Abstract
Key message
A major locus for spontaneous haploid genome doubling was detected by a case–control GWAS in an exotic maize germplasm. The combination of double haploid breeding method with this locus leads to segregation distortion on genomic regions of chromosome five.
Abstract
Temperate maize (Zea mays L.) breeding programs often rely on limited genetic diversity, which can be expanded by incorporating exotic germplasm. The aims of this study were to perform characterization of inbred lines derived from the tropical BS39 population using different breeding methods, to identify genomic regions showing segregation distortion in lines derived by the DH process using spontaneous haploid genome doubling (SHGD), and use case–control association mapping to identify loci controlling SHGD. Four different sets were used: BS39_DH and BS39_SSD were derived from the BS39 population by DH and single-seed descendent (SSD) methods, and BS39 × A427_DH and BS39 × A427_SSD from the cross between BS39 and A427. A total of 663 inbred lines were genotyped. The analyses of gene diversity and genetic differentiation for the DH sets provided evidence of the presence of a SHGD locus near the centromere of chromosome 5. The case–control GWAS for the DH set also pinpointed this locus. Haplotype sharing analysis showed almost 100% exclusive contribution of the A427 genome in the same region on chromosome 5 of BS39 × A427_DH, presumably due to an allele in this region affecting SHGD. This locus enables DH line production in exotic populations without colchicine or other artificial haploid genome doubling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize (Zea mays L.) breeding contributed to significant yield gains in the past several decades (Andorf et al. 2019), while its germplasm base narrowed (Mikel 2011). Incorporation of exotic germplasm broadens the genetic base of temperate breeding programs, and its use has risen over the past several years (Cruz-Cárdenas et al. 2019). For example, lowland tropical landraces such as Cuban Flint, Suwan, Tusón, and Tuxpeño (Goodman 1999) have all been introgressed into temperate materials. Among exotic germplasm sources, maize breeders prefer adapted inbred lines instead of heterozygous plants from populations of tropical germplasm. The synthetic population BS39 represents tropical Tusón germplasm, photoperiod adapted to temperate environments (Hallauer and Carena 2016), and could serve as a unique source of genetic diversity for US Corn Belt breeding programs.
Traditionally, inbred lines in maize breeding programs have been produced through pedigree selection. The single-seed descent (SSD) method has been used for developing inbred lines to be used in quantitative genetic studies of maize populations (Hallauer and Carena 2016). The SSD method requires 6–7 generations to obtain lines with minimal residual heterozygosity (Adamski et al. 2014). The doubled haploid (DH) approach has almost completely replaced traditional self-pollination for inbred line development, primarily because it decreases the time to obtain homozygous lines. Application of DH technology has been shown to be suitable for exploring the variability within landraces (Strigens et al. 2013) and for quantitative genetic studies such as linkage map construction and quantitative trait locus (QTL) identification (Trampe et al. 2020).
DH line production in maize requires the induction of haploid kernels, identification of haploid seed, and genome doubling of haploids (Wu et al. 2017). While tools and methods for induction and identification of haploids have improved over time, haploid genome doubling remains a challenge for successful application of DH technology at a large-scale (Boerman et al. 2020). Genome doubling in haploids derived from exotic germplasm is even more challenging due to the presence of deleterious recessive alleles that are expressed in haploids (Smelser et al. 2016). Hence, direct application of DH technology for exotic germplasm is not as effective as in temperate and elite germplasm (Prigge et al. 2011).
Genome doubling rates can be increased through spontaneous haploid genome doubling (SHGD) (Wu et al. 2014). SHGD may also help to reduce the exposure of humans to chemicals (e.g., colchicine) necessary for artificial genome doubling. Haploids derived by SHGD can be directly sown in field nurseries, removing associated costs with greenhouses, chemical treatment of haploids, and transplanting (Boerman et al. 2020).
Public line A427 was found to have high rates of haploid male fertile (HMF) exceeding 78% (De la Fuente et al. 2020) and to carry a major QTL on chromosome 5 (Ren et al. 2020; Trampe et al. 2020). De La Fuente et al. (2020) derived haploid plants from a full diallel cross, scoring for HMF. A427 provided positive and significant general combining ability (GCA) for HMF, suggesting that it carries alleles that are additive in nature and work in different genetic backgrounds.
Genome-wide association (GWAS) studies under a case–control scenario can be a powerful approach to identify loci controlling SHGD. Case–control GWAS has been widely applied in human genetics for investigating associations between SNPs and dichotomous disease traits (Thomas and Witte 2002; Yu et al. 2017). The most important factors in this analysis are the accurate definition of phenotypes (cases and controls) and trait heritability (Zondervan and Cardon 2007). In plant breeding, the only studies that used binary case–control GWAS addressed disease resistance. Rincker et al. (2016) identified SNPs related to brown stem rot using a case–control GWAS in soybean, Chang et al. (2016) characterized disease resistance loci in the USDA soybean germplasm collection, and Hart and Griffiths (2015) screened viral resistance in common bean.
In this study, we derived lines from BS39, a temperate-adapted synthetic population, and from a cross between BS39 and A427, used as SHGD donor, by DH and SSD methods. Four sets of inbreds were created (BS39_DH, BS39_SSD, BS39 × A427_DH, BS39 × A427_SSD), and a total 663 inbred lines were genotyped to understand the impact of the breeding method and SHGD in exploiting exotic germplasm. The objectives of this study were (1) to compare the four sets of inbred lines derived from BS39 at the genotype level in order to investigate the impact of different breeding methods and SHGD genes on developing inbred lines from an exotic population, (2) to map genomic regions showing segregation distortion in inbred lines derived by the DH process using SHGD, and (3) to use a case–control association mapping to identify loci controlling SHGD.
Materials and methods
Plant materials and inbred line development
A total of 663 inbred lines were derived from BS39 or from the cross between BS39 and A427 through DH and SSD breeding methods. BS39 is a temperate-adapted germplasm serving as a source to expand the genetic base in maize breeding programs (Hallauer and Carena 2016). A427 is a public non-stiff stalk inbred line developed by the University of Minnesota (Gerdes et al. 1993) that shows a high rate of HMF (~ 78%) and is used as a source of SHGD alleles (De la Fuente et al. 2020). Maternal haploid inducer BHI201 (http://isurftech.technologypublisher.com/technology/19126) was used to develop DH lines (DHLs). DHLs were produced by both artificial haploid genome doubling (AHGD) and SHGD. To develop AHGD lines, 648 BS39 plants were crossed with BHI201. After haploid selection—made manually based on embryo coloration (R1-nj)—colchicine was injected in haploid seedlings following the protocol of Vanous et al. (2017). Outliers were removed in the field based on plant vigor. Putative haploid plants shedding pollen were self-pollinated. At physiological maturity, 153 DHLs were harvested and coded as BS39_DH lines (Fig. 1). To develop SHGD lines, 648 BS39 plants were crossed with A427. The resulting F1 population was crossed with BHI201. Since F1 plants received the SHGD trait from A427, haploids were not treated with colchicine or any other chemical for genome doubling. After selection based on embryo coloration (R1-nj), haploid kernels were directly sown into the field. Haploid plants shedding pollen were self-pollinated. In total, 318 DHLs were obtained and coded as BS39 × A427_DH lines (Fig. 1). In parallel with developing DHLs, inbred lines were also derived by SSD from 648 BS39 plants and from the cross between 750 BS39 plants and A427 (BS39_SSD, BS39 × A427_SSD; Fig. 1). Six generations of self-pollination were carried out, generating 96 inbred lines for each of the two SSD sets. Agronomic traits such as maturity, plant and ear height, tassel size, foliar diseases, ear size, kernel texture, ear diseases, stalk, and root lodging were considered for mild selection during the six generations of self-pollination.
Genotyping and SNP calling
Genotyping of DHLs (153 BS39_DH and 318 BS39 × A427_DH lines) and 310 individuals from the BS39 population was performed using genotyping by sequencing (GBS) (Elshire et al. 2011). Plant tissue was collected at the seedling stage from 10 plants of each DHL and from 310 individual plants of the BS39 population. Freeze-dried tissue samples were sent to Cornell University Genomic Diversity Facility for DNA extraction and genotyping. GBS was performed as described by Elshire et al. (2011). Briefly, libraries were constructed in a 96-plex and genomic DNA was digested with the ApeKI restriction enzyme. DNA fragments were sequenced using Illumina Inc. next-generation sequencing platforms. The raw sequence was processed into SNP genotypes, as described by Glaubitz et al. (2014) using the B73 reference genome version 2 (AGPv2) as a reference. In total, 955,690 SNPs were generated by GBS. Filtering was conducted using TASSEL 5.2.58 (Glaubitz et al. 2014). SNPs with minor allele frequency (MAF) below 5% and call rate below 0.50 (50%) were removed. Additionally, any DHL with more than 5% heterozygosity was discarded. The remaining heterozygous loci were considered missing data. After filtering, 282,034 SNPs were retained in 118 BS39_DH and 317 BS39 × A427_DH lines. Beagle 5.0 (Browning et al. 2018) was used for imputation of missing data. For SSD lines (96 BS39_SSD and BS39 × A427_SSD), Diversity Arrays Technology sequencing was used (DArtseq) (Jaccoud et al. 2001). Kernels from 120 BS39_SSD and 120 BS39 × A427_SSD inbred lines were sent to the Genetic Analysis Service for Agriculture (SAGA) at the International Maize and Wheat Improvement Center (CIMMYT) for genotyping. SNPs were obtained using DArtseq and were called using the DArtsoft analytical pipeline (https://www.diversityarrays.com), using the B73 reference genome version 4 (AGPv4) as a reference. A total of 32,930 SNPs were generated by DArtseq. Quality control and imputation of Dartseq SNPs were similar to the GBS procedures described for DHLs. After correction, 17,366 SNPs were retained in 51 BS39_SSD and 72 BS39 × A427_SSD lines.
Gene diversity and genetic differentiation
Estimates of gene diversity (HS) were calculated according to Nei (1987), based on the identities of two randomly chosen loci within and between populations, independently of the number of alleles. The assumption was that there are n alleles at a locus and the frequency of the kth allele is xk in a population. In order to evaluate the impact of A427 and the breeding method on gene diversity, BS39_DH lines were compared with BS39 × A427_DH lines and BS39_SSD lines compared with BS39 × A427_SSD lines. The degree of genetic differentiation (FST) between BS39_DH versus BS39 × A427_DH lines and BS39_SSD versus BS39 × A427_SSD was calculated as described by Weir and Cockerham (1984) as a ratio of the variance between populations to the total variance in the parental population. Both HS and FST analyses were obtained using the R package hierfstat (Goudet 2005).
In order to answer whether the genetic diversity present in BS39 from tropical germplasm was represented in the four sets of inbred lines, we compared the allelic frequencies at each locus of the 310 BS39 plants with each of the BS39-derived sets using a Chi-squared test with one degree of freedom. The comparison between the 310 BS39 and the DH sets (118 BS39_DH and 317 BS39 × A427_DH) considered the 282,034 SNPs. As BS39 was originally genotyped based on B73 reference genome version 2, we converted it to version 4 for comparison with SSD sets (for which the B73 reference genome version 4 was used). The conversion was made based on the assembly Converter tool found on the Gramene website (http://ensembl.gramene.org/Oryza_sativa/Tools/AssemblyConverter?db=core). After conversion, BS39 and SSD sets were merged in TASSEL (Bradbury et al. 2007) and additional filtering was used to discard unmatched markers. In total, 3,401 markers were used to compare the 310 BS39 with 51 BS39_SSD and 71 BS39 × A427_SSD lines.
Linkage disequilibrium
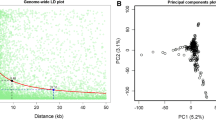
Linkage disequilibrium (LD) analysis was performed for all pairwise combinations of SNPs by computing the squared correlation (r2) of marker genotypes using the software TASSEL (Bradbury et al. 2007). The rate of LD decay with r2 threshold set at 0.2 was calculated for each of the BS39 derived sets based on a marker matrix and a map with distances between markers in base pairs using a nonlinear regression based on Hill and Weir (1988) using the nls function in R software (R Core Team 2020).
Case–control GWAS
A case–control GWAS was performed to map distorted segregation differences between subsets of BS39-derived lines. We contrasted inbred lines with the same phenotype (successful haploid genome doubling), obtained with different mechanisms. BS39 × A427_DH lines utilized a genetic mechanism: spontaneous haploid genome doubling without application of colchicine or similar treatment. In contrast, BS39_DH lines were obtained after a colchicine treatment. Although similar to case–control GWAS to detect disease resistance loci by contrasting “cases” with non-afflicted individuals, all individuals surveyed in our approach showed the same phenotype (haploid genome doubling), attained by either a genetic or a non-genetic mechanism. By using this contrast, we intended to identify genetic loci responsible for haploid genome doubling. Based on prior information of a major QTL for SHGD on chromosome 5 contributed by A427 (Ren et al. 2020; Trampe et al. 2020), our hypothesis was that we would be able to detect this locus using the case–control GWAS. Since the only difference between these sets was the presence of A427 alleles, the 317 BS39 × A427_DHLs were scored as “1” (cases) and the 118 BS39_DH were scored as “0” (controls). GWAS was performed by using the fixed and random model circulating probability unification (FarmCPU) method in the R package GAPIT (Liu et al. 2016). The first five principal components, obtained by GAPIT, were included as covariates in the model. The kinship matrix was automatically estimated in FarmCPU. To determine a significance threshold to account for multiple testing, the false discovery rate (FDR) control (Benjamini and Hochberg 1995) is implemented in the procedure. Because FarmCPU model was developed to fit quantitative variables, statistical assumptions such as normality of residuals were violated. In order to confirm the associations detected by the model, all significant SNPs from the FarmCPU analysis were included into a logistic regression model using SAS PROC LOGISTIC (SAS Institute 2013).
Haplotype sharing—segregation distortion
Analyses of haplotype sharing between A427 and both BS39 × A427_DH and BS39 × A427_SSD sets were conducted using the software Genetic Error-Tolerant Regional Matching with Linear-Time Extension (GERMLINE) (Gusev et al. 2009). Shared haplotypes were identified with a seed of identical genotypes at 10 neighboring SNPs that were extended until up to two homozygous mismatches were encountered. Analyses were based on segments with a minimum size of 2 cM using B73 reference genome version 2 for the comparison between A427 and BS39 × A427_DH, and B73 reference genome version 4 for A427 and BS39 × A427_SSD. The comparisons were made between IBS-SNPs on a site-by-site basis. As we had previous information about a QTL for SHGD on chromosome 5 (Trampe et al. 2020) and we wanted to know whether there was a significant difference in A427 haplotype contribution caused by DH method, we performed a nonparametric Mann–Whitney statistical test for assessing the significance in the median of BS39 × A427_DH and BS39 × A427_SSD within the region of the SHGD QTL shown by Trampe et al. (2020). We used the percentage of A427 haplotype on chromosome 5 from 87 to 130 Mb and compared both sets of BS39 × A427 derived lines using a significance level of \(\alpha\) \(=\) 0.05 using the wilcox.test function in R software (R Core Team 2020).
Results
Gene diversity and genetic differentiation between BS39-derived sets
BS39_DH, BS39 × A427_DH, and BS39 × A427_SSD had very similar allele frequencies compared to a sample of BS39 for most loci. 57.5% of BS39_DH loci did not statistically differ from BS39, BS39 × A427_DH had 62.1% loci that did not differ, BS39 × A427_SSD 52%, and BS39_SSD 31.9%.
The FST values from the comparison between BS39_SSD and BS39 × A427_SSD reached values up to 0.064 (Fig. 2B), which means that up to 6.5% of genetic variation observed among genotypes is due to the difference between sets, and 93.5% of genetic variation is within sets. The overall mean for the comparison between BS39_DH and BS39 × A427_DH was 0.0095 (Fig. 2a). A clear distortion on chromosome 5 was observed with FST values close to 0.120 in the region close to the centromere (S5.89156625-S5.117624647).
A substantial loss of HS on chromosomes 3, 4, and 5 (Fig. 3a) was observed in BS39 × A427_DH compared to BS39_DH. The highest HS loss was observed in the region S5.1874148-S5.216538534 on chromosome 5, followed by chromosomes 3 and 4. The largest difference between the two sets was 0.218 at S5.143957693. HS losses were smaller between SSD sets (Fig. 3b). The highest HS loss of BS39xA427_SSD compared to BS39_SSD was 0.120 on chromosome 5, in the region flanked by markers S5.48032093-S5.174692242.
Gene diversity (HS) comparison by chromosome between (A) BS39_DH versus BS39 × A427_DH and (B) BS39_SSD versus BS39 × A427_SSD. BS39_DH and BS39_SSD are baselines (with their HS values adjusted to zero). The differences between baseline sets and their respective pairs are represented by blue dots. Dots above zero represent a higher HS in the baseline’s pair for the chromosomal region. Dots below zero represent a lower HS in the baseline’s pair
Linkage disequilibrium
An average r2 of 0.2 was reached among BS39_DH individuals within about ~ 94 kb (Fig. 4). Further reduced LD decay was found among BS39 × A427_DH lines with an average r2 of 0.2 at 150 kb. The same pattern was observed among inbred lines derived by the SSD method. BS39_SSD lines reached an average r2 of 0.2 within about 4 kb, and BS39 × A427_SSD lines reached an average r2 of 0.2 within about ~ 51 kb.
Case–control approach to identify loci controlling SHGD
A strong signal for haploid genome doubling was detected on chromosome 5 using a case–control GWAS approach when comparing BS39_DH and BS39 × A427_DH lines (Fig. 5). The strongest association was located at S5.90859140 bp (p value 4.27 × 10–23) on chromosome 5 based on the B73 reference genome version 2 (AGPv2), which corresponds to S5.93191130 on the version 4 (AGPv4). In addition, significant SNPs were found on chromosomes 1 (S1.115866538, p value 0.00079272) and 7 (S7.1286028, p value 8.16 × 10–5). However, the results from the logistic regression model of these three significant SNPs revealed a weak association for S1.115866538, with a p value of 0.2035 (Table 1).
Haplotype sharing—segregation distortion
Haplotype sharing analysis between A427 and BS39 × A427_SSD (Fig. 6) showed A427 average percentages varying from 37% on chromosome 9 to 61% on chromosome 2 (Table S1). Overall, all chromosomes had A427 contributions close to the expected 50% in this set of inbred lines.
The comparison between haplotypes of A427 and BS39 × A427_DH (Fig. 7) revealed a lower contribution of A427 genome-wide, especially on chromosomes 5 and 6, where only 21% and 17% of the BS39 × A427_DH genome matched with A427 haplotypes on average, respectively (Table S1). Segregation distortion on chromosome 5 revealed a peak of approximately 90% exclusive contribution of the A427 genome in the region close to the centromere (~ 88–130 Mb). This region includes the significant SNP identified by the case–control GWAS (S5.90859140) and it is in the same region pinpointed by FST analysis (S5.89156625-S5.117624647) (Fig. 6).
The comparison between peaks of A427 haplotype within the region 88–130 Mb on chromosome 5 in BS39 × A427_DH and BS39 × A427_SSD showed a significant difference (P = 0.005507) according to the Mann–Whitney test that indicates that DH and SSD methods acted differently to keep SGHD alleles in the genome of its respective lines.
Discussion
Genotypic characterization of BS39-derived inbred lines
BS39 is a unique source of tropical alleles for inbred line development, distinct from current US elite germplasm, and thus an option to expand the genetic base in maize breeding programs (Hallauer and Carena 2016). A fundamental question was how well the different BS39-derived sets represent the original BS39 population. Since more than 50% of BS39_DH, BS39 × A427_DH, and BS39 × A427_SSD loci did not differ from BS39 in their allele frequencies, we can infer that these sets represent BS39 sufficiently well. However, allele frequencies for most loci in BS39_SSD were significantly different from BS39, which may be due to small sample size. The 51 lines in BS39_SSD and 71 lines in BS39 × A427_SSD likely led to greater deviation from BS39 (31.9% and 52% for BS39_SSD and BS39 × A427_SSD, respectively), when compared to the DH sets (57.5% and 62.1% for 118 BS39_DH and 317 BS39 × A427_DH, respectively).
Based on HS and FST values, both SSD and DH breeding methods appear promising for capturing genetic variability from the base population. In addition, all sets displayed significant genotypic variance for agronomic traits (Verzegnazzi et al. in preparation). Application of DH technology can help to purge the genetic load present in exotic germplasm without strongly affecting diversity (Strigens et al. 2013). However, segregation distortion observed in BS39 × A427_DH suggests that selective neutrality of the in vivo DH method can be affected by SHGD genes in particular genome regions. The SSD method seems to be more suitable to retain genetic diversity of the BS39 population across the genome (Figs. 3, 6, and 7). However, the trade-off for the observed modest increase in capturing diversity across the genome is the time-consuming nature of the SSD process. While it is economic to use isolation fields for a large-scale haploid seed production using haploid inducers as male followed by self-pollination of haploid plants, producing inbred lines by SSD requires selfing of multiple individuals for each of at least six generations.
Mapping genomic regions for SHGD based on segregation distortion
A427 was shown to carry a major QTL for SHGD on chromosome 5 as well as a few minor QTL on chromosomes 1, 6, 7, and 10 (Trampe et al. 2020). Since all BS39 × A427_DH lines were obtained by spontaneously haploid genome doubling, selection of the A427 haplotype was expected for genome regions affecting SHGD. The impact of the known major QTL for SHGD for developing exotic lines was confirmed by the combined results of HS, FST, and LD decay analyses. As expected, LD decay among DH lines was reduced compared to SSD lines. Even though SSD lines were genotyped by using Dartseq and DH lines by GBS, the LD decay pattern did not change because SSD inbred lines had six opportunities of recombination, while DH inbreds had one. The extensive HS loss on chromosome 5 in the region flanked by markers S5.1874148-S5.216538534, when comparing BS39_DH and BS39 × A427_DH, suggests that the presence of SHGD alleles using the DH breeding method reduced allelic diversity in this region. The smaller HS loss for the contrast between BS39_SSD and BS39 × A427_SSD indicates that the inheritance of SHGD genes over generations of self-pollination has less impact in these specific regions than in DH line production. The peak of FST values on chromosome 5 within the region of higher HS loss (S5.89156625-S5.117624647) indicates a major contribution of SHGD alleles in this region in the DH set (Fig. 2). We did observe a peak of A427 haplotypes on chromosome 5 in the same region highlighted by FST and HS analyses. Moreover, a highly significant SNP coincided with this region in the case–control GWAS. Taken together, our findings are consistent with the presence of a major SHGD QTL from A427 identified in this region (Ren et al., 2020; Trampe et al. 2020). QTL analysis showed pleiotropic effects of a major QTL on chromosome 5 that explained 51.3% of the phenotypic variation for anther emergence, 55.9% for pollen production, 48.5% for tassel size, and 45.7% for haploid male fertility (Trampe et al. 2020).
However, segregation distortion did not generally favor A427 haplotypes. Reasons for segregation distortion were discussed by Murigneux et al. (1993). They observed a higher segregation distortion in DH when compared to SSD inbred lines as a consequence of either sampling effect, selection, or difference in the viability of some genetic combinations. On chromosomes 5 and 6, small regions showed complete absence of A427 haplotypes in BS39 × A427_DH (Fig. 6). This finding suggests that A427 may carry regions in chromosome 5 that have adverse effects on the DH process, given that regions with a high contribution of A427 were next to regions where the A427 haplotypes were absent. Thus, selection of recombinant SHGD donor genotypes on chromosome 5 should be possible, with even stronger benefits for the DH process. This should increase the efficiency of DH line development based on SHGD even further.
Differences in A427 haplotype frequencies between BS39 × A427_DH versus BS39 × A427_SSD were helpful to study the impact of the two breeding methods (DH and SSD) on genomic composition and genetic diversity in the respective populations. Our results confirmed selection of particular A427-derived SHGD alleles using the DH method, not selected for by the SSD method (Figs. 6 and 7). If we consider the region between 88 and 140 Mb on chromosome 5, 65% of the BS39 × A427_DH genome has more than 70% of A427 haplotype while for BS39 × A427_SSD, 83% of this region has 50% or less of A427 haplotype. Moreover, since SSD inbred lines had multiple recombination events due to six self-pollinations, linkage blocks and A427 haplotypes were smaller on average compared to the DH lines. In conclusion, haplotype analysis can help to monitor genetic diversity in breeding populations at the genome level, to avoid specific regions of being unintentionally fixed, and to identify regions of selection and variation in the genome (Coffman et al. 2020).
Case–control approach to identify loci controlling SHGD
Case–control GWAS is a common approach in human genetics but not in plant breeding. The validity of this methodology relies on how well population structure and sample size are modeled to avoid false positives (Hirschhorn and Daly 2005; Wang et al. 2005). In total, 6000 cases and 6000 controls provided approximately 43% and 94% power to detect disease susceptibility variants with MAF of 0.05 and 0.01, respectively, in a study conducted by Wang et al. (2005). Hauer et al. (2017) studied genetic risk loci for ischemic stroke in a Dutch population based on 1375 cases and 1533 controls. However, cases and controls in human studies cannot be replicated, in contrast to entries of agronomic experiments. By using experimental designs with replications, it is possible to improve the heritability of the traits (e.g., heritability on an entry mean basis) by reducing the residual variation. Moreover, successful studies in humans were reported with smaller population size. Samarani et al. (2019) found associations between killer cell immunoglobin-like receptors in three groups of Canadian patients using a case–control population ranging from 93 to 245 individuals. Ozaki et al. (2002) performed a study to investigate the susceptibility to myocardial infarction using 94 cases and 658 controls. A candidate locus was identified, and the result was further supported by an additional haplotype structure and LD analysis.
Case–control GWAS applied in a plant breeding scenario has the same issues regarding population structure and sample size as in human studies. However, large effect loci can be reliably detected with smaller population sizes. Hart and Griffiths (2015) used 84 recombinant inbred lines and identified 44 SNPs strongly associated with virus resistance. To identify minor effect QTL, a case-to-control ratio higher than 1:1 is desirable, but it should not exceed 1:4 as shown in a study with 248 cases (Hong and Park 2012). Despite our limited number of cases and controls (317 and 118, respectively), we were able to identify a highly significant SNP but not the minor effect QTL from Trampe et al. (2020). The strong association at S5.90859140 bp (p value 4.27 × 10–23) within the chromosome 5 genomic region confirms the large genetic effect of this particular locus on SHGD in exotic background. Thus, case–control GWAS seems to be suitable to identify major loci, and small sample size may limit identification of minor effect QTL, as we only found one additional QTL (Fig. 5, Table 1). As we had the previous information that A427 carries a major QTL with strong effect on SHGD, our primary interest was to determine whether we can detect this QTL in an exotic genetic background. The A427 haplotype on chromosome 5 was enriched to near fixation. Based on all results in our study, we conclude that the SHGD QTL is transferable to genotypes with an exotic background like BS39.
Outlook
The region flanked by markers S5.89156625-S5.11762464788 on chromosome 5 is useful for deriving DH lines from exotic germplasm using SHGD. The major SHGD QTL identified by Trampe et al. (2020) between positions 91–93 Mb is within this region (S5.86261290-S5.92805032). No obvious linkage drag was found for this SHGD QTL (Verzegnazzi et al. in preparation), which is important for using the target region to develop high performing inbred lines. Fine mapping would be desirable to determine the location of this major QTL in more detail. However, since this region is close to the centromere, where recombination is usually suppressed, this is challenging. Moreover, Schneider et al. (2016) reported neocentromere formation on chromosome 5, which is another complicating factor. The reduced contribution of A427 genome to some chromosomes (e.g., chromosomes 5 or 6) of DH versus SSD lines suggest the presence of unfavorable alleles of A427 in those regions interfering with the inbred line development via the DH process. Consequently, SSD lines seem to capture allelic diversity of the parental genotypes better than DH lines in those chromosome regions in this population.
Different from improving haploid inducers (Trentin et al. 2020), genes controlling SHGD needs to be present in breeding populations (Boerman et al. 2020). The first step for applying SHGD in breeding programs will be introgression of these genes into elite germplasm. This requires initial crosses with a SHGD donor. Second cycle selection of DH lines should already benefit from increased efficiencies of DH line development due to SHGD. A recurrent selection approach to introgress haploid male fertility was presented by Molenaar et al. (2019). Recurrent selection for haploid male fertility resulted in a substantial improvement in SHGD. The identification of the major SHGD loci in A427 and the information about the absence of linkage drag with the SHGD QTL makes the introgression of it in breeding populations even more straightforward (Boerman et al. 2020), with or without using marker-assisted selection.
Producing DH lines with SHGD means that all lines would carry the alleles because just the lines that shed pollen will produce seeds. The exclusive use of a SHGD system to develop inbred lines increases the risk of fixing genome regions such as on chromosome 5 identified in this study. However, being able to accomplish SHGD with alleles at one or a few QTL makes this approach feasible in combination with marker-assisted backcrossing for efficient introgression into elite material, in contrast to relying on minor QTL reported in other studies (Yang et al. 2019).
References
Adamski T, Krystkowiak K, Kuczyńska A, Mikołajczak K, Ogrodowicz P, Ponitka A, Surma M, Ślusarkiewicz-Jarzina A (2014) Segregation distortion in homozygous lines obtained via anther culture and maize doubled haploid methods in comparison to single seed descent in wheat (Triticum aestivum L.). Electron J Biotechnol 17:6–13. https://doi.org/10.1016/j.ejbt.2013.12.002
Andorf C, Beavis WD, Hufford M, Smith S, Suza WP, Wang K, Woodhouse M, Yu J, Lübberstedt T (2019) Technological advances in maize breeding: past, present and future. Theor Appl Genet 132:817–849. https://doi.org/10.1007/s00122-019-03306-3
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Boerman NA, Frei UK, Lübberstedt T (2020) Impact of spontaneous haploid genome doubling in maize breeding. Plants 9:369. https://doi.org/10.3390/plants9030369
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Browning BL, Zhou Y, Browning SR (2018) A One-Penny imputed genome from next-generation reference panels. Am J Hum Genet 103:338–348. https://doi.org/10.1016/j.ajhg.2018.07.015
Chang H, Lipka AE, Domier LL, Hartman GL (2016) Characterization of disease resistance Loci in the USDA Soybean germplasm collection using genome-wide association studies. Phytopathology 106:1139–1151. https://doi.org/10.1094/PHYTO-01-16-0042-FI
Coffman SM, Hufford MB, Andorf CM, Lübberstedt T (2020) Haplotype structure in commercial maize breeding programs in relation to key founder lines. Theor Appl Genet 133:547–561. https://doi.org/10.1007/s00122-019-03486-y
Cruz-Cárdenas CI, Cortés-Cruz M, Gardner CA, Costich DE (2019) Wild relatives of maize. In: Greene SL, Williams KA, Khoury CK, Kantar MB, Marek LF (eds) North American Crop Wild Relatives. Springer International Publishing, Berlin. https://doi.org/10.1007/978-3-319-97121-6_1
De la Fuente GN, Frei UK, Trampe B, Ren J, Bohn MO, Yana N, Verzegnazzi AL, Murray SC, Lübberstedt T (2020) A diallel analysis of a maize donor population response to in vivo maternal haploid induction. II: Spontaneous Haploid Genome Doubling. Crop Sci 60:873–882. https://doi.org/10.1002/csc2.20021
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379. https://doi.org/10.1371/journal.pone.0019379
Gerdes JT, Behr CF, Coors JG, Tracy WL (1993) Compilation of North American maize breeding germplasm. Crop Sci Soc Am Madison. https://doi.org/10.2135/1993.compilationofnorthamerican.frontmatter
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE 9:e90346. https://doi.org/10.1371/journal.pone.0090346
Goodman MM (1999) Broadening the genetic diversity in maize breeding by use of exotic germplasm. In: Coors JG, Pandey S (eds) Genetics and Explotaition of Heterosis in Crops. Crop Science Society of America, Madison. https://doi.org/10.2134/1999.geneticsandexploitation.c13
Goudet J (2005) Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186. https://doi.org/10.1111/j.1471-8286.2004.00828.x
Gusev A, Lowe JK, Stoffel M, Daly MJ, Altshuler D, Breslow JL, Friedman JM, Pe’er I (2009) Whole population, genome-wide mapping of hidden relatedness. Genome Res 19:318–326. https://doi.org/10.1101/gr.081398.108
Hallauer AR, Carena MJ (2016) Registration of BS39 Maize Germplasm. J Plant Regist 10:296–300. https://doi.org/10.3198/jpr2015.02.0008crg
Hart JP, Griffiths PD (2015) Genotyping-by-sequencing enabled mapping and marker development for the By-2 potyvirus resistance allele in common bean. Plant Genome 8:1–14. https://doi.org/10.3835/plantgenome2014.09.0058
Hauer AJ, Pulit SL, van den Berg LH, de Bakker PIW, Veldink JH, Ruigrok YM (2017) A replication study of genetic risk loci for ischemic stroke in a Dutch population: a case-control study. Sci Rep 7:12175. https://doi.org/10.1038/s41598-017-07404-4
Hill WG, Weir BS (1988) Variances and covariances of squared linkage disequilibria in finite populations. Theor Popul Biol 33:54–78
Hirschhorn JN, Daly MJ (2005) Genome-wide association studies for common diseases and complex traits. Nat Rev 6:95–108. https://doi.org/10.1038/nrg1521
Hong EP, Park JW (2012) Sample size and statistical power calculation in genetic association studies. Genom Inf 10:117–122. https://doi.org/10.5808/GI.2012.10.2.117
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res 29:E25. https://doi.org/10.1093/nar/29.4.e25
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12:e1005767. https://doi.org/10.1371/journal.pgen.1005767
Mikel MA (2011) Genetic composition of contemporary U.S. commercial dent corn germplasm. Crop Sci 51:592–599. https://doi.org/10.2135/cropsci2010.06.0332
Molenaar WS, Schipprack W, Brauner PC, Melchinger AE (2019) Haploid male fertility and spontaneous chromosome doubling evaluated in a diallel and recurrent selection experiment in maize. Theor Appl Genet 132:2273–2284. https://doi.org/10.1007/s00122-019-03353-w
Murigneux A, Baud S, Beckert M (1993) Molecular and morphological evaluation of doubled-haploid lines in maize. 2. comparison with single-seed-descent lines. Theor Appl Genet 8:278–287. https://doi.org/10.1007/BF00223777
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y, Tanaka T (2002) Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat Gen 32:650–654. https://doi.org/10.1038/ng1047
Prigge V, Sánchez C, Dhillon BS, Schipprack W, Araus JL, Bänziger M, Melchinger AE (2011) Doubled haploids in tropical maize: I. effects of inducers and source germplasm on in vivo haploid induction rates. Crop Sci 51:1498–1506. https://doi.org/10.2135/cropsci2010.10.0568
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/
Ren J, Boerman N, Liu R, Wu P, Vanous K, Trampe B, Frei UK, Chen S, Lübberstedt T (2020) Mapping of QTL and identification of candidate genes conferring spontaneous haploid genome doubling in maize (Zea mays L.). Plant Sci 293:110337. https://doi.org/10.1016/j.plantsci.2019.110337
Rincker K, Lipka AE, Diers BW (2016) Genome-wide association study of brown stem rot resistance in soybean across multiple populations. Plant Genome 9:1–11. https://doi.org/10.3835/plantgenome2015.08.0064
Samarani S, Mack DR, Bernstein CN, Iannello A, Debbeche O, Jantchou P, Faure C, Deslandres C, Amre DK, Ahmad A (2019) Activating killer-cell immunoglobulin-like receptor genes confer risk for crohn’s disease in children and adults of the western european descent: findings based on case-control studies. PLoS ONE 14:e0217767. https://doi.org/10.1371/journal.pone.0217767
SAS Institute Inc (2013) SAS 9.4. Cary, United States of America. https://www.sas.com/en_us/software/sas9.html
Schneider KL, Xie Z, Wolfgruber TK, Presting GG (2016) Inbreeding drives maize centromere evolution. PNAS USA 113:E987–E996. https://doi.org/10.1073/pnas.1522008113113
Smelser A, Gardner C, Blanco M, Lübberstedt T, Frei UK (2016) Germplasm enhancement of maize: a look into haploid induction and chromosomal doubling of haploids from temperate-adapted tropical sources. Plant Breed 135:593–597. https://doi.org/10.1111/pbr.12397
Strigens A, Schipprack W, Reif JC, Melchinger AE (2013) Unlocking the genetic diversity of maize landraces with doubled haploids opens new avenues for breeding. PLoS ONE 8:e57234. https://doi.org/10.1371/journal.pone.0057234
Thomas DC, Witte JS (2002) Point: population stratification: a problem for case-control studies of candidate-gene associations? Cancer Epidemiol Biomark Prev 11:505–512
Trampe B, Santos IG, Frei UK, Ren J, Chen S, Lübberstedt T (2020) QTL mapping of spontaneous haploid genome doubling using genotyping-by-sequencing in maize (Zea mays L.). Theor Appl Genet 133:2131–2140. https://doi.org/10.1007/s00122-020-03585-1
Trentin UH, Frei UK, Lübberstedt T (2020) Breeding maize maternal haploid inducers. Plants 9:614. https://doi.org/10.3390/plants9050614
Vanous K, Vanous A, Frei UK, Lübberstedt T (2017) Generation of Maize (Zea mays) Doubled Haploids via Traditional Methods. Curr Protoc Plant Biol 2:147–157. https://doi.org/10.1002/cppb.20050
Verzegnazzi AL, Santos IG, Edwards J, Frei UK, Boerman N, Zuffo LT, Pires LPM, De La Fuente GN, and Lubberstedt T (2021) Usefulness of adapted exotic maize lines developed by doubled haploid and single seed descent methods. In preparation
Wang WYS, Barratt BJ, Clayton DG, Todd JA (2005) Genome-wide association studies: theoretical and practical concerns. Nat Rev 6:109–118. https://doi.org/10.1038/nrg1522
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358. https://doi.org/10.2307/2408641
Wu P, Ren J, Li L, Chen S (2014) Early spontaneous diploidization of maternal maize haploids generated by in vivo haploid induction. Euphytica 200:127–138. https://doi.org/10.1007/s10681-014-1166-5
Wu P, Ren J, Tian X, Lübberstedt T, Li W, Li G, Li X, Chen S (2017) New insights into the genetics of haploid male fertility in maize. Crop Sci 57:637–647. https://doi.org/10.2135/cropsci2016.01.0017
Yang J, Li H, Qu Y, Chen Q, Tang J, Lübberstedt T, Liu Z (2019) Genetic dissection of haploid male fertility in maize (Zea mays L.). Plant Breed 138:259–265. https://doi.org/10.1111/pbr.12688
Yu X, Sun NR, Jang HT, Guo SW, Lian MX (2017) Associations between EGFR gene polymorphisms and susceptibility to glioma a systematic review and meta-analysis from GWAS and case-control studies. Oncotarget 8:86877–86885. https://doi.org/10.18632/oncotarget.21011
Zondervan K, Cardon L (2007) Designing candidate gene and genome-wide case–control association studies. Nat Protoc 2:2492–2501. https://doi.org/10.1038/nprot
Funding
This work was supported by USDA’s National Institute of Food and Agriculture (grant numbers: IOW04314, IOW01018, IOW05520; IOW05510; NIFA award 2018–51181-28419), the Plant Sciences Institute, Crop Bioengineering Center, R.F. Baker Center for Plant Breeding, K.J. Frey Chair in Agronomy at Iowa State University.
Author information
Authors and Affiliations
Contributions
ALV, UKF and TL design the project and performed the experiments. ALV, IGS, MDK, MH, JC, VCA, LTZ and NB analyzed the data. ALV and IGS wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Yunbi Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
About this article
Cite this article
Verzegnazzi, A.L., dos Santos, I.G., Krause, M.D. et al. Major locus for spontaneous haploid genome doubling detected by a case–control GWAS in exotic maize germplasm. Theor Appl Genet 134, 1423–1434 (2021). https://doi.org/10.1007/s00122-021-03780-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03780-8