\), \(\beta _{1}(\cdot ,\cdot )\) and \(\beta _{2}(\cdot ,\cdot )\) is similar to the choices in Goldsmith et al. (2011), whereas \(X_{i2}(r)\) is a modification of a Brownian bridge. For the scalar covariates, we considered a binary variable \(W_{1}=1\{\text{ Unif }[0,1]\ge .75\}\), and a continuous variable \(W_{2} \sim N(10,5^2)\). A univariate cubic B-spline basis with \(\kappa _{0}= 10\) basis functions with second-order difference penalty (Eilers and Marx 1996) was used to fit \(\beta _{0}(\cdot )\). Tensor products of cubic B-splines with \(\kappa _{1}=\kappa _{2}=25\) basis functions and second-order difference penalties in both directions were used to fit \(\beta _{1}(\cdot ,\cdot )\) and \(\beta _{2}(\cdot ,\cdot )\). For more complex functional parameters, increasing the number of basis functions may be necessary to capture the increased complexity. In our setting, increasing the number of basis functions for the bivariate smoothers from \(25\) to \(100\) did not significantly affect the fit or computation times.
We considered all possible combinations of the following choices:
-
1.
Number of subjects: (a) \(n=100\) and (b) \(n=200\).
-
2.
Functional predictors: (a) noiseless \(\sigma _{X}=0\) and (b) noisy \(\sigma _{X}=0.2\).
-
3.
Standard deviation for the error: \(\sigma _{\epsilon }=1\).
-
4.
Effects of the scalar covariates:
-
(a)
no scalar covariates, and
-
(b)
scalar covariates \(W_{1}\) and \(W_{2}\) with \(\gamma _{1}=1\) and \(\gamma _{2}=-0.5\).
-
(a)
The combination of the various choices provides eight different scenarios and for each scenario we simulated \(500\) data sets. For illustration, Fig. 1 displays one simulated data set for scenario 4(a). Curves for three subjects are highlighted in color, with each color representing one subject across the three panels. PFFR uses two steps for case 2(b). First, the functional predictors were estimated as \(\widehat{X}_{i1}(s)\) and \(\widehat{X}_{i2}(r)\) using the smoothing approach previously used in Goldsmith et al. (2011). Second, these estimated functions were used instead of \(X_{i1}(s)\) and \(X_{i2}(r)\) when fitting model (1).
The left panel displays a sample of 200 simulated functional responses \(Y_{i}(t)\) with \(\sigma _{\epsilon }=1\). The middle and right panels display 200 simulated functions from \(X_{i1}(s)\) and \(X_{i2}(r)\), respectively, highlighting three examples of the functional predictors with no error (solid) and with measurement error \(\sigma _{X}=0.2\) (dashed)
We computed the integrated mean squared error (IMSE), integrated squared bias \((\hbox {IBIAS}^{2})\), and integrated variance (IVAR), where IMSE\(\{ \widehat{\beta }(t,s)\} = \int _{\mathcal {T}} \int _{\mathcal {S}} {E} [ \{ \widehat{\beta }(t,s) -\beta (t,s) \}^{2} ] dt ds,\hbox { IBIAS}^{2}\{ \widehat{\beta }(t,s)\} = \int _{\mathcal {T}} \int _{\mathcal {S}} [{E}\{ \widehat{\beta }(t,s)\} - \beta (t,s) ]^{2} dt ds\), and \(\hbox {IVAR}\{\widehat{\beta }(t,s)\} = \int _{\mathcal {T}} \int _{\mathcal {S}} {Var}\{ \widehat{\beta }(t,s)\} dt ds\). Here \({E}\{ \widehat{\beta }(t,s)\}\) and \({Var}\{ \widehat{\beta }(t,s)\}\) were estimated by the empirical mean and variance of \(\widehat{\beta }(t,s)\) in \(500\) simulations. To characterize the properties of the pointwise confidence intervals we report the integrated actual pointwise coverage (IAC) and integrated actual width (IAW), where IAC = \({E}[\int _{\mathcal {T}} \int _{\mathcal {S}} 1 \{ \beta (t,s) \in CI_{p}(t,s) \} dt ds]\), where \(CI_{p}\) is the pointwise approximate confidence interval for the model parameter \(\beta (t,s)\), based on an independence assumption of the errors. We implemented approximate pointwise confidence intervals for the nominal level 0.95. For example, an approximate 95 % pointwise confidence interval \(CI_{p}(t_{0},s_{0})\) for \(\beta _{1}(t_{0},s_{0})\) can be constructed as \(\widehat{\beta }_{1}(t_{0},s_{0}) \pm 1.96 \ \widehat{\text{ sd }}\{\widehat{\beta }_{1}(t_{0},s_{0})\}\). As \(\widehat{\beta }_{1}(t_{0},s_{0})= \sum _{l=1}^{\kappa _{1}} a_{1,l}(t_{0},s_{0})\widehat{\beta }_{1,l}\), for every \((t_{0}, s_{0})\), \(\widehat{\text{ sd }}\{ \widehat{\beta }_{1}(t_{0},s_{0})\}=\sqrt{{\varvec{a_{1}(t_{0},s_{0})}}\widehat{{\varvec{\varSigma }}}_1 {\varvec{a_{1}^{t}(t_{0},s_{0})}}}\), where \(\widehat{{\varvec{\varSigma }}}_1\) is the estimated covariance matrix of \(\widehat{{\varvec{\beta }}}_{1}\), and \({\varvec{a_{1}(t_{0},s_{0})}} = \{ a_{1,l}(t_{0},s_{0}) \}_{l}\). We use the Bayesian posterior covariance matrix, see Ruppert et al. (2003), available under the Vp specification for gam; alternatively, for REML estimation, a Vc option that additionally accounts for smoothing parameter uncertainty is available in the mgcv R-package (Wood 2014) and can in some cases lead to slightly improved coverage. The width of this confidence interval is \(3.92\ \widehat{\text{ sd }}\{\widehat{\beta }_{1}(t_{0},s_{0})\}\) and \(\text{ IAW }= 3.92 {E}[\int _{\mathcal {T}}\int _{\mathcal {S}}\widehat{\text{ sd }}\{\widehat{\beta }_{1}(t,s)\}dtds].\) As a measure of accuracy of the fit we provide the functional \(R^2\), denoted by \(\hbox {fR}^2\), and computed as fR\(^2=1- \{ \sum _{i} \sum _{j} [Y_{i}(t_j)-{\widehat{\mu }_{i}(t_j)}]^2 \} / \{ \sum _{i}\sum _j [Y_{i}(t_j)-\widehat{\mu }_{Y}(t_j)]^2\}\), where \(\widehat{\mu }_Y(t_{j})=\sum _{i=1}^{n}Y_i(t_{j})/n\), and \({\widehat{\mu }}_{i}(t_{j})\) are the estimated \({\mu }_{i}(t_j)\) using the fitted model. Table 1 compares the averages of these measures over simulations, whereas Online Appendix B displays boxplots of these measures for the bivariate functional parameters calculated for each data set. Overall, our results indicate that PFFR outperforms the modified-PFR in terms of estimation precision. To provide the intuition behind these results, Fig. 2 displays the fits using PFFR and PFR obtained in one simulation for the setting \(n=200, \sigma _{\epsilon }=1, \sigma _{X}=0\) and case 4(a). Both methods capture the general features of the true parameter surfaces well, but the modified-PFR method does not borrow strength between the neighboring values of the functional responses since it is derived from PFR. As our results show, this causes unnecessary roughness of the estimates along the \(t\) dimension. In contrast, PFFR provides a much smoother surface that better approximates the shape of the true underlying functions.
Displayed are the bivariate functional parameters \(\beta _{1}(t,s) \) (left panels) and \(\beta _{2}(t,r)\) (right panels): true values (top panels), estimates for one simulation iteration via PFFR (middle panels), and estimates via PFR (bottom panels). Scenario: \(n=200\), \(\sigma _{\epsilon }=1\), \(\sigma _{X}=0\) and case 4(a)
Results in Table 1 can be summarized as follows. PFFR performs better than modified-PFR in terms of IMSE for almost all the estimated parameters, and gives comparable performance in a few cases, irrespective of the noise level in the functional predictors and the presence of additional non-functional covariates; see the columns labeled IMSE. For the bivariate functional parameters, the variability of the MSEs of PFFR and modified-PFR is comparable, and the median MSE of PFFR is smaller than that of modified-PFR; see Online Appendix B. As the number of subjects increases, IMSE for both methods decreases, confirming that in the settings considered, the methodology yields consistent estimators; this is expected to hold more generally, see, for example Claeskens et al. (2009), for the asymptotics of penalized splines. In terms of the performance of the pointwise approximate confidence intervals, the PFFR intervals are reasonably narrow and have a coverage probability that is relatively close to the nominal level; see the columns corresponding to IAW and IAC in Table 1. By contrast, confidence intervals for modified-PFR are constructed based on models for each \(t\) separately, which do not make use of the full information available, and intervals are thus excessively wide. While this results in coverage close to 1 for some settings, in other settings the point estimates have such high IMSE and IBIAS values that intervals are incorrectly centered and give very low coverage despite the large width. Overall, confidence intervals based on PFFR are narrower, more properly centered and give coverage more reliably close to the nominal level, despite some amount of undercoverage in some of the settings. For PFFR confidence intervals, as the number of subjects increases, IAW decreases, as expected, while maintaining IAC close to the nominal level. The approximate confidence intervals discussed in this section provide results that overall perform rather close to the 0.95 nominal level. Alternative bootstrap confidence intervals are discussed in Online Appendix A. Overall, when functional predictors are measured with error, the estimation of parameters and coverage of confidence intervals tends to deteriorate slightly. Otherwise, results are similar to the setting without noise. The accuracy of the fit seems similar for PFFR and modified-PFR, as illustrated by \(\hbox {fR}^2\).
Compared to PFFR, the modified-PFR alternative yields inferior results. First, fit results tend to be rougher and may require additional and specialized smoothing. Second, running a large number of scalar on function regressions can lead to longer computation times. Third, confidence intervals are not easy to obtain, which may further affect computation times.
4.2 Sparsely sampled functional response
Consider again scenario 4(a) described in Sect. 4.1 and assume that for each subject \(i\), the functional response is observed at randomly sampled \(m_{i}\) points from \(t \in \{ j/10, j=1,2,...,60\}\). Estimation of the parameter functions was carried out using our PFFR approach with \(\kappa _{0}=10\) basis functions for the univariate spline basis, and \(\kappa _{1}=\kappa _{2}=25\) for the bivariate spline bases. The estimates are evaluated using the same measures as described in Sect. 4.1. In the situation of sparsely sampled functional response, our method does not have any competitors.
Table 2 shows the results for two sparsity levels, \(m_i=20\) and \(m_i=6\). As expected, the sparsity of the functional response affects both the bias and the variance of the parameter estimators, as well as the width of the confidence intervals; compare Table 2 with the PFFR results of Table 1 corresponding to scenario 4(a) and \(n=200\). Nevertheless PFFR continues to show very good performance for many levels of missingness in the functional response with coverage of the confidence intervals similar to before.
4.3 Functional predictors sampled with moderate sparsity
The sparse design for the functional predictors was generated by starting with the scenario 4(a) described in Sect. 4.1 with few changes. The number of eigenfunctions for the two functional predictors was set to \(E_{s}=2\) and \(E_{r}=4\) respectively. For each functional predictor curve \(i\), \(1\le i \le n\), we randomly sampled \(K_{i}\) points from \(s \in \{ k/10, k=1,2,...,50\}\) and \(Q_{i}\) points from \(r \in \{ q/10, q=1,2,..., 70\}\). We apply the same smoothing approach used in Goldsmith et al. (2011) to reconstruct the trajectories for each functional predictor; see the digitally available R program for Sec 4.3 at the first author’s website.
For illustration, Fig. 3 displays the simulated data and predicted values for four curves, two for process \(X_{i1}(s)\) displayed in the two leftmost columns, and two for process \(X_{i2}(r)\) displayed in the two rightmost columns. The true underlying curves are shown as black dotted lines, the actual observed data are the red dots and the predicted curves are the solid red lines. Visual inspection of these plots indicates that the smoothing procedure recovers the underlying signal quite well. This is one of the main reasons PFFR continues to perform well for these scenarios.
Prediction of trajectories for functional predictors in simulation settings with \(K_{i}=12\), \(Q_{i}=25\) points per curve, and \(\sigma _{X}=0.20\). Shown are: \(X_{i1}(s_{ik})\) for two subjects (leftmost two panels) and \(X_{i2}(r_{iq})\) for the same subjects (rightmost two panels). For each panel we have the true signal (dotted lines), the observed signal (points), and the predicted signal (solid lines)
Table 3 displays the results for our model parameters for the case of functional predictors observed with moderate sparsity, \(n= 200\) subjects and different sparsity levels. Both the IMSE and the coverage performance of the PFFR are affected by the sparsity of the predictor function. Due to the sparsity of the functional predictors, a reduced number of basis functions is used for estimating the bivariate parameters. Throughout this simulation exercise we used \(\kappa _{0}=10\) basis functions for the univariate basis, and \(\kappa _{1}=\kappa _{2}=20\) basis functions for the two bivariate bases.
4.4 Simulations: a single functional predictor
In this section we compare pffr to the fully functional regression model of Ramsay and Silverman (2005, Ch. 16), which is implemented in the linmod function in the R-package fda (Ramsay et al. 2009, Sects. 10.3–4). As linmod cannot handle more than one functional covariate and sparsely sampled functional responses and/or covariates, we consider a model with a single functional predictor and densely observed functions. Our simulation design for this section corresponds to that in Sect. 4.1, Scenario 4(a), \(\sigma _{X}=0\), with the exception that now the model has a single functional predictor, \(X_1(s)\), instead of two functional predictors. We generate \(n\) realizations of functional responses from the model:
The linmod function requires the construction of functional data objects for the functional response and functional predictor prior to the model fitting step. We first outline the requirements for setting up the functional data objects, and then provide details for using the linmod function. One first has to choose the basis for object generation (Ramsay et al. 2009, Sect. 5.2.4), which must be the same as the basis used for the representation of the regression parameters: the functional intercept \(\beta _{0}(t)\), and the bivariate regression slope \(\beta _{1}(t,s)\). In addition to the choice of basis for functional data representation, linmod requires the manual specification of the smoothing parameter. We use generalized cross validation search across 41 values ranging from \(10^{-5}\) to \(10^5\) in steps of \(10^{0.25}\) in a manner similar to the implementation described in Sect. 5.3. of Ramsay et al. (2009). Separate cross validations are performed for obtaining functional data objects for the functional response and the functional predictor, respectively.
The linmod function requires the specification of three additional smoothing parameters: one for the functional intercept, and one each for the \(s\) and \(t\) directions of the regression coefficient \(\beta _1(s,t)\). The specification of these three parameters, necessary for fitting the linmod function, is determined with the help of a leave-one-curve-out cross-validation. Section 10.1.3 of Ramsay et al. (2009) presents implementation examples for a leave-one-curve-out cross validation. We use the choices \(\lambda _{\beta _{0}}=\{10^{-4}, 10^{-5}, 10^{-6}\}\) for the smoothing parameter for \(\beta _{0}\), and \(\lambda _{\beta _{1}^{(s)}} = \{10^{2}, 10^{3}, 10^{4}\}\) for the smoothing parameter in the \(s\) direction for \(\beta _{1}(t,s)\), \(\lambda _{\beta _{1}^{(t)}} = \{10^{2}, 10^{3}, 10^{4}\}\) for the smoothing parameter in the \(t\) direction for \(\beta _{1}(t,s)\). These correspond to extended choices compared to the implementation of the linmod function introduced in Sect. 10.4 of Ramsay et al. (2009). This constructed grid consists of 27 options available for the leave-one-curve-out cross validation. We executed our search using the cross-validated integrated squared error \(CVISE (\lambda _{\beta _{0}}, \lambda _{\beta _{1}^{(s)}}, \lambda _{\beta _{1}^{(t)}}) = \sum _{i=1}^{n} \int _{\mathcal {T}} (Y_i(t) - \widehat{Y}^{(-i)}_i(t) ) ^{2} dt \), where \(\widehat{Y}^{(-i)}_i(t) = \widehat{\beta }^{(-i)}_{0}(t) + \int _\mathcal {S} \widehat{\beta }^{(-i)}_{1}(t,s) X_{1i}(s) ds\) was computed as the predicted value for \(Y_{i}(t)\) when it was omitted from estimation. We tracked computation time for the linmod model fitting (that includes the generalized cross validations, and the leave-one-curve-out cross validation), and for sample size \(n=100\) the time was approximately 20 minutes on a MacBook 2GHz. Table 4 shows the computation time for all considered approaches and shows that our approach is faster than linmod by several orders of magnitude. This is likely due to the need for cross-validation for linmod as opposed to REML estimation in our case.
Table 4 also shows that our approach improves on linmod in terms of mean squared error, bias, variance and fR\(^2\). This finding might be related to the fact that REML can determine the optimal smoothing parameters more precisely than a grid search using cross-validation can do within a reasonable computation time, especially for multiple smoothing parameters. Another benefit of our method might be that not having to smooth the outcome functions before estimation might allow for a more accurate incorporation of uncertainty into the model fit. The current linmod function does not permit to obtain confidence intervals for the regression parameters, whereas pffr enables the estimation of lower and upper bounds for pointwise confidence intervals.
5 Application to DTI-MRI tractography
We consider a DTI brain tractography study of multiple sclerosis (MS) patients; cf. Greven et al. (2010), Goldsmith et al. (2011), Staicu et al. (2012), McLean et al. (2014). Diffusion tensor imaging is a magnetic resonance imaging (MRI) technique that quantifies water diffusion, whose properties we will use as a proxy for demyelination. DTI, at some level of output complexity, estimates the water diffusion at every voxel using its first three directions of variation (Basser et al. 1994, 2000). Multiple sclerosis is a demyelinating autoimmune-mediated disease that is associated with brain lesions and results in severe disability. Little is known about in-vivo demyelination including whether it is a global or local phenomenon, whether certain areas of the brain demyelinate faster, or whether lesion formation in certain areas is associated with demyelination in other areas of the brain. Also, it is unclear how these patterns in the demyelination process differ in MS patients from the normal aging process in healthy subjects. Here we attempt to provide an answer to some of these questions using function-on-function regression.
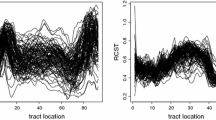
We focus on three major tracts that are easy to recognize and identify on the MRI scans: the corpus callosum (CCA), corticospinal (CTS) and optical radiation (OPR) tracts. The study comprises 160 MS patients and 42 healthy controls, who are observed at multiple visits. At each visit, functional anisotropy (FA) is extracted along these three major tracts for each subject, resulting in three functional measurements per observation. For illustration, the panels in Fig. 4 display the FA in the MS and control groups for the three tracts—CCA tract (left), CTS tract (middle) and OPR tract (right)—at the baseline visit. Depicted in red/blue/green are the FA measurements corresponding to three subjects in each group (subjects are color coded).
FA in the CCA tract (left panels), FA in the left CTS tract (middle) and FA in the left OPR tract (right panels) for the MS patients (top panels) and controls (bottom panels) observed at the baseline visit. In each of the top and bottom panels, depicted are the FA measurements for three subjects, with each mark representing a subject
Our goal here is mainly exploratory, as we are trying to further understand the spatial and temporal course of the disease. Following Tievsky et al. (1999) and Song et al. (2002), we use FA as our proxy variable for demyelination of the white matter tracts; larger FA values are closely associated with less demyelination and fewer lesions. MS is typically associated with lesions and axon demyelination in the corpus callosum. In advanced stages of MS, there is evidence for significant neuronal loss in the corpus callosum (e.g. Evangelou et al. 2000; Ozturk et al. 2010). To study the spatial association between demyelination along the CCA, CTS and OPR, we focus in our first regression model on the CCA—one of the main structures in the brain—as the response, and CTS and OPR as the covariates. It would also be possible, and potentially of interest, to look at regression models with each of the other two white matter tracts as the response. Interpretation of the associations between demyelination in certain areas of the different tracts can potentially inform medical hypotheses on spatial progression of the disease. In addition, models for each of the three responses could also be thought of as potential future building blocks in a mediation analysis (Lindquist 2012) with functional variables, to investigate the causal pathway of the effect of changes in the brain during MS on disability outcomes (e.g. Huang et al. 2013). This would involve both extension of the Lindquist (2012) method to the fully functional setting and expert clinical knowledge to interpret the neurological findings.
Consider that the response of interest is the FA along the CCA tract, and the functional predictors are the FA along the left CTS and left OPR tracts, at the baseline visit. The first step is to de-noise and deal with missing data in the functional predictors, as discussed in Sect. 2. Following the notation in Sect. 2, \(Y_i(t)\) denotes the FA along the CCA tract at location \(t\), \(X_{i1}(s)\) and \(X_{i2}(r)\) denote the centered de-noised FA along the left CTS tract at location \(s\), and the left OPR tract at location \(r\), respectively, of subject \(i\). We use regression model (1), where \({\varvec{W}}_i\) is the vector of age and gender of subject \(i\). The proposed methods are employed to estimate the regression functions \(\beta _0(\cdot )\), \(\beta _1(\cdot , \cdot )\) and \(\beta _2(\cdot , \cdot )\) as well as the regression parameters \({\varvec{\gamma }}\).
Figure 5 shows the regression function estimates along with information on their pointwise approximate 95 % confidence intervals for each of the two groups: MS subjects (left column) and controls (right column). We begin with \(\widehat{\beta }_0 (t)\), which accounts for a relative \(R^{2}\) of 28.9 % for the MS group and 38 % for the control group. For the MS group, the estimated overall mean of the FA for the CCA tract has a wavy shape, with two main peaks, one near the beginning of the tract, around location 10, and one at the end, around 90. For the control group, the estimated mean function shows similar patterns, but we observe lower FA values for the MS group than for the control; this is biologically plausible, as FA values tend to decrease in MS-affected subjects due to lesions or demyelination. Next, we consider the effect of the FA for the left CTS tract; the relative \(R^{2}\) corresponding to this predictor is 10.8 % for the MS group and 15.9 % for the control group. The estimated function \(\widehat{\beta }_1(t,s)\), displayed in the middle row of Fig. 5, shows the association between the FA for the CTS tract at location \(s\) and the FA of the CCA tract at location \(t\). Pointwise 95 % confidence intervals for the coefficient functions are constructed under independence assumption of the errors, and are displayed using different colors of the facets of the surface mesh. Specifically, red represents positive values, \(\widehat{\beta }_1(t,s)>0\), that are found to be significant, blue represents significant negative values, \(\widehat{\beta }_1(t,s)<0\); light red and light blue correspond to the remaining positive and negative values, respectively.
Estimates of the regression functions \(\widehat{\beta }_0\) (top panels), \(\widehat{\beta }_1\) (middle panels) and \(\widehat{\beta }_2\) (bottom panels) corresponding to the MS group (left column) and the control group (right column). The dashed lines (top panels) correspond to the pointwise 95 % confidence intervals. The facets of the surface mesh are marked: darker shades for more than approximately two standard errors above/below zero, and lighter shades for less than approximately two standard errors above/below zero
For the control group, most of the CTS–FA values (corresponding to locations below 40) are positively associated with FA values along the CCA, indicating that demyelination occurs simultaneously in those tracts. The coefficient surface for the control group is mostly constant, indicating a spatially homogeneous association for most regions of these two tracts. In MS patients, on the other hand, there are areas (30 to 45, and around 10) of the CTS for which FA measures show negative associations with the FA values in the CCA. This might indicate regions of those tracts where demyelination due to lesions typically occurs only in one tract at a time, while stronger positive associations may suggest that lesions often occur simultaneously in those areas. Our results support the expected pattern that the pathological demyelination in MS patients is a more localized phenomenon than demyelination in control patients due to aging. These findings, while exploratory, may yield new insights into the motivating question of whether lesion formation in certain areas of the brain is associated with demyelination in other areas of the brain and can lead to hypotheses for further studies.
We examine the effect of the FA of the left OPR tract, as estimated by \(\widehat{\beta }_2(t,r)\). The profile of the left OPR tract seems to be less predictive of the response for the control group (relative \(R^{2}\) is 12.6 %) and more predictive for the MS group (relative \(R^{2}\) is 25.7 %). Here, the associations between the FA of the OPR and the FA of the CCA seem somewhat similar between the two groups, but more pronounced for the MS patients, especially for the inferior OPR; see Fig. 5, bottom panels.
The estimates of the additional covariates, age and gender (reference group: female), are \(0.0002^*(0.00004)\) and \(0.0008(0.0009)\), for the MS group, and \(-0.0003^*(0.00005)\) and \(-0.0052^*(0.0018)\) for the control group, respectively, where the asterisks indicate significance at level \(0.01\). Standard errors are displayed within brackets. Negative age effects as seen in the controls are expected, as myelination and FA values tend to decrease with age even in healthy subjects.
6 Discussion
We develop a novel method for function-on-function regression that: (1) is applicable to one or multiple functional predictors observed on the same or different domains than the functional response; (2) accommodates sparsely and/or noisily observed functional predictors as well as a sparsely observed functional response, (3) accommodates linear or non-linear effects of additional scalar predictors; (4) produces likelihood-based approximate confidence intervals based on a working independence assumption of the errors, as a by-product of the associated mixed model framework; (5) is accompanied by open-source and fully documented software. In Online Appendix A we briefly discuss and numerically investigate bootstrap-based confidence intervals to account for non-i.i.d. error structure.
PFFR was applied and tested in a variety of scenarios and showed good results in both simulations and a medical application. Equally important PFFR is an easy to use automatic fitting procedure implemented in the pffr function of the R package refund.
Recent work by Scheipl et al. (2014) develops a framework of regression models for correlated functional responses, allowing for multiple partially nested or crossed functional random effects with flexible correlation structures for, e.g., spatial, temporal, or longitudinal functional data. While the focus is on modeling of the covariance structure for correlated functional responses, Scheipl et al. (2014) successfully use our PFFR methods for modeling the dependence of the functional mean on functional and scalar covariates within their framework. This shows that methods introduced in this paper are potentially useful in a much wider range of settings than considered here and can be seen as an important building block in the development of flexible regression models for functional responses.
7 Electronic supplementary material
Additional material is available with our article and consists of the following online appendices available as supplementary material online. Material is organized as Online Appendix A, B, and C. Online Appendix A outlines our pffr bootstrap confidence intervals. Online Appendix B consists of box plots corresponding to numerical measures on mean squared error, coverage of the approximate confidence intervals, and width for \(\beta _1\) and\(\beta _2\) parameters and simulation settings presented in Section 4.1. Online Appendix C consists of an additional exploratory study of our data for the temporal progression of demyelination in MS patients across two consecutive visits, both within the same location and with spatial propagation.
References
Aguilera A, Ocaña F, Valderrama M (1999) Forecasting with unequally spaced data by a functional principal component approach. Test 8(1):233–253
Aneiros-Pérez G, Vieu P (2008) Nonparametric time series prediction: a semi-functional partial linear modeling. J Multivar Anal 99:834–857
Basser P, Mattiello J, LeBihan D (1994) MR diffusion tensor spectroscopy and imaging. Biophys J 66:259–267
Basser P, Pajevic S, Pierpaoli C, Duda J (2000) In vivo fiber tractography using DT-MRI data. Magn Reson Med 44:625–632
Bunea F, Ivanescu AE, Wegkamp MH (2011) Adaptive inference for the mean of a Gaussian process in functional data. J R Stat Soc Ser B 73(4):531–558
Claeskens G, Krivobokova T, Opsomer JD (2009) Asymptotic properties of penalized splines estimators. Biometrika 96(3):529–544
Crainiceanu C, Reiss P, Goldsmith J, Huang L, Huo L, Scheipl F, Swihart B, Greven S, Harezlak J, Kundu M, G, Zhao Y, McLean M, Xiao L (2014) Refund: regression with functional data, Website: http://CRAN.R-project.org/package=refund
Crainiceanu CM, Ruppert D (2004) Likelihood ratio tests in linear mixed models with one variance component. J R Stat Soc Ser B 66(1):165–185
Crainiceanu CM, Staicu A-M, Di C (2009) Generalized multilevel functional regression. J Am Stat Assoc 104(488):177–194
Eilers P, Marx B (1996) Flexible smoothing with B-splines and penalties. Stat Sci 11(2):89–121
Evangelou N, Konz D, Esiri MM, Smith S, Palace J, Matthews PM (2000) Regional axonal loss in the corpus callosum correlates with cerebral white matter lesion volume and distribution in multiple sclerosis. Brain 123(9):1845–1849
Fan Y, Foutz N, James GM, Jank W (2014) Functional response additive model estimation with online virtual stock markets. Ann Appl Stat, To appear
Ferraty F, Laksaci A, Tadj A, Vieu P (2011) Kernel regression with functional response. Electron J Stat 5:159–171
Ferraty F, Van Keilegom I, Vieu P (2012) Regression when both response and predictor are functions. J Multivar Anal 109:10–28
Ferraty F, Vieu P (2006) Nonparametric functional data analysis. Springer, New York
Ferraty F, Vieu P (2009) Additive prediction and boosting for functional data. Comput Stat Data Anal 53:1400–1413
Goldsmith J, Bobb J, Crainiceanu CM, Caffo BS, Reich D (2011) Penalized functional regression. J Comput Graph Stat 20(4):830–851
Greven S, Crainiceanu CM, Caffo BS, Reich D (2010) Longitudinal functional principal component analysis. Electron J Stat 4:1022–1054
Greven S, Crainiceanu CM, Küchenhoff H, Peters A (2008) Restricted likelihood ratio testing for zero variance components in linear mixed models. J Comput Graph Stat 17(4):870–891
He G, Müller H-G, Wang J-L, Wang W (2010) Functional linear regression via canonical analysis. Bernoulli 16(3):705–729
Horváth L, Kokoszka P (2012) Inference for functional data with applications. Springer, New York
Huang L, Goldsmith J, Reiss PT, Reich DS, Crainiceanu CM (2013) Bayesian scalar-on-image regression with application to association between intracranial DTI and cognitive outcomes. NeuroImage 83:210–223
Kadri H, Preux P, Duflos E, Canu S (2011) Multiple functional regression with both discrete and continuous covariates. In: Ferraty F (ed) Recent advances in functional data analysis and related topics, contributions to statistics. Physica-Verlag, Heidelberg, pp 189–195
Krivobokova T, Kauermann G (2007) A note on penalized spline smoothing with correlated errors. JASA 102(480):1328–1337
Lindquist MA (2012) Functional causal mediation analysis with an application to brain connectivity. J Am Stat Assoc 107(500):1297–1309
Matsui H, Kawano S, Konishi S (2009) Regularized functional regression modeling for functional response and predictors. J Math Ind 1(3):17–25
McLean MW, Hooker G, Staicu A-M, Scheipl F, Ruppert D (2014) Functional generalized additive models. J Comput Graph Stat 23(1):249–269
Matlab, The MathWorks Inc. (2014) Natick. Massachusetts, United States
Nychka D (1988) Confidence intervals for smoothing splines. J Am Stat Assoc 83:1134–1143
Ozturk A, Smith SA, Gordon-Lipkin EM, Harrison DM, Shiee N, Pham DL, Caffo BS, Calabresi PA, Reich DS (2010) MRI of the corpus callosum in multiple sclerosis: association with disability. Mult Scler 16(2):166–177
R Development Core Team (2014) R: a language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria. www.R-project.org
Ramsay JO, Wickham H, Graves S, Hooker G (2014) FDA: functional data analysis. Website: http://CRAN.R-project.org/package=fda
Ramsay JO, Hooker G, Graves S (2009) Functional data analysis with R and Matlab. Springer, New York
Ramsay JO, Silverman BW (2005) Functional data analysis. Springer, New York
Reiss P, Ogden T (2009) Smoothing parameter selection for a class of semiparametric linear models. J R Stat Soc Ser B 71(2):505–523
Ruppert D, Wand MP, Caroll RJ (2003) Semiparametric regression. Cambridge University Press, Cambridge
Scheipl F, Greven S, Küchenhoff H (2008) Size and power of tests for a zero random effect variance or polynomial regression in additive and linear mixed models. Comput Stat Data Anal 52(7):3283–3299
Scheipl F, Staicu A-M, Greven S (2014) Functional additive mixed models. J Comput Graph Stat. doi:10.1080/10618600.2014.901914
Song SK, Sun SW, Ramsbottom MJ, Chang C, Russell J, Cross AH (2002) Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage 17(3):1429–1436
Staicu A-M, Crainiceanu CM, Reich DS, Ruppert D (2012) Modeling functional data with spatially heterogeneous shape characteristics. Biometrics 68(2):331–343
Tievsky AL, Ptak T, Farkas J (1999) Investigation of apparent diffusion coefficient and diffusion tensor anisotropy in acute and chronic multiple sclerosis lesions. Am J Neuroradiol 20(8):1491–1499
Valderrama MJ, Ocaña FA, Aguilera AM, Ocaña-Peinado FM (2010) Forecasting pollen concentration by a two-step functional model. Biometrics 66(2):578–585
Wahba G (1983) Bayesian ‘confidence intervals’ for the cross-validated smoothing spline. J R Stat Soc Ser B 45:133–150
Wood SN (2006) Generalized additive models: an introduction with R. Chapman & Hall/CRC, New York
Wood SN (2011) Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J R Stat Soc Ser B 73(1):3–36
Wood SN (2014) MGCV: mixed GAM computation vehicle with GCV/AIC/REML smoothness estimation. Website: http://CRAN.R-project.org/package=mgcv
Wu Y, Fan J, Müller H-G (2010) Varying-coefficient functional linear regression. Bernoulli 16(3):730–758
Yao F, Müller H-G, Wang J-L (2005a) Functional data analysis for sparse longitudinal data. J Am Stat Assoc 100(740):577–590
Yao F, Müller H-G, Wang J-L (2005b) Functional linear regression analysis for longitudinal data. Ann Stat 33(6):2873–2903
Zhang JT, Chen J (2007) Statistical inferences for functional data. Ann Stat 35(3):1052–1079
Acknowledgments
Staicu’s research was supported by U.S. National Science Foundation Grant Number DMS 1007466 and by the NCSU Faculty Research and Professional Development Grant. Fabian Scheipl and Sonja Greven were funded by Emmy Noether Grant GR 3793/1-1 from the German Research Foundation. We thank Daniel Reich and Peter Calabresi for the DTI tractography data. We would like to thank Ciprian Crainiceanu for helpful discussions and comments on earlier versions of the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
The refund package is available from CRAN at the following website: http://CRAN.R-project.org/package=refund.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ivanescu, A.E., Staicu, AM., Scheipl, F. et al. Penalized function-on-function regression. Comput Stat 30, 539–568 (2015). https://doi.org/10.1007/s00180-014-0548-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00180-014-0548-4