Abstract
Key message
Two key barley genes independently control anthesis and senescence timing, enabling the manipulation of grain fill duration, grain size/plumpness, and grain protein concentration.
Abstract
Plant developmental processes such as flowering and senescence have direct effects on cereal yield and quality. Previous work highlighted the importance of two tightly linked genes encoding a glycine-rich RNA-binding protein (HvGR-RBP1) and a NAC transcription factor (HvNAM1), controlling barley anthesis timing, senescence, and percent grain protein. Varieties that differ in HvGR-RBP1 expression, ‘Karl’(low) and ‘Lewis’(high), also differ in sequence 1 KB upstream of translation start site, including an ~ 400 bp G rich insertion in the 5′-flanking region of the ‘Karl’ allele, which could disrupt gene expression. To improve malt quality, the (low-grain protein, delayed-senescence) ‘Karl’ HvNAM1 allele was introgressed into Montana germplasm. After several seasons of selection, the resulting germplasm was screened for the allelic combinations of HvGR-RBP1 and HvNAM1, finding lines combining ‘Karl’ alleles for both genes (−/−), lines combining ‘Lewis’ (functional, expressed) HvGR-RBP1 with ‘Karl’ HvNAM1 alleles ( ±), and lines combining ‘Lewis’ alleles for both genes (+ / +). Field experiments indicate that the functional (‘Lewis,’ +) HvGR-RBP1 allele is associated with earlier anthesis and with slightly shorter plants, while the ‘Karl’ (−) HvNAM1 allele delays maturation. Genotypes carrying the ± allele combination therefore had a significantly (3 days) extended grain fill duration, leading to a higher percentage of plump kernels, slightly enhanced test weight, and lower grain protein concentration when compared to the other allele combinations. Overall, our data suggest an important function for HvGR-RBP1 in the control of barley reproductive development and set the stage for a more detailed functional analysis of this gene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant developmental processes such as flowering transition and whole plant senescence impact crop yield and quality. Fine-tuning flowering control can affect biomass production during vegetative growth, thereby substantially improving grain yield and its components (Alqudah and Schnurbusch 2017; Blümel et al. 2015; Mathan et al. 2016). A better understanding of senescence regulation has the potential for advancing the nutritional quality of cereal grains (Distelfeld et al. 2014; Gregersen 2011) and reducing fertilizer use (Yang and Udvardi 2018), thus reducing environmental impacts (Omara et al. 2019). Considering the food production needs of the twenty-first century, research has focused on determining the genetic control of flowering transition and whole-plant senescence. Integrated studies combining classical breeding, association mapping, and -omics techniques have improved our understanding of mechanisms controlling flowering and senescence in both model species and crops (Borràs-Gelonch et al. 2012; Greenup et al. 2009; Ibrahim et al. 2018; Kim et al. 2018; Rangan et al. 2017; Woo et al. 2013).
NAC genes, a large, plant-specific family of transcription factors, are among the genes identified that regulate senescence in different species. Several NAC genes induce leaf senescence so that nutrients are remobilized to seeds or grains. In Arabidopsis thaliana, T-DNA insertions in the AtNAP gene lead to significantly delayed leaf senescence, while overexpression of this gene causes precocious senescence (Guo and Gan 2006). In wheat, presence of an allele coding for a functional NAC protein (TtNAM-B1) on chromosome arm 6BS leads to faster leaf and whole-plant senescence and more efficient nutrient remobilization to developing kernels, increasing grain protein, iron, and zinc contents (Uauy et al. 2006; Waters et al. 2009). Reducing the transcript levels of all NAM copies in hexaploid wheat using RNA interference leads to a strong stay-green phenotype (delayed senescence) accompanied by a 30% reduction in percent grain protein (Uauy et al. 2006). A barley gene orthologous to TtNAM-B1, HvNAM1, is located in a co-linear (with wheat chromosome arm 6BS) region of chromosome six (Distelfeld et al. 2008). Alleles of this gene (GenBank accessions EU368851 and EU368852) found in two varieties (‘Lewis’ and ‘Karl,’ respectively) (Burger et al. 1979; Hockett et al. 1985; Wesenberg et al. 1976) code for proteins that differ in three positions (W78C, A102P, A357T). Previous work from our laboratory, comparing near-isogenic germplasm carrying either the ‘Lewis’ or ‘Karl’ HvNAM1 alleles, demonstrated that the ‘Karl’ allele is associated with delayed senescence and lower percent grain protein, suggesting that its function is impaired (Heidlebaugh et al. 2008; Jukanti et al. 2008).
Our laboratory has previously derived near-isogenic lines varying in the allelic state of HvNAM1 from a ‘Lewis’ x’Karl’ mapping population (See et al. 2002; Mickelson et al. 2003). Transcriptomic comparison of flag leaves from variety ‘Karl’ and near-isogenic line ‘10_11′ (containing the ‘Lewis’ HvNAM1 allele) identified an AtGRP7 ortholog (HvGR-RBP1) as one of the most strongly upregulated (in line ‘10_11’ vs. ‘Karl’) genes. For all time points from 7 to 28 days past anthesis, more than 100-fold differences in HvGR-RBP1 expression also were observed (Jukanti et al. 2008). Similar differences in expression were found in leaves of younger (pre-anthesis) plants (Lacerenza et al. 2010) with HvGR-RBP1 expressed at very low levels (detectable by quantitative real-time RT-PCR), in variety ‘Karl.’ Intriguingly, comparison of plant development in ‘Karl’ and ‘10_11’ indicated that, in addition to differences in leaf and whole-plant senescence (faster in line ‘10_11,’ attributed to the ‘Lewis’ HvNAM1 allele), pre-anthesis plant development was different as well. Starting with leaf eight, and throughout the rest of plant development, leaf development was significantly delayed in ‘Karl.’ A majority of ‘Karl’ and ‘10_11’ shoots had 13 leaves (i.e., leaf 13 was the flag leaf), but production of a 14th leaf was about twice as frequent in ‘Karl’ as in ‘10_11’ and only (very few) ‘Karl’ plants developed 15 leaves on their main shoots, emphasizing their slower development. This behavior was explained by slower development of the shoot apex and led to delayed anthesis (~ 5 days) in ‘Karl’ (Lacerenza et al. 2010). The relationship between HvNAM-1 and HvGR-RBP1 is unclear; however, recent barley genome information (Mascher et al. 2017) indicates that both are located on chromosome six, with a genetic distance of ~ 5 cM.
Glycine-rich RNA-binding proteins (GR-RBPs) are small (molecular weight < 20 kD) proteins with an N-terminal RNA-binding domain (~ 90 amino acids; also known as an RNA Recognition Motif or RRM) and a C-terminal glycine-rich domain (~ 70 amino acids) which is intrinsically disordered in the free protein (Ciuzan et al. 2015; Tripet et al. 2014). They comprise a subgroup of the large family of glycine-rich proteins, designated as subfamily IVa (Czolpinska and Rurek 2018; Mangeon et al. 2010). The best-understood plant GR-RBP is Arabidopsis thaliana glycine-rich RNA-binding protein 7 (AtGRP7). This protein binds both RNA and DNA, with a preference for single-stranded nucleic acids (Schüttpelz et al. 2008). AtGRP7 affects plant stress tolerance under high salt and dehydration conditions; the protein is involved in the regulation of stomata and also confers tolerance to low temperatures, most likely through its RNA chaperone activity (Cao et al. 2006; Hecht et al. 2005; Kim et al. 2008; Yang et al. 2014). AtGRP7 is a component of the flowering autonomous (or earliness per se) pathway which promotes floral transition, as demonstrated by the late-flowering phenotype of knockout mutants (Steffen et al. 2019; Streitner et al. 2008). Individual nucleotide resolution cross-linking and immunoprecipitation (iCLIP) has identified several hundred potential AtGRP7 RNA targets, besides its own pre-mRNA, suggesting its involvement in numerous functions besides flowering time control (Meyer et al. 2017).
Information outlined above suggests that developmental differences observed between variety ‘Karl’ and its near-isogenic line ‘10_11’ are due to both HvNAM1 and HvGR-RBP1 function, with HvNAM-1 regulating senescence and HvGR-RBP1 influencing pre-anthesis development and anthesis date. Using a wild barley nested association mapping population, Maurer et al. (2016) identified a quantitative trait locus (QTL) influencing flowering in the same region of chromosome six, closely linked to HvNAM1, and have suggested HvGR-RBP1 as a candidate gene. In this context, the purpose of research presented here was to (1) characterize differences in HvGR-RBP1 function between ‘Karl’ and ‘Lewis’; (2) use this information for the development of a molecular marker; and (3) apply this marker and previously developed markers for HvNAM1 (Distelfeld et al. 2008) to malt barley breeding germplasm, in order to dissect the effects of these genes on plant development and agronomic parameters.
Materials and methods
Sequence analysis, molecular marker design and genotyping
DNA isolation
Tissue from young barley leaves (~ 0.5 to 1 g) was finely ground in liquid nitrogen, using mortar and pestle, and mixed with 750 µl of extraction buffer containing 0.1 M Tris/HCl (pH 7.5), 50 mM EDTA, and 1.25% SDS (preheated to 65 °C). After incubation at 65 °C for 30 min, samples were cooled on ice and 300 µl of cold (4 °C) 6 mM ammonium acetate was added. Samples were incubated for 15 min at 4 °C, followed by centrifugation at 13,300 × g for 15 min. The supernatant was transferred to a new tube and mixed gently with 2 volumes of cold isopropanol. After incubation at 4 °C for 5 min, samples were centrifuged at 17,500 × g for 15 min to pellet the DNA. The pellet was washed twice with 75% ethanol, dried and suspended in 20 µl of H2O. Subsequently, DNA quantification was performed using a NanoDrop ND-2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Quantified DNA was diluted to 100 ng/µl for genotyping via PCR and restriction digest assays.
Primer design
To dissect the allelic state of HvGR-RBP1 in different barley genotypes, we designed a versatile molecular marker. The HvGR-RBP1 sequence was obtained from NCBI (https://www.ncbi.nlm.nih.gov/) (ID: JX126694.1) and BLASTed against the reference barley genome (variety ‘Morex’) (Mascher et al. 2017), identifying HORVU6Hr1G055440. Subsequently, primers (listed in Table S1) were designed, allowing amplification of an approximately 5 kb region encompassing the HvGR-RBP1 coding, flanking 5′- and flanking 3′-regions. PCR amplification of HvGR-RBP1 was performed using genomic DNA from barley varieties ‘Karl’ (Burger et al. 1979; Wesenberg et al. 1976) and ‘Lewis’ (Hockett et al. 1985). PCR reactions were performed using GoTaq DNA polymerase (Promega, Madison, WI, USA) following the manufacturer’s guidelines. An Eppendorf Mastercycler 5333 PCR Thermal Cycler (Eppendorf, Hauppauge, NY, USA) was used for PCR with the following cycling profile: 94 °C for initial denaturation, 30 cycles of 94 °C for 1 min, Tm for 1 min (Table S1), 72 °C for 1 min per kb, and 7 min of final elongation at 72 °C. PCR products were then visualized on 1% agarose gels, and Sanger sequencing was performed by Genewiz (South Plainfield, NJ, USA) with PCR samples cleaned using a ZR-96 DNA Clean-Up Kit (Zymo Research, Irvine, CA, USA).
Sequence analysis
Differences in the sequence of HvGR-RBP1 between barley varieties ‘Karl,’ ‘Lewis,’ and ‘Morex’ (reference genome) (Mascher et al. 2017) were analyzed using NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and pairwise and multiple sequence alignment tools from EBI (https://www.ebi.ac.uk/Tools/psa/, https://www.ebi.ac.uk/Tools/msa/), and Ensembl Plant (https://plants.ensembl.org/index.html/) with default settings. Single nucleotide polymorphisms (SNPs) between varieties were detected manually. PCR amplification and sequencing results identified differences between ‘Karl’ and ‘Lewis’ in the promoter region of the HvGR-RBP1; hence, this region was inspected in more detail. Two kb upstream of the HvGR-RBP1 start codon were scanned for potential transcription factor binding motifs via an online tool, PlantPAN 3.0 (Chow et al. 2019) (https://plantpan.itps.ncku.edu.tw/index.html) using the reference sequence from variety ‘Morex.’ The analysis was performed with default settings chosen for Brachypodium distachyon, and transcription factor motifs with a similarity score of 1 were selected as potential candidates.
Analysis of the allelic state of HvNAM1 was performed using the molecular markers described by Distelfeld et al. (2008). Two marker sequences (UHB6 and UHB7) were amplified with previously described PCR reactions. PCR products were digested with MwoI (for UHB6; New England Biolabs, Ipswich, MA, USA) and HpyCH4III (for UHB7; New England Biolabs) (Distelfeld et al. 2008). For a few barley lines, restriction enzyme cleavage analysis of amplified sequences was complemented with Sanger sequencing to confirm HvNAM1 allelic state.
In silico mapping
To determine genetic distance between HvNAM1 and HvGR-RBP1, the HvNAM1 sequence was obtained from NCBI (https://www.ncbi.nlm.nih.gov/) (ID: DQ869678). The sequence was BLASTed against the reference barley genome (Mascher et al. 2017) using the EnsemblPlant tool (Bolser et al. 2015) (https://plants.ensembl.org/Hordeum_vulgare/Tools/Blast), identifying HORVU6Hr1G019380 as a perfect match. Both HvGR-RBP1/HORVU6Hr1G055440 (see materials and methods, ‘primer design’) and HvNAM1/HORVU6Hr1G019380 were further characterized using the BARLEX Barley Genome Explorer (https://apex.ipk-gatersleben.de/apex/f?p=284:10::::::) (Colmsee et al. 2015) ‘gene list’ function. This allowed location of HORVU6Hr1G055440 on cluster 130, and of HORVU6Hr1G019380 on cluster 926 within the barley reference genome (Mascher et al. 2013, 2017; Beier et al. 2017). The BARLEX ‘cluster list’ function positions cluster 130 at 49.6 cM and cluster 926 at 55.0 cM on chromosome six, indicating a genetic distance of ~ 5 cM between the two genes.
Plant material, field studies and phenotypic data collection
To understand the impact of different HvGR-RBP1 and HvNAM1 alleles, we genotyped a subset (95 lines) of the material described as the ‘Malt Panel’ by Pauli et al. (2015), resulting from a set of crosses aimed at introgressing the ‘Karl’ HvNAM1 allele into malt barley breeding germplasm (Table 1; see Table S2 for pedigrees). Subsequent to genotyping, we re-analyzed agronomic traits that had been collected as described in Pauli et al. (2015) and results from the re-assessment are listed as experiment 1 throughout this manuscript. In order to test the interaction of HvGR-RBP1 and HvNAM1 allelic states with nitrogen fertilization and irrigation, we also performed a smaller experiment (13 varieties and lines; Tables 1 and S2) which is referred to as experiment 2 throughout this manuscript. Experiment 2 was grown for three different location-years, namely Bozeman 2016, Bozeman 2017 (Arthur Post Research Farm, Bozeman, MT, USA; 45°40′40.78 N, 111°09′07.14 W), and Conrad 2017 (Western Triangle Agricultural Research Station, Conrad, MT, USA; 48°18′26.05 N, 111°55′29.24 W), with three replicates for each location-year and treatment. For each location-year, plant material was grown under two water treatments in separate but adjacent blocks. Post Farm plots received 119 mm of rainfall from May to July during the 2016 season and 124.5 mm during 2017, while rainfall (2017) amounted to 89.9 mm for the Western Triangle location. All plots had adequate pre-season rainfall to reach field capacity. Approximately 150 mm water was added to the irrigated plots over three events throughout the season to ensure that plots did not dry below the barley wilting point; irrigation was stopped in the first week of July. Each location-year and water treatment also consisted of two nitrogen treatments in a split-plot randomized complete block design where the main plots were nitrogen treatment and the subplots were the genotypes. Prior to treatment, field soil samples were analyzed for total nitrogen content by AGVISE Laboratories (Benson, MN, USA) on a field level in 2016 and a replication level in 2017. This information was used to adjust the field nitrogen to the desired level in each individual split-plot, which was calculated by using the Montana Barley Production Guide (McVay et al. 2017) for malting barley nitrogen application (61.77 g nitrogen * expected yield in kg ha−1). The exact amount of nitrogen (applied as urea) used in this experiment for each location and treatment is listed in Table S3.
Taking advantage of the smaller experiment size, we collected more data from experiment 2, particularly for developmental parameters. Plants from this experiment were analyzed for developmental traits including plant height, anthesis date, maturity date, grain fill duration, and tiller numbers. Agronomic traits included percentage of plump kernels, test weight, grain protein concentration, yield, and harvest index. Grain protein concentration and kernel plumpness were measured and reported in the same way as experiment 1 (Pauli et al. 2015). Plant height was measured at maturity and was determined by extending the tip of the barley ear upward in order to record the maximum height including awns. Two measurements per plot were performed and the averages were recorded. Tiller count was measured at maturity by counting the number of productive tillers per 30.5 cm of row length. Again, two measurements were performed per plot (replicate), and the average was recorded. Anthesis date was recorded when approximately 50% of plants in each plot had reached Zadoks growth stage 49 (‘awn tipping’) (Alqudah and Schnurbusch 2017; Zadoks et al. 1974). Maturity dates were recorded only at the Post Farm (2016, 2017) location-years as the date at which approximately 50% of barley ears had reached Zadoks stage 92. The grain filling period was estimated by subtracting anthesis from maturity dates for each plot/replicate. Harvest index was calculated as grain weight divided by total row biomass weight (only for Post Farm location-years). Test weights were determined using a GAC2500-UGMA (Dickey-john Corporation, Auburn, IL, USA) and reported as kg hL−1.
Screening of HvGR-RBP1 allelic state in the USDA barley core collection
The USDA Barley Core Collection is a genetically diverse group of 2465 6-row and 2-row lines consisting of advanced breeding lines, cultivars, and landraces collected from different geographical locations around the world (Muñoz-Amatriaín et al. 2014). PCA analysis of 2-row genotypes consisting of 6906 polymorphic markers from a 9 K Infinium SNP chip (https://triticeaetoolbox.org/) was used to select a subset of 138 lines representing the genetic diversity of the population. To determine the frequency of the nonfunctional (‘Karl’) allele in a more genetically diverse population than the breeding population, the allelic state of HvGR-RPB1 was analyzed in 138 accessions using the molecular marker described in Fig. 2.
Data analysis
In this study, R software (v.3.5.3) was used to investigate the statistical significance of collected agronomic and developmental data (R Core Team 2018). Experiment 1 was treated as a randomized incomplete block design with a 2-level irrigation treatment (Pauli et al. 2015) (Table S4). Experiment 2 was treated as a randomized complete block design with 2-level irrigation and 2-level fertilizer treatments (Table S5). In both experiments, due to the non-independence of the barley lines with shared pedigrees, the genetic relatedness of lines used in the study was taken into consideration during statistical analysis. For this purpose, we performed analysis of variance (ANOVA) with the R package lme4qtl. This package considers the genetic relatedness of individual lines as a random factor (Ziyatdinov et al. 2018).
For experiment 1, a relationship matrix was constructed from the SNP genotypes (Table S6) consisting of 333 markers obtained by Pauli et al. (2015). Method-of-moments estimators were used to construct a kinship matrix, Gn × n where n is the number of genotypes and the kinship estimate between the most distant subpopulations is zero on average using the R package popkin (Ochoa and Storey 2019). The following linear mixed model was fit to these data
where \(Y_{ijk}\) is a single phenotypic observation, \(\mu\) is the grand mean, \({\text{alleleCombination}}_{i}\) is a dummy variable capturing the effect of the allele combinations at the HvNAM1 and HvGR-RBP1 loci, \({\text{irrigation}}_{j}\) is the effect of the jth irrigation treatment, \(\left( {{\text{alleleCombination}} \times {\text{irrigation}}} \right)_{ij}\) is the effect of the interaction between the ith alleleCombination and the jth irrigation treatment, \({\text{genotype}}_{k}\) is the effect of the kth genotype following \({\mathcal{N}}\)(0,Gn × n), and \(\varepsilon_{ijk}\) is the random error following \({\mathcal{N}}\)(0,\(\sigma^{2}\)). The model terms allelic combination and irrigation were modeled as fixed effects, and the other terms were modeled as random effects.
In experiment 2, the pedigree of the 13 utilized varieties and lines was obtained from T3/Barley (https://triticeaetoolbox.org/barley/) with manual extraction until there was an ‘unknown’ ancestor for the variety (Table S7). The genetic relatedness matrix Gn × n where n is the number of genotypes was then built using this information with the R package synbreed with the kin function (Wimmer et al. 2012). The following linear mixed model was fit to these data
where \(Y_{ijk}\) is a single phenotypic observation, \(\mu\) is the grand mean, \({\text{alleleCombination}}_{i}\) is a dummy variable capturing the effect of the allele combinations at the HvNAM1 and HvGR-RBP1 loci, \({\text{irrigation}}_{j}\) is the effect of the jth irrigation treatment, \(\left( {{\text{alleleCombination}} \times {\text{irrigation}}} \right)_{ij}\) is the effect of the interaction between the ith alleleCombination and the jth irrigation treatment, \({\text{genotype}}_{k}\) is the effect of the kth genotype following \({\mathcal{N}}\)(0,Gn × n), and \(\varepsilon_{ijk}\) is the random error following \({\mathcal{N}}\)(0,\(\sigma^{2}\)).
The model terms allelic combination and irrigation were modeled as fixed effects, and the other terms were modeled as random effects. The normality of model residuals was tested with the R package ‘fitdistrplus’ (Delignette-Muller and Dutang 2015), and datasets that did not fit a normal distribution were normalized via the R package ‘bestNormalize’ (Peterson and Cavanaugh 2019). Comparisons between different allelic groups and treatments were made with the Wilcoxon test, with the significance cutoff value p < 0.05; since no multiple testing corrections were made, p values are recorded in Supplemental Table S13.
Results
A G-rich insertion in the promoter region disrupts HvGR-RBP function in barley variety ‘Karl’
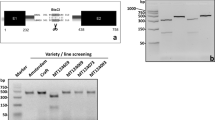
Previous studies from our laboratory have indicated that HvGR-RBP1 is expressed at a very low level in barley variety ‘Karl’ (see introduction). In order to discover the reason for this finding, a ~ 5 kb region, including ~ 2 kb of 5′ UTR, the 500 bp coding region and ~ 3 kb of 3′ UTR, was amplified from genomic DNA of varieties ‘Karl’ and ‘Lewis’ (with high HvGR-RBP1 expression) and sequenced (Fig. 1a). The primer combinations PM, P1, and P2 amplified the 5′ UTR, while P5 and P6 amplified the 3′ UTR (Fig. 1a, Table S1). The coding region is captured by P3 and P4, with P4 including both part of the coding region and 3′ UTR. A gene model, which is based on previous information about this gene (Tripet et al. 2014), the reference barley (var. ‘Morex’) genome (Mascher et al. 2017), and novel sequence information from this study, is presented in Fig. 1b. Gel electrophoretic analysis of PCR products showed a prominent difference in the DNA sequence amplified with the PM primer combination, with the fragment from ‘Karl’ ~ 400 bp longer than the ‘Lewis’ fragment (Fig. 2a). Sanger sequencing of this PCR amplicon identified a G-rich insertion in ‘Karl,’ which is absent in the reference genome, and in variety ‘Lewis’ (Supplementary File 1). This insertion may be responsible for the low gene transcription that was reported in var. ‘Karl’ (Jukanti et al. 2008; Lacerenza et al. 2010). In addition to the insertion, sequencing identified three single nucleotide differences that resulted in three amino acid substitutions, asparagine to serine at the 59th position, alanine to glycine at the 114th position, and tyrosine to histidine at the 115th position, between ‘Lewis’ and ‘Karl’ (Fig. 1b).
Amplification of HvGR-RBP1 and potential gene model. a The amplified PCR products of HvGR-RPB1 from variety ‘Lewis’ are shown on %1 agarose gel. PM is the amplicon from ‘Lewis’ that varied for the insertion with ‘Karl.’ b A potential gene model for HvGR-RBP1 is represented. The promoter region of the gene possesses several potential transcription factor binding motifs. The G-rich insertion in ‘Karl’ may disrupt gene expression. HvGR-RPB1 consist of two exons and an intron, which are highly similar between ‘Karl’ and ‘Lewis’ with only three single nucleotide polymorphisms (A261G, C426G, T428C) that are located in Exon-2 and result in missense mutations Aspargine to Serine at 59th, Alanine to Glycine at 114th and Tyrosine to Histidine at 115th positions in ‘Karl.’ The locations for forward and reverse primers used for amplification of PCR products from Fig. 1a are also shown as PM, P1-P6
HvGR-RBP1 Marker Development and Genotyping. a The G-rich insertion in variety ‘Karl’ is used as a marker for genotyping of HvGR-RBP1 gene. Here, four different varieties are represented: Karl, Lewis, MT124017, and 10_11. The difference in amplification length in the marker region is ~ 300 to 400 bp differentiates ‘Karl’ and ‘Lewis’ genotypes, as well as the heterozygote ‘MT124017.’ b A pie chart representing the distribution of each allelic group in the screened MSU breeding population. + / + indicating functioning alleles for both HvGR-RPB1 and HvNAM1. −/− indicating non-functioning alleles for both genes. ± indicating functional ‘Lewis’ allele for HvGR-RBP1 and non-functioning ‘Karl’ allele for HvNAM1. −/ + indicating non-functioning ‘Karl’ allele for HvGR-RBP1 and functional ‘Lewis’ allele for HvNAM1 were not observed in this subset that were advanced from the original larger population. 2c. A figure representing the genetic map distance between HvGR-RBP1 and HvNAM-1 gene. Two genes are 5.4 cM apart, resulting in linkage
The promoter region of HvGR-RBP1 was scanned for potential transcription factor binding motifs. Our analysis indicated that this region is almost identical in varieties ‘Lewis’ and ‘Morex’; hence, the ‘Morex’ (reference) sequence was used for this part of the study. Several transcription factor binding motifs were identified using the PlantPAN (v. 3.0) tool (https://plantpan.itps.ncku.edu.tw/) (Chow et al. 2019). Motifs included those which may be recognized by members of the MADF, WRKY, Myb/SANT, and NAC families, suggesting roles for HvGR-RBP1 in plant developmental and stress response processes (Fig. 1b, Table S8).
Screening for HvGR-RBP1 allelic state in breeding germplasm suggests a phenotypic selection for the functional allele
To understand the allelic effect of HvGR-RBP1 on agronomic traits in malt barley, we screened germplasm from the Montana State University malt barley breeding program included in experiments 1 and 2. The molecular marker exploited the amplicon length difference in the 5′ UTR of HvGR-RBP1, enabling easy differentiation of homozygous and heterozygous genotypes (Fig. 2a). The germplasm screened represents efforts to control grain protein concentration by introgressing the ‘Karl’ (low grain protein; delayed senescence) HvNAM1 allele; lines were advanced and phenotypically selected for yield, low grain protein, and a high percentage of plump kernels. As expected, the ‘Karl’ HvNAM1 allele was present in a majority of tested lines. Interestingly, the functional (high gene expression; ‘Lewis’) HvGR-RBP1 allele was present in a considerably larger percentage of lines than expected based on genetic distance between the two genes (~ 5 cM; Fig. 2c) (Table 1, Fig. 2b, Table S2). Phenotypic selection may have favored lines carrying the functional HvGR-RBP1 allele.
Barley plants carrying a functional HvGR-RBP1 allele head earlier and are slightly reduced in height
To understand whether the combination of the functional ‘Lewis’ allele of HvGR-RBP1 and a ‘Karl’ HvNAM1 allele was due to phenotypic selection during breeding, we compared agronomic traits in lines with different allele combinations. Lines carrying a ‘Karl’ allele of HvGR-RBP1 and a ‘Lewis’ allele of HvNAM1 were rare after selection and have therefore not been included in our analyses. Data from experiment 1 (re-analysis of agronomic data from Pauli et al. (2015) based on new molecular marker analyses) indicate that germplasm homozygous for the functional (‘Lewis’) HvGR-RBP1 allele reaches the heading stage 0.5 days earlier (Table 2). The effect was stronger in experiment 2, with 1.2 days of difference in anthesis date (Table 3), with measured differences significant in both experiments. Lines combining functional (‘Lewis’) HvGR-RBP1 with ‘Karl’ HvNAM1 alleles had the earliest heading or anthesis dates (Tables 2 and 3). Lines with this allele combination were also reduced in height; separate analysis of HvGR-RBP1 and HvNAM1 effects suggests a small but consistent effect of HvGR-RBP1 on height, while the influence of HvNAM1 was not consistent between the two experiments (Tables 2 and 3).
Experiment 2 also analyzed maturity dates. Based on these, plants carrying the ‘Karl’ HvNAM1 allele matured 2 days later (Table 3). This finding agrees with previous research from our laboratory comparing barley lines with ‘Lewis’ vs. ‘Karl’ HvNAM1 alleles (Heidlebaugh et al. 2008; Jukanti and Fischer 2008; Jukanti et al. 2008; Lacerenza et al. 2010), and with the recent literature regarding the function of NAC genes in senescence regulation (Distelfeld et al. 2014; Podzimska-Sroka et al. 2015). Interestingly, as the functional (‘Lewis’) HvGR-RBP1 allele accelerates flowering and heading while the ‘Karl’ HvNAM1 allele delays plant maturity, the combination of these two alleles (± group in Table 3) resulted in a significantly increased (~ 3 days longer) grain fill duration.
Experiments also analyzed the effect of irrigation and N fertilizer treatments on flowering/heading dates, maturity dates, and plant height (Tables 2, 3 and S5). The statistical model used indicates that both genes contribute significantly to the control of grain fill duration. The model also suggests that the effects of HvGR-RBP1 alleles are independent of the environmental conditions tested, while HvNAM1 allelic effects are slightly affected by irrigation treatments. As expected, both irrigation and N treatments had significant effects on plant height, with irrigated plants ~ 20 cm taller in experiment 1 and ~ 13 cm taller in experiment 2 (Table S9). Significant interactions between allele combinations and irrigation treatments occurred in both experiments (Tables 2 and 3). In contrast to heading, maturity, and grain fill duration, tillering (i.e., numbers of tillers per meter) was not influenced by the genes studied here (Table 3), while both nitrogen fertilization and irrigation significantly influenced this parameter (Table S9).
Germplasm combining functional (‘Lewis’) HvGR-RBP1 with ‘Karl’ HvNAM1 alleles has a lower grain protein concentration and a higher percentage of plump kernels
The agronomic traits most important to malt quality, grain protein, and percentage of plump kernels, were also examined to determine their regulation by HvGR-RBP1 and HvNAM1 allelic states. In both experiments, germplasm homozygous for ‘Lewis’ HvGR-RBP1 and ‘Karl’ HvNAM1 alleles presented a strongly enhanced percentage of plump kernels when compared to the other allele combinations (Tables 2 and 3). The statistical model indicates that, in both experiments, the influence of alleles and allele combinations on this trait are highly significant (Tables 2 and 3). Lines with this allele combination also had the lowest grain protein concentrations (Tables 2 and 3), with the influence of HvNAM1 consistent between the two experiments. The ‘Karl’ HvNAM1 allele lowered grain protein from 13.4 to 12.4% in experiment 1 and from 13.1 to 11.8% in experiment 2, confirming previous studies in barley and wheat (Heidlebaugh et al. 2008; Jukanti et al. 2008; Uauy et al. 2006).
Irrigation treatments applied in this study significantly enhanced the percentage of plump kernels and lowered percent grain protein in experiment 1, while applied N fertilization (experiment 2) slightly enhanced grain protein concentration (Fig. 3, Table S9). Germplasm homozygous for ‘Lewis’ HvGR-RBP1 and ‘Karl’ HvNAM1 alleles had the highest percentage of plump kernels and the lowest grain protein concentration, irrespective of irrigation treatment, in both experiments 1 (Fig. 3) and 2 (Tables S10 and S11). This allele combination also maintained its advantage for malt barley production under the two N levels tested in experiment 2 (Table S11), suggesting that observed effects are robust under a range of growing conditions.
Changes in agronomic traits under irrigation treatment in re-analysis of Pauli et al (2015). Data collected on lines segregating for HvGR-RBP1 and HvNAM1 and for traits from dry and irrigated conditions. Lines intersect data from the two treatments at the mean value for each trait for each allelic combination. Agronomic traits are represented as (a) kernel plumpness (%), (b) grain protein (%) (c) test weight (kg/L), (d) yield (kg/ha). Note that change in mean between dry and irrigated is less for allelic combo ± except for in the case of yield
Germplasm combining functional (‘Lewis’) HvGR-RBP1 with ‘Karl’ HvNAM1 alleles has slightly enhanced test weight, but yield is not increased
Based on data presented in Tables 2 and 3, lines homozygous for the functional (‘Lewis’) HvGR-RBP1 and ‘Karl’ HvNAM1 alleles have slightly enhanced test weight; this effect can be mostly attributed to the functional HvGR-RBP1 allele. In experiment 1, this allele combination appears associated with a slight yield penalty, while essentially no yield effects associated with alleles and allele combinations were observed in experiment 2.
As expected, tested environmental parameters had a significant influence on test weight and yield in both experiments. Irrigation increased test weight by 1–3 kg hL−1 and yield by 2000 to 3000 kg ha−1 (Fig. 3, Tables S6 and S7), while higher N levels were associated with slightly (but not significantly) enhanced yield in experiment 2 (Table S11). The statistical model confirms that effects of irrigation on test weight and yield were significant in both experiments (Tables 2 and 3), while no significant effect of applied N treatments was observed in experiment 2 (Table 3). Some combinations between studied genes/alleles and irrigation affected test weight, but not yield (Table 3).
The HvGR-RBP1 promoter insertion is not unique to variety ‘Karl’
In our analysis, we studied the influence of the functional (‘Lewis’) vs. nonfunctional (‘Karl’) HvGR-RBP1 alleles on plant developmental and agronomic parameters. To determine if the nonfunctional HvGR-RBP1 allele is unique to ‘Karl,’ we used the newly developed marker (Fig. 2a) to screen a subset (138 accessions) from the USDA Barley Core Collection (Muñoz-Amatriaín et al. 2014). Based on this analysis (Table S12), 22 accessions (15.9%) possessed an upstream insertion identical or similar to the one found in var. ‘Karl.’ Thirteen (13) of the accessions carrying the insertion were landraces, three were breeding lines, and two were cultivars, while the status of the remaining four accessions is unclear (Table S12). The presence of the upstream insertion in landraces with a geographically diverse origin (including Asia, Africa, and South America) indicates that this variation is not particularly rare. However, it stands as an intriguing question whether the functional vs. nonfunctional allele provides a different selective advantage depending on the environment.
Discussion
Flowering and senescence are important phases of a plant’s life cycle, particularly in annual species with monocarpic senescence. They define adaptation to a particular environment; in annual crops including cereals, timing of flowering and senescence control yield and quality (Distelfeld et al. 2014; Hill and Li 2016; Woo et al. 2018). Interactions between flowering and senescence control appear particularly relevant, and both flowering time-dependent and -independent inputs into the control of senescence have been identified (Bogard et al. 2011; Hensel et al. 1993; Kim et al. 2004; Miryeganeh et al. 2018; Parrott et al. 2012; Wingler et al. 2010; Wu et al. 2008).
Through sequencing of HvGR-RBP1, an amplicon size difference was identified that is directly applicable as a molecular marker, which has been utilized here to screen malt barley breeding germplasm (Table S2) and a geographically diverse subset of the USDA Barley Core Collection, finding that lines with the insertion are not rare (Table S12) (Muñoz-Amatriaín et al. 2014). Data shown in Tables 1 and 2 indicate that germplasm with a strongly expressed HvGR-RBP1 allele reaches anthesis earlier and is slightly reduced in height when compared with germplasm carrying the ‘Karl’ allele, which is very weakly expressed. These findings fit with previous analyses comparing variety ‘Karl’ with a near-isogenic line (‘10_11’) carrying the functional allele from variety ‘Lewis’ (Lacerenza et al. 2010; Parrott et al. 2012). Furthermore, a homologous gene in A. thaliana (AtGRP7) is important for the autonomous (or earliness per se) flowering pathway; knockout mutants exhibit substantially delayed flowering (Steffen et al. 2019; Streitner et al. 2008). Together, these data indicate that AtGRP7 and HvGR-RBP1 have similar or identical functions in A. thaliana and barley flowering time control. Although, at this point, we cannot exclude the possibility that allelic differences in genes linked to HvNAM1 and HvGR-RBP1 contribute to the observed phenotype. Future research will focus on dissecting the molecular interactions through which HvGR-RBP1 modulates barley flowering, considering known differences between flowering time control in A. thaliana and cereals (Blümel et al. 2015; Greenup et al. 2009). The fact that vernalization treatments eliminate developmental differences between germplasm differing in GR-RBP function (Parrott et al. 2012; Streitner et al. 2008) may be helpful in this context.
The ‘Lewis’ allele of HvNAM1 leads to plants reaching maturity 2 days earlier than the ‘Karl’ allele in experiment 2 (Table 3). This finding from field experiments agrees with our previous (greenhouse) based comparison of flag leaf senescence in near-isogenic germplasm varying in HvNAM1 allelic state (Heidlebaugh et al. 2008; Jukanti et al. 2008; Mason et al. 2016). Importantly, based on experiment 2, effects of HvGR-RBP1 and HvNAM1 are additive; genotypes combining functional (‘Lewis’) HvGR-RBP1 with ‘Karl’ HvNAM1 alleles have a grain fill duration that is ~ 3 days longer. While this difference may not appear large, it corresponds to an ~ 9% extension of this developmental phase. This extension in grain fill duration may explain the substantial and significant increase in the percentage of plump kernels, and the slight (but significant) increase in test weight and decrease in grain protein concentration seen in germplasm with this allele combination (Table 3). While maturity was not measured in experiment 1, the same effects on kernel plumpness, test weight, and grain protein as in experiment 2 were observed when comparing lines combining functional (‘Lewis’) HvGR-RBP1 with ‘Karl’ HvNAM1 alleles with the other allele combinations (Table 2), confirming conclusions drawn from experiment 2. Analysis of seed samples from the Bozeman 2017 (non-irrigated) location indicates that enhanced kernel plumpness is associated with a slight (~ 3%) increase in kernel diameter, an ~ 10% increase in single kernel weight, but no change in seed length (data not shown).
Maltsters can reject barley for malt if grain protein is too high and/or the percentage of plump kernels too low, reducing the farmer’s profit by half. A variety of conditions can increase grain protein and reduce plump seed, including too much soil nitrogen, lack of rainfall, and heat during grain fill. In the current study, the positive effects on kernel plumpness and protein content contributed by the combination of the ‘Lewis’ HvGR-RBP1 and the ‘Karl’ HvNAM1 alleles were stable under the different environments (irrigation, nitrogen) tested (Fig. 3, Tables S6 and S7). Weston et al. (1993) found that low grain protein lines from ‘Karl’ had lower protein across nitrogen treatments when compared with types not carrying low protein gene. However, the low protein types also had lower plumps and kernel weight across treatments, making them undesirable. Our data suggest that this is due to linkage between the ‘Karl’ HvGR-RBP1 and HvNAM1 alleles. Recombinants between the genes studied here satisfy a breeding goal that benefits growers and end-users—varieties with stable low protein and high plumps.
It might be expected that a genotype with increased grain fill duration leads to a yield increase. Extended grain fill duration (known as a ‘stay-green phenotype’ if maturation is delayed) has been extensively discussed in the literature and is often suggested to increase yield (Distelfeld et al. 2014; Thomas and Howarth 2000; Thomas and Ougham 2014), yet no significant yield increase was seen in our experiments when considering lines carrying the ‘Karl’ HvNAM1 allele (delayed maturity) combined with either the ‘Lewis’ or ‘Karl’ HvGR-RBP1 alleles (Tables 2, 3). Significantly increased kernel plumpness and test weight, combined with no or marginal yield effects, suggest that lines combining functional (‘Lewis’) HvGR-RBP1 with ‘Karl’ HvNAM1 alleles produce fewer but larger kernels. While this is still desirable in malting barley, the system may be sink-limited. As tillering is not affected (Table 3), one possible explanation is that the early-flowering (‘Lewis’ allele for HvGR-RBP1) genotype is associated with the production of fewer fertile spikelets, i.e., faster exhaustion of the meristem, and/or enhanced seed abortion. This is clearly an issue that warrants further analysis.
The observation that HvGR-RBP1 expression is strongly reduced in barley variety ‘Karl’ (Jukanti et al. 2008; Lacerenza et al. 2010) could be due to changes in the 5′ flanking sequence about 1 kb upstream of the translation start site, which includes an ~ 400 bp G-rich insertion (Fig. 2a). Efforts to determine differences in HvGR-RBP1 protein levels between ‘Karl’ and ‘Lewis’ types have so far been unsuccessful, and effects of the identified amino acid sequence differences remain unexplored. The insertion could disrupt transcription for a variety of reasons, including disruption of enhancers, or attraction of silencers. The fact that the insertion includes repeated G sequence could be important as such sequences are known to interact with interfering RNA (Pernitzsch et al. 2014) or form secondary structures that impede expression (Yang et al. 2018). A more detailed analysis of HvGR-RBP1 sequence and function will allow discrimination between these possibilities.
Overall, data presented here combined with past work suggest an important function for HvGR-RBP1 in the control of barley reproductive development, similar or identical to AtGRP7 function in Arabidopsis development. This research sets the stage for a detailed functional analysis of HvGR-RBP1; furthermore, our data indicate that screening for the allelic state of HvGR-RBP1, particularly in combination with HvNAM1, is of high practical value in malt barley breeding.
Code availability
Not applicable.
Availability of data and material
All data utilized are contained within the manuscript, or within Supplementary Material. Data are shown in Tables.
References
Alqudah AM, Schnurbusch T (2017) Heading date is not flowering time in spring barley. Front Plant Sci 8:896. https://doi.org/10.3389/fpls.2017.00896
Beier S, Himmelbach A, Colmsee C, Zhang XQ, Barrero RA, Zhang Q, Li L, Bayer M, Bolser D, Taudien S, Groth M, Felder M, Hastie A, Simkova H, Stankova H, Vrana J, Chan S, Munoz-Amatriain M, Ounit R, Wanamaker S, Schmutzer T, Aliyeva-Schnorr L, Grasso S, Tanskanen J, Sampath D, Heavens D, Cao S, Chapman B, Dai F, Han Y, Li H, Li X, Lin C, McCooke JK, Tan C, Wang S, Yin S, Zhou G, Poland JA, Bellgard MI, Houben A, Dolezel J, Ayling S, Lonardi S, Langridge P, Muehlbauer GJ, Kersey P, Clark MD, Caccamo M, Schulman AH, Platzer M, Close TJ, Hansson M, Zhang G, Braumann I, Li C, Waugh R, Scholz U, Stein N, Mascher M (2017) Construction of a map-based reference genome sequence for barley. Hordeum vulgare L Sci Data 4:170044. https://doi.org/10.1038/sdata.2017.44
Blümel M, Dally N, Jung C (2015) Flowering time regulation in crops-what did we learn from Arabidopsis? Curr Opin Biotechnol 32:121–129. https://doi.org/10.1016/j.copbio.2014.11.023
Bogard M, Jourdan M, Allard V, Martre P, Perretant MR, Ravel C, Heumez E, Orford S, Snape J, Griffiths S, Gaju O, Foulkes J, Le Gouis J (2011) Anthesis date mainly explained correlations between post-anthesis leaf senescence, grain yield, and grain protein concentration in a winter wheat population segregating for flowering time QTLs. J Exp Bot 62:3621–3636. https://doi.org/10.1093/jxb/err061
Bolser DM, Kerhornou A, Walts B, Kersey P (2015) Triticeae resources in Ensembl plants. Plant Cell Physiol 56:e3. https://doi.org/10.1093/pcp/pcu183
Borràs-Gelonch G, Rebetzke GJ, Richards RA, Romagosa I (2012) Genetic control of duration of pre-anthesis phases in wheat (Triticum aestivum L.) and relationships to leaf appearance, tillering, and dry matter accumulation. J Exp Bot 63:69–89. https://doi.org/10.1093/jxb/err230
Burger WC, Wesenberg DM, Carden JE III, Pawlisch PE (1979) Protein content and composition of Karl and related barleys. Crop Sci 19:235–238. https://doi.org/10.2135/cropsci1979.0011183X001900020016x
Cao S, Jiang L, Song S, Jing R, Xu G (2006) AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis. Cell Mol Biol Lett 11:526–535. https://doi.org/10.2478/s11658-006-0042-2
Chow C-N, Lee T-Y, Hung Y-C, Li G-Z, Tseng K-C, Liu Y-H, Kuo P-L, Zheng H-Q, Chang W-C (2019) PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucl Acids Res 47:D1155–D1163. https://doi.org/10.1093/nar/gky1081
Ciuzan O, Hancock J, Pamfil D, Wilson I, Ladomery M (2015) The evolutionarily conserved multifunctional glycine-rich RNA-binding proteins play key roles in development and stress adaptation. Physiol Plant 153:1–11. https://doi.org/10.1111/ppl.12286
Colmsee C, Beier S, Himmelbach A, Schmutzer T, Stein N, Scholz U, Mascher M (2015) BARLEX - the barley draft genome explorer. Mol Plant 8:964–966. https://doi.org/10.1016/j.molp.2015.03.009
Czolpinska M, Rurek M (2018) Plant glycine-rich proteins in stress response: an emerging, still prospective story. Front Plant Sci 9:302. https://doi.org/10.3389/fpls.2018.00302
Delignette-Muller ML, Dutang C (2015) fitdistrplus: An R package for fitting distributions. J Stat Softw. https://doi.org/10.18637/jss.v064.i04
Distelfeld A, Korol A, Dubcovsky J, Uauy C, Blake T, Fahima T (2008) Colinearity between the barley grain protein content (GPC) QTL on chromosome arm 6HS and the wheat Gpc-B1 region. Mol Breed 22:25–38. https://doi.org/10.1007/s11032-007-9153-3
Distelfeld A, Avni R, Fischer AM (2014) Senescence, nutrient remobilization, and yield in wheat and barley. J Exp Bot 65:3783–3798. https://doi.org/10.1093/jxb/ert477
Greenup A, Peacock WJ, Dennis ES, Trevaskis B (2009) The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann Bot 103:1165–1172. https://doi.org/10.1093/aob/mcp063
Gregersen PL (2011) Senescence and nutrient remobilization in crop plants. In: Hawkesford MJ, Barraclough PB (eds) The molecular and physiological basis of nutrient use efficiency in crops. Wiley, Oxford, pp 83–102
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612. https://doi.org/10.1111/j.1365-313X.2006.02723.x
Hecht V, Foucher F, Ferrandiz C, Macknight R, Navarro C, Morin J, Vardy ME, Ellis N, Beltran JP, Rameau C, Weller JL (2005) Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol 137:1420–1434. https://doi.org/10.1104/pp.104.057018
Heidlebaugh NM, Trethewey BR, Jukanti AK, Parrott DL, Martin JM, Fischer AM (2008) Effects of a barley (Hordeum vulgare) chromosome 6 grain protein content locus on whole-plant nitrogen reallocation under two different fertilisation regimes. Funct Plant Biol 35:619–632. https://doi.org/10.1071/FP07247
Hensel LL, Grbic V, Baumgarten DA, Bleecker AB (1993) Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5:553–564. https://doi.org/10.1105/tpc.5.5.553
Hill CB, Li C (2016) Genetic architecture of flowering phenology in cereals and opportunities for crop improvement. Front Plant Sci 7:1906. https://doi.org/10.3389/fpls.2016.01906
Hockett EA, Gilbertson KM, McGuire CF, Bergman LE, Wiesner LE, Robbins GS (1985) Registration of ‘Lewis’ barley. Crop Sci 25:570–571. https://doi.org/10.2135/cropsci1985.0011183X002500030036x
Ibrahim A, Harrison M, Meinke H, Fan Y, Johnson P, Zhou M (2018) A regulator of early flowering in barley (Hordeum vulgare L.). PLoS One 13:e0200722. https://doi.org/10.1371/journal.pone.0200722
Jukanti AK, Fischer AM (2008) A high-grain protein content locus on barley (Hordeum vulgare) chromosome 6 is associated with increased flag leaf proteolysis and nitrogen remobilization. Physiol Plant 132:426–439. https://doi.org/10.1111/j.1399-3054.2007.01044.x
Jukanti AK, Heidlebaugh NM, Parrott DL, Fischer IA, McInnerney K, Fischer AM (2008) Comparative transcriptome profiling of near-isogenic barley (Hordeum vulgare) lines differing in the allelic state of a major grain protein content locus identifies genes with possible roles in leaf senescence and nitrogen reallocation. New Phytol 177:333–349. https://doi.org/10.1111/j.1469-8137.2007.02270.x
Kim JH, Durrett TP, Last RL, Jander G (2004) Characterization of the Arabidopsis TU8 glucosinolate mutation, an allele of TERMINAL FLOWER2. Plant Mol Biol 54:671–682. https://doi.org/10.1023/B:PLAN.0000040897.49151.98
Kim JS, Jung HJ, Lee HJ, Kim KA, Goh C-H, Woo Y, Oh SH, Han YS, Kang H (2008) Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 55:455–466. https://doi.org/10.1111/j.1365-313X.2008.03518.x
Kim J, Kim JH, Lyu JI, Woo HR, Lim PO (2018) New insights into the regulation of leaf senescence in Arabidopsis. J Exp Bot 69:787–799. https://doi.org/10.1093/jxb/erx287
Lacerenza JA, Parrott DL, Fischer AM (2010) A major grain protein content locus on barley (Hordeum vulgare L.) chromosome 6 influences flowering time and sequential leaf senescence. J Exp Bot 61:3137–3149. https://doi.org/10.1093/jxb/erq139
Mangeon A, Junqueira RM, Sachetto-Martins G (2010) Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav 5:99–104
Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Munoz-Amatriain M, Close TJ, Wise RP, Schulman AH, Himmelbach A, Mayer KF, Scholz U, Poland JA, Stein N, Waugh R (2013) Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J 76:718–727. https://doi.org/10.1111/tpj.12319
Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, Bayer M, Ramsay L, Liu H, Haberer G, Zhang X-Q, Zhang Q, Barrero RA, Li L, Taudien S, Groth M, Felder M, Hastie A, Šimková H, Staňková H, Vrána J, Chan S, Muñoz-Amatriaín M, Ounit R, Wanamaker S, Bolser D, Colmsee C, Schmutzer T, Aliyeva-Schnorr L, Grasso S, Tanskanen J, Chailyan A, Sampath D, Heavens D, Clissold L, Cao S, Chapman B, Dai F, Han Y, Li H, Li X, Lin C, McCooke JK, Tan C, Wang P, Wang S, Yin S, Zhou G, Poland JA, Bellgard MI, Borisjuk L, Houben A, Doležel J, Ayling S, Lonardi S, Kersey P, Langridge P, Muehlbauer GJ, Clark MD, Caccamo M, Schulman AH, Mayer KFX, Platzer M, Close TJ, Scholz U, Hansson M, Zhang G, Braumann I, Spannagl M, Li C, Waugh R, Stein N (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544:427–433. https://doi.org/10.1038/nature22043
Mason KE, Hilmer JK, Maaty WS, Reeves BD, Grieco PA, Bothner B, Fischer AM (2016) Proteomic comparison of near-isogenic barley (Hordeum vulgare L.) germplasm differing in the allelic state of a major senescence QTL identifies numerous proteins involved in plant pathogen defense. Plant Physiol Biochem 109:114–127. https://doi.org/10.1016/j.plaphy.2016.09.008
Mathan J, Bhattacharya J, Ranjan A (2016) Enhancing crop yield by optimizing plant developmental features. Development 143:3283–3294. https://doi.org/10.1242/dev.134072
Maurer A, Draba V, Pillen K (2016) Genomic dissection of plant development and its impact on thousand grain weight in barley through nested association mapping. J Exp Bot 67:2507–2518. https://doi.org/10.1093/jxb/erw070
McVay K, Burrows M, Jones C, Wanner K, Menalled F (2017) Montana barley production guide. Montana State University Extension Service, Bozeman
Meyer K, Köster T, Nolte C, Weinholdt C, Lewinski M, Grosse I, Staiger D (2017) Adaptation of iCLIP to plants determines the binding landscape of the clock-regulated RNA-binding protein AtGRP7. Genome Biol 18:204. https://doi.org/10.1186/s13059-017-1332-x
Mickelson S, See D, Meyer FD, Garner JP, Foster CR, Blake TK, Fischer AM (2003) Mapping of QTL associated with nitrogen storage and remobilization in barley (Hordeum vulgare L.) leaves. J Exp Bot 54:801–812. https://doi.org/10.1093/jxb/erg084
Miryeganeh M, Yamaguchi M, Kudoh H (2018) Synchronisation of Arabidopsis flowering time and whole-plant senescence in seasonal environments. Sci Rep 8:10282. https://doi.org/10.1038/s41598-018-28580-x
Muñoz-Amatriaín M, Cuesta-Marcos A, Endelman JB, Comadran J, Bonman JM, Bockelman HE, Chao S, Russell J, Waugh R, Hayes PM, Muehlbauer GJ (2014) The USDA barley core collection: genetic diversity, population structure, and potential for genome-wide association studies. PLoS ONE 9:e94688. https://doi.org/10.1371/journal.pone.0094688
Ochoa A, Storey JD (2019) FST and kinship for arbitrary population structures II: method-of-moments estimators. bioRxiv. https://doi.org/10.1101/083923
Omara P, Aula L, Oyebiyi F, Raun WR (2019) World cereal nitrogen use efficiency trends: review and current knowledge. Agrosyst Geosci Environ 2:180045. https://doi.org/10.2134/age2018.10.0045
Parrott DL, Downs EP, Fischer AM (2012) Control of barley (Hordeum vulgare L.) development and senescence by the interaction between a chromosome six grain protein content locus, day length, and vernalization. J Exp Bot 63:1329–1339. https://doi.org/10.1093/jxb/err360
Pauli D, Brown-Guedira G, Blake TK (2015) Identification of Malting Quality QTLs in Advanced Generation Breeding Germplasm. J Am Soc Brew Chem 73(1):29–40
Pernitzsch SR, Tirier SM, Beier D, Sharma CM (2014) A variable homopolymeric G-repeat defines small RNA-mediated posttranscriptional regulation of a chemotaxis receptor in Helicobacter pylori. Proc Natl Acad Sci USA 111(4):E501–E510. https://doi.org/10.1073/pnas.1315152111
Peterson RA, Cavanaugh JE (2019) Ordered quantile normalization: a semiparametric transformation built for the cross-validation era. J Appl Stat. https://doi.org/10.1080/02664763.2019.1630372
Podzimska-Sroka D, O’Shea C, Gregersen P, Skriver K (2015) NAC transcription factors in senescence: From molecular structure to function in crops. Plants 4:412–448. https://doi.org/10.3390/plants4030412
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rangan P, Furtado A, Henry RJ (2017) The transcriptome of the developing grain: a resource for understanding seed development and the molecular control of the functional and nutritional properties of wheat. BMC Genomics 18:766. https://doi.org/10.1186/s12864-017-4154-z
Schüttpelz M, Schöning JC, Doose S, Neuweiler H, Peters E, Staiger D, Sauer M (2008) Changes in conformational dynamics of mRNA upon AtGRP7 binding studied by fluorescence correlation spectroscopy. J Am Chem Soc 130:9507–9513. https://doi.org/10.1021/ja801994z
See D, Kanazin V, Kephart K, Blake T (2002) Mapping genes controlling variation in barley grain protein concentration. Crop Sci 42:680–685. https://doi.org/10.2135/cropsci2002.6800
Steffen A, Elgner M, Staiger D (2019) Regulation of flowering time by the RNA-binding proteins AtGRP7 and AtGRP8. Plant Cell Physiol 60:2040–2050. https://doi.org/10.1093/pcp/pcz124
Streitner C, Danisman S, Wehrle F, Schöning JC, Alfano JR, Staiger D (2008) The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. Plant J 56:239–250. https://doi.org/10.1111/j.1365-313X.2008.03591.x
Thomas H, Howarth CJ (2000) Five ways to stay green. J Exp Bot 51:329–337. https://doi.org/10.1093/jexbot/51.suppl_1.329
Thomas H, Ougham H (2014) The stay-green trait. J Exp Bot 65:3889–3900. https://doi.org/10.1093/jxb/eru037
Tripet BP, Mason KE, Eilers BJ, Burns J, Powell P, Fischer AM, Copié V (2014) Structural and biochemical analysis of the Hordeum vulgare L. HvGR-RBP1 protein, a glycine-rich RNA-binding protein involved in the regulation of barley plant development and stress response. Biochemistry 53:7945–7960. https://doi.org/10.1021/bi5007223
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314:1298–1301. https://doi.org/10.1126/science.1133649
Waters BM, Uauy C, Dubcovsky J, Grusak MA (2009) Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J Exp Bot 60:4263–4274. https://doi.org/10.1093/jxb/erp257
Wesenberg DM, Hayes RM, Standridge NN, Burger WC, Goplin ED, Petr FC (1976) Registration of Karl barley (Reg. No. 147). Crop Sci 16:737. https://doi.org/10.2135/cropsci1976.0011183X001600050039x
Weston DT, Horsley RD, Schwarz PB, Goos RJ (1993) Nitrogen and planting date effects on low-protein spring barley. Agron J 85:1170–1175
Wimmer V, Albrecht T, Auinger H-J, Schön C-C (2012) synbreed: a framework for the analysis of genomic prediction data using R. Bioinformatics 28:2086–2087. https://doi.org/10.1093/bioinformatics/bts335
Wingler A, Purdy SJ, Edwards S-A, Chardon F, Masclaux-Daubresse C (2010) QTL analysis for sugar-regulated leaf senescence supports flowering-dependent and -independent senescence pathways. New Phytol 185:420–433. https://doi.org/10.1111/j.1469-8137.2009.03072.x
Woo HR, Kim HJ, Nam HG, Lim PO (2013) Plant leaf senescence and death - regulation by multiple layers of control and implications for aging in general. J Cell Sci 126:4823–4833. https://doi.org/10.1242/jcs.109116
Woo HR, Masclaux-Daubresse C, Lim PO (2018) Plant senescence: how plants know when and how to die. J Exp Bot 69:715–718. https://doi.org/10.1093/jxb/ery011
Wu K, Zhang L, Zhou C, Yu C-W, Chaikam V (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59:225–234. https://doi.org/10.1093/jxb/erm300
Yang J, Udvardi M (2018) Senescence and nitrogen use efficiency in perennial grasses for forage and biofuel production. J Exp Bot 69:855–865. https://doi.org/10.1093/jxb/erx241
Yang DH, Kwak KJ, Kim MK, Park SJ, Yang KY, Kang H (2014) Expression of Arabidopsis glycine-rich RNA-binding protein AtGRP2 or AtGRP7 improves grain yield of rice (Oryza sativa) under drought stress conditions. Plant Sci 214:106–112. https://doi.org/10.1016/j.plantsci.2013.10.006
Yang X, Yang M, Deng H, Ding Y (2018) New era of studying rna secondary structure and its influence on gene regulation in plants. Front Plant Sci 9:671. https://doi.org/10.3389/fpls.2018.00671
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421. https://doi.org/10.1111/j.1365-3180.1974.tb01084.x
Ziyatdinov A, Vázquez-Santiago M, Brunel H, Martinez-Perez A, Aschard H, Soria JM (2018) lme4qtl: linear mixed models with flexible covariance structure for genetic studies of related individuals. BMC Bioinform 19:68. https://doi.org/10.1186/s12859-018-2057-x
Acknowledgements
The authors thank Huseyin Yavas for technical assistance.
Funding
This project was supported by funding from the Montana Agricultural Experiment Station (MAES) and the Montana Wheat and Barley Committee.
Author information
Authors and Affiliations
Contributions
BA performed laboratory experiments, analyzed data, and wrote the manuscript; DM performed field studies for experiment 2 and collected agronomic data; DP and TB contributed agronomic data for experiment 1; JL guided and assisted BA in statistical analyses of both experiments; TH provided USDA barley core; JS and AF designed and guided the research and assisted BA with manuscript preparation. The manuscript was reviewed and approved by all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors have read the manuscript and approved its publication.
Additional information
Communicated by Andreas Graner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Alptekin, B., Mangel, D., Pauli, D. et al. Combined effects of a glycine-rich RNA-binding protein and a NAC transcription factor extend grain fill duration and improve malt barley agronomic performance. Theor Appl Genet 134, 351–366 (2021). https://doi.org/10.1007/s00122-020-03701-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03701-1