Abstract
Key message
Multi-environmental characterization of flag leaf morphology traits in the US winter wheat revealed nine stable genomic regions for different flag leaf-related traits including a major region governing flag leaf angle.
Abstract
Flag leaf in wheat is the primary contributor to accumulating photosynthetic assimilates. Flag leaf morphology (FLM) traits determine the overall canopy structure and capacity to intercept the light, thus influencing photosynthetic efficiency. Hence, understanding the genetic control of these traits could be useful for breeding desirable ideotypes in wheat. We used a panel of 272 accessions from the hard winter wheat (HWW) region of the USA to investigate the genetic architecture of five FLM traits including flag leaf length (FLL), width (FLW), angle (FLANG), length–width ratio, and area using multilocation field experiments. Multi-environment GWAS using 14,537 single-nucleotide polymorphisms identified 36 marker-trait associations for different traits, with nine being stable across environments. A novel and major stable region for FLANG (qFLANG.1A) was identified on chromosome 1A accounting for 9–13% variation. Analysis of spatial distribution for qFLANG.1A in a set of 2354 breeding lines from the HWW region showed a higher frequency of allele associated with narrow leaf angle. A KASP assay was developed for allelic discrimination of qFLANG.1A and was used for its independent validation in a diverse set of spring wheat accessions. Furthermore, candidate gene analysis for two regions associated with FLANG identified seven putative genes of interest for each of the two regions. The present study enhances our understanding of the genetic control of FLM in wheat, particularly FLANG, and these results will be useful for dissecting the genes underlying canopy architecture in wheat facilitating the development of climate-resilient wheat varieties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bread wheat (Triticum aestivum) is the most extensively grown staple cereal crop for ensuring the food security of the growing human population worldwide. It provides approximately 19% of the total dietary calories and 21% of proteins globally along with a wealth of additional nutrients that promote human health in everyday diets (Tadesse et al. 2019). Given that the global human population is projected to reach 10 billion by 2050 and that farmland will decline steadily, there will inevitably be a need to increase food production. However, current trends indicate that progress toward higher-yielding wheat cultivars may fall short of meeting the impending demand (Tilman et al. 2011; Ray et al. 2013). Therefore, enhancing the genetic potential of wheat productivity remains the primary objective for all wheat breeding programs.
Many studies have shown that the photosynthetic capacity of crop species will play a major role in improving crop yield (Zhu et al. 2008; Ort et al. 2015). Canopy architecture and photosynthesis efficiency together play a pivotal role in determining photosynthetic capacity; therefore, it is imperative to discover genes that control canopy architecture and sustained flow of nutrients that lead to optimal spike development and yield (Mantilla-Perez and Salas Fernandez 2017; Ma et al. 2020). The flag leaf plays an important role in wheat as the primary site for accumulating photo-assimilates contributing 40–60% of photosynthetic performance and over 40% of assimilates during the grain-filling stage (Sharma et al. 2003; Ba et al. 2020; Du et al. 2022). Various characteristics of flag leaf morphology including the size and area of the flag leaf have been found to be positively associated with various yield-related traits and ultimately contribute to wheat productivity (Fan et al. 2015; Liu et al. 2018a). Additionally, high yields have been associated with the size and area of the flag leaf in rice and have become a breeding target to achieve an ideal phenotype (Li et al. 1998; Zhang et al. 2014).
Flag leaf morphology (FLM) is characterized by the flag leaf length (FLL), width (FLW), angle (FLANG), area (FLA), and ratio (FLR), which together determine the canopy architecture. Understanding the genetic mechanisms governing these quantitative traits influenced by environmental factors will enable the development of genotypes with stable and enhanced photosynthetic efficiency (Kobayashi et al. 2003). Photosynthesis is affected by many factors that determine the distribution of light over the leaves in the canopy. Among the flag leaf traits, leaf posture, or more specifically leaf inclination (angle), plays an important role in terms of light interception over the canopy. The flag leaf angle (FLANG), defined as the angle between the leaf blade midrib and the stem, directly influences the amount of light that plants receive, which ultimately affects yield (Mantilla-Perez and Salas Fernandez 2017). Modeling approaches have been employed for decades to predict the optimal plant characteristics that would optimize yield with leaf inclination being a key factor (Mantilla-Perez and Salas Fernandez 2017). Many researchers have proposed that upright leaf angles in the upper canopy, less erect leaves in the middle canopy, and more horizontally oriented leaves in the lower canopy provide an ideal plant architecture (Long et al. 2006; Zhu et al. 2010; Ku et al. 2011; Ort et al. 2015). The impact of leaf angle on photosynthetic efficiency has been extensively studied in various cereal crops including maize (Hammer et al. 2009; Fischer and Edmeades 2010; Edwards 2011; Tian et al. 2011), rice (Sinclair and Sheehy 1999; Sakamoto et al. 2006; Kumagai et al. 2009), barley, sorghum, and oats (Tanner et al. 1966; Shearman et al. 2005; Fischer and Edmeades 2010; Truong et al. 2015). The findings of these studies suggested that cultivars with erect leaves exhibit better light interception, higher photosynthesis, greater crop growth, less photoinhibition, and higher yield in comparison with those with horizontally positioned leaves. The plant model for 'super' high-yielding hybrid rice, also known as the 'second generation of new plant types (NPTs),’ incorporates a reduced leaf angle as one of its suggested morphological characteristics (Li et al. 2002; Peng et al. 2008). In wheat, Donald (1968) also considered upright leaves as the desired 'ideotype.' Over the past five decades, studies in wheat have consistently suggested that genotypes with small and erect leaves exhibit higher exposure of leaf area to sunlight resulting in higher Leaf Area Index (LAI) and a higher rate of dry matter production, which ultimately translates to superior yields (Donald 1968; Choudhury 2000; Parry et al. 2011). In rice, narrow flag leaf angles have been reported to result in 13% higher photosynthetic rates, a reduction in photoinhibition, and 15% higher yields (Chen et al. 2020; Mantilla-Perez and Salas Fernandez 2017). Thus, understanding the underlying genetics of leaf angle is crucial for the development of photosynthetically efficient wheat cultivars.
Several studies have been conducted to characterize the FLM traits in wheat. Most of these studies used linkage mapping to identify QTLs governing FLM traits (Xue et al. 2013; Fan et al. 2015; Yang et al. 2016; Hussain et al. 2017; Zhao et al. 2018; Liu et al. 2018a, b; Khanna-Chopra et al. 2020; Ma et al. 2020; Yan et al. 2020a, b, c; Wang et al. 2022). However, the majority of these studies have focused on the FLL and FLW, and few studies have focused on the FLANG (Yang et al. 2016; Sun et al. 2017; Liu et al. 2018a; Chen et al. 2021). Liu et al. (2019) used map-based cloning to identify the first gene TaSPL8 that regulates leaf angle in wheat. TaSPL8 encodes a SQUAMOSA PROMOTER BINDING-LIKE (SPL) protein that regulates leaf angle in wheat through auxin signaling and brassinosteroid biosynthetic pathway. Recently, Zhang et al. (2024) fine-mapped and validated a major quantitative trait locus on chromosome 4B for flag leaf angle in wheat and identified candidate genes underlying this QTL using expression analysis. It is also noteworthy that only a handful of studies have used genome-wide association studies (GWAS) to characterize FLM traits in wheat, especially FLANG. Most of these studies have focused on the characterization of yield-related traits using FLM traits as secondary traits ( Sun et al. 2017; Liu et al. 2017; Chen et al. 2021; Li et al. 2021; Muhammad et al. 2021a, b; Wang et al. 2022). Furthermore, the majority of reported studies are from China, whereas such studies have not been reported in the US winter wheat. In the above context, there is an opportunity to explore the phenotypic and genetic diversity associated with FLM traits in US winter wheat that will provide insights into the genetic mechanism governing FLM traits and enable its utilization in wheat breeding programs.
The majority of GWAS reports in various crop species commonly utilize assembled diversity panels or landraces (Sidhu et al. 2020; AlTameemi et al. 2021; Gudi et al. 2024), rather than using the breeding materials (Ward et al. 2019). The existing genotypic variation in various germplasm and diversity panels provides an opportunity to exploit photosynthetic efficiency through characterization and subsequent integration into breeding programs (Fischer and Edmeades 2010; Lawson et al. 2012; Singh et al. 2022). However, the utilization of elite breeding lines within these panels not only assists in uncovering novel genomic regions associated with the desired trait(s) but also enables the successful integration of beneficial alleles into the cultivar development process without any linkage drag (Gill et al. 2022).
To the best of our knowledge, this is the first study on the characterization of FLM traits in US wheat germplasm in multilocation field experiments. We assembled a population comprising diverse lines from the South Dakota State University (SDSU) breeding program and a set of released cultivars/breeding lines from different breeding programs in the US Great Plains region. The assembled panel was genotyped using genotyping-by-sequencing (GBS) and evaluated for FLM traits in field trials at multiple locations. The objectives of the current study were to (a) assess the phenotypic and genetic variation for the FLM traits in the US hard winter wheat, (b) dissect the genetic architecture of these traits using GWAS to identify genomic regions and putative candidate genes for the FLM using a wheat reference genome, and (c) develop and validate Kompetitive Allele Specific Polymerase Chain Reaction (KASP) assays for important genomic regions to facilitate marker-assisted selection (MAS) and fine mapping.
Materials and methods
Plant materials and experimental setup
In this study, we assembled a new association mapping panel named the Winter Wheat Association Mapping (WWAM) panel, which includes 272 diverse winter wheat accessions (Supplementary Table S1). The WWAM panel primarily comprised breeding lines developed over the last 10 years at the South Dakota State University (SDSU) winter wheat breeding program, selected based on their diverse pedigrees. To enhance mapping resolution, the panel also included historical and recently released cultivars and elite lines from the Hard Winter Wheat (HWW) region that have been widely used in various breeding programs. Additionally, a small set of winter wheat accessions from the global collection of wheat lines, which were recently exome-captured (He et al. 2019), were included in the panel based on their previous evaluation in the South Dakota environment. The panel also included ‘Kharkof’ and ‘Scout-66’ which have been used as historical checks in regional HWW nurseries. The metadata for the WWAM panel is provided in Supplementary Table S1.
The WWAM panel was evaluated for FLM traits in three field environments including SDSU experimental stations at Aurora, Brookings, and Felt farm, South Dakota (referred to as E1, E2, and E3) in the 2022–2023 winter wheat growing season. Each experiment was planted using a randomized complete block design (RCBD) with two replications (complete blocks), resulting in six independent field replicates for the whole experiment. The experimental unit within each replicate was a four-row plot that was 1.25 m long with an inter-row spacing of 20 cm. All the trials were managed following regional standard cultural practices for proper growth and development of wheat.
Phenotyping for FLM and agronomic traits
After flowering, disease-free flag leaves from the 10 main tillers in the two middle rows of each plot were selected for measuring the flag leaf length (FLL), flag leaf width (FLW), and flag leaf angle (FLANG). Two additional traits, namely flag leaf area (FLA) and the length-to-width ratio of the flag leaf (FLR), were estimated as described in previous studies (Ma et al. 2020; Chen et al. 2021). The FLL was determined by measuring the distance from the base to the tip of the leaf, while flag leaf width (FLW) represented the width at the broadest section of the leaf. The FLANG was measured as the angle between the stem and midrib of the blade. FLM measurements were taken after flowering (Feekes 10.5.1), with the identification and tagging of ten plants 4–5 days post-flowering using spray paint to ensure consistency in the crop stage in each plot. The flag leaf area was measured as the multiplication of FLL and FLW by a factor of 0.75 (FLL × FLW × 0.75) (Yang et al. 2016; Liu et al. 2018b). Plant height (PH) was measured as the distance from the base of the stem to the top of the spike, excluding the awns after reaching physiological maturity. Days to heading (DH) were recorded as the Julian days when 50% of the heads were emerged in each plot.
Statistical analysis of phenotype data
The statistical analyses were performed using various packages in R environment (Core R Team 2019). Initially, we analyzed the three experiments independently to obtain the best linear unbiased estimates (BLUE) for various traits using the following model:
where \({y}_{ij}\) is the trait of interest, \(\mu\) represents the overall mean, \({R}_{i}\) denotes the random effect of the ith replicate in each location, \({G}_{j}\) refers to the fixed effect of the jth genotype, and \({e}_{ij}\) refers to the residual error effect of the ith replication and jth genotype.
Secondly, BLUEs were calculated from all three environments using the following equation:
where \({y}_{ijk}\) is the trait of interest, \(\mu\) is overall mean, \({E}_{i}\) refers to the random effect of the ith environment, \({R}_{j(i)}\) refers to the random effect of the jth replicate nested in the ith environment, \({G}_{k}\) refers to the fixed effect of the kth genotype, \({GE}_{ik}\) denotes the genotype by environment (G × E) interaction, and \({e}_{ijk}\) refers to the residual error. The environment corresponds to the individual locations, and replicates correspond to the complete blocks within each location.
The broad-sense heritability (H2) was estimated by fitting the genotypic effect from the above equation as random using the following formula:
where \({\sigma }_{g}^{2}\) and \({\sigma }_{e}^{2}\), are the genotype and error variance components, respectively; \({\sigma }_{ge}^{2}\) is the variance component for G × E, nRep refers to the number of replicates, and nLoc refers to the number of locations. The above analysis was performed with the META-R tool (Alvarado et al. 2019), which uses ‘lme4’ package (Bates et al. 2015). The correlations among traits were obtained using the BLUEs from across environment analysis and visualized using ‘psych’ library in R (William 2023). All the model comparisons were visualized using ‘ggplot2’ in a R package (Wickham 2016).
Genotyping, population structure, and linkage disequilibrium
The WWAM panel was genotyped using GBS as previously described (Gill et al. 2022) in different sequencing runs as a limited number of lines are sequenced each year for the breeding program. DNA was isolated from fresh leaf tissue collected at the three-leaf stage using the hexadecyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1987), and GBS libraries were prepared using the PstI and MspI restriction enzymes as described by Poland et al. (2012). The libraries were sequenced using Ion Proton sequencer (Thermo Fisher Scientific, Waltham, MA, USA) or NextSeq2000 sequencer (Illumina, San Diego) at the USDA Central Small Grain Genotyping Lab, Manhattan, KS. The raw GBS reads were used to call single-nucleotide polymorphisms (SNPs) for the complete set of 272 accessions using GBS discovery pipeline v2.0 in TASSEL v5.0 (Trait Analysis by Association, Evolution and Linkage) (Bradbury et al. 2007), and Chinese Spring (CS) wheat genome RefSeq v2.1 (Zhu et al. 2020) was used as the reference. The SNPs with > 30% missing data points, > 10% heterozygotes, < 5% minor allele frequency (MAF), or unmapped to any chromosome were removed for quality control, followed by imputation for missing data using BEAGLE v4.1 (Browning and Browning 2007). In addition, two accessions were removed due to the high frequency of missing data. After filtration, 14,537 high-quality SNPs for 270 accessions were retained and used for further analysis.
The WWAM panel was examined for population stratification using principal component analysis (PCA) and STRUCTURE analysis. PCA was performed based on the genotypic data using ‘SNPRelate’ package (Zheng et al. 2012) and visualized using ‘ggplot2’ in R (Wickham 2016). A Bayesian model-based clustering program, STRUCTURE v2.3.4, was also used assuming an Admixture model (Pritchard et al. 2000). We performed ten independent runs with a burn-in period of 20,000 iterations followed by 20,000 Monte Carlo iterations assuming ten subpopulations (K = 1–10). The analysis was performed in parallel using StrAuto v1.0 on the SDSU high-performance computing (HPC) cluster (Chhatre and Emerson 2017; Tange 2018). The most likely number of subpopulations within the WWAM panel was inferred using an ad hoc statistic (DeltaK) that used the rate of change in the log probability between runs using successive K-values using STRUCTURE HARVESTER (Evanno et al. 2005; Earl and vonHoldt 2012). Further, a neighbor-joining-based phylogenetic tree was constructed in TASSEL using the filtered set of SNP markers. Linkage disequilibrium (LD) analysis was performed using the complete set of SNPs with all pairwise marker comparisons and a sliding window size of 50 SNPs in TASSEL v5.0 (Bradbury et al. 2007). The LD decay for the whole genome as well as individual sub-genomes was obtained by fitting a nonlinear model using the modified Hill and Weir method (Hill and Weir 1988) as described in Gill et al. (2022) and visualized in the R.
GWAS and candidate gene analysis
We performed GWAS using 14,537 SNPs and trait BLUEs obtained by analyzing individual experiments (E1, E2, and E3) as well as the BLUEs from combined analysis (CEnv). A multi-locus model, Fixed and random model Circulating Probability Unification (FarmCPU), was used to perform GWAS for all the traits (Liu et al. 2016). The FarmCPU model used four principal components to account for the population structure and was implemented in Genomic Association and Prediction Integrated Tool (GAPIT) version 3.0 in the R (Wang and Zhang 2021). The fit of the model was observed by visualizing the quantile–quantile (QQ) plots. A stringent criterion was set to report the marker-trait associations (MTAs) that were stable and observed in different environments. According to this criterion, only those stable MTAs were reported that were identified in at least three GWAS analyses (E1, E2, E3, or CEnv) at an exploratory threshold of − log10(P) = 3.50, as well as surpassed the Bonferroni-corrected (P < 0.05) threshold of − log10(P) = 5.40 in one of those three environments. The proportion of the phenotypic variance explained by a major genomic region on chromosome 1A associated with FLANG (qFLANG.1A) was estimated by fitting a mixed-linear model (MLM) in TASSEL v5.0 (Bradbury et al. 2007). We performed a pairwise comparison of two alleles of qFLANG.1A identified on chromosome 1A, for differences in FLANG, days to heading, and plant height. For this purpose, mean trait values (BLUEs from combined analysis) for two groups of alleles were compared using a t-test and visualized using boxplots in R.
We performed candidate gene analyses for the selected regions based on the local LD, rather than relying on a random upstream/downstream search around the significant SNP. For candidate gene analyses of identified genomic regions, we used rTASSEL (Monier et al. 2021) to visualize LD blocks in the region harboring associated SNP. Further, we used CS RefSeq v2.1 (Zhu et al. 2021) to extract all high confidence (HC) genes within the LD block harboring respective SNP. The coding sequences for HC genes were retrieved from the CS RefSeq v2.1 database and were used to manually annotate the HC genes using ‘Blast2GO’ (Conesa et al. 2005). The functional annotation, comparison with reported FLM-related genes, and publicly available expression databases were used to select the putative candidate genes of interest.
Development of KASP and independent validation of qFLANG.1A on chromosome 1A
For the utilization of stable and novel genomic region for FLANG (qFLANG.1A) identified on chromosome 1A, we designed primers for the KASP assay. The 200 bp sequences from upstream and downstream of significant SNP associated with FLANG were identified in IWGSC CS RefSeq v2.1 (Zhu et al. 2021) to design KASP primers using PolyMarker (Ramirez-Gonzalez et al. 2015). The tail sequences were added to 5’-ends of the two forward primers and the tail sequences matched with FAM (6-carboxy-fluorescein) and HEX (Hexachlorofluorescein) fluorescence-labelled sequences in KASP mix (Supplementary Table S2). The total reaction volume for each KASP assay was 10.14 µL comprising 5 µL of genomic DNA (~ 50 ng), 5 µL of 2X KASP-TF master mix, and 0.14 µL of KASP assay mix. The details of KASP genotyping thermal cycle protocol are provided in Supplementary Table S3. The PCR and fluorescence signal detection were performed in the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, USA). The KASP primers were first tested using a set of 158 lines selected from the WWAM panel representing two alleles.
Further, the developed KASP marker was used for independent validation of the qFLANG.1A in spring wheat to broaden the scope of findings from the current study. We used an independent natural population of 200 diverse spring wheat accessions randomly selected from the National Small Grains Collection (Supplementary Table S4). Importantly, the 200 accessions were part of a larger core collection that represents considerable diversity for spring wheat germplasm. The selected 200 accessions were grown in the greenhouse conditions using DEEPOT D40 CELLS (Stuewe & Sons, Tangent, OR, USA) filled with Sunshine R 360 potting soil (Sun Gro Horticulture, Agawam, MA, USA). The cells were arranged in trays (Stuewe & Sons, Tangent, OR, USA) following a randomized complete block design with three replications. The independent set was phenotyped for FLANG as described in the previous section. Further, the 200 accessions were subjected to KASP assay to determine the qFLANG.1A allele, and the average phenotypic data for FLANG were compared between two allelic groups of qFLANG.1A using a t-test and visualized using boxplots in R.
Results
Phenotypic variation, correlations, and heritability
A significant variation was observed for FLM traits in all tested environments (Table 1). The distribution for FLM traits was almost consistent in all three environments, with slightly lower FLW and FLA measurements in E2 compared to other environments (Figs. 1c-1f). A high broad-sense heritability (H2) was observed for FLM traits, ranging from 0.80 to 0.94 (Table 1) with the highest H2 (0.94) for FLANG. There was no significant correlation among three major FLM traits, including FLL, FLW, and FLANG (Fig. 1g). As expected, significant correlations were observed for FLA and FLR with FLL and FLW owing to the mathematical relationship between these traits (Fig. 1g). Plant height showed a significant positive correlation with FLL (r = 0.40, P ≤ 0.001), a nonsignificant correlation with FLANG (0.09), but a significant negative correlation with FLW (r = − 0.16, P ≤ 0.01). Days to heading showed lower but significant positive correlations with both FLL (r = 0.17, P ≤ 0.01) and FLW (r = 0.28, P ≤ 0.001), and a significant negative correlation with FLANG (r = − 0.19, P ≤ 0.01).
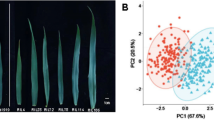
Phenotypic variability and correlation among various flag leaf morphology (FLM) traits in 272 genotypes of WWAM panel evaluated in three different environments (E1, E2, and E3) and combined over environments (CEnv). a Representative images of diverse phenotypes of FLANG, flag leaf angle; b FLL, flag leaf length. Density plots showing the phenotypic distribution of (c) FLL, flag leaf length; d FLW, flag leaf width; e FLA, flag leaf area; f FLANG, flag leaf angle. g Correlation coefficients among investigated traits were calculated using the best linear unbiased estimates (BLUEs) from a combined analysis of three environments. The pair plot shows bivariate scatter plots below the diagonal, histograms on the diagonal, and Pearson correlation between given traits above the diagonal. Statistically significant correlations are denoted by an asterisk (*) where * P ≤ 0.05, ** P ≤ 0.01, and *** P ≤ 0.001
Population structure and linkage disequilibrium
A set of 14,537 high-quality SNPs was used for analyzing the population structure and LD. Among the three sub-genomes, the highest number of SNPs were mapped to the B sub-genome (n = 6409), followed by A (n = 5641) and D (n = 2487) (Supplementary Table S5). Among chromosomes, chromosome 7A had the most SNPs (n = 1186), while chromosome 4D had the least SNPs (n = 117) (Supplementary Table S5). The STRUCTURE analysis showed a distinct peak at K = 4, suggesting the presence of four subgroups within the WWAM panel (Supplementary Figure S1). The principal component analysis also showed stratification in the WWAM panel, with the first two principal components explaining only 4.5% and 3.7% variance, respectively (Fig. 2a). To better explain the clustering within the WWAM panel, we used a neighbor-joining tree based on the genotypic data of the 270 lines (Supplementary Figure S2). Overall, the sub-groups within the WWAM panel were not very distinct. Two smaller sub-groups were identified with the first comprising of the germplasm derived from CIMMYT or Kansas material, and the second was dominated by material from Eastern Kansas and Nebraska. A third large sub-group comprised of majority SDSU releases and breeding lines, and it can be further divided into two groups with one having lines derived from Western Great Plains material, while the other one dominated by lines derived from Northern Plains material. The average LD decay distance was approximately 3 Mbp for the whole wheat genome (Fig. 2b) with a considerable variation in LD decay distance among the three sub-genomes (Figs. 2c-2e), with 1.7, 2.2, and 7 Mbp for sub-genomes A, B, and D, respectively.
Association mapping for FLM traits
The GWAS was conducted for five FLM traits (FLL, FLW, FLANG, FLA, and FLR) using BLUEs for each trait obtained from individual environments (namely E1, E2, and E3), as well as for the combined analysis (CEnv). In total, we identified 36 MTAs (Supplementary Table S6) for different FLM traits based on the Bonferroni-corrected threshold in individual GWAS analyses (Fig. 3). Nevertheless, we were interested in reporting only those MTAs that could be considered reliable based on stability in multiple environments. Hence, we focused on MTAs that were identified in at least three GWAS analyses (E1, E2, E3, or CEnv) at an exploratory threshold of − log10(P) = 3.50, as well surpassed the Bonferroni-corrected (P < 0.05) threshold of − log10(P) = 5.40 in one of those three environments. Based on these criteria, a total of nine MTAs were identified representing nine QTLs for FLM traits (Table 2), with six for FLL and three for FLANG. Stable MTAs were not identified for FLW, FLA, and FLR (Table 2, Supplementary Table S7).
Manhattan plots presenting the results of genome-wide association studies for (a) flag leaf angle (FLANG) and (b) flag leaf length (FLL). The positions of SNPs on chromosomes are indicated on the x-axis relative to their -log10(P) values on the y-axis. The red dotted line refers to the exploratory threshold of -log10(P) = 3.5, and the blue dotted line refers to the Bonferroni-corrected threshold of -log10(P) = 5.40
For FLL, six stable MTAs were identified on chromosomes 1B, 2B, 3A, 4A, 4B, and 5A, with all six MTAs being detected in three GWAS analyses (Table 2, Fig. 3b). The most significant association for FLL was detected on chromosome 1B (represented by SNP S1B_653531690) with − log10(P) values ranging from 4.81 to 8.96. Based on a comparison with previous reports, three of the six MTAs for FLL mapped on chromosomes 1B, 4A, and 4B represented novel genomic regions for this trait. Furthermore, three novel stable MTAs were detected for FLANG located on chromosomes 1A, 5A, and 5B (Table 2, Fig. 3a). Among the three MTAs for FLANG, a major genomic region (represented by SNP S1A_581955638) on the long arm of chromosome 1A (referred to as qFLANG.1A) was of particular interest owing to its consistency in all environments and strong signal. The qFLANG.1A (mapped at 581.9 Mbp on chromosome 1A) was detected in all four GWAS analyses with a strong signal, with − log10(P) ranging from 7.64 in E1 to 16.13 in CEnv. Moreover, the percentage of variance explained by qFLANG.1A (S1A_581955638) was estimated by fitting an MLM model and it ranged from 9 to 13% in different environments.
Further, we performed a pairwise comparison of two alleles of SNP S1A_581955638, which represents qFLANG.1A to evaluate the allelic effects on FLANG, PH, and HD. The 270 accessions in the WWAM panel were grouped based on the S1A_581955638 allele (C/G), and the two groups were compared for mean trait values using a t-test. The two groups significantly differed (P < 0.001) in FLANG, but not in plant height or days to heading (Fig. 4a-4c).
Candidate gene analysis
Candidate gene analysis was performed for only two genomic regions for FLANG (qFLANG.1A and qFLANG.5A), as LD blocks were notably larger (> 7 Mbp) for the other seven regions associated with FLL and FLANG. The two genomic regions were mapped on IWGSC CS RefSeq v2.1, and high-confidence genes were identified from the candidate region based on the available wheat expression data and previous literature. For qFLANG.1A, we visualized the LD around the associated SNP and did not observe a clear single LD block (Fig. 4d). Hence, we extracted the high-confidence (HC) genes from the ~ 581 to ~ 587 Mbp region on chromosome 1A. Out of a total of 79 HC genes from this region, seven putative candidate genes were identified encoding various types of proteins including a F-box family protein, kinase domains, and a glucan endo-1,3-beta-glucosidase 3-like protein (Table 3). Furthermore, the significant SNP (S1A_581955638) for qFLANG.1A was mapped within the gene TraesCS1A03G1030200 encoding glucan endo-1,3-beta-glucosidase 3. Although we did not find any predicted protein in the candidate region that was previously reported to regulate FLM in the literature, it would be valuable to further investigate the gene underlying qFLANG.1A.
Further, 64 HC genes were retrieved in the qFLANG.5A region (473–479 Mb) on chromosome 5A, and seven were identified as putative candidates based on their annotations (Table 3). Among these seven genes, TraesCS5A03G0658100 and TraesCS5A03G0644800 encoding SHAGGY-like kinase and a squamosa promoter-binding like (SPL) protein, respectively, are of particular interest because of their involvement in brassinosteroid (BR) pathway or relatedness to previously characterized genes for FLANG.
Spatial distribution of qFLANG.1A alleles in HWW germplasm
The frequency of allele ‘C' for narrow and ‘G’ for wide flag leaf angle for marker S1A_581955638 in the WWAM panel was 68.6% (177) and 31.4% (81), respectively. To study the spatial distribution of the two alleles in the HWW region, we used a large set of 2354 HWW lines consisting of 2203 breeding lines from SDSU preliminary or advanced trials and 151 Elite materials consisting of released HWW cultivars or lines tested in regional nurseries from different breeding programs. Among the 2203 breeding lines, 1,494 (68%) carried the ‘C’ allele, while 709 (32%) carried the ‘G’ allele (Fig. 5a). However, a slightly higher frequency of the ‘C’ allele was observed in Elite materials (74%) compared to breeding lines, whereas only 26% of lines carrying the ‘G’ allele (Fig. 5b).
Pie charts showing the frequency distribution of two alleles of S1A_581955638, the SNP representing qFLANG.1A. (a) The frequency distribution in breeding lines from the South Dakota State University winter wheat breeding program, and (b) in a set of released cultivars and elite material from various HWW breeding programs of the US Great Plains region
Development of a KASP assay to validate qFLANG.1A in spring wheat population
Considering the large effect of qFLANG.1A on flag leaf angle, a KASP assay was developed to facilitate marker-assisted selection of qFLANG.1A in the breeding programs. The KASP assay was tested on a set of 158 accessions from the WWAM panel, and the marker correctly discriminated the two alleles at SNP S1A_581955638 (Fig. 6a). Further, the KASP assay was used to screen an independent set of 200 diverse spring wheat accessions described in materials (Fig. 6b; Supplementary Table S4). Subsequently, the FLANG data for these 200 accessions were compared for different allelic groups (Supplementary Table S4). The mean FLANG values were significantly different (P = 0.00013) with an average angle of 50.03 for the ‘C’ group and 61.47 for the ‘G’ group (Fig. 6c), confirming the significance of qFLANG.1A and the effectiveness of the KASP assay in spring wheat germplasm.
Allele discrimination plot for SNP S1A_581955638 using KASP assay evaluated on (a) a set of genotypes from the WWAM panel and b on a set of diverse spring wheat genotypes used for independent validation. c Boxplot comparing the FLANG among allelic groups of qFLANG.1A in an independent natural population
Discussion
Flag leaf and its posture determine plant photosynthetic capacity and in turn wheat productivity (Zanella et al. 2023). Altering the flag leaf architecture to maximize photosynthate assimilation at the grain-filling stage is a desirable goal for wheat breeding programs. Various photosynthetic component traits including FLM showed genetic variation and were consistently heritable despite their quantitative nature. This makes them suitable for phenotypic selection, as required for breeding superior wheat cultivars (Poland 2015). We observed a wide range of variations in FLM traits (FLL, FLW, FLANG, FLR, FLA), plant height, days to heading (Table 1), and high heritability, consistent with previous reports (Chen et al. 2021; Wang et al. 2022). The high heritability for FLANG (0.94) in the current study was similar to or higher as compared to previous studies, indicating a significant potential for wheat canopy improvement (Chen et al. 2020, 2021).
Similar to previous studies, we also observed significant positive correlations (Fig. 1g) between plant height and FLL (0.40), FLA (0.16), and FLR (0.40), respectively (Hu et al. 2020; Gao et al. 2021; Sun et al. 2017; Liu et al. 2017; Chen et al. 2021; Li et al. 2021; Muhammad et al. 2021a, b). In addition to other factors, the observed correlations between height and FLL might be attributed to the effect of various genes involved in the brassinosteroid (BR) signaling pathway. Previous studies have shown BR pathway genes have a direct impact on leaf architecture and exhibit pleiotropic effects on plant height (Makarevitch et al. 2012; Hirano et al. 2017; Mantilla-Perez and Salas Fernandez 2017). BR-deficient (BRASSINOSTEROID INSENSITIVE 1 gene) or mutant phenotypes tend to exhibit small leaf angles with reduced height (Morinaka et al. 2006; Sakamoto et al. 2006). Mutants of the BRASSINOSTEROID-SIGNALING KINASE1 (BSK1) gene in rice also resulted in dwarf plants with erect leaves (Bai et al. 2007; Zhang et al. 2012). Sorghum plants with a mutated DWARF1 (DW1) are similar to BR-deficient rice mutants, with phenotypes that include a reduced plant height and leaf angle (Hirano et al. 2017). However, surprisingly we did not observe a significant correlation between FLANG and plant height, suggesting alternate pathways (phytohormones, auxin, ethylene, strigolactone, and gibberellin) may also affect the FLANG in wheat (Shimada et al. 2006; Zhao et al. 2013; Li et al. 2014, 2020a, b). Wang et al. (2020) have also suggested other signals, including circadian rhythm, biotic stresses, MAPK signaling, phosphatidylinositol signaling, and calcium-mediated signaling, may play important roles in regulating leaf angle plasticity; however, it needs further investigation.
The GWAS identified 36 MTAs (Supplementary Table S6) for FLM traits; however, we focused on nine stable MTAs that were consistently identified in multiple environments, making them more reliable and valuable for wheat breeding (Table 2, Supplementary Table S7). Out of six stable MTAs for FLL identified in the present study, three MTAs (S2B_44931079, S3A_688732686, and S5A_570625938) on chromosome 2B, 3A, and 5A are located in the vicinity of previously reported QTLs for FLL in recent studies (Wu et al. 2016; Liu et al. 2018a; Li et al. 2019; Yan et al. 2020c; Du et al. 2022). The other three genomic regions (MTAs, S1B_653531690, S4A_665013137, and S4B_459361891) associated with FLL are novel with no QTL previously reported in these regions. The identification of known and new genomic regions affecting FLL shows the robustness and consistency of our findings and validates the importance of these regions in controlling FLL.
In this study, we identified three stable genomic regions/MTAs for FLANG, namely S1A_581955638, S5A_473723888, and S5B_247555050, on chromosomes 1A, 5A, and 5B, respectively. Notably, all three genomic regions seem to be novel as no genes/QTLs affecting FLANG have been documented in their vicinity in wheat. The understanding of the genetic architecture of leaf inclination is limited in wheat ( Sun et al. 2017; Liu et al. 2018a; Chen et al. 2021). To date, only one gene, TaSPL8, has been cloned for FLANG in wheat using map-based cloning. It encodes a SQUAMOSA PROMOTER BINDING-LIKE (SPL) protein, which regulates lamina joint development through auxin signaling and the brassinosteroid biosynthesis pathway, resulting in the exhibition of erect leaves. Conversely, the genetics of flag leaf angle have been extensively studied in various cereal crop species (Mantilla-Perez and Salas Fernandez 2017). In maize, numerous genes associated with leaf angle including LIGULELESS (Bauer et al. 2004; Buescher et al. 2014), ZmCLA4 (Zhang et al. 2014), and BRASSINOSTEROIDDEFICIENT DWARF1 (Makarevitch et al. 2012) have been cloned. Similarly in rice, genes like OsDWARF (Hong et al. 2002), OsBRI1 (Li and Chory 1997; Yamamuro et al. 2000), LAZY1 (LA1) (Li et al. 2007), LEAF AND TILLER ANGLE INCREASED CONTROLLER (OsLIC) (Sinclair and Sheehy 1999; Tanaka et al. 2009; Zhang et al. 2012), and BRASSINOSTEROID UPREGULATED 1-LIKE1 (OsBUL1) (Jang et al. 2017), and in sorghum, DWARF3 (DW3) (Knöller et al. 2010; Truong et al. 2015) and DWARF1 (DW1) (Yamaguchi et al. 2016; Hirano et al. 2017) have been associated with the leaf inclination.
Among three MTAs, we identified a major and novel region (qFLANG.1A) with a consistently strong signal on chromosome 1A that explained 9–13% of the variation across different environments. Comparative synteny with rice and maize revealed no orthologs associated with canopy architecture in the region (581–587 Mb) harboring qFLANG.1A (Mantilla-Perez and Salas Fernandez 2017). To further validate the significance of this major region and facilitate MAS, we developed a KASP assay specifically for qFLANG.1A to discriminate the two alleles (Fig. 6a). Moreover, we were interested to validate the effect of qFLANG.1A beyond hard winter wheat to other classes of wheat. Hence, we used the KASP assay to discriminate the alleles for qFLANG.1A in a diverse set of spring wheat accessions that was randomly selected from a global core set of spring wheat. The spring wheat set was phenotyped for FLANG, and the KASP assay was robust for identifying the narrow and broad FLANG genotypes in an independent set, and the two allelic groups differed significantly (P = 0.00013) for FLANG. The independent validation in a diverse set provides strong evidence supporting the importance of this genomic region in regulating FLANG not only in winter wheat but also in spring wheat germplasm. Therefore, it becomes compelling to delve deeper into this region and identify the putative gene underlying this important and novel QTL.
Building upon this validation, the spatial distribution of qFLANG.1A alleles across a substantial dataset comprising 2354 HWW lines, including SDSU breeding lines and elite material from various breeding programs evaluated in regional nurseries across the Great Plains, revealed a notable pattern. We observed a higher frequency of alleles associated with narrow leaf angle in all breeding lines similar to the allele distribution observed in our WWAM panel. Remarkably, this consistent trend was also evident in the elite material/varieties, indicating a widespread distribution of these alleles across the Great Plains. This suggests that narrow leaf angle traits have been indirectly selected during the breeding process, likely as part of efforts to enhance yield potential. However, it is noteworthy that despite the strong association with narrow leaf angle traits, we found only one report of QTL (QYld.crc.1A) affecting yield and yield-related traits mapped close to qFLANG.1A (Cuthbert et al. 2008).
We identified seven high-confidence genes of interest for qFLANG.1A. The most interesting among these seven genes is TraesCS1A03G1030200 encoding glucan endo-1,3-beta-glucosidase 3. The most significant and stable SNP for qFLANG.1A (S1A_581955638) was mapped within the gene TraesCS1A03G1030200. Although we did not find the role of this predicted protein in the regulation of flag leaf angle, a recent study on mapping flag leaf angle has delimited an important QTL for FLANG on chromosome 4B to a 5 Mb region and identified seven candidate genes based on differential expression and one of the seven genes encodes a beta-glucosidase protein (Zhang et al. 2024). This suggests a putative role of this gene family in the regulation of FLANG and supports further investigation. Another gene of interest is the F-box family protein coding gene (TraesCS1A03G1026000) identified in this region, which has been reported to be responsible for plant growth and development by regulating leaf growth and cell division, influencing cell numbers and consequently impacting leaf size (Yan et al. 2020a, b, c; Kong et al. 2023). Another gene identified encodes typical receptor-like protein kinases (RLKs) proteins that regulate plant growth, development, and stress responses (Cui et al. 2022). However, some reports have shown that RLKs are involved in the BRASSINOSTEROID INSENSITIVE 1 (BRI1) gene pathway, which mediates the BR signal to regulate cell elongation required for normal growth and development of the plant (Hothorn et al. 2011; She et al. 2011; Cui et al. 2022). We did not identify any annotated gene in the candidate region that was directly related to the BR pathway, which regulates leaf angle in many crops (Mantilla-Perez and Salas Fernandez 2017). The lack of BR pathway genes in the candidate region suggests the role of alternate pathways in regulating leaf angle. Furthermore, the nonsignificant correlation between FLANG and plant height in our study also suggests the possibility of alternate mechanisms affecting FLANG. Various phytohormones, including auxin, ethylene, and gibberellin, may play a role in the regulation of FLANG and need to be further investigated.
By studying the genomic region spanning qFLANG.5A, we identified several genes that may potentially affect FLANG. The gene TraesCS5A03G0658100 encodes a squamosa promoter-binding-like protein, which is similar to SPL protein exhibiting an erect leaf phenotype and regulates lamina joint development by affecting the auxin response and BR biogenesis pathway in wheat (Liu et al. 2019). Additionally, the SQUAMOSA PROMOTER BINDING-LIKE (SPL) protein is related to an erect leaf phenotype in maize and rice (Moreno et al. 1997; Lee et al. 2007). Another gene, TraesCS5A03G0644800, encodes a SHAGGY-like kinase protein, which is known to be involved in hormonal signaling networks and required for growth and stress responses. This protein is a key player in the brassinosteroid (BR) signaling pathway and affects various developmental processes and responses to environmental factors (Bittner et al. 2015; Kloc et al. 2020). Additionally, gene TraesCS5A03G0657600 encodes the ETHYLENE INSENSITIVE 3-like protein which plays a crucial role in regulating a range of functions including leaf development, senescence, and germination stimulation (Dubois et al. 2018). Further studies to validate these candidate genes will help unravel their precise roles and mechanisms regulating FLANG.
In conclusion, this is the first major study dissecting the genetic architecture of FLM traits in US winter wheat where multi-environment GWAS identified 36 MTAs for FLM traits. Nine genomic regions were stable across environments, and three each for FLL and FLANG are novel and have not been previously reported. Further investigation and independent validation of qFLANG.1A, a major QTL associated with FLANG using KASP markers, suggested its importance in both winter and spring wheat panels and its potential use in marker-assisted selection (MAS) in the wheat breeding programs. Fine mapping of this region will be useful to gain a more detailed understanding of the genetic regulations of FLANG in wheat and its potential implications in wheat breeding programs. Further, the SNP markers linked to the stable regions identified in this study could also be incorporated into multivariate genomic selection models for the enhancement of the photosynthetic efficiency of wheat. This research opens avenues for both practical breeding applications and further exploration of genomic regions influencing FLM traits in winter wheat.
Data availability
The datasets generated in the study are either presented in the supplementary information associated with the article or can be provided on request to the corresponding author.
References
AlTameemi R, Gill HS, Ali S et al (2021) Genome-wide association analysis permits characterization of Stagonospora nodorum blotch (SNB) resistance in hard winter wheat. Sci Rep. https://doi.org/10.1038/s41598-021-91515-6
Alvarado G, López M, Vargas M, et al (2019) META-R (Multi Environment Trail Analysis with R for Windows) Version 6.04
Ba Q, Zhang L, Chen S et al (2020) Effects of foliar application of magnesium sulfate on photosynthetic characteristics, dry matter accumulation and its translocation, and carbohydrate metabolism in grain during wheat grain filling. Cereal Res Commun 48:157–163. https://doi.org/10.1007/s42976-020-00026-z
Bai MY, Zhang LY, Gampala SS et al (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.0706386104
Bates D, Mächler M, Bolker BM, Walker SC (2015) Fitting linear mixed-effects models using lme4. J Stat Softw. https://doi.org/10.18637/jss.v067.i01
Bauer P, Lubkowitz M, Tyers R et al (2004) Regulation and a conserved intron sequence of liguleless3/4 knox class-I homeobox genes in grasses. Planta 219:359–368. https://doi.org/10.1007/s00425-004-1233-6
Bittner T, Nadler S, Schulze E, Fischer-Iglesias C (2015) Two homolog wheat Glycogen Synthase Kinase 3/SHAGGY—like kinases are involved in brassinosteroid signaling. BMC Plant Biol 15:1–20. https://doi.org/10.1186/s12870-015-0617-z
Bradbury PJ, Zhang Z, Kroon DE et al (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81:1084–1097. https://doi.org/10.1086/521987
Buescher EM, Moon J, Runkel A et al (2014) Natural variation at sympathy for the ligule controls penetrance of the semidominant liguleless narrow-R mutation in zea mays. G3 Genes Genomes Genet 4:2297. https://doi.org/10.1534/g3.114.014183
Chen S, Cheng X, Yu K et al (2020) Genome-wide association study of differences in 14 agronomic traits under low- and high-density planting models based on the 660k SNP array for common wheat. Plant Breed 139:272. https://doi.org/10.1111/pbr.12774
Chen S, Liu F, Wu W et al (2021) A SNP-based GWAS and functional haplotype-based GWAS of flag leaf-related traits and their influence on the yield of bread wheat (Triticum aestivum L.). Theor Appl Genet 134:3895–3909. https://doi.org/10.1007/s00122-021-03935-7
Chhatre VE, Emerson KJ (2017) StrAuto: automation and parallelization of STRUCTURE analysis. BMC Bioinform 18:192. https://doi.org/10.1186/s12859-017-1593-0
Choudhury BJ (2000) A sensitivity analysis of the radiation use efficiency for gross photosynthesis and net carbon accumulation by wheat. Agric for Meteorol 101:217. https://doi.org/10.1016/S0168-1923(99)00156-2
Conesa A, Gotz S, Garcia-Gomez JM et al (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Core R Team (2019) A Language and Environment for Statistical Computing. R Found Stat Comput 2
Cui Y, Lu X, Gou X (2022) Receptor-like protein kinases in plant reproduction: current understanding and future perspectives. Plant Commun 3:100273
Cuthbert JL, Somers DJ, Brûlé-Babel AL et al (2008) Molecular mapping of quantitative trait loci for yield and yield components in spring wheat (Triticum aestivum L.). Theor Appl Genet 117:595. https://doi.org/10.1007/s00122-008-0804-5
Donald CM (1968) The breeding of crop ideotypes. Euphytica 17:385. https://doi.org/10.1007/BF00056241
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Du B, Wu J, Islam MS et al (2022) Genome-wide meta-analysis of QTL for morphological related traits of flag leaf in bread wheat. PLoS ONE 17:1–19. https://doi.org/10.1371/journal.pone.0276602
Dubois M, Van den Broeck L, Inzé D (2018) The pivotal role of ethylene in plant growth. Trends Plant Sci 23:311
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Edwards J (2011) Changes in plant morphology in response to recurrent selection in the Iowa StiffStalk Synthetic maize population. Crop Sci 51:2352. https://doi.org/10.2135/cropsci2010.09.0564
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Fan X, Cui F, Zhao C et al (2015) QTLs for flag leaf size and their influence on yield-related traits in wheat (Triticum aestivum L.). Mol Breed. https://doi.org/10.1007/s11032-015-0205-9
Fischer RA, Edmeades GO (2010) Breeding and cereal yield progress. Crop Sci. https://doi.org/10.2135/cropsci2009.10.0564
Gao L, Meng C, Yi T et al (2021) Genome-wide association study reveals the genetic basis of yield- and quality-related traits in wheat. BMC Plant Biol 21:1. https://doi.org/10.1186/s12870-021-02925-7
Gill HS, Halder J, Zhang J et al (2022) Whole-genome analysis of hard winter wheat germplasm identifies genomic regions associated with spike and kernel traits. Theor Appl Genet 135:2953–2967. https://doi.org/10.1007/s00122-022-04160-6
Gudi S, Halladakeri P, Singh G et al (2024) Deciphering the genetic landscape of seedling drought stress tolerance in wheat (Triticum aestivum L.) through genome-wide association studies. Front Plant Sci. https://doi.org/10.3389/fpls.2024.1351075
Hammer GL, Dong Z, McLean G et al (2009) Can changes in canopy and/or root system architecture explain historical maize yield trends in the U.S. corn belt? Crop Sci 49:299. https://doi.org/10.2135/cropsci2008.03.0152
He F, Pasam R, Shi F et al (2019) Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat Genet 51:896–904. https://doi.org/10.1038/s41588-019-0382-2
Hill WG, Weir BS (1988) Variances and covariances of squared linkage disequilibria in finite populations. Theor Popul Biol 33:54–78. https://doi.org/10.1016/0040-5809(88)90004-4
Hirano K, Kawamura M, Araki-Nakamura S et al (2017) Sorghum DW1 positively regulates brassinosteroid signaling by inhibiting the nuclear localization of BRASSINOSTEROID INSENSITIVE 2. Sci Rep 7:126. https://doi.org/10.1038/s41598-017-00096-w
Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S et al (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32:495. https://doi.org/10.1046/j.1365-313X.2002.01438.x
Hothorn M, Belkhadir Y, Dreux M et al (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474:467. https://doi.org/10.1038/nature10153
Hu J, Wang X, Zhang G et al (2020) QTL mapping for yield-related traits in wheat based on four RIL populations. Theor Appl Genet 133:917. https://doi.org/10.1007/s00122-019-03515-w
Hussain W, Stephen Baenziger P, Belamkar V et al (2017) Genotyping-by-sequencing derived high-density linkage map and its application to QTL mapping of flag leaf traits in bread wheat. Sci Rep 7:1–15. https://doi.org/10.1038/s41598-017-16006-z
IWGSC (2018) International Wheat Genome SequencingConsortium(IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science (80) 361
Jang S, An G, Li HY (2017) Rice leaf angle and grain size are affected by the OsBUL1 transcriptional activator complex. Plant Physiol 173:688. https://doi.org/10.1104/pp.16.01653
Khanna-Chopra R, Singh K, Shukla S et al (2020) QTLs for cell membrane stability and flag leaf area under drought stress in a wheat RIL population. J Plant Biochem Biotechnol 29:276–286. https://doi.org/10.1007/s13562-019-00534-y
Kloc Y, Dmochowska-Boguta M, Zielezinski A et al (2020) Silencing of hvgsk1.1—a gsk3/shaggy-like kinase–enhances barley (hordeum vulgare L.) growth in normal and in salt stress conditions. Int J Mol Sci 21:6616. https://doi.org/10.3390/ijms21186616
Knöller AS, Blakeslee JJ, Richards EL et al (2010) Brachytic2/ZmABCB1 functions in IAA export from intercalary meristems. J Exp Bot 61:3689. https://doi.org/10.1093/jxb/erq180
Kobayashi S, Fukuta Y, Morita S et al (2003) Quantitative trait loci affecting flag leaf development in rice (Oryza sativa L.). Breed Sci 53:255–262. https://doi.org/10.1270/jsbbs.53.255
Kong B, Ma J, Zhang P et al (2023) Deciphering key genomic regions controlling flag leaf size in wheat via integration of meta-QTL and in silico transcriptome assessment. BMC Genomics 24:33. https://doi.org/10.1186/s12864-023-09119-5
Ku L, Wei X, Zhang S et al (2011) Cloning and characterization of a putative tac1 ortholog associated with leaf angle in maize (Zea mays L.). PLoS ONE 6:1–7. https://doi.org/10.1371/journal.pone.0020621
Kumagai E, Araki T, Ueno O (2009) Effect of nitrogen-deficiency on midday photoinhibition in flag leaves of different rice (Oryza sativa L.) cultivars. Photosynthetica 47:241. https://doi.org/10.1007/s11099-009-0038-z
Lawson T, Kramer DM, Raines CA (2012) Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Curr Opin Biotechnol 23:215
Lee J, Park JJ, Kim SL et al (2007) Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65:487. https://doi.org/10.1007/s11103-007-9196-1
Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90:929. https://doi.org/10.1016/s0092-8674(00)80357-8
Li Z, Pinson SRM, Stansel JW, Paterson AH (1998) Genetic dissection of the source-sink relationship affecting fecundity and yield in rice (shape Oryza sativa L.). Mol Breed 4:419–426
Li X, Jiao DM, Liu YL, Huang XQ (2002) Chlorophyll fluorescence and membrane lipid peroxidation in the flag leaves of different high yield rice variety at late stage of development under natural condition. Acta Bot Sin 44:413
Li P, Wang Y, Qian Q et al (2007) LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res 17:402. https://doi.org/10.1038/cr.2007.38
Li X, Sun S, Li C et al (2014) The strigolactone-related mutants have enhanced lamina joint inclination phenotype at the seedling stage. J Genet Genomics 41:605
Li F, Wen W, Liu J et al (2019) Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol 19:1–9. https://doi.org/10.1186/s12870-019-1781-3
Li H, Wang L, Liu M et al (2020a) Maize plant architecture is regulated by the ethylene biosynthetic gene ZmACS7. Plant Physiol 183:1184. https://doi.org/10.1104/pp.19.01421
Li X, Wu P, Lu Y et al (2020b) Synergistic interaction of phytohormones in determining leaf angle in crops. Int J Mol Sci 21:5052
Li Y, Tang J, Liu W et al (2021) The genetic architecture of grain yield in spring wheat based on genome-wide association study. Front Genet 12:1–19. https://doi.org/10.3389/fgene.2021.728472
Liu X, Huang M, Fan B et al (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. https://doi.org/10.1371/journal.pgen.1005767
Liu Y, Lin Y, Gao S et al (2017) A genome-wide association study of 23 agronomic traits in Chinese wheat landraces. Plant J 91:861–873. https://doi.org/10.1111/tpj.13614
Liu K, Xu H, Liu G et al (2018a) QTL mapping of flag leaf-related traits in wheat (Triticum aestivum L.). Theor Appl Genet 131:839–849. https://doi.org/10.1007/s00122-017-3040-z
Liu Y, Tao Y, Wang Z et al (2018b) Identification of QTL for flag leaf length in common wheat and their pleiotropic effects. Mol Breed 38:1. https://doi.org/10.1007/s11032-017-0766-x
Liu K, Cao J, Yu K et al (2019) Wheat TaSPL8 modulates leaf angle through auxin and brassinosteroid signaling. Plant Physiol 181:179–194. https://doi.org/10.1104/pp.19.00248
Long SP, Zhu XG, Naidu SL, Ort DR (2006) Can improvement in photosynthesis increase crop yields? Plant. Cell Environ 29:315
Ma J, Tu Y, Zhu J et al (2020) Flag leaf size and posture of bread wheat: genetic dissection, QTL validation and their relationships with yield-related traits. Theor Appl Genet 133:297–315. https://doi.org/10.1007/s00122-019-03458-2
Makarevitch I, Thompson A, Muehlbauer GJ, Springer NM (2012) Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS ONE 7:e30798. https://doi.org/10.1371/journal.pone.0030798
Mantilla-Perez MB, Salas Fernandez MG (2017) Differential manipulation of leaf angle throughout the canopy: current status and prospects. J Exp Bot 68:5699–5717. https://doi.org/10.1093/jxb/erx378
Monier B, Casstevens TM, Bradbury PJ, Buckler ES (2021) rTASSEL: an R interface to TASSEL for association mapping of complex traits. BioRxiv. https://doi.org/10.1101/2020.07.21.209114
Moreno MA, Harper LC, Krueger RW et al (1997) liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11:616. https://doi.org/10.1101/gad.11.5.616
Morinaka Y, Sakamoto T, Inukai Y et al (2006) Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol 141:924. https://doi.org/10.1104/pp.106.077081
Muhammad A, Li J, Hu W et al (2021a) Uncovering genomic regions controlling plant architectural traits in hexaploid wheat using different GWAS models. Sci Rep 11:1–14. https://doi.org/10.1038/s41598-021-86127-z
Muhammad A, Li J, Hu W et al (2021b) Uncovering genomic regions controlling plant architectural traits in hexaploid wheat using different GWAS models. Sci Rep 11:6767. https://doi.org/10.1038/s41598-021-86127-z
Ort DR, Merchant SS, Alric J et al (2015) Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci USA 112:8529–8536. https://doi.org/10.1073/pnas.1424031112
Parry MAJ, Reynolds M, Salvucci ME et al (2011) Raising yield potential of wheat. II. Increasing photosynthetic capacity and efficiency. J Exp Bot 62:453. https://doi.org/10.1093/jxb/erq304
Peng S, Khush GS, Virk P et al (2008) Progress in ideotype breeding to increase rice yield potential. F Crop Res 108:32
Poland J (2015) Breeding-assisted genomics. Curr Opin Plant Biol 24:119–124. https://doi.org/10.1016/j.pbi.2015.02.009
Poland J, Endelman J, Dawson J et al (2012) Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome. https://doi.org/10.3835/plantgenome2012.06.0006
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ramirez-Gonzalez RH, Uauy C, Caccamo M (2015) PolyMarker: a fast polyploid primer design pipeline. Bioinformatics 31:2038. https://doi.org/10.1093/bioinformatics/btv069
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428. https://doi.org/10.1371/journal.pone.0066428
Sakamoto T, Morinaka Y, Ohnishi T et al (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24:105. https://doi.org/10.1038/nbt1173
Sharma SN, Sain RS, Sharma RK (2003) The genetic control of flag leaf length in normal and late sown durum wheat. J Agric Sci 141:323–331. https://doi.org/10.1017/S0021859603003642
She J, Han Z, Kim TW et al (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474:472. https://doi.org/10.1038/nature10178
Shearman VJ, Sylvester-Bradley R, Scott RK, Foulkes MJ (2005) Physiological processes associated with wheat yield progress in the UK. Crop Sci 45:175. https://doi.org/10.2135/cropsci2005.0175a
Shimada A, Ueguchi-Tanaka M, Sakamoto T et al (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48:390. https://doi.org/10.1111/j.1365-313X.2006.02875.x
Sidhu JS, Singh D, Gill HS et al (2020) Genome-wide association study uncovers novel genomic regions associated with coleoptile length in hard winter wheat. Front Genet 10:1345. https://doi.org/10.3389/fgene.2019.01345
Sinclair TR, Sheehy JE (1999) Erect leaves and photosynthesis in rice. Science 283:1455. https://doi.org/10.1126/science.283.5407.1455c
Singh G, Gudi S, Amandeep, et al (2022) Unlocking the hidden variation from wild repository for accelerating genetic gain in legumes. Front Plant Sci. https://doi.org/10.3389/fpls.2022.1035878
Sun C, Zhang F, Yan X et al (2017) Genome-wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China. Plant Biotechnol J 15:953–969. https://doi.org/10.1111/pbi.12690
Tadesse W, Sanchez-garcia M, Assefa SG (2019) Genetic gains in wheat breeding and its role in feeding the world. Crop Breed Genet Genom. https://doi.org/10.20900/cbgg20190005
Tanaka A, Nakagawa H, Tomita C et al (2009) Brassinosteroid Upregulated1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol 151:669. https://doi.org/10.1104/pp.109.140806
Tange O (2018) GNU Parallel 2018. https://doi.org/10.5281/ZENODO.1146014
Tanner JW, Gardener CJ, Stoskopf NC, Reinbergs K (1966) Some observations on upright-leaf-type small grains. Can J Plant Sci 46:690. https://doi.org/10.4141/cjps66-115
Tian F, Bradbury PJ, Brown PJ et al (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43:159
Tilman D, Balzer C, Hill J, Befort BL (2011) Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci U S A 108:20260–20264. https://doi.org/10.1073/pnas.1116437108
Truong SK, McCormick RF, Rooney WL, Mullet JE (2015) Harnessing genetic variation in leaf angle to increase productivity of sorghum bicolor. Genetics 201:1229. https://doi.org/10.1534/genetics.115.178608
Wang J, Zhang Z (2021) GAPIT Version 3: boosting power and accuracy for genomic association and prediction. Genom Proteom Bioinform. https://doi.org/10.1016/j.gpb.2021.08.005
Wang R, Liu C, Li Q et al (2020) Spatiotemporal resolved leaf angle establishment improves rice grain yield via controlling population density. iScience. https://doi.org/10.1016/j.isci.2020.101489
Wang Y, Qiao L, Yang C et al (2022) Identification of genetic loci for flag-leaf-related traits in wheat (Triticum aestivum L.) and their effects on grain yield. Front Plant Sci 13:990287. https://doi.org/10.3389/fpls.2022.990287
Ward BP, Brown-Guedira G, Kolb FL et al (2019) Genome-wide association studies for yield-related traits in soft red winter wheat grown in Virginia. PLoS ONE. https://doi.org/10.1371/journal.pone.0208217
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer, New York
William R (2023) psych: Procedures for psychological, psychometric, and personality research. Northwestern University, Evanston, Illinois, USA. https://CRAN.R-project.org/package=psych
Wu Q, Chen Y, Fu L et al (2016) QTL mapping of flag leaf traits in common wheat using an integrated high-density SSR and SNP genetic linkage map. Euphytica 208:337–351. https://doi.org/10.1007/s10681-015-1603-0
Xue S, Xu F, Li G et al (2013) Fine mapping TaFLW1, a major QTL controlling flag leaf width in bread wheat (Triticum aestivum L.). Theor Appl Genet 126:1941–1949. https://doi.org/10.1007/s00122-013-2108-7
Yamaguchi M, Fujimoto H, Hirano K et al (2016) Sorghum Dw1, an agronomically important gene for lodging resistance, encodes a novel protein involved in cell proliferation. Sci Rep 6:28366. https://doi.org/10.1038/srep28366
Yamamuro C, Ihara Y, Wu X et al (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12:1591. https://doi.org/10.1105/tpc.12.9.1591
Yan X, Wang S, Yang B et al (2020a) QTL mapping for flag leaf-related traits and genetic effect of QFLW-6A on flag leaf width using two related introgression line populations in wheat. PLoS ONE 15:1–15. https://doi.org/10.1371/journal.pone.0229912
Yan X, Wang S, Yang B et al (2020b) QTL mapping for flag leaf-related traits and genetic effect of QFLW-6A on flag leaf width using two related introgression line populations in wheat. PLoS ONE 15:e0229912. https://doi.org/10.1371/journal.pone.0229912
Yan X, Zhao L, Ren Y et al (2020c) Identification of genetic loci and a candidate gene related to flag leaf traits in common wheat by genome-wide association study and linkage mapping. Mol Breed 40:1–5. https://doi.org/10.1007/s11032-020-01135-7
Yang D, Liu Y, Cheng H et al (2016) Genetic dissection of flag leaf morphology in wheat (Triticum aestivum L.) under diverse water regimes. BMC Genet 17:1–15. https://doi.org/10.1186/s12863-016-0399-9
Zanella CM, Rotondo M, McCormick-Barnes C et al (2023) Longer epidermal cells underlie a quantitative source of variation in wheat flag leaf size. New Phytol 237:1558. https://doi.org/10.1111/nph.18676
Zhang C, Xu Y, Guo S et al (2012) Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet 8:e1002686. https://doi.org/10.1371/journal.pgen.1002686
Zhang J, Ku LX, Han ZP et al (2014) The ZmCLA4 gene in the qLA4–1 QTL controls leaf angle in maize (Zea mays L.). J Exp Bot 65:5063. https://doi.org/10.1093/jxb/eru271
Zhang B, Ye W, Ren D, Tian P, Peng Y, Gao Y, Ruan B et al (2015) Genetic analysis of flag leaf size and candidate genes determination of a major QTL for flag leaf width in rice. Rice 8:1–10
Zhang W, Chen X, Yang K et al (2024) Fine-mapping and validation of the major quantitative trait locus QFlANG-4B for flag leaf angle in wheat. Theor Appl Genet 137:121. https://doi.org/10.1007/s00122-024-04629-6
Zhao SQ, Xiang JJ, Xue HW (2013) Studies on the rice leaf inclination1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol Plant 6:174. https://doi.org/10.1093/mp/sss064
Zhao C, Bao Y, Wang X et al (2018) QTL for flag leaf size and their influence on yield-related traits in wheat. Euphytica 214:1–15. https://doi.org/10.1007/s10681-018-2288-y
Zheng X, Levine D, Shen J et al (2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28:3326–3328. https://doi.org/10.1093/bioinformatics/bts606
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome. https://doi.org/10.3835/plantgenome2008.02.0089
Zhu XG, Long SP, Ort DR (2010) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61:235–261. https://doi.org/10.1146/annurev-arplant-042809-112206
Zhu Z, Chen L, Zhang W et al (2020) Genome-wide association analysis of fusarium head blight resistance in Chinese elite wheat lines. Front Plant Sci 11:206. https://doi.org/10.3389/fpls.2020.00206
Zhu T, Wang L, Rimbert H et al (2021) Optical maps refine the bread wheat Triticum aestivum cv. Chinese spring genome assembly. Plant J 107:303–314. https://doi.org/10.1111/tpj.15289
Acknowledgements
The authors would like to thank the South Dakota Agriculture Experimental Station (Brookings, SD, USA) and the South Dakota State University High-Performance Computing (HPC) team for providing the resources to conduct and analyze the experiments. The mention of trade names or commercial products in this publication is solely to provide specific information and does not imply recommendation or endorsement by the United States Department of Agriculture. The USDA is an equal opportunity provider and employer.
Funding
This project was collectively funded by the USDA hatch projects SD00H695-20, the Agriculture and Food Research Initiative Competitive Grants 2022-68013-36439 (Wheat-CAP) from the USDA National Institute of Food and Agriculture, and South Dakota Wheat Commission grant 3X3079. The funders had no role in the study design, data collection, analysis, decision to publish, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
SS and PK conceptualized the experiment and designed the methodology. PK, MS, KK, DK, HG, and SS performed the field trials and data collection. PK, HG, and SS performed the data curation, performed data analysis, software implementation, and visualization. PS, AB, and GB performed genotyping and SNP discovery. PK, HG, and SS wrote the original manuscript. GB, PS, AB, ST, PK, MS, KK, and DK contributed to the interpretation of results and revision of the manuscript. SS obtained the funding and supervised the project. All authors approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Communicated by Lingrang Kong.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kumar, P., Gill, H.S., Singh, M. et al. Characterization of flag leaf morphology identifies a major genomic region controlling flag leaf angle in the US winter wheat (Triticum aestivum L.). Theor Appl Genet 137, 205 (2024). https://doi.org/10.1007/s00122-024-04701-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-024-04701-1