Abstract
Chromosome analysis is an important approach to detecting genetic diseases. However, the process of identifying chromosomes in metaphase images can be challenging and time-consuming. Therefore, it is important to use automatic methods for detecting chromosomes to aid diagnosis. This work proposes a study of deep learning approaches for classification and detection of chromosome in metaphase images. Furthermore, we propose a method for detecting chromosomes, which includes new stages for preprocessing and reducing false positives and false negatives. The proposed method is evaluated using 74 chromosome images in the metaphase stage, which were obtained from the CRCN-NE database, resulting in 2174 chromosome regions. We undertake three types of evaluation: segmentation; classification of cropped regions of chromosomes; and detection of chromosomes in the original images. For the segmentation analysis, we evaluated the Otsu, adaptive, fuzzy and fuzzy-adaptive methods. For classification and detection, we evaluated the following state-of-the-art algorithms: VGG16, VGG19, Inception v3, MobileNet, Xception, Sharma and MiniVGG. The classification results showed that the proposed approach, using segmented images, obtained better results than using RGB images. Furthermore, when analyzing deep learning approaches, the VGG16 algorithm obtained the best results, using fine tuning, with a sensitivity of 0.98, specificity of 0.99 and AUC of 0.955. The results also showed that the proposed negative reduction method increased sensitivity by 18%, while maintaining the specificity value. Deep learning methods have been proved to be efficient at detecting chromosomes, but preprocessing and post-processing are important to avoid false negatives. Therefore, using binary images and adding stages for reducing false positives and false negatives are necessary in order to increase the quality of the images of the chromosomes detected.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Chromosomes are present in each of the nucleus cells of all living organisms, their function being to carry genetic information to reproduction cells and organisms [1]. Chromosome analysis and classification can identify several anomalies associated with changes in the structure of chromosomes, such as in Down Syndrome, Turner Syndrome [2] and when seeking to identify several types of cancer [3]. However, this process is very time-consuming for the cytogeneticist and for the patient, who has to wait for the proper treatment. To make a diagnosis, a cytogeneticist needs to analyze hundreds of images and select those with chromosomes in the metaphase stage for later analysis [4]. Next, cytogeneticists analyze each image which shows chromosomes, which they count, and then they identify each one. This analysis, which is usually performed manually in many hospitals, is a laborious procedure, and very time-consuming [5]. Furthermore, this process usually results in divergences between the diagnosis of the same image [6, 7]. Also, the skill and experience of the cytogeneticist is a relevant aspect of the correctly analyzing chromosomes [1]. That happens because some images, obtained by using microscopes, are usually of a poor quality, which makes the manual process inaccurate [8]. In Fig. 1, note the difference in quality obtained from different slides, using the same microscope.
According to Matta et al. [9], 3–4 days are needed to prepare the samples and produce slides, and at least 5 days to evaluate each individual. Besides the time needed to analyze a single individual, chromosomes present several types of structure, e.g., they may overlap, be curved, or have some abnormality. Other factors that challenge the process of chromosomic analysis include chromosomes that do not have a rigid structure and may overlap or be connected to others [10]. Many studies have been developed that allow chromosomes to be analyzed automatically. However, the segmentation stage and automatic classification of chromosomes is still an open problem [5]. Consequently, many cytogenetic laboratories still conduct analysis manually, and as the demand is frequently very high, this process is slow and susceptible to errors.
Developing an automatic solution for classifying and detecting chromosomes has two primary impacts: (i) processing and analyzing chromosome images take less time to do (ii) it aids the cytogeneticist by reducing misdiagnosis. The aim of the proposed work is to provide a method to detect chromosomes in a noisy environment, which can help to identify chromosome anomalies. The task of automatically counting chromosomes can speed up the process of detecting anomalies, such as Down syndrome, which produces an extra copy of chromosome 21, resulting in 47 chromosomes. Turner’s syndrome, on the other hand, arises from the absence of an X chromosome, meaning only 45 chromosomes are present. Furthermore, the detection method can be used to find structural chromosomal abnormalities..
This paper analyzes several solutions for segmenting and classifying chromosomes and proposes an approach for detecting and reducing false positives and false negatives in the image. Thus, this method provides a solution that can be used by cytogeneticists to give a faster and more accurate diagnosis. Our main contributions can be summarized as follows:
-
Construction of a public labeled database of chromosomes in metaphase images.
-
A study of segmentation methods for segmenting chromosomes.
-
An analysis of CNN architectures to classify chromosomes.
-
A novel approach to detecting chromosomes, with proposals for reducing false positives and false negatives.
This paper is structured as follows: Sect. 2 discusses related studies in the area, Sect. 3 describes the methodology we used in this work, Sect. 4 presents the results of the analysis, Sect. 5 discusses the results obtained and Sect. 6 draws some conclusions and indicates some future lines of research.
2 Related work
According to Wang et al. [11], since 1980, several research groups have been developing solutions to improve chromosome analysis from images by separating overlapped chromosomes and classifying them in order to detect anomalies.
Arora and Dhir [5] conducted a survey of the main segmentation algorithms applied to chromosome images. Arora and Dhir analyzed six algorithms that proposed solutions to the segmentation problem in metaphase chromosomes. Based on 12 characteristics, they found that the method proposed by Uttamatanin et al. [12] is the one that is most recommended for segmenting chromosomes.
One of the most cited works on chromosome segmentation is MetaSel [12], which has an accuracy of 90% for chromosome classification. Uttamatanin et al. [12] segment groups of chromosomes by subdividing them in four main groups, based on their shapes. The first group consists of individual chromosomes with a well-defined shape, whereas the second group has a curved structure. The third group is categorized by overlapped chromosomes, and the last group is defined as non-chromosomes. Their study provides a software that is available for download and test. The algorithm proposed by Uttamatanin et al. [12] is based on the Otsu technique [13]. It achieves high accuracy for the first and second group, this being 99.42% and 90.67%, respectively. In some cases, chromosomes from the second group are classified as the third group, because of the similarity between edges and some residual objects. In their study, chromosomes are not segmented individually, but as groups of chromosomes. Therefore, the cytogeneticist still has to spend time verifying the results and counting and identifying chromosomes.
Gagula-Palalic and Can [14] use competitive neural network teams (CNNTs) along with the nearest neighbor technique (NNT). The perceptrons of NNT are trained to identify two classes of chromosomes, with the training done in pairs. Next, the algorithm defines to which pair the chromosome belongs. The experiments in this study were performed on the Copenhagen and Saravejo database, which contains more than 3300 individual chromosomes, and achieved an accuracy rate of 95.7% with CNNTs, and a rate of 98.27% with CNNTs + NNT.
Andrade et al. [15] proposed a hybrid segmentation technique based on fuzzy and adaptive methods, applied to chromosome images. It is compared with state-of-the-art approaches, and their approach obtained better segmentation results, with sensitivity and specificity values of 91% and 92%, respectively. However, their approach is only used for segmentation and no classification task using the segmented images are evaluated.
Recent papers have used deep learning approaches, using convolutional neural networks (CNN) to classify chromosomes. Qin et al. [16] use two CNN architectures to perform classification: the first architecture being a Global Net (G-Net) and the second a Local Net (L-Net). The G-Net extracts the global characteristics so as to find the regions where the chromosomes are located. With their method, they obtained 97% accuracy, which was more efficient than the techniques that it was compared to. However, the images used in their study have neither many noises nor overlapped chromosomes.
Sharma et al. [17] use a crowdsourcing platform, called CrowdFlower, to segment chromosomes. On this platform, the users annotate the edges of the chromosomes and are rewarded for doing so. However, some users used incorrect labeling, usually rectangular shape. To avoid this, Sharma uses a verification step to filter the best annotations in order to segment the metaphases. Sharma also proposes a method to deal with curved and folded chromosomes. Folded chromosomes are corrected using an algorithm based on Javan-Roshtkhari and Setarehdan [18]. This post-processing resulted in an improvement in accuracy from 68.5 to 86.7%. Sharma also made an interface to cytogeneticists which they can use to correct the mistakes produced during segmentation.
Swati et al. [19] use Siamese Networks to classify chromosomes, and using a dataset of 1740 images, obtained 85.6% of accuracy in their best configuration. Comparing these with state-of-the-art algorithms, Swatti obtained better results. Although they combined different types of machine learning techniques to reduce the computational cost, 124 h for training were needed to find the best architecture. This can be unfeasible when considering that new data may well be inserted frequently in order to improve training.
Table 1 shows a comparative summary of the related work. Most of the works focus either on segmentation or classification types of chromosomes. This works in focused chromosome detection on images with many non-chromosome elements. It differs from the literature because it combines segmentation with deep learning methods to provide chromosome regions with high sensitivity and specificity.
There are different approaches for identifying chromosomes, and they generate different types of images, with distinct challenges [20]. Most of the works in the literature use a method called G-banding, where the chromosomes appear as a succession of dark and light bands [21], as illustrated in Fig. 2a. This type of image is easier to segment, once there are no noises in the images. In this paper, images are slightly different from those described in related works, mainly because these images are obtained by using a cheaper method, which is used by many laboratories. In this methodology, healthy samples of images with chromosomes in metaphase stage are submitted to radiation at various levels. After that, a drop of Giemsa pigment impregnates chromosome regions and other noises [9]. The type of image used in this work is shown in Fig. 2b. The problems found in these images have similarities in terms of chromosomes shape, overlapped, curved, but the images from our dataset have a larger amount of noise than those found in the literature. Therefore, the task of detecting chromosomes is more challenging and requires additional steps. Based on this, in our work, there is a preprocessing step to reduce these noises and a post-processing step to decrease false positives and false negatives. We also evaluated different segmentation and classification models from the literature applied to this kind of image, besides providing our dataset for public access.
3 Materials and methods
3.1 Analyzed approaches
In this paper, we evaluate deep learning models to classify chromosome images in the metaphase stage using two different approaches: using colored images as input and using binary images obtained from a segmentation step. For training the model, we compare the use of several state-of-the-art CNN architectures. Finally, we conduct two types of classification analysis.
3.1.1 CNN applied to colored regions
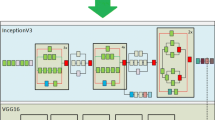
This is a classical end-to-end approach, where the input that the network receives is the region to be classified, and the output of the network is the class that the input belongs to. For this analysis, we used the RGB image provided by the database and identified whether or not the input image is a chromosome. Figure 3 shows an example of the architecture of this model. The CNN architecture used in Fig. 3 is just to illustrate one general CNN approach, but the architecture of the model will vary depending on the network used.
3.1.2 CNN applied to segmented regions
In this approach, we used segmentation followed by classification, as illustrated in Fig. 4. To perform segmentation, we extended the analysis made in our previous work [15]. This work uses the following segmentation algorithms: Otsu, adaptive thresholding, fuzzy and fuzzy adaptive. We extend the analysis conducted in the previous study, by increasing the database and adding a classification analysis. Although Otsu and adaptive thresholding are classical thresholding methods, they are still applied by segmentation methods in state-of-the-art techniques [12, 23, 24]. In the result Sect. 4, we analyze the segmentation method that performed best for chromosome segmentation. Other methods can be applied for segmentation without impact on the architecture.
3.2 Database
The images used during the experiments were acquired from the Centro Regional de Ciências Nucleares do Nordeste (CRCN-NE), in the city of Recife, Brazil. The CRCN-NE provided the dataset with chromosome images for developing the algorithm, and specialists helped to label all images. The acquired database has 74 images of chromosomes in the metaphase stage, including their labeling. The images were digitized using an optical microscope (Leica DM 500), using a resolution of 2028 \(\times \) 1536 pixels. Figure 5 shows examples of images of chromosomes acquired from the database.
Note that the images obtained from this database are quite complex, with some images with noises, chromosomes overlapped, connected or incomplete. These are real cases of images, where cytogeneticists have to analyze the chromosomes in order to count and identify them. This database and labeling are publicly available on the Zenodo platform [22].
3.3 Methodology
Figure 6 shows a flowchart of the methodology used to train the models. First, the image regions were cropped into chromosomes and non-chromosomes, based on the annotations of the CRCN-NE database. After cropping them, preprocessing is performed to remove noises and resize the images. Finally, the model is trained with these regions, followed by test and analysis of results. The details of each step are described next.
3.3.1 Creation of the cropped database
The CRCN-NE database is divided in 60% for training, 20% for validation and 20% for tests. This resulted in 48 metaphases images for training, 11 for validation and 15 for tests. Thereafter, we extracted the regions as being chromosome and non-chromosome. To obtain the cropped areas, it was used the LabelMe online tool [25], using the annotations provided by the specialists. The non-chromosome regions were generated randomly. Then, it was obtained 2174 cutouts for each class, chromosome and non-chromosome, in the training stage, and 501 cutouts in the validation stage. For test, it had 641 cutouts for chromosome and 579 for non-chromosome. The training, validation and test cropped regions were extracted from their respective images of training, validation and test sets. Figure 7 shows the size and proportion of cropped chromosomes and non-chromosomes of the database.
The constructed cropped database is also available on the Zenodo platform [22].
3.3.2 Preprocessing
After constructing the cropped dataset, the following preprocessing was done: orientation correction, resizing and padding. The orientation correction for the cropped region is corrected to put the chromosome ones closer to a vertical alignment. For this purpose, the image was rotated in the region whenever the width of an image is greater than its height.
Resizing is necessary because the CNNs used require as input squared images and a minimum size for each architecture. Therefore, we tried two approaches: simple resizing, and resizing with padding. In simple resizing, the image is resized to the final dimensions. In the resize with padding, it preserves the aspect of ratio and fills the squared area with zeros or replicates the edges. Figure 8 shows simple resizing, and resizing with padding.
In the experiments, it was used simple resizing, resizing, padding with black and edge replicate filing, and no padding. Some models had better results without the use of padding, while others worked better with padding. The result section shows the best configuration for each approach.
We also constructed a binary version of the cropped database, to be used as input to the models, according to the approach described in Sect. 3.1.2. To generate the regions, we segmented the original images and cropped the regions based on the annotations provided.
After creating the cropped database and conducting the preprocessing step, the model is trained using the training and validation cropped sets. Next, we evaluate the model by using the cropped test images in the test stage. Finally, we analyze the model using the evaluation metrics.
3.4 Chromosome detection
Chromosomes are detected by generating the candidates, by segmentation, and using the trained model to classify the candidates. Figure 9 shows each step of the proposed approach to detecting chromosomes.
In the proposed model, segmentation algorithms are used to generate the candidate regions, which will be classified as chromosome or non-chromosome. For this task, we prioritized approaches that did not remove any chromosome region and generated few noise regions. To this purpose, classical segmentation algorithms were applied: Otsu, adaptive thresholding, fuzzy and fuzzy adaptive [22] segmentation algorithms.
After the segmentation process, all the resulting contours of the image are selected as chromosome candidates. A bounding box area defines the candidate region. However, as the segmentation process generates many noise regions, a filtering stage is required. This stage is based on the size of the chromosomes of the database, as shown in Fig. 7. We eliminate all small regions that are smaller than the minimum chromosome width and height found in the database, that is equal to 5. A filtering related to large areas can also be applied.
After, the candidate regions are generated and filtered. The candidates are processed using the same operations as was done in the training process: orientation alignment, resizing and padding. Each candidate is used as input to the previously trained model, and each candidate is predicted to be a chromosome or non-chromosome. In the classification step, some false positive and false negatives cases may be found in the image. To avoid this problem, this work proposes a false positive and a false negative reduction step. These steps are described below.
3.4.1 False positive reduction
After the detection obtained by classifying the candidate regions, a small number of false positives were generated for most of the images. Figure 10 shows an example of detection, where the green areas are the true positives detected, the red areas are the false negatives, and the blue regions are the false positives.
Many false positive areas are isolated from other regions classified as chromosome, as shown in Fig. 10. Based on the fact that chromosomes are usually located next to each other, if a candidate predicted as chromosome is located isolated from other candidates predicted to be in the same class, it has a high chance that it was labeled incorrectly. Based on this argument, the proposed method updates the label of these candidates, that are located far from other candidates identified as chromosome. This approach is illustrated in Fig. 11.
In Fig. 11, the green triangles represent the candidates classified as chromosome and the gray squares those classified as non-chromosomes. For every candidate classified as chromosome, its neighborhood is checked in a radius r. If fewer than k candidates are found in the area of radius r, then the candidate changes its label to non-chromosome. That would mean that there are no other candidates classified as chromosome in the neighborhood, and it is probable that this candidate was mislabeled. Therefore, its label is updated to non-chromosome. In Fig. 11a, a k value of 1 was defined, so the chromosome in the red circle has its label changed, as shown in Fig. 11b. To this task, the values of r and k defined empirically to 100 and 2, respectively. The false positive reduction algorithm is described in Algorithm 1.

3.4.2 False negative reduction
It is expected that false negatives will be avoided, as all the chromosomes should be detected in each image. However, they are frequently present after a simple classification process, as represented by the red squares in Fig. 10.
As the chromosomes in the image are usually grouped in the same region, it is rare to find non-chromosome regions surrounded by other chromosomes, after the segmentation stage. Based on this, we verify the neighborhood of each non-chromosome candidate in a radius r, and if the candidate is surrounded by k candidates classified as chromosome, it changes its label to chromosome. This method is illustrated in Fig. 12.
It is true that a candidate is not always surrounded by other chromosome candidates is, in fact, a chromosome. Moreover, this can lead to generate a false positive being generated in some cases. However, it was verified that most of the time it more often corrects candidate labels, thus reducing false negatives, rather than generating false positives, as will be shown in Results section. For this task, we used a radius value of 60 and a k value of 2, defined empirically.
Figure 13 shows all steps of the proposed method, performed on an image of the test database. Figure 13a shows the original image from the test database. After applying the segmentation algorithm, it is obtained the image in Fig. 13b is obtained. The green square is the area shown in Fig. 13c. The black boxes are the candidates generated from the segmentation step. Next, the filtering process is applied, thus reducing the small regions, resulting in Fig. 13d. Then, each candidate is submitted to the trained classifier, thereby generating the prediction for each candidate, as illustrated in Fig. 13e. The green areas are the true positive detected, in which the blue and red are the false positive and false negative detections. The true negative regions are not shown in the image. To reduce the false negative regions, each candidate classified as chromosome that is isolated from the others is removed. Therefore, the blue candidate, in Fig. 13f, has its label changed to non-chromosome, producing a true negative, and generating the image from Fig. 13g. Finally, false positive candidates are reduced by changing the label of regions classified as non-chromosome and that are surrounded by candidates classified as chromosome. With this method, all the red regions, in Fig. 13g, have their labels changed, thereby generating the final detection in Fig. 13h. The false negative reduction algorithm is described in Algorithm 2.

3.5 Implemented methods
3.5.1 Models
In order to classify a selected region of the image as chromosome, this study uses the following deep learning state-of-the-art methods: VGG16 [26], VGG19 [26], Inception_v3 [27], MobileNet [28], Xception [29], MiniVGG and Sharma Model [17]. To initialize the weights of the model, is used the transfer learning approach [30]. Transfer learning is a technique to reuse a model trained in a Domain D1, and to use it in a similar or different Domain D2. The fundamentals that support transfer learning are that the basic features, such as edges and curves, are present even in different domains, so the feature extraction learned from a model can be reused in another domain. Models trained in large databases, such as Imagenet [31], which has millions of images, have been used as a base model to classify images in medical analysis [32].
When using transfer learning, we have the option to freeze the model and just train the fully connected (FC) layers, or we can fine-tune the previous layers to adjust their weights to the new database. In this work, both approaches are analyzed , using the initial weights from the model trained on Imagenet. The models in which we apply the transfer learning approach and train only the FC layers are defined with a ’TL’ suffix. For example, the VGG16 model using this approach is defined in this document as VGG16_TL.
We also conduct transfer learning with fine tuning on the first layers. In this approach, only the first layer is frozen and all the consequent ones are trained. For the models using this fine-tuning approach, we use the ’FT’ suffix. For example, the VGG16 model, using this approach, is defined in this document as VGG16_FT.
These models are also evaluated training all layers, without transfer learning and fine-tuning. However, they were not able to learn well, their accuracy being around 50%. These architecture models may be too complex for the size of CRCN database, and it might therefore be that they do not learn the features properly.
The CNNs described also are compared with simpler models, which we designed for this problem. The first model, defined as MiniVGG, is a sequence of blocks of Convolution (CONV), Batch Normalization (BN), Dropout (DO), Max Pooling (MP), and Softmax (SM) layers. The MiniVGG is a simpler version of the VGG model, consisting of two blocks of CONV\(\rightarrow \)BN\(\rightarrow \)CONV\(\rightarrow \)BN\(\rightarrow \)MP\(\rightarrow \)DO layers, followed by FC \(\rightarrow \)BN\(\rightarrow \)DO\(\rightarrow \)SM layers. The size of the filters was defined as 32 for the first block and 64 for the second block, with dropout values of 0.25 and filter size of 3 for both blocks. The FC layer has 512 nodes and the dropout value of 0.5.
The model used by Sharma et al. [17] is also implemented, which is named the Sharma model in this work.
We also compared the deep models with a simple classifier. For this purpose, we used Zernike moments [33] to extract the features, and a multilayer perceptron (MLP). For the MLP, we used 256 neurons in the hidden layer and we added a dropout layer with value 0.2. The MLP was trained using Adam optimizer, with learning rate 0.001, for 50 epochs. The best parameters used for the MLP were found empirically.
The algorithms implemented in this study were developed using Python language, Keras [34] and the OpenCV library [35]. The algorithms were executed using a computer with an i7 processor and 8GB RAM memory.
3.6 Metrics
The metrics used to evaluate the results are divided into segmentation and classification metrics.
3.6.1 Segmentation
To perform the analysis, we implemented the following metrics used in the literature to analyze medical images: Sensibility, Specificity, the Jaccard Index, Matthews coefficient correlation and positive prediction value. These metrics are calculated from the true positive (TP), true negative (TN), false positive (FP) and false negative (FN) rates found in the segmented images. Next, each metric to be analyzed is described.
Sensibility (SE) represents the effectiveness of the algorithm at correctly classifying the pixels from the object in the image. The sensibility can be described by means of Eq. 1:
Specificity (SP) represents the effectiveness of the algorithm at correctly classifying the pixels from the background in the image. The specificity can be described by means of Eq. 2:
The Jaccard index (J) represents the similarity index between a segmented image and its respective ground truth. The Jaccard index is calculated by means of Eq. 3:
The Matthews Correlation Coefficient (MCC) is considered a useful metric to evaluate the similarity between binary classifications [36]. This metric shows if the prediction values are being random or not. Values closest to 0 are considered a random result. Value -1 indicates a total divergence between the images, while value 1 shows total similarity. The metric MCC is calculated by means of Eq. 4:
The value of the positive prediction value (PPV) shows the total number of pixels classified as an object, divided by the total number of pixels of the object, as shown in Eq. 5:
3.6.2 Classification
To evaluate the classification and detection of the models, we use, besides SE and SP, the metrics for accuracy (ACC), F1-score (F1), average precision (AP) and area under curve (AUC).
Accuracy is a common metric used for classification, where it considers the fraction of TP and TN, under the total sum of the samples (positive and negative). In resume, it is the number of correct classifications divided by the total number of samples. The accuracy equation is defined by Eq. 6
The F1-score calculates the harmonic mean between precision and recall, where recall is the same as the SE metric. A high precision value means that the system does not produce a large number of false alarms, while high recall means that most of the subjects are being detected. Since both measures are important, we also consider the harmonic mean, as described in Equation
The Average Precision (AP) metric summarizes the weighted increase in precision with each change in recall for different values of thresholds.
The AUC metric evaluates the area under the receiver operating characteristic curve, which measures the trade-off between the false positive rate and the true positive rate.
4 Results
The algorithms evaluation is evaluated using the 74 images from the database, with the models described in Sect. 3.5.1. First, the segmentation methods are evaluated to find the best model to use by the CNN with the segmentation approach, described in Sect. 3.1.2, and also to generate the candidates in the detection stage. Next, the models to classify the cropped dataset are evaluated in the proposed detection approach.
4.1 Segmentation algorithm analysis
The following segmentation algorithms are applied to CRCN-NE database: Otsu, adaptive thresholding, fuzzy and fuzzy-adaptive (FA). Table 2 shows the parameters explored for each technique, followed by the best configuration found for it.
The first column shows the segmentation method, the second column shows the parameters of each method, the third column shows the range of values explored for each parameter, and the last column shows the best value obtained.
Figure 14 shows the segmentation results of each technique, for an image of the database. Figure 14a shows the original image, while Fig. 14b shows the ground truth image. Figure 14d–f shows the segmentation results of the Otsu, fuzzy, fuzzy-adaptive and adaptive techniques, respectively. Note that all techniques generate significant noise in the image. At this stage, we are interested in not missing any chromosome region, while having the least noise possible.
The results obtained by computing the segmentation metrics described in Sect. 3.6.1 are showed in Fig. 15. The results are the metrics average to all images of the database. Note from Fig. 15 that the fuzzy adaptive technique obtained higher values for Jaccard, MCC and PPV metrics, while had high values of SE and SP. Next, note the results from Otsu and adaptive techniques are close for most metrics. Therefore, based on these results, we would tend to use FA segmentation in the detection approach. However, despite these results, are identified some problems in FA and Otsu segmentation, that led us to use the adaptive method.
The problem we found in the FA technique is that the chromosomes are very close to each other in segmentation, and in some images it is hard separate them without removing small chromosome areas. Figure 16 illustrates the problem. The first column of Fig. 16a shows the original image, and the green rectangle shows the zoom region, that is shown in Fig. 16b, followed by its ground truth, in Fig. 16c. Figure 16d shows the segmented region using FA, and Fig. 16e shows the results of the adaptive method. As presented in Fig. 16d, the FA segmentation sometimes produces regions with connected chromosomes, making the classification task harder. The same problem does not happen using the method adaptive thresholding.
Looking at Otsu segmentation, it can also generate undesirable scenarios, as shown in Fig. 17. Figure 17a shows the original image, Fig. 17b shows the zoom region, and Fig. 17c shows its ground truth. Figures 17d, e show the segmentation results of the Otsu and adaptive methods, respectively. Note from Fig. 17d that Otsu segmentation can generate incorrect segmentation, thus missing the individual chromosomes of the regions. This occurs in some images of the database yet while using the adaptive method this problem was not found.
Based on these observations, the segmentation made by the adaptive thresholding algorithm does not lose chromosome areas. The only problem of adaptive method is the amount of noise generated in the image, but this is solved in the classification stage. Therefore, the adaptive method was selected as segmentation method for the following analysis.
4.2 Classification analysis
The classification algorithms were applied to the cropped database. For this analysis, are compared some recent deep learning techniques, described in Sect. 3.5.1: VGG16_TL, VGG16_FT, VGG19_TL, VGG19_FT, Inception_v3_TL, Inception_v3_FT, MobileNet_TL, MobileNet_FT, Xception_TL, Xception_FT, Sharma and miniVGG.
Different parameters values of the input image were used to perform the classification, as shown in Table 3:
The architectures based on VGG, Inception, MobileNet and Xception require a minimum size of input image of 64\(\times \) 64, 192\(\times \) 192, 128\(\times \) 128 and 128\(\times \) 128, respectively. Was evaluated higher input sizes for VGG, but as the chromosome region is small, when resizing, it loses quality, which impacts the final classification. The padding preprocessing operation on input images was evaluated, and also, using colored and segmented images, as described in Sect. 3.1. The best configurations for each technique are described in Table 4.
All models were executed using the Adam optimizer, with a learning rate of 0.001, using 50 epochs and a batch size value of 32. For this analysis, the models were trained on the cropped training and validation datasets and tested on the cropped testing dataset. Each architecture was executed 10 times, and average metrics were calculated.
The results of accuracy obtained for each technique are shown in Table 5, where the highest value obtained is represented by the bold value. From Table 5, note that all models obtained better results using the binary approach, using the segmented chromosome regions. The best models were the ones based on VGG architecture. The results based on VGG architecture have a similar accuracy, there being no significant statistical difference between them. The accuracy of other approaches, such as the Inception, MobileNet and Xception, was lower, probably because they required a bigger input size image, which impacted the accuracy. There is not any significant difference between the models defined as FT and TL, thus indicating that the initial weights of the first layer are enough to provide a good classification. The Sharma model, although using a 64\(\times \)64 input image, did not obtain good results.
4.3 Detection analysis
An analysis to detect chromosomes in the test images was also conducted. The experiments were performed using the detection approach described in Fig. 9, for the test images. Table 6 shows the results using the metrics of SE, SP, F1 and AP, where the highest values for each metric are showed in bold.
As can be seen in Table 6, the VGG16_FT model achieved the best results for F1 and AP metric, while having the highest accuracy. The other models based on VGG16 and VGG19 obtained similar performance. The Sharma model presented poor results and is therefore not recommended for detection. Figure 18 shows the results of segmentation of the VGG16_FT model for some figures of the database. As can be seen in Fig. 18, the proposed approach, using VGG16_FT, can detect all the chromosomes and generates few false positives per image. These results are supported by the high values of the metrics shown in Table 6.
After the classification, it was analyzed the impact of the false positive reduction and false negative reduction stages on the metrics of sensitivity and specificity, to the VGG_FT Algorithm. The results are shown in Fig. 19 where it can be seen that when the false positive reduction is applied, it does not affect the SE metric, because the metric is not based on false positives. But it increases the SE metric, from 0.98, which is already high, to 0.99. When applying the FN reduction, it recovers many misclassified candidates, reducing the false positives and significantly increasing the SE metric.
Finally, it evaluates the precision-recall curve and the AUC metrics. To this analysis, it was taken the best execution results found by each architecture. The results are shown in Fig. 20. It can be observed that the VGG16_FT model had better results than the other approaches, having higher AUC value.
5 Discussion
The segmentation analysis performed in Sect. 4.1 extends the analysis conducted by Andrade et al. [15], thus adding more images to the database. Although the metrics used suggested the use of fuzzy-adaptive method as the best method to perform segmentation, Fig. 16 shows some problems were found in its segmented images. Similar problems were found with the Otsu technique, as shown in Fig. 17. These results show that using classical segmentation metrics, without a qualitative verification of the images, can lead to bad project decisions. For example, the Jaccard metric promotes segmentations with high amounts of TP, while having low amounts of FN and FP. While the Jaccard metric promotes the expansion of the segmented area by having a higher value when an image has high TP and low FN values, it controls the expansion by adding a FP term in the equation, as shown in Eq. 3. However, the equation does not take into consideration if there are connected chromosomes in final segmentation, which is not desirable for chromosome detection. A small expansion in the segmented area, generating false positives and connected chromosomes, has a more negative impact for the classifier than having a medium retraction in the segmented area, thereby generating less TP and more FN, but still preserving the chromosome structure. However, as there are more terms promoting expansion (TP and FN), the metric benefits the results that overextend the ground truth area rather than reducing its area. A similar scenario occurs when applying the other metrics. The segmentation of adaptive, on the other hand, avoids cases of connected chromosomes, which can help to detect chromosomes.
In this study, we also proposed evaluating the classification using two approaches described in Sect. 3.1, RGB and binary segmented images. From Tables 4 and 5, are observed that using a binary image as input provided better results for all the techniques analyzed. This in an interesting observation, since most of the methods used in state-of-the-art to classify chromosomes use RGB images, as described in Sect. 2. The better results of binary images may occur because the colored images from the database have more noise and a slighter variance, which can affect in the classification process.
Results in classification analysis show that the models based on VGG architecture obtained the best values, and achieved 93% for the accuracy metric. One of the aspects that benefits VGG architecture is that it accepts input images with smaller sizes, such as 64 \(\times \) 64. When we resized the images to bigger sizes, the values obtained were lower. However, Sharma architecture also used 64 \(\times \) 64 images as input and obtained lower values.
When analyzing the detection approach, we found that VGG16_FT has obtained the highest values for the metrics SE, SP, F1 and AP, as shown in Table 6. Results presented in Fig. 18 show that the proposed approach is able to detect the chromosomes even when there are cells and noise in the image. The high values of SE and SP also show that most chromosomes were detected and few false positives were generated. We noticed that when the original image did not have much noise and the chromosomes are clear in the image, the algorithm achieves the best results, obtaining SE and SP values of 1.0 in some cases. Therefore, the quality of the input image can impact the final detection, but, as illustrated, the method presented can work well even with noisy images.
As the number of images in our dataset is small, the models trained from scratch had lower performance compared to the pretrained models. This is expected, because deep learning model requires a large number of data to be trained. Even using a simpler model, such as the combination of Zernike moments and MLP, the pretrained models still had better results, because they were pretrained with millions of images, from ImageNet. However, this is an interesting results, because it shows that the transfer learning can be used to classify chromosome candidates, even with reduced training sets. This strategy has not been explored in the literature to classify chromosomes, but our study shows its potential and how it can outperforms other models trained from scratch. Although the transfer learning performed well for the small training set, results from Table 6 shows that a fine tuning stage is important to have better results.
Results presented in Fig. 19 show the impact of false positive and false negative reduction stages, on average, for the VGG16_FT model. For the false positive reduction stage, which affects only the specificity metric, the proposed method contributes to reducing the number of false positives cases. As most of the databases used in the literature do not have the presence of noise or cells in the image, no post-processing steps have been proposed to deal with false positive reduction. The same can be considered for the false negative reduction step. Adding this step increased the value of sensitivity metric by 18%. As reported in Sect. 3.4.2, this process can add few false positive detections in some images, but on average, it did not affect the specificity metric.
The last analysis, illustrated in Fig. 20, it shows using the proposed method, with VGG16_FT CNN architecture, obtained 0.955 of AUC accuracy, for its best model run, indicating that its use for chromosome detection is recommended and can help cytogeneticists to analyze chromosome images.
6 Conclusion
This paper is set out to analyze deep learning approaches in order to classify and detect chromosomes in metaphase images. For this purpose, we evaluated state-of-the-art deep learning algorithms, thereby analyzing the processes of segmentation, classification, detection and post-processing. We constructed a database of metaphase chromosome images, with the aid of the CRCN-NE lab, which is available on the Zenodo platform [22] and consists of 74 labeled images. From the original labeled images, we also constructed a cropped database, which has 2174 regions of each class: chromosome and non-chromosome. Finally, we proposed a chromosome detection approach, with false positive and false negative reduction stages, which was evaluated with SE, SP, F1 and AP metrics.
Results showed that adaptive thresholding obtained better results for segmenting chromosome images. It was also reported that relying only on the metric values can lead to choosing segmentations with connected chromosomes. Furthermore, we proposed to add a segmentation step before submitting the image to the classifier, which provided better results than using RGB images, as most of the approaches in the literature do.
From the classification analysis, we showed that CNN models based on VGG architecture, using fine tuning, could obtain 93.19% accuracy. For detection results, we showed that the proposed approach could obtain values of 0.983 and 0.989 for the metrics of sensibility and specificity, respectively, when used VGG16 with fine tuning was used. Moreover, qualitative results indicate the detection of all chromosomes in most of the images, while generating few false positives per image.
In this paper, we also proposed false positive and false negative post-processing stages, which increase the sensitivity value by 18%, obtaining final results of 0.98 and 0.99, on average, for the metrics of sensitivity and specificity.
Future research should include applying solutions for overlapped chromosomes found and improving the detection process.
References
Pantaleão, C.H.Z., et al.: Contribuição à análise e classificação citogenética baseada no processamento digital de imagens e no enfoque lógico-combinatório, 168 p. Doctoral dissertation, UFSC - Universidade Federal de Santa Catarina, Florianópolis (2003)
Cunha, D.M.d.C., et al.: Análise cromossômica por microarranjos em probandos com indicação clínica de síndrome de down sem alterações cariotípicas, 54 p, Masters dissertation, Pontifícia Universidade Católica de Goiás, Goiânia (2015)
Nussbaum, R.L., Mcinnes, R.R., Willard, H.: Thompson & Thompson: Genética médica. Elsevier Brasil (2008)
Grisan, E., Poletti, E., Ruggeri, A.: Automatic segmentation and disentangling of chromosomes in q-band prometaphase images. IEEE Trans. Inf. Technol. Biomed. 13, 575–581 (2009)
Arora, T., Dhir, R.: A review of metaphase chromosome image selection techniques for automatic karyotype generation. Med. Biol. Eng. Comput. 54, 1147–1157 (2016)
Organization, W.H., et al.: Cytogenetic dosimetry: applications in preparedness for and response to radiation emergencies, Technical Report, International Atomic Energy Agency (2011)
Roemer, E., Zenzen, V., Conroy, L.L., Luedemann, K., Dempsey, R., Schunck, C., Sticken, E.T.: Automation of the in vitro micronucleus and chromosome aberration assay for the assessment of the genotoxicity of the particulate and gas-vapor phase of cigarette smoke. Toxicol. Mech. Methods 25, 320–333 (2015)
Kurtz, G.C.: Metodologias para detecção do centrômero no processo de identificação de cromossomos, 116 p. Masters Dissertation, Universidade Federal de Santa Maria, Santa Maria (2011)
Da Matta, M., Dümpelmann, M., Lemos-Pinto, M., Fernandes, T., Amaral, A.: Processamento de imagens em biodosimetria: influência da qualidade das preparações cromossômicas. Sci. Lena 9 (2013)
Grisan, E., Poletti, E., Ruggeri, A.: An improved segmentation of chromosomes in q-band prometaphase images using a region based level set. In: World Congress on Medical Physics and Biomedical Engineering, September 7–2, Munich, Germany, Springer, pp. 748–751 (2009)
Wang, X., Li, S., Liu, H., Wood, M., Chen, W.R., Zheng, B.: Automated identification of analyzable metaphase chromosomes depicted on microscopic digital images. J. Biomed. Informatics 41, 264–271 (2008)
Uttamatanin, R., Yuvapoositanon, P., Intarapanich, A., Kaewkamnerd, S., Phuksaritanon, R., Assawamakin, A., Tongsima, S.: Metasel: a metaphase selection tool using a Gaussian-based classification technique. BMC Bioinformatics 14, S13 (2013)
Vala, H.J., Baxi, A.: A review on otsu image segmentation algorithm. Int. J. Adv. Res. Comput. Eng. Technol. (IJARCET) 2, 387–389 (2013)
Gagula-Palalic, S., Can, M.: Human chromosome classification using competitive neural network teams (cnnt) and nearest neighbor. In: IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI), pp. 626–629. IEEE (2014)
Andrade, M.F., Cordeiro, F.R., Macário, V., Lima, F.F., Hwang, S.F., Mendonça, J.C.: A fuzzy-adaptive approach to segment metaphase chromosome images. In: 2018 7th Brazilian Conference on Intelligent Systems (BRACIS). IEEE, pp. 290–295
Qin, Y., Wen, J., Zheng, H., Huang, X., Yang, J., Wu, L., Song, N., Zhu, Y.-M., Yang, G.-Z.: Varifocal-net: a chromosome classification approach using deep convolutional networks. IEEE Trans. Med. Imaging 38, 2569–2581 (2019)
Sharma, M., Saha, O., Sriraman, A., Hebbalaguppe, R., Vig, L., Karande, S.: Crowdsourcing for chromosome segmentation and deep classification. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW). IEEE, pp. 786–793
Javan-Roshtkhari, M., Setarehdan, S.K.: A new approach to automatic classification of the curved chromosomes. In: 5th International Symposium on Image and Signal Processing and Analysis, (2007). ISPA 2007. IEEE, pp. 19–24
Swati, G.G., Yadav, M., Sharma, M., Vig, L., et al.: Siamese networks for chromosome classification. In: ICCV Workshop, pp. 72–81
Estandarte, A.K.C.: A Review of the Different Staining Techniques for Human Metaphase Chromosomes. Department of Chemistry, University College London, University of London, London (2012)
Abid, F., Hamami, L.: A survey of neural network based automated systems for human chromosome classification. Artif. Intell. Rev. 49, 41–56 (2018)
Andrade, M.F.S., Cordeiro, F.R., Silva, J.J.G., Lima, F.F., Hwang, S., Macario, V.: CRCN-NE Chromosomes DataSet (Version 1.3) [Data set]. Zenodo. (2019) https://doi.org/10.5281/zenodo.3229434
Arora, T., Dhir, R.: Segmentation approaches for human metaspread chromosome images using level set methods. In: International Conference on Mass Data Analysis of Images and Signals MDA 2016 in New York
Dougherty, A.W., You, J.: A kernel-based adaptive fuzzy c-means algorithm for m-fish image segmentation. In: 2017 International Joint Conference on Neural Networks (IJCNN). IEEE, pp. 198–205
Russell, B.C., Torralba, A., Murphy, K.P., Freeman, W.T.: LabelMe: a database and web-based tool for image annotation. Int. J. Comput. Vis. 77, 157–173 (2008)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition (2014). arXiv:1409.1556
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826
Howard, A.G., Zhu, M., Chen, B., Kalenichenko, D., Wang, W., Weyand, T., Andreetto, M., Adam, H.: Mobilenets: efficient convolutional neural networks for mobile vision applications (2017). arXiv:1704.04861
Chollet, F.: Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1251–1258
Huh, M., Agrawal, P., Efros, A.A.: What makes imagenet good for transfer learning? (2016). arXiv:1608.08614
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S., Ma, S., Huang, Z., Karpathy, A., Khosla, A., Bernstein, M., et al.: Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 115, 211–252 (2015)
Litjens, G., Kooi, T., Bejnordi, B.E., Setio, A.A.A., Ciompi, F., Ghafoorian, M., Van Der Laak, J.A., Van Ginneken, B., Sánchez, C.I.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Khotanzad, A., Hong, Y.H.: Invariant image recognition by zernike moments. IEEE Trans. Pattern Anal. Mach. Intell. 12, 489–497 (1990)
Chollet, F. & others, 2015. Keras. Available at: https://github.com/fchollet/keras
Bradski, G.: The OpenCV Library. Dr. Dobb’s J. Softw. Tools (2000)
Poletti, E., Zappelli, F., Ruggeri, A., Grisan, E.: A review of thresholding strategies applied to human chromosome segmentation. Comput. Methods Programs Biomed. 108, 679–688 (2012)
Acknowledgements
The authors would like to thank FACEPE and CRCN-NE for supporting this project. We also gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp used for this research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Andrade, M.F.S., Dias, L.V., Macario, V. et al. A study of deep learning approaches for classification and detection chromosomes in metaphase images. Machine Vision and Applications 31, 65 (2020). https://doi.org/10.1007/s00138-020-01115-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00138-020-01115-z