Abstract
Key message
A novel recessive bacterial blight resistance locus designated as a xa-45(t) was identified from Oryza glaberrima accession IRGC 102600B, transferred to O. sativa and mapped to the long arm of chromosome 8 using ddRAD sequencing approach. The identified QTL spans 80 kb region on Nipponbare reference genome IRGSP-1.0 and contains 9 candidate genes. An STS marker developed from the locus LOC_Os08g42410 was found co-segregating with the trait and will be useful for marker-assisted transfer of this recessive resistance gene in breeding programs.
Abstract
Bacterial blight, caused by Xanthomonas oryzae pv. oryzae, is one of the major constraints of rice productivity in Southeast Asia. In spite of having 44 bacterial blight resistance genes from cultivated rice and wild species, the durability of resistance is always at stake due to the continually evolving nature of the pathogen and lack of suitable chemical control. Here, we report high-resolution genetic mapping of a novel bacterial blight resistance gene tentatively designated as a xa-45(t) from an introgression line derived from Oryza glaberrima accession IRGC 102600B. This introgression line was crossed with the susceptible rice indica cultivar cv. Pusa 44 to generate F2 and F2:3 populations for inheritance and mapping studies. The inheritance studies revealed the presence of single recessive locus controlling resistance to the Xanthomonas pathotype seven. A high-density linkage map was constructed using double-digest restriction-associated DNA sequencing of 96 F2 populations along with the parents. The QTL mapping identified a major locus on the long arm of rice chromosome 8 with a LOD score of 33.22 between the SNP markers C8.26737175 and C8.26818765. The peak marker, C8.26810477, explains 49.8% of the total phenotypic variance and was positioned at 202.90 cM on the linkage map. This major locus spans 80 kb region on Nipponbare reference genome IRGSP-1.0 and contains 9 candidate genes. A co-segregating STS marker was developed from the LOC_Os08g42410 for efficient transfer of this novel gene to elite cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacterial blight (BB), caused by Xanthomonas oryzae pv. Oryzae (Xoo), is an important disease of rice (Oryza sativa L.) causing significant yield losses annually in irrigated and rain-fed lowland ecologies throughout Asia (Mew 1987; Chen et al. 2011). In general, the yield losses due to bacterial blight disease range from 20 to 30%, but in epidemic years, it may cause up to 50% reduction in yield (Ou 1985; Noh et al. 2007; Gu 2008). Breeding for bacterial blight resistance with deployment of R genes in high-yielding rice cultivars is the key economical and environment-friendly approach for the management of this disease. Globally, 44 different R genes (Xa/xa) conferring resistance against Xoo have been identified (Nino-Liu et al. 2006; Cheema et al. 2008; Wang et al. 2009; Kim et al. 2015; Zhang et al. 2015; Kim 2018; Kim and Reinke 2019). Out of these, 17 genes, namely xa5, xa8, xa13, xa15, xa19, xa20, xa24(t), xa25, xa26(t), xa28(t), xa31(t), xa32(t), xa33(t), xa34, xa41(t), xa42 and a 44 (t), are recessive and the rest is dominant (Chen et al. 2011; Liu et al. 2011; Busungu et al. 2016; Kim and Reinke 2019). A total of nine genes have been cloned, of which six (Xa1, Xa3/Xa26, Xa10, Xa21, Xa23 and Xa27) are dominant and three are (xa5, xa13 and xa25) recessive (Iyer and McCouch 2004; Kim et al. 2015). High-resolution mapping and candidate gene analysis approach plays a crucial role in map-based cloning projects in rice (Ashikari et al. 1999; Blair et al. 2003). The bacterial blight resistance genes are mostly nucleotide-binding site–leucine-rich repeat (NBS-LRR) genes and the cell surface pattern recognition receptors (Song et al. 1995; Yoshimura et al. 1998; Sun et al. 2004). Out of three cloned recessive genes, the xa5 encodes a mutated subunit of the basal transcription factor IIA on chromosome 5 (TFIIAg5) (Iyer and McCouch 2004; Jiang et al. 2006), whereas xa13 and xa25 belong to the SWEET multiple gene family (Chu et al. 2006; Liu et al. 2011). Of the dominant genes, Xa1 encodes an NB-LRR-type protein (Yoshimura et al. 1998), whereas Xa3/Xa26 and Xa21 encodes an LRR receptor kinase-like protein (Song et al. 1995; Sun et al. 2004; Xiang et al. 2006). The other three dominant genes, viz. Xa 10, Xa23 and Xa27, encode for executor R protein (Gu et al. 2005; Tian et al. 2014).
The genus Oryza comprises two distinct cultivated, viz. O. sativa L. (Asian rice) and O. glaberrima Steud. (African rice) and 22 wild species (Brar and Singh 2011). The O. glaberrima is the native cultivated rice of economic importance in West Africa and has many useful traits such as tolerance to drought, soil acidity, iron toxicity, phosphorus deficiency, as well as to biotic stresses such as resistance to African gall midge, nematodes, bacterial blight, blast, etc. (Sanchez et al. 2013; Tanaka et al. 2014; Sikirou et al. 2018), but are very poor yielder. The major obstacle in the transfer of useful variability from O. glaberrima to O. sativa is the presence of reproductive barriers which cause interspecific hybrid sterility (Sano 1986; Heuer and Miézan 2003; Li et al. 2018). The phenomenon of segregation distortion (SD) was well documented in wide crosses of many crop species (Zhang 2007; Li et al. 2012; Yang et al. 2012; Wang et al. 2013; Xu et al. 2004). In rice, SD is frequently observed in inter-subspecies crosses (Maekawa et al. 1981; Lin et al. 1992; Xu et al. 1997; Zhao et al. 2006; Reflinur et al. 2014) and inter-species crosses (Causse et al. 1994; McCouch et al. 1988; Lorieux et al. 2000; Koide et al. 2012). However, with the advancement in genomics and phenomics facility, it became possible to utilize these species in rice improvement programs. One such example is the development of NERICA (New Rice for Africa) varieties with increased rice productivity in Africa by combining the high yielding quality of Asian rice with locally adapted African rice (Sie 2008; Somado et al. 2008).
Though many bacterial blight resistance genes have been deployed in resistance breeding programs, breakdown of resistance in many rice cultivars and in near isogenic lines containing bacterial blight resistance genes either singly or in combination was observed. This could be either due to a narrow resistance spectrum of deployed gene or rapid evolution in the virulence behavior of Xoo pathotypes prevalent in a particular region. Mega rice cultivars of Punjab like PR114, PR116, PR120, PAU 201 and many others released variety became susceptible to one or the other virulent strain of Xoo. The probable explanation could be the presence of a high degree of pathogenic variation in Xanthomonas, often leading to the breakdown of resistance (Vera Cruz et al. 2000; Suh et al. 2013). In the Punjab State of India alone, 10 pathotypes have evolved over a very short period (Lore et al. 2011; Neelam et al. 2016). None of the previously reported genes currently exhibit resistance to all the prevalent pathotypes in India. Thus, expanding genetic diversity by identifying novel resistance genes from both cultivated rice gene pool as well as wild relatives of rice and subsequently their deployment in breeding programs can help in developing rice cultivars with durable BB resistance to Xoo (Suh et al. 2009; Kumar et al. 2012).
Punjab Agricultural University (PAU), India, is maintaining an active collection of around 1600 wild species germplasm received from the International Rice Research Institute, Los Baños, Philippines, and ICAR-National Rice Research Institute (ICAR-NRRI), Cuttack, India (formerly CRRI, Central Rice Research Institute). These accessions were screened against Xoo pathotypes prevalent in Punjab State of India, over several years, starting from 2001 (Vikal et al. 2007; Neelam et al. 2016). Among these, thirteen accessions of O. glaberrima were found resistant to seven Xoo pathotypes (Vikal et al. 2007), prevalent in Punjab State of India (Lore et al. 2011), and one accession was used for the development of mapping population. Here, we report high-resolution mapping of bacterial blight resistance gene designated as xa-45(t) and development of markers for marker-assisted transfer of this recessive resistance gene into other backgrounds.
Materials and methods
Plant material and population development
The O. glaberrima accession IRGC102600B (single plant selection from the original accession IRGC102600) from Liberia procured from IRRI, Philippines, showed a high level of resistance to all the seven Xoo pathotypes prevalent in Punjab (Vikal et al. 2007). The accession was crossed as male to O. sativa cv. Pusa44, a widely adapted genotype in Punjab, which is high yielding, lodging tolerant, but susceptible to bacterial blight (Fig. 1). The F1 was made by crossing Pusa44 with O. glaberrima. The F1’s were completely male sterile; therefore, the BC1F1 seeds were generated by mass pollination of F1 plants with Pusa44 pollen, in isolation, without clipping of florets. This was done because clipping of the florets of the F1 plants leads to 100% shattering of florets within 24 h of pollination. Pollination in each F1 plant was continued for 4–5 days to ensure sufficient seed set. All the BC1F1 showed partial to complete pollen sterility. All the BC1F1 plant was bagged to generate BC1F2 seeds. Out of more than 100 BC1F1 plants, only eight BC1F1 plants showed partial fertility and set seeds upon selfing. The BC1F2 populations of the eight plants were inoculated with Xoo pathotype PbXo7 and two progenies showed segregation for BB resistance and all others were susceptible. Selfed seeds from each BC1F2 plant were collected from one population and again screened against Xoo pathotype PbXo7 to confirm the reaction of the BC1F2 plants and ascertaining their genotype. Since pollen sterility was still a concern, the progenies homozygous for resistance to BB were further backcrossed two times to Pusa 44 and selection continued to generate a stable and agronomically desirable BC3F8 introgression line (Pusa 44/O. glaberrima IRGC102600B//3* Pusa 44), designated as IL 274 (Vikas 2010). The IL 274 was again crossed to Pusa 44 to generate F2 and F2:3 population for mapping and inheritance studies.
Screening with X. oryzae pv. oryzae pathotype PbXo-7
Bacterial culture of Xanthomonas pathotype PbXo7 (Lore et al. 2011), originally isolated from a single colony, was streaked on Walkimoto medium (10 g sucrose, 5 g peptone and 20 g agar, total volume made up to 1000 ml using distilled water) slants in test tubes under aseptic conditions. After streaking, the cultures were incubated for 72 h at 28 °C. The bacterial growth was scrapped and homogenized by vigorous shaking. The bacterial suspension was standardized to the inoculum density of 109 cells/ml. This virulent culture was used for inoculation of the parents (IL 274 and Pusa 44), F1 plants, F2 and F2:3 progenies at maximum tillering stage following ‘clip inoculation technique’ (Kauffman et al. 1973) during the kharif crop season 2014 to 2016.
Disease assessment
The disease data were recorded 14 days after inoculation (DAI). The disease reaction on the basis of lesion length (cm) was classified according to the scale given by Cottyn and Mew (2004). The lesion length up to 5.0 cm (disease score 1–3) was classified as resistant, between 5 and 10 cm (disease score 3–5) as moderately resistant, 11–15 cm (disease score 5–7) as moderately susceptible and greater than 15 cm (disease score 7–9) as susceptible. For the inheritance study, at least five leaves from each of 312 F2 individual plants were inoculated and the disease score was recorded using the standard scoring system. Seeds were harvested from individual F2 plants and planted as F3 progeny with 15 plants per progeny. Five leaves of each plant were inoculated with pathotype PbXo-7 and lesion length recorded after 14 days of inoculation. The average of the lesion length of five leaves was used for analysis. The Chi-square test was used to test the goodness of fit for ascertaining the number of genes governing the BB resistance.
DNA extraction and SNP genotyping
Large-scale DNA extraction protocol was followed to isolate high-molecular-weight DNA from 25- to 30-day-old field-grown F2 plants using CTAB (cetyl trimethyl ammonium bromide) method. DNA was quantified using a spectrophotometer (Eppendorf Biophotometer). The quality and integrity of genomic DNA were also checked on 0.8% agarose gel and normalized by adding Tris EDTA buffer. 1000 ng DNA from each F2 plant and the parents was used for double-digest restriction site-associated DNA sequencing (ddRAD-seq). The ddRAD sequencing was outsourced to SciGenom (www.scigenom.com). Briefly, the Illumina HiSeq 2000/2500 sequencing technology was used for fetching out SNPs. The enzymes used for double digestion of the samples are SphI and MlucI. Approximately, 100–120 MB data per sample was obtained for further analysis.
Linkage map construction
High-quality SNP markers with less than 5% missing data were retained for the construction of a genetic map. The Chi-square test was performed to exclude markers with segregation distortion or χ2 > 10. Linkage groups (LGs) were generated with RECORD (REcombination Counting and ORDering) software (Van Os et al. 2005), using the Kosambi mapping function. Linkage groups (LGs) were drawn using MapChart V2.2 (Voorrips 2002). The markers with similar segregation patterns (based on a similarity of ≥ 0.95) were placed in the same genetic bin and only one of the markers was retained.
The composite interval mapping (CIM) function of winQTL cartographer software (Wang et al. 2012) was used for mapping QTL(s) for BB resistance with a window size of 10 cM. The stepwise threshold logarithm of the odds (LOD) scores for the QTL was calculated based on 1000 permutations at P ≤ 0.05 (Churchill and Doerge 1994). Markers used as cofactors in the CIM model were selected by forward and backward regression analyses. The proportion of observed phenotypic variance attributable to the QTL was estimated by the coefficient of determination (R2) using maximum likelihood for composite interval mapping.
Markers development
To enrich the QTL region, we have used available SSR markers within the defined region (International Rice Genome sequencing Project 2005). Further, we have designed new primers harboring SSRs in the whole 80 kb region on chromosome 8 for getting more number of polymorphic markers. In addition to this, gene specific primers were also designed from the nine putative candidate genes within the QTL region. In total, 39 primer pairs were designed for saturating the identified region. The markers were amplified in a set of 10 μl PCR reactions containing 60 ng of genomic DNA, 10 pmol of each primer, 2.5 mM, each of the four dNTPs, 1 units of Taq DNA polymerase, 1X PCR buffer and 1.5 mM MgCl2. Reactions were performed in a thermal cycler (Eppendorf), programmed at 94 °C for 5 min, followed by 35 cycles of 1 min at 94 °C, 1 min at 55 °C and 1 min at 72 °C with a final extension of 7 min at 72 °C. PCR amplified products were resolved in 2.5% agarose gel, and individual alleles were scored by comparing to parental alleles.
Sequencing of xa13 gene (SWEET11) in IRBB13, IL274 and Pusa44
The Oryza sativa Japonica sequence (LOC4346153) was used for designing the overlapping primers from position 26,725,954–26,728,807 bp covering a total length of 2853 bp on reference sequence. The amplicons were purified using Wizard® SV 96 PCR clean-up/Gel extraction kit from Promega, USA, as per manufacturer’s protocol. Sequencing reaction was performed using ABI Big-dye Terminator v3.1 chemistry and sequenced using ABI Sequencer 3730XL. Hi-fidelity long-read DNA polymerase (Phusion Taq) from Promega, USA, was employed to obtain the required amplicon size. A minimum of three replications was carried out for the confirmation of single nucleotide polymorphism (SNPs).
Results
Inheritance of bacterial blight resistance using BC1F2 population
The disease reaction of Pusa 44, O. glaberrima 102600B and F1 of the cross Pusa44/O. glaberrima 102600B is shown in Fig. 2. The susceptible reaction of the F1 plants indicated the recessive nature of BB resistance gene in O. glaberrima 102600B. Further, the BC1F1 progeny was inoculated with Xoo pathotype PbXo7. All the BC1F1 plants were susceptible, thereby confirming the recessive nature of BB resistance. Only two of the BC1F2 progeny designated as 532 and 535 with normal segregation for BB was used for inheritance studies. In population 532, out of 129 plants, only 27 were resistant and 102 susceptible, whereas in population 535, out of 191 plants, 51 were resistant and 140 susceptible (Table 1). Assuming single gene segregation, the Chi-square values were nonsignificant for both the populations with P values < 0.29 and < 0.59 for population 532 and 535, respectively. The population of 535 was tall and leafy and had a high level of sterility and shattering; hence, BC1F3 progenies of this population could not be tested, but the majority of the plants in a population 532 set seed. A total of 129 BC1F3 progenies of this population were screened against Xoo pathotype PbXo7 during 2006. In each progeny, 29 plants were inoculated and data recorded as homozygous resistant (HR), homozygous susceptible (HS) or segregating (Seg). Of the 129 progenies tested, 27 were homozygous resistant, 35 were homozygous susceptible and 67 showed segregation. The Chi-square value fitted to the expected for 1:2:1 HR/Seg/HS (Table 1) with P value < 0.55. This confirmed that the bacterial blight resistance in O. glaberrima 102600B was governed by a single recessive gene.
Mapping of bacterial blight resistance using BC1F2 population
Population of 535 was initially used for parental polymorphism and bulked segregant analysis. From the rice linkage map (Temnykh et al. 2001), parental polymorphism and BSA were carried out using 159 SSR markers spanning all the twelve linkage groups. Out of 159 primers tested, 86 showed polymorphism between the parents. The level of polymorphism between Pusa44 and O. glaberrima 102600B varied from 20.0% in chromosome 10–71.4% in chromosome 5, with an overall polymorphism of 54.0% (Table S1). Segregation distortion was observed for several markers throughout the genome where only one parent band was observed in both resistant and susceptible bulks (Figure S1); hence, this population was discontinued for gene mapping studies. The introgression of O. glaberrima allele was observed on chromosome 5 and 6 but was unable to map the gene, primarily due to segregation distortion and a smaller number of markers (Figure S2). We continued further selfing and backcrossing to generate several populations, including one BC1F6:BC2F10 lines.
Inheritance of bacterial blight resistance using IL 274
In order to map the BB resistance gene, we attempted a new cross between O. sativa cv. Pusa 44 with the BC3F8 introgression line IL 274 (Pusa 44/O. glaberrima IRGC102600B//3* Pusa 44). The F1 plant of the above-said cross was susceptible when inoculated with Xanthomonas pathotype PbXo7. Out of 312 F2 individuals, 221 plants were found susceptible and 91 plants resistant (Table 2). The segregation ratio of susceptible to resistant plants agreed with 3: 1 segregation ratio of the single recessive gene for BB resistance in the introgression line. Out of 312 F2:3 progeny, 74 were homozygous resistant, 158 segregating and 80 homozygous susceptible, which fitted well to the expected 1:2:1 segregation (χ 2c = 0.271, χ 20.75,2 = 0.575). Thus, it could be deduced that the gene governing BB resistance in the introgression line IL274 is governed by a single recessive gene. The phenotypic data on bacterial blight were also recorded on F4 and F5 progenies in consecutive years 2017 and 2018 (Fig. 3).
Construction of linkage map and segregation distortion
In total, 19,458 SNP markers were obtained with ddRAD-sequencing (Table 3). The SNP markers with indel or heterozygote SNP in either of the parents were removed, and only SNPs in the homozygous state (AA, CC, TT and GG) were used for scoring the polymorphism between O. sativa cv. Pusa 44 and the introgression line IL274. With this approach, we were left with only 1754 polymorphic SNPs between the parents for further linkage analysis. The SNPs markers with distorted segregation were removed using Chi-square test. A total of 652 markers were thus used for construction of the linkage map using RECORD. The genetic map spanned a total of 2426.17 cM on 12 chromosomes of rice except for 7, 10 and 12, where all the identified polymorphic markers showed a complete deviation from the expected Mendelian frequencies. Only two markers mapped on rice chromosome 1 and 3 after the Chi-square test. In most of the cases, the markers were skewed toward the introgression line allele (Table 4). The genetic length of the smallest LG was for chromosome 3 (6.14 cM), and the largest was on chromosome 11 (847.86 cM).
QTL analysis and predicted genes
Composite interval mapping with 652 SNP markers revealed a sharp peak at the SNP marker interval C8.26737175 and C8.26818765 at LOD score 33.22 indicating major effect QTL (Figs. 4, 5) on chromosome 8. The peak marker, C8.26810477, was positioned at 202.90 cM on linkage map and explains 49.8% of the total phenotypic variance. This marker interval spans a physical distance of 80 kb on the Nipponbare reference genome IRGSP v1 (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/). The QTL region flanked by the two SNPs markers C8.26737175 and C8.26818765 contains nine predicted genes (Table 5) including seven genes with assisted functions, one hypothetical protein and one expressed protein.
Markers development
Out of 39 primer pairs designed within the identified QTL region, only one of the primer pair designed from the LOC_Os08g42410 showed polymorphism between the parents. This marker was analyzed on F5 population (273 plants). This marker is co-segregating with the phenotype (Fig. 6, Table 6). Five recombinant plants (3581-40-3-3, 3581-72-3-3, 3581-93-3-3 3581-193-3-3 and 3581-272-3-3) were observed with this marker. Our observation indicated that the location of the xa-45(t) (QTL location, chr8: 26737175…26818765) is fairly close to the position of xa13 (LOC_Os084346153, chr8:26725954…26728807), another cloned recessive bacterial blight resistance gene on chromosome 8. Therefore, we also applied the STS marker for xa13 (F-GGCCATGGCTCAGTGTTTAT, R-GAGCTCCAGCTCTCCAAATG) to the same F5 population. This marker is also found as co-segregating with the trait (Figure S3). However, 11 recombination points were observed between the xa-45(t) marker and the xa13 marker on the whole population. For the individuals 3581-13-3-3, 3581-93-3-3, 3581-193-3-3, 3581-269-3-3, 3581-272-3-3 and 3581-273-3-3 both of the markers showed opposite results, i.e., with one marker they are showing resistant genotype scores and with another it is the susceptible genotype score (Table S2). The individuals 3581-129-3-3, 3581-235-3-3, 3581-250-3-3, 3581-267-3-3 and 3581-268-3-3 had a resistant genotype score (B) converted to heterozygote (H) with either of the markers. This also provides an indication that both of the genes are different.
Comparative sequence analysis of xa13 gene with other diploid wild species of rice
We performed a comparative sequence analysis of the xa13 gene from nine diploid wild Oryza species and three of the cultivated Oryza species (Fig. 7). The genomic sequence of the cloned xa13 gene, i.e., LOC4346153 (SWEET11), was used as reference sequence for the analysis. A number of polymorphic sites were found at the nucleotide positions 232, 358, 359, 442, 694 and 728 between the reference sequence and O. glaberrima assembly. Phylogenetic analysis demonstrated high gene collinearity across the AA genome Oryza species. The O. rufipogon, O. barthii and O. nivara share a common clade with O. sativa japonica group, whereas O. glumaepatula and O. glaberrima fall closer to O. sativa. Two of the AA genome species, O. longistaminata and O. meridionalis, showed less similarity with the reference sequence as well as among themselves. The O. punctata (BB) genome and O. brachyantha (FF) genome did not grouped, indicating non-collinearity of xa13 gene across other diploid genome species. On contrary, the comparative analysis of 80 kb region containing a xa-45(t) candidate genes in Nipponbare reference sequence aligned with the Chr8 of O. glaberrima genome (nucleotide position 26737175–26818765) showed a higher degree of genomic rearrangement. This was evident by several polymorphic sites, including several INDELS of 3-5 bp (Figure S4).
Comparison of xa13 gene sequence in IRBB13, IL274 and Pusa 44
We analyzed the xa13 gene sequence (from two of the resistant lines, viz. a NIL containing xa13 (IRBB13), introgression line (IL274) and the susceptible check, Pusa44. To our surprise, this gene is having similar sequences in all the three lines except at six nucleotide positions when compared to the reference sequence (Figure S4). At the nucleotide positions 1148 and 1586, the reference sequence and Pusa44 had the nucleotide C as compared to G in IL274 and IRBB13, whereas at the nucleotide position 2223, G/A transition was seen between these pairs. The IRBB13 and IL274 differ at the nucleotide position 2238(A/C), whereas singletons in Pusa44 were observed at nucleotide positions 1964 (G/C) and 2001(T/C). However, having similar genomic sequence does not always correlate well with the expression of the gene. There might be a difference in the expression level or it may be induced after inoculation with the Xanthomonas in resistant genotypes. Another possible explanation could be differences in the promoter region of susceptible allele as compared to the resistant allele. Similar findings were also reported in the case of the bacterial blight resistance gene Xa23 and Xa27. In the case of Xa23, the susceptible allele has an identical open reading frame as that of resistant allele but it lacks the TALE binding elements in the promoter region for Avrxa23, whereas the Xa27-resistant allele (IRBB27) and the susceptible allele xa27 in IR24 contain the identical coding sequences but the resistant allele expression induced only after inoculation with Xanthomonas (Wang et al. 2015). Further, investigation is needed to decipher the mechanism governing resistance in the xa-45(t) to conclude that if it’s only an allele of the xa13 or a new gene.
Phenotypic evaluation of xa45(t) and xa13 against 10 key pathotypes of X. oryzae pv. oryzae
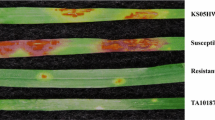
To date, 10 pathotypes of XOO oryzae pv. oryzae have been identified in Northern India (Lore et al. 2011, 2013). Since xa13 locus is co-located with xa-45(t) on chromosome 8, we investigated the disease reaction of the introgression line (IL274) carrying bacterial blight resistance gene xa45(t), IRBB13 carrying xa13 gene, resistant donor O. glaberrima acc. 102600B, susceptible recipient parent Pusa 44 and susceptible check IR 24 against 10 key pathotypes of Xoo (Table 7, Fig. 8). The IL 274 and the donor O. glaberrima acc. 102600B showed resistant reaction to all the 10 pathotypes of Xoo; however, IRBB 13 (xa13) showed susceptible reaction to the pathotype PbXo-8. The cultivar Pusa 44 and IR24 (Xa18) were susceptible to all the tested pathotypes, thus confirming the novelty of the new gene.
Discussion
The durability of resistance depends upon the prevalence of the pathogen population in a particular area. Pathogen Xanthomonas is highly variable and showed diverse reaction to rice cultivars possessing genetic resistance to bacterial blight under field conditions. The pathogen population has been divided into many groups in different countries based on local rice cultivars/near isogenic lines (Adhikari et al. 1999; Noda et al. 2001; Lore et al. 2011). The pathogen population from Northern India are comparatively more aggressive and became virulent to most of the single BB resistance genes such as Xa1, Xa3, Xa4, xa5, Xa7, xa8, Xa10, Xa11, xa13, Xa21, Xa23, Xa38 and even some of the Xanthomonas strains showing virulence on pyramided genes such as Xa4 + xa5, Xa4 + Xa21, xa5 + Xa21 and xa13 + Xa21 in India (Lore et al. 2011). In the present study, we identified and mapped a new bacterial blight resistance gene from O. glaberrima on the long arm of chromosome 8. None of the bacterial blight resistance gene is reported from O. glaberrima previously. It is important to note that the most widely used resistance gene xa13 shows susceptibility to Xoo pathotype PbXo-8, whereas xa-45(t) showed resistance against all the 10 pathotypes present in North India. This infers that the addition of this new gene or allele to the bacterial blight resistance gene pool will definitely broader the genetic base and hence will be helpful in the creation of durable resistance.
Segregation distortion
In our analysis, we observed that the markers were distorted either toward one of the alleles from O. sativa/O. glaberrima. The presence of sterility barriers between O. sativa and O. glaberrima in early hybrid generations is evident in many earlier studies (Sano et al. 1979; Sano 1986; Ikehashi and Araki 1986; Pham and Bougerol 1993; Doi et al. 1998; Lorieux et al. 2000; Li et al. 2004; Reflinur et al. 2014). Till date, approximately ten hybrid sterility loci as gamete eliminator or pollen killer between O. sativa and O. glaberrima were identified (Li et al. 2018). The introgression line alleles were over represented on chromosome 2, 3, 6 and 10. This may be attributed to the presence of gametophytic and sterility genes on these chromosomes (Lin et al. 1992; Xu et al. 1997) that might lead to the partial female gamete abortion. Approximately, 60% of the markers were skewed toward introgression line allele on chromosome 2. Maekawa et al. (1981) reported the presence of the gametophyte gene (ga-6) on chromosome 2 causing significant deviation of the markers from the normal Mendelian ratio. Another hybrid sterility gene S29 (t) on chromosome 2 was also observed to cause female gamete abortion in remote cross of rice (Zhu et al. 2005). Liu et al. (1997) mapped two of the minor genes on chromosome 2 and 12 which jointly cause reduction in the female fertility even in the presence of wide- compatibility genes. Segregation distortion of the marker locus due to the presence of gametophyte genes on chromosomes 6 (ga-1) was reported way back by Iwata et al. (1964) and on chromosome 3 (ga-2 and ga-3) by Nakagahra et al. (1972, 1986). It is noteworthy that the region around waxy locus on chromosome 6 was reported as distortion hotspot showing one-locus sporo-gametophytic distortion (Sano 1990; Lorieux et al. 2000). According to this model, the Sa gametes (O. sativa genotype) will abort in heterozygous F1 plants and only S gametes (O. glaberrima genotype) will be represented in F2 population (Sano et al. 1979; Heuer and Miézan 2003). Another pollen killer gene S1 is also located on chromosome 6. This gene could behave as gamete eliminator or pollen killer depending on the activity of modifiers and genetic background (Sano 1990). They observed higher female sterility in plants carrying distal segments of chromosome 6 of O. glaberrima due to preferential abortion of female gametes (O. sativa alleles). Aluko et al. (2004) also observed skewness of the markers toward O. glaberrima allele specifically on chromosomes 3 and 6 while studying grain quality traits in the interspecific cross between O. sativa and O. glaberrima. Similarly, transgressive segregation has been reported in crosses of O. sativa with O. rufipogon as well, due to a gene block of sterility genes on chromosome 6 (Matsubara et al. 2003). Lin et al. (1992) also observed strong distortion of the markers on chromosomes 3, 7, 8, 11 and 12 in a hybrid between indica and japonica crosses. However, as per our observation, the O. sativa alleles were in higher frequency of chromosome 11. This could owe to the fact that the sterility/gametophytic gene’s behavior modifies due to the existence of multiple alleles or interaction among them including the presence of neutral alleles.
Putative candidate genes
Within the 80 kb region, 9 putative genes were predicted (Table 5) using rice genome annotation project database (http://rice.plantbiology.msu.edu). The probable function of these genes is discussed here. The Zinc finger, DHHC domain-containing protein has been reported in brachypodium, maize, sorghum to be involved in palmitotransferase function. This gene family has been reported to play a role in reversible posttranslational lipid modification of long chain fatty acid, usually the 16-carbon palmitate, covalently attaches to a cysteine residue(s) throughout the protein via a thioester bond. They regulate various biological processes during growth and development and reproduction (Gleason et al. 2006; Xiang et al. 2010; Yuan et al. 2013; Li and Qi 2017). The genome-wide transcriptome analysis revealed enhanced expression of this gene against bacterial blight disease (Jung et al. 2014). Mitochondrial import inner membrane translocase subunit Tim is intermembrane chaperone that participates in the import and insertion of some multi-pass transmembrane proteins into the mitochondrial inner membrane in arabidopsis, brachypodium, maize, sorghum and rice (Werhahn et al. 2001; Lister et al. 2004).
The glycerophosphoryl diester phosphodiesterase family protein is basically responsible for maintaining cellular phosphate homeostasis under phosphate starvation and organization of primary cell wall (Rest et al. 2004; Cheng et al. 2011). The no apical meristem protein is a NAC4 domain-containing protein in O. sativa japonica group, whereas it is putatively un characterized protein in O. sativa indica group. The NAC family transcription factors play an important role in the regulation of the various plant physiological process and response to stress. Their involvement in activating defense responses in rice, innate immune responses and host recognition of pathogens and further recognition triggered early signaling pathways is well characterized (Skamnioti and Gurr 2009; Valent and Khang 2010). The transketolase is involved in biological processes such as acetyl-CoA biosynthetic process from pyruvate, protein phosphorylation and also act as precursors for producing thymine in C3 cycle. The thymine biosynthesis is activated during plant response to abiotic and biotic stresses, including inoculation with Xanthomonas (Wang et al. 2006). Its molecular function includes ATP binding, protein serine/threonine kinase activity and pyruvate dehydrogenase activity. The protein serine/threonine kinases are well known to be involved in signal transduction and defense mechanism either directly as resistance genes or downstream components of defense pathways (Martin et al. 1993; Meskiene and Hirt 2000; Frye et al. 2001; Afzal et al. 2008). The membrane-associated DUF588 domain-containing protein may have catalytic activity, and these proteins are predicted to contain 3 or 4 transmembrane helices. In rice, only three BBX genes (CCT/B-box Zinc finger) have been identified and known to be involved in the regulation of flowering time (Huang et al. 2012). Based on the above-described functions of putative candidate genes in the QTL region, LOC_Os08g42370, LOC_Os08g42400 and LOC_Os08g42410 are worth further investigation in their role in response to bacterial blight infection.
PCR-based markers for marker-assisted selection
A PCR-based marker was developed from the LOC_Os08g42410 for marker-aided transfer of this recessive R gene. This can be of immediate use in rice bacterial blight breeding programs as being the recessive gene; desirable phenotypes are not expressed in heterozygotes. Being closely located to xa13, an STS marker for xa13 could also be used for MAS of a xa-45(t). However, the bacterial blight resistance provided by the xa-45(t) has a wider spectrum than the xa13 gene which indicates presence of superior alleles from O. glaberrima. This new gene was incorporated in mega rice variety Pusa 44 and it was released as new variety PR127 in Northern India during the year 2018 (POP 2018).
Conclusion
In summary, we identified a novel gene or an allele from O. glaberrima of the existing xa13 gene from O. sativa, conferring broad spectrum resistance to all the ten Xoo pathotypes of Northern states of India. Though, further confirmation is needed with the allelic test. A cross between NIL carrying xa13 (IRBB 13) and IL274 will be carried out in the next kharif season to validate the concept. In addition, we also developed co-dominant marker for the transfer of this recessive gene to the elite cultivar for the deployment of bacterial blight resistance in breeding programs.
References
Adhikari TB, Basnyat RC, Mew TW (1999) Virulence of Xanthomonas oryzae pv. Oryzae on rice lines containing single resistance genes and gene combinations. Plant Dis 83:46–50. https://doi.org/10.1094/PDIS.1999.83.1.46
Afzal AJ, Wood AJ, Lightfoot DA (2008) Plant receptor-like serine threonine kinases: roles in signalling and plant défense. Mol Plant Microbe Interact 21:507–517. https://doi.org/10.1094/MPMI-21-5-0507
Aluko G, Martinez C, Tohme J, Castano C, Bergman C, Oard JH (2004) QTL mapping of grain quality traits from the interspecific cross Oryza sativa × Oryza glaberrima. Theor Appl Genet 109:630–639. https://doi.org/10.1007/s00122-004-1668-y
Ashikari M, Wu JZ, Yano M, Sasaki T, Yoshimura A (1999) Ricegibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc Natl Acad Sci USA 96:10284–10289
Blair MW, Garris AJ, Iyer AS, Chapman B, Kresovich S, McCouch SR (2003) High resolution genetic mapping and candidate gene identification at the xa5 locus for bacterial blight resistance in rice (Oryza sativa L.). Theor Appl Genet 107:62–73. https://doi.org/10.1007/s00122-003-1231-2
Brar DS, Singh K (2011) Rice. In: Kole C (ed) Wild crop relatives: genomics and breeding resources. Springer, Berlin, pp 321–365
Busungu C, Taura S, Sakagami JI, Ichitani K (2016) Identification and linkage analysis of a new rice bacterial blight resistance gene from XM14, a mutant line from IR24. Breed Sci 66:636–645. https://doi.org/10.1270/jsbbs.16062
Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu KS, Xiao JH, Yu ZH, Ronald PC, Harrington SE, Second G, McCouch SR, Tanksley SD (1994) Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138:1251–1274
Cheema K, Grewal N, Vikal Y, Sharma R, Lore J, Das A, Singh K (2008) A novel bacterial blight resistance gene from O. nivara mapped to 38 kb region on chromosome 4L and transferred to O. sativa L. Genet Res 90(5):397–407. https://doi.org/10.1017/S0016672308009786
Chen S, Liu X, Zeng L, Ouyang D, Yang J, Zhu X (2011) Genetic analysis and molecular mapping of a novel recessive gene. Theor Appl Genet 122:1331–1338. https://doi.org/10.1007/s00122-011-1534-7
Cheng Y, Zhou W, Sheery NI, Peters C, Li M, Wang X, Huang J (2011) Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J 66:781–795. https://doi.org/10.1111/j.1365-313XOO2011.04538.x
Chu Z, Fu B, Yang H, Xu C, Li Z, Sanchez A, Park YJ, Bennetzen JL, Zhang Q, Wang S (2006) Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor Appl Genet 112:455–461
Churchill G, Doerge R (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cottyn B, Mew TW (2004) Bacterial blight of rice. In: Goodman RM (ed) Encyclopedia of plant and crop science. Marcel Dekker, New York, pp 79–83. https://doi.org/10.1111/jph.12338
Doi K, Taguchi K, Yoshimura A (1998) A new locus affecting high F1 pollen sterility found in backcross progenies of japonica rice and African rice. Rice Genet News Lett 15:146–148. https://doi.org/10.1007/978-3-319-71997-9_20
Frye CA, Tang D, Innes RW (2001) Negative regulation of defense response in plants by a conserved MAPKK kinase. Proc Natl Acad Sci 98:373–378. https://doi.org/10.1073/pnas.98.1.373
Gleason EJ, Lindsey WC, Kroft TL, Singson AW, L’hernault SW (2006) Spe-10 encodes a DHHC–CRD zinc-finger membrane protein required for endoplasmic reticulum/Golgi membrane morphogenesis during Caenorhabditis elegans spermatogenesis. Genetics 172:145–158. https://doi.org/10.1534/genetics.105.047340
Gu K (2008) High-resolution genetic mapping of bacterial blight resistance gene Xa10. Theor Appl Genet 116:155–163. https://doi.org/10.1007/s00122-007-0655-5
Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, Yin Z (2005) R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435:1122–1125
Heuer S, Miézan KM (2003) Assessing hybrid sterility in Oryza glaberrima × O. sativa hybrid progenies by PCR marker analysis and crossing with wide compatibility varieties. Theor Appl Genet 107:902–909. https://doi.org/10.1007/s00122-003-1325-x
Huang J, Zhao X, Weng X, Wang L, Xie W (2012) The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 77(10):e48242. https://doi.org/10.1371/journal.pone.0048242
Ikehashi H, Araki H (1986) Genetics of F1 sterility in remote crosses of rice. In: Rice genetics. Proceedings of the international rice genetics symposium. IRRI, Manila, Philippines, pp 119–130
Iwata N, Nagamatsu T, Omura T (1964) Abnormal segregation of waxy and apoculus coloration by a gametophyte gene belonging to the first linkage group in rice. Jpn J Breed 14:33–39. https://doi.org/10.1270/jsbbs1951.14.33
Iyer AS, McCouch SR (2004) The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol Plant Microbe Interact 17:1348–1354. https://doi.org/10.1094/MPMI.2004.17.12.1348
Jiang GH, Xia ZH, Zhou YL, Wan J, Li DY, Chen RS, Zhai WX, Zhu LH (2006) Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAg1. Mol Genet Genom 275:354–366
Jung KH, Han M, Nguyen VNT, Yoo Y, Nguyen MP, Lee C, Lee SW, Jeon JS (2014) Development of defense signaling pathways against bacterial blight disease in rice using genome-wide transcriptome data. J Agric Sci 6:48–111. https://doi.org/10.5539/jas.v6n7p48
Kauffman HE, Reddy APK, Hsieh SPY, Merca SD (1973) An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep 57:537–541
Kim SM (2018) Identification of novel recessive gene Xa44(t) conferring resistance to bacterial blight races in rice by QTL linkage analysis using an SNP chip. Theor Appl Genet 131:2733–2743. https://doi.org/10.1007/s00122-018-3187-2
Kim SM, Reinke RF (2019) A novel resistance gene for bacterial blight in rice, Xa43(t) identified by GWAS, confirmed by QTL mapping using a bi-parental population. PLoS ONE 14(2):e0211775. https://doi.org/10.1371/journal.pone.0211775
Kim SM, Suh JP, Qin Y, Noh TH, Reinke RF, Jena KK (2015) Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (O. sativa L.). Theor Appl Genet 128:1933–1943. https://doi.org/10.1007/s00122-015-2557-2
Koide Y, Shinya Y, Ikenaga M, Sawamura N, Matsubara K, Onishi K, Kanazawa A, Sano Y (2012) Complex genetic nature of sex-independent transmission ratio distortion in Asian rice species: the involvement of unlinked modifiers and sex-specific mechanisms. Hered 108:242–247. https://doi.org/10.1038/hdy.2011.64
Kumar PN, Sujatha K, Laha GS, Rao KS, Mishra B, Viraktamath BC, Hari Y, Reddy CS, Balachandran SM, Ram T, Madhav MS, Rani NS, Neeraja CN, Reddy GA, Shaik H, Sundaram RM (2012) Identification and fine-mapping of Xa33, a novel gene for resistance to Xanthomonas oryzae pv. oryzae. Phytopathology 102:222–228. https://doi.org/10.1094/PHYTO-03-11-0075
Li Y, Qi B (2017) Progress toward understanding protein S-acylation: prospective in plants. Front Plant Sci. https://doi.org/10.3389/fpls.2017.00346
Li JM, Xiao JH, Grandillo S, Jiang LY, Wan YZ, Deng QY, Yuan LP, McCouch SR (2004) QTL detection for rice grain quality traits using an interspecific backcross population derived from cultivated Asian (O. sativa L.) and African (O. glaberrima S.) rice. Genome 47:697–704. https://doi.org/10.1139/g04-029
Li JQ, Shahid MQ, Feng JH, Liu XD, Zhao XJ, Lu YG (2012) Identification of neutral alleles at pollen sterility gene loci of cultivated rice (Oryza sativa L.) from wild rice (O. rufipogon Griff.). Plant Syst Evol 298:33–42
Li J, Zhou J, Xu P, Deng X, Deng W, He M, Yang Y, Zhang Y, Tao D (2018) Neutral alleles at hybrid sterility loci of Oryza glaberrima from AA genome relatives in Genus Oryza. Breed Sci 68:343–351. https://doi.org/10.1270/jsbbs.18006
Lin SY, Ikehashi H, Yanagihara S, Kawashima A (1992) Segregation distortion via male gametes in hybrids between Indica and Japonica or wide-compatibility varieties of rice (Oryza sativa L.). Theor Appl Genet 84:812–818. https://doi.org/10.1007/BF00227389
Lister R, Chew O, Lee MN, Heazlewood JL, Clifton R, Parker KL, Millar AH, Whelan J (2004) A transcriptomic and proteomic characterization of the arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol 134:777–789. https://doi.org/10.1104/pp.103.033910
Liu KD, Wang J, Li HB, Xu CG, Liu AM, Li XH, Zhang Q (1997) A genome-wide analysis of wide compatibility in rice and the precise location of the S5 locus in the molecular map. Theor Appl Genet 95:809–814. https://doi.org/10.1007/s001220050629
Liu Q, Yuan M, Zhou Y, Li X, Xiao J, Wang S (2011) A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ 34:1958–1969. https://doi.org/10.1111/j.1365-3040.2011.02391.x
Lore JS, Vikal Y, Hunjan MS, Goel RK, Bharaj TS, Raina GL (2011) Genotypic and pathotypic diversity of Xanthomonas oryzae pv. oryzae, the cause of bacterial blight of rice in Punjab state of India. J Phytopathol 159:479–487
Lore JS, Jyoti J, Mangat GS (2013) Evaluation of rice germplasm for multiple disease resistance under artificial inoculation conditions. Indian J Genet Plant Breed 74:670–673. https://doi.org/10.5958/0975-6906.2014.00908.0
Lorieux M, Ndjiondjop MN, Ghesquiere A (2000) A first interspecific Oryza sativa × O. glaberrima microsatellite genetic linkage map. Theor Appl Genet 100:593–601. https://doi.org/10.1007/s001229900061
Maekawa M, Kinoshita T, Takahashi M (1981) Genetical studies on the rice plant. LXXVI. A new gametophyte gene in the second linkage group of rice. Jpn Fac Agric Hokkaido Univ 60:107–114
Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262:1432–1436. https://doi.org/10.1126/science.7902614
Matsubara K, Thidar K, Sano Y (2003) A gene block causing cross-incompatibility hidden in wild and cultivated rice. Genetics 165:343–352
McCouch SR, Kochert G, Yu ZH, Wang ZY, Khush GS, Coffman WR, Tanksley SD (1988) Molecular mapping of rice chromosomes. Theor Appl Genet 76:815–829. https://doi.org/10.1007/BF00273666
Meskiene I, Hirt H (2000) MAP kinase pathways: molecular plug and play chips for the cell. Plant Mol Biol 42:791–806
Mew TW (1987) Current status of future prospects of research on bacterial blight of rice. Annu Rev Phytopathol 25:359–382. https://doi.org/10.1146/annurev.py.25.090187.002043
Nakagahra M (1986) Geographic distribution of gametophyte genes in wide crosses of rice cultivars. In: Rice genetics. Proceedings of the international rice genetics symposium. IRRI, Manila, Philippines pp 73–82. https://doi.org/10.1142/9789812814265_0007
Nakagahra M, Omura T, Iwata N (1972) Gametophyte genes and their loci on the eleventh linkage group of cultivated rice. Jpn J Breed 22:305–312
Neelam K, Lore JS, Kaur K, Pathania S, Kumar K, Sahi G, Mangat G, Singh K (2016) Identification of resistance sources in wild species of rice against two recently evolved pathotypes of Xanthomonas oryzae pv. oryzae. Plant Genet Resour 10:1–5. https://doi.org/10.1017/S1479262116000149
Nino-Liu DO, Ronald PC, Bogdanove AJ (2006) Xanthomonas oryzae pv. oryzae: model pathogen of a model crop. Mol Plant Pathol 7:303–324. https://doi.org/10.1111/j.1364-3703.2006.00344.x
Noda T, Li C, Li J, Ise HO, Kaku H (2001) Pathogenic diversity of Xanthomonas oryzae pv. oryzae strains from Yunnan province, China. Jap Agric Res 35:97–103
Noh TH, Lee DK, Park JC, Shim HK, Choi MY, Kang MH, Park YJ (2007) Effect of bacterial leaf blight occurrence on rice yield and grain quality in different rice growth stage. Plant Dis 13:2023
Ou SH (1985) Rice diseases, 2nd edn. Commonwealth Mycological Institute, Kew
Pham JL, Bougerol B (1993) Abnormal segregations in crosses between two cultivated rice species. Hered 70:466–471
Reflinur KB, Kim B, Jang SM, Chu SH, Bordiya Y, Akter MB, Lee J, Chin JH, Koh HJ (2014) Analysis of segregation distortion and its relationship to hybrid barriers in rice. Rice 7:3. https://doi.org/10.1186/s12284-014-0003-8
Rest B, Rolland N, Boisson AM, Ferro M, Blingy R, Douce R (2004) Identification and characterization of plant glycerophosphodiester phosphodiesterase. Biochem J 379:601–607. https://doi.org/10.1042/BJ20031489
Sanchez P, Wing R, Brar D (2013) The wild relative of rice: genomes and genomics: crops and models. In: Zhang Q, Wing RA (eds) Genetics and genomics of rice. Plant genetics and genomics, vol 5. Springer, New York, pp 9–25. https://doi.org/10.1007/978-1-4614-7903-1_2
Sano Y (1986) Sterility barriers between Oryza sativa and O. glaberrima. In: Kush GS (ed) Rice genetics. Proceedings of the international rice genetics symposium. International Rice Research Institute, Manila, Philippines, pp 109–118
Sano Y (1990) The genic nature of gamete eliminator in rice. Genetics 125:183–191
Sano Y, Chu YE, Oka HI (1979) Genetic studies of speciation in cultivated rice, 1. Genic analysis for the F1 sterility between O. sativa L. and O. glaberrima Steud. Jpn J Genet 54:121–132
Sie M (2008) NERICA for the high-potential irrigated and rain-fed lowlands. In: Somado EA, Guei RG, Keya SO (eds) NERICA: the new rice for Africa—a compendium. Africa Rice Center (WARDA), Cotonou, pp 19–26
Sikirou M, Shittu A, Konaté KA, Maji AT, Ngaujah AS, Sanni KA, Ogunbayo SA, Akintayo I, Saito K, Dramé KN, Ahanchédé A, Venuprasad R (2018) Screening African rice (O. glaberrima) for tolerance to abiotic stresses: I. Fe toxicity. Field Crops Res 220:3–9. https://doi.org/10.1016/j.fcr.2016.04.016
Skamnioti P, Gurr SJ (2009) Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol 27:141–150. https://doi.org/10.1016/j.tibtech.2008.12.002
Somado EA, Guei RG, Keya SO (2008) NERICA: origins, nomenclature and identification characteristics. In: Somado EA, Guei RG, Keya SO (eds) NERICA: the new rice for Africa—a compendium. Africa Rice Center (WARDA), Cotonou, pp 10–18
Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804–1806
Suh JP, Noh TH, Kim KY, Kim JJ, Kim YG, Jena KK (2009) Expression levels of three bacterial blight resistance genes against K3a race of Korea by molecular and phenotype analysis in japonica rice (O. sativa L.). J Crop Sci Biotechnol 12(3):103–108. https://doi.org/10.1007/s12892-009-0103-y
Suh JP, Jeung JU, Noh TH, Cho YC, Park SH, Park HS, Shin MS, Kim CK, Jena KK (2013) Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 6:5. https://doi.org/10.1186/1939-8433-6-5
Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J 37:517–527
Tanaka JP, Chin JH, Drame KN, Dalid C, Heuer S, Wissuwa M (2014) A novel allele of the P starvation tolerance gene OsPSTOL1 from African rice (O. glaberrima Steud) and its distribution in the genus Oryza. Theor Appl Genet 127:1387–1398. https://doi.org/10.1007/s00122-014-2306-y
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation and utilization as genetic markers. Genome Res 11:1441–1452
Tian D, Wang J, Zeng X, Gu K, Qiu C, Yang X, Zhou Z, Goh M, Luo Y, Murata-Hori M, White FF, Yin Z (2014) The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 26:497–515
Valent B, Khang CH (2010) Recent advances in rice blast effector research. Curr Opin Plant Biol 13:434–441. https://doi.org/10.1016/j.pbi.2010.04.012
Van Os H, Stam P, Visser RGF, Van Eck HJ (2005) RECORD: a novel method for ordering loci on a genetic linkage map. Theor Appl Genet 112:30–40. https://doi.org/10.1007/s00122-005-0097-x
Vera Cruz CM, Bai J, Ona I, Leung H, Nelson RJ, Mew TW, Leach JE (2000) Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc Natl Acad Sci USA 97:13500–13505. https://doi.org/10.1073/pnas.250271997
Vikal Y, Das A, Patra B, Goel RK, Sidhu JS, Singh K (2007) Identification of new sources of bacterial blight (Xanthomonas oryzae pv. oryzae) resistance in wild Oryza species and O. glaberrima. Plant Genet Resour 5(2):108–112. https://doi.org/10.1017/S147926210777661X
Vikas G (2010) Fine mapping of bacterial blight resistance genes from O. glaberrima (Steud.) and O. barthii (A. Chev.). Punjab Agricultural University, Ludhiana
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang YS, Pi LY, Chen X, Chakrabarty PK, Jiang J, De Leon AL, Liu GZ, Li L, Benny U, Oard J, Ronald PC, Song WY (2006) Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18:3635–3646. https://doi.org/10.1105/tpc.106.046730
Wang C, Wen G, Lin X, Liu X, Zhang D (2009) Identification and fine mapping of the new bacterial blight resistance gene, Xa31(t), rice. J Plant Pathol 123:235–240. https://doi.org/10.1007/s10658-008-9356-4
Wang S, Basten CJ, Zeng ZB (2012) Windows QTL cartographer 2.5. Department of Statistics, North Carolina state University, Raleigh
Wang CS, Wang AZ, Lin DG (2013) The application of mutants in breeding disease resistance in rice. Paper presented at the special issue or the symposium on important crop pathogen detection and management, Taichung
Wang C, Zhang X, Fan Y, Gao Y, Zhu Q, Zheng C, Qin T, Li Y, Che J, Zhang M, Yang B, Liu Y, Zhao K (2015) XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol Plant 8(2):290–302. https://doi.org/10.1016/j.molp.2014.10.010
Werhahn W, Niemeyer A, Jansch L, Kruft V, Schmitz UK, Braun H-P (2001) Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis: identification of multiple forms of Tom 20. Plant Physiol 125:943–954. https://doi.org/10.1104/pp.125.2.943
Xiang Y, Cao Y, Xu C, Li X, Wang S (2006) Xa3 conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor Appl Genet 113:1347–1355
Xiang J, Lin J, Tang D, Zhou B, Guo M, He R, Huang X, Zhao X, Liu X (2010) DHHC-type zinc finger protein gene regulates shoot branching in Arabidopsis. Afr J Biotechnol 9(45):7759–7766. https://doi.org/10.5897/AJB10.650
Xu Y, Zhu L, Xiao J, Huang N, McCouch SR (1997) Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (O. sativa L.). Mol Gen Genet 253:535–545. https://doi.org/10.1007/s004380050355
Xu PJ, Zhou JL, Hu F, Deng X, Feng S, Ren G, Zhang Z, Deng WD, Yan CQ, Qian KX, Yan QS, Zhang XQ, Xue GP, Huangfu WG, Wu YF, Zhao YZ, Xue ZY, Huang J, Xu GZ, Wu P (2004) Use of asymmetric somatic hybridization for transfer of the bacterial resistance trait from O. meyeriana L. to O. sativa L. ssp. japonica. Plant Cell Rep 22:569–575
Yang J, Zhao X, Cheng K, Du H, Ouyang Y, Chen J, Qiu S, Huang J, Jiang Y, Jiang L, Ding J, Wang J, Xu C, Li X, Zhang Q (2012) A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337:1336–1340. https://doi.org/10.1126/science.1223702
Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, Kono I, Kurata N, Yano M, Iwata N, Sasaki T (1998) Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci 95:1663–1668
Yuan X, Shizhong ZS, Sun M, Liu S, Qi B, Li X (2013) Putative DHHC-cysteine-rich domain S-acyltransferase in plants. PLoS ONE. https://doi.org/10.1371/journal.pone.0075985
Zhang Q (2007) Genetics of quality resistance and identification of major resistance genes to rice bacterial blight. In: Zhang Q (ed) Genetics and improvement of resistance to bacterial blight in rice. Science Press, Beijing, pp 130–177
Zhang F, Zhuo DL, Zhang F, Huang LY, Wang WS, Xu JL, Cruz CV, Li ZK, Zhou YL (2015) Xa39, a novel dominant gene conferring broad-spectrum resistance to Xanthomonas oryzae pv. oryzae in rice. Plant Pathol 64:568–575. https://doi.org/10.1111/ppa.12283
Zhao Z, Wang C, Jiang L, Zhu S, Ikehashi H, Wan J (2006) Identification of a new hybrid sterility gene in rice (Oryza sativa L.). Euphytica 151:331–337. https://doi.org/10.1007/s10681-006-9154-z
Zhu S, Wang C, Zheng T, Zhao Z, Ikehashi H, Wan J (2005) A new gene located on chromosome 2 causing hybrid sterility in a remote cross of rice. Plant Breed 124:440–445. https://doi.org/10.1111/j.1439-0523.2005.01143.x
Acknowledgements
The authors gratefully acknowledge the financial support from the Department of Biotechnology, Ministry of Science and Technology, Government of India, vide Grant Nos. BT/AB/03/FG-2/2003 and BT/AB/FG-2(PH-2) (2-B)/2009 for carrying out research. The grant provided by the Indian Agricultural Research Institute (ICAR), New Delhi, under Niche Area of Excellence vide Project Number F. No. 10(9)2011-EPD is also acknowledged for providing additional financial support for carrying out extension of the research work. We are thankful to Dr. Amit Kishore and Mr. Inderjit Singh Yadav for their suggestions during the revision of the manuscript.
Author information
Authors and Affiliations
Contributions
KS and KN designed the experiment and performed the phenotyping and genetic mapping using F2 population. JSL provided Xanthomonas cultures and assisted in phenotyping of mapping population. KS, RM, VG, DB, BKG and RK contributed in the phenotyping and genotyping of backcrossed derivatives. KN and KS analyzed the data and wrote the manuscript. KS and GSM involved in the field trials and the release of the rice variety PR127 with xa-45(t).
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Dr. Takuji Sasaki.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2019_3501_MOESM1_ESM.pdf
Figure S1: Bulk segregant analysis showing the presence of Pusa 44 allele in homozygous conditions in both resistant and susceptible bulks, Pusa 44 (P1), O. glaberrima 102600B (P2), resistant bulk (RB), Susceptible bulk (SB), C-negative control, M- 50bp ladder (Fermentas). Figure S2: Bulk segregant analysis showing association of SSR markers RM 267, RM 593 and RM 13 of rice chromosome 5 with BB resistance in O. glaberrima 102600B, Pusa 44 (P1), O. glaberrima 102600B (P2), resistant bulk (RB), Susceptible bulk (SB), C-negative control, M-50bp ladder (Fermentas). Figure S3: Genotyping of 94 F5 progenies along with susceptible and resistant parents using xa13 marker on agarose gel (2.5%). Figure S4: Multiple sequence alignment showing polymorphic sites at SWEET11 locus among IL274, IRBB13 and Pusa44 as compared to the reference sequenceLOC4346153. Arrow indicates polymorphic sites (PDF 1432 kb)
Rights and permissions
About this article
Cite this article
Neelam, K., Mahajan, R., Gupta, V. et al. High-resolution genetic mapping of a novel bacterial blight resistance gene xa-45(t) identified from Oryza glaberrima and transferred to Oryza sativa. Theor Appl Genet 133, 689–705 (2020). https://doi.org/10.1007/s00122-019-03501-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03501-2