Abstract
In recent years, histone deacetylase (HDAC) has become one of the hottest and most effective targets for the treatment of cancer. In this work, we designed and synthesized a series of novel o-aminobenzamide based HDAC inhibitors and evaluated their antitumor properties in vitro. All 23 compounds obtained showed micromolar IC50 values against A549 cells proliferation, and the most effective compound was 8 u (IC50 = 0.165 μM). In vitro, 8 u showed potent antiproliferative activity against another three cancer cell lines, outperforming the approved drug Chidamide. Enzyme inhibition and western blot assays confirmed that 8 u was a selective inhibitor of HDAC1-3 isoforms. 8 u was able to induce apoptosis of A549 cells and arrest the tumor cells in G2/M phase. Moreover, 8 u significantly mitigated the migration A549 cells. All these results suggest that 8 u deserves further biological studies.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Loss of cellular identity triggered by epigenetic abnormalities has been reported to be responsible for the progression of intractable human diseases including cancers [1]. These epigenetic changes can provoke abnormal activation or silencing of certain genes, which leads to uncontrolled cell growth [2, 3]. Among all enzymes in the field of epigenetic, histone acetyltransferases (HATs) and histone deacetylases (HDACs) are responsible for maintaining dynamic balance of the acetylation level of core histones and precisely regulate gene transcription and expression [4]. Past studies have revealed that HDACs are overexpressed in kinds of malignant tumors and are key regulators of cell proliferation [5], differentiation [6], and regulation of cell death in hematological malignancies and a variety of solid tumors [7]; and it has been shown that HDACs are directly correlated with the degree of undifferentiated tumors, hyperproliferation, invasiveness, disease staging, and prognosis [8].
The human HDACs family contains 18 members [9] which are divided into class I (HDACs 1, 2, 3 and 8), class II a (HDACs 4, 5, 7 and 9), class II b (HDACs 6 and 10), class III (named sirtuins 1–7) [10] and class IV (HDAC 11). The classical HDACs refer to class I, II and IV isoforms which require zinc ion as an indispensable cofactor for deacetylation activity, while class III isoforms are nicotine adenine dinucleotide dependent. Among them, HDAC1-3 play particularly important roles in apoptosis, cell division and cell cycle control in cancer [11]. HDAC1-3 are predominantly found in the nucleus, and their overexpression has been associated with a wide range of malignant tumors, including those of the stomach, esophagus, colon, breast, prostate, and lungs, and has been demonstrated to be a potential therapeutic targets for cancer therapy [12, 13].
HDAC inhibitors, as a class of sought-after antitumor agents, can hamper tumor cell proliferation via inducing tumor cell growth arrest, differentiation, or apoptosis, and thus achieve the effect of inhibition of tumor cell proliferation and lower toxicity to normal cells [7, 14]. More and more efforts have been dedicated to rationally design and synthesized HDAC inhibitors with diverse skeletons [15]. The majority of the novel HDAC inhibitors reported could be divided into three classes: isoform selective HDAC inhibitors which could avoid unwanted side effects, multi-targeting HDAC inhibitors which could suppress more than one pathway or targets associated with cancers and HDAC degraders such as PROTAC which could address the problems of drug resistance [16]. Hitherto, five HDAC inhibitors: Vorinostat [17], Belinostat [18], Panobinostat [19] and Romidepsin [20], have been approved by the FDA for the treatment of cutaneous T-cell lymphoma, peripheral T-cell lymphoma, and multiple myeloma. Chidamide [21] as the only benzamide-based HDAC inhibitor was approved by the NMPA in 2015 for the treatment of relapsed refractory peripheral T-cell lymphoma (Fig. 1).
The canonical pharmacophore model of HDAC inhibitors consists of three components: Cap which interact with the entrance of the active pocket, zinc binding group (ZBG), and a Linker. The model enables the rational and high effective design of HDAC inhibitors [22]. Till now, the ZBG of majority of HDAC inhibitors ether approved or undergoing clinical trials is hydroxamate due to its high affinity to zinc ion. However, the drawbacks of hydroxamate are also conspicuous, including off-target effects to metalloproteinase, poor pharmacokinetic profiles, and unwanted toxicities [23]. As alternative, o-aminobenzamide based HDAC inhibitors generally exhibit selectivity for HDAC 1–3 isoforms, desirable in vitro and in vivo antitumor activity as well as fewer side effects [24]. In addition to Chidamide which has been marketed, several o-aminobenzamide HDAC inhibitors are in clinical trials, such as CI994 for inhibition of hemolymphatic proliferation [25], Entinostat for the treatment of metastatic melanoma [26], Mocetinostat for the treatment of myelogenous leukemia [27], Domatinostat with the potential to ameliorate refractory cancers by combining with immunotherapeutic therapy [28], and CXD101 which shows effectiveness against Hodgkin’s lymphoma, T-cell lymphoma and follicular lymphoma with stable activity [29] (Fig. 2).
On the other hand, piperazine as an important heterocycle [30] is frequently used as a building block in drug design due to its capacity of optimizing the biological activity of the parent scaffold and improving the water solubility. Drugs containing the structure of piperazine displayed abundant therapeutic effects including antitumor (such as Entrectinib, Abemaciclib for breast cancer, Olmutinib for lung cancer) (Fig. 3), anti-inflammatory, anti-psychotic, anti-Alzheimer’s disease, antibacterial and anti-diabetic [31, 32]. Piperazine also widely exists in the structure of reported HDAC inhibitors. For instance, Xing et al designed and synthesized indole-piperazine derivative 1 with better affinity for class I HDACs, especially for HDAC1-3 [33]. Ruzic et al used 1-benzylpiperazine as a Cap to design novel benzohydroxycarbamic acid derivative 2 as HDAC6 selective inhibitor possessing anti-metastatic effect against breast cancer [34]. Trivedi’s team designed, synthesized compound 3 with alkyl piperazine as a linking group, which exerted inhibitory activity toward HDAC8 [35] (Fig. 3).
In present work, we designed and synthesized a series of novel o-aminobenzamides HDAC inhibitors containing piperazine fragment which functioned as a linker (Fig. 4), and evaluated their antitumor activities in vitro with the aim of digging out novel and potent HDAC inhibitors as candidate for cancer treatment.
Results and discussion
Chemistry
The synthetic route for compounds 8a–8w was shown in Scheme 1. The condensation reaction was carried out between commercially available methyl 4-(bromomethyl) benzoate and tert-butyl piperazine-1-carboxylate to obtain the compound 4 whose Boc group was removed by adding HCl-ethylacetate solution to give intermediate 5. Compound 5 reacted with different acy-lchlorides or sulfonyl chlorides under the treatment of triethylamine to obtain all compounds 6. The methyl-ester in all compounds 6 were hydrolyzed under strong basic environment. And the free carboxyl group in compound 7 condensed with much excessive o-phenylenediamine to afford all target compounds 8a–8w (Table 1).
Reagents and reaction conditions, A: CH3CN, K2CO3, 0 °C, 30 min then R.T. 16 h, 89%. B: 2 M HCl in EA, R.T., 24 h, 100%. C: acyl chloride or sulfonyl chloride or phenyl isocyanate, TEA, 0 °C, DCM, 30 min, then R.T. 16 h, 87–90%. D: NaOH, MeOH: H2O = 8:1, R.T., 24 h, 100%. E: HOBT, EDCI, DIPEA, R.T., 24 h, 59–66%
Biological activity
To assess the biological activity of the target compounds, we initially tested the antiproliferative activity of all 23 target compounds against A549 lung tumor cells by MTT assay, and from the results, the compound 8u displayed the best inhibitory effect with the IC50 value of 0.165 μM, which was much more potent than the positive control Chidamide (IC50 = 9.521 μM) (Table 2). Therefore, further studies were focused on compound 8u.
HDAC isoforms inhibitory activity
The inhibitory activity of compound 8 u against five HDAC isoforms (HDAC1, 2, 3, 6, and 8) were tested still with Chidamide as the positive control. Just as shown in Table 3, 8 u exhibited potent activity to HDAC 1 to 3 with the IC50 of 226.3, 263.1 and 133.4 nM, less potent than Chidamide. Regarding for HDAC6 and 8, no noticeable inhibitory activity of 8 u was detected under the concentration of 5000 nM, which validated that 8 u as a HDAC1-3 selective inhibitor.
Antiproliferation assay of 8 u for other cells
To further confirm the cytotoxicity of 8 u, three more tumor cell lines of MDA-MB-231, Hela and PC-3 were tested with the results showed in Table 4. Compound 8 u still possessed robust antiproliferative activity against threes tumor cell lines with IC50 values at the sub-μM or μM level, better than the positive control Chidamide.
Effect of compound 8 u on the cell cycle distribution of A549 cells
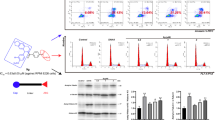
Flow Cytometry was utilized to study the effect of compound 8 u on A549 cell cycle distribution. As shown in Fig. 5, compound 8 u was able to arrest A549 cells in the G2/M phase from 7.04% in blank group to 69.34% after treatment of 8 u at 3 μM compared, much more significantly than Chidamide at the same concentration.
Effect of compound 8 u on apoptosis of A549 cells
Annexin V-FITC/7-AAD double staining was utilized to detect the ability of compound 8 u to induce apoptosis in A549 cells. Compound 8 u was able to significantly induce apoptosis in A549 cells compared to the DMSO blank control group (Fig. 6).
Effect of compound 8 u on the migration of A549 cancer cells
Tumor cell invasion and migration are fatal features of tumors which directly result in the death of patients. HDAC2 was reported to be associated with the migration and invasion of non-small cell lung cancer cells [36]. Accordingly, cell scratch assay was used to test the anti-tumor cell migration ability of compound 8 u. A549 cells were treated with 0.06 μM and 0.2 μM concentrations of compound 8 u, respectively, and the wound healing was photographed and recorded after 48 h. It could be observed that compound 8 u could effectively inhibit cell migration (Fig. 7).
Western blot assay
In order to assess 8 u as a cell active compound. Western blotting was employed to monitor the expression change of four proteins in A549 cells. As shown in Fig. 8, the level of cleaved-PARP increased significantly after the treatment 8 u at 500 n M which implied the occurrence of apoptosis of A549 cells. In addition, to explore whether compound 8 u inhibited HDACs in A549 cells, the levels of acetylated histones H3 (Ac-HH3) and H4 (Ac-HH4), which are well documented biomarkers associated with intracellular HDAC1-3 inhibition, were determined. Consistent with its enzyme inhibition activity, 8 u could remarkably elevated the level of two substrate proteins in a dose dependent manner. All results above confirmed that 8 u was a cell permeable HDAC inhibitor and cell active.
Expression change of cleaved-PARP, Ac-tubulin, Ac-HH3 and Ac-HH4 after A549 cells were treated with 8 u and Chidamide at various concentrations. A The western blot results of 8 u. B 8 u could upregulate the expression of Cleaved-PARP at 500 nM. C 8 u could upregulate the expression of Ac-tubulin at 500 nM. D 8 u significantly upregulated the expression of Ac-HH3 at both 100 nM and 500 nM. E 8 u significantly upregulated the expression of Ac-HH4 at 500 nM. Values represent the mean ± SD, n = 3. **** (P < 0.0001)
Molecular docking study
With the aim of validating the inhibitory activity of 8 u to HDACs and figuring out the plausible binding mode, docking simulation work was performed with compound 8 u acting on the catalytic pocket of the human HDAC2 using the available crystal structure (PDB code: 4LXZ). As can be seen from the in Fig. 9, the hydrogen atom on the amino group in the structure of compound 8 u forms a hydrogen bond with ASP181, the nitrogen atom on the amide bond coordinated with the zinc ion. These hydrogen bonds forced 8 u in a suitable spatial conformation to effectively contact with HDAC2. Moreover, the hydrogen atom on the amide bond forms a hydrogen bond with GLY154, the oxygen atom on the amide bond forms a hydrogen bond with HIS183, and the benzene ring of 8 u forms an extra π-π staking interaction.
Conclusion
Herein, we rationally designed and obtained a series of novel HDAC inhibitory with piperazine and benzamide as core skeletons. All 23 compounds showed moderate to potent antiproliferative potency towards the A549 tumor cells, out of which, compound 8 u stood out as the leading compound. 8 u could effectively and selectively inhibit the enzymatic activities of HDAC1-3 with IC50 values all in sub-μM range. Furthermore, 8 u also exhibited potent antiproliferative activity against MDA-MB-231, Hela, and PC-3 cells, more potent than Chidamide. Further mechanistic studies showed that 8 u significantly inhibited the migration of A549 cells, and the results of DAPI staining and flow cytometry showed that 8 u possessed a dose-dependent effect on apoptosis and G2/M cell cycle block in A549 cells, cell scratch assay shows that 8 u can effectively inhibit the migration of A549 tumor cells and western blot experiments indicated that 8 u could effectively and dose dependently attenuate the expression level of proteins which were relevant with apoptosis and deacetylation activity. Finally, the results of molecular docking showed that 8 u could well accommodated with the active pocket of HDAC2 and formed multiple hydrogen bonds with each other. Taken together, all these results indicate that 8 u has the potential to be a valuable lead compound for further antitumor activity evaluation.
Materials and methods
Materials
All compounds were prepared in DMSO (cell culture grade). DMEM medium and MTT solution were purchased from Basal Mide (Shanghai, China), and fetal bovine serum was from Zeta Life (California, America). Annexin V-FITC/PI Apoptosis Detection Kit was purchased from MBL (Beijing, China). All other reagents used were of analytical grade.
HDAC isoforms inhibitory activity
All enzymatic reactions were carried out at 37 °C for 30 min. 25 mM Tris, 1 mM MgCl2, 0.1 mg/mL BSA, 137 mM NaCl, 2.7 mM KCI, and HDAC enzyme substrate were mixed to form a 50 uL mixture. The compounds were diluted with 10% DMSO, and 5 u L of the diluted compounds were added to 50 μL of the reaction so that the final concentration of DMSO in all reactions was 1%. The amount of fluorescent product in the solution was determined by quantifying it after the enzyme reaction. Fluorescence intensity was then analyzed on a SpectraMax M5 reader at 350–360 nm. Normalized dose/response fitting was performed using GraphPad Prism 8.0 software, and IC50 values were calculated using nonlinear regression.
Antiproliferative activity
The in vitro anti-A549, MDA-MB-231, Hela, PC-3 cells proliferative activity of the target compounds and positive control Chidamide was assayed by MTT assay. Cells were cultured in DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C, 5% CO2. Cells were inoculated in 96-well plates at a density of 4 × 103 cells/well. After 12 h, cells were treated with different concentrations of target compounds for 72 h. MTT solution was added for 4 h of incubation followed by addition of a termination solution in overnight incubation. Absorbance at 490 nm was measured using an enzyme meter. All experiments were repeated three times independently. IC50 was calculated by GraphPad Prism 8.0 based on the ratio of absorbance of compound-treated cells to non-compound-treated cells.
Cell apoptosis assay
A549 cells in logarithmic growth phase were taken and inoculated into six-well plates with 2 × 105 cells per well. After cell attachment, different concentrations of compound 8 u and Chidamide were added to each well and incubated for 48 h. Cells were rinsed twice with PBS buffer, digested with trypsin, collected by centrifugation and washed with PBS buffer and the supernatant was discarded. Annexin V-FITC and Propidium Iodide (PI) were added to the cell precipitates, and the cells were incubated for 15 min in the dark. Flow cytometry was performed on the machine and apoptosis was analyzed by FlowJo V10.
Cell cycle assay
Logarithmic growth phase cells were taken and inoculated in six-well plates with 2 × 105 cells per well. After the cells were attached to the wall, different concentrations of compound 8 u and Chidamide were added to each well and incubated for 48 h. The medium was discarded and the cells were washed with PBS buffer, the cells were moderately digested with trypsin, centrifuged to collect the cells, the supernatant was discarded, the cells were washed with PBS buffer two times, and the supernatant was discarded. 300 μL of PBS buffer was added to resuspend the cells, and the cells were blown apart to avoid clumping. Subsequently, the cell suspension was added drop by drop into 700 uL of anhydrous ethanol (pre-cooled) and then fixed at −20 °C overnight. On the following day, cells were collected by centrifugation and supernatants were aspirated with a pipette gun, followed by resuspension of the cells with 1 mL of PBS buffer and centrifugation for 2–3 washes. Under the condition of light avoidance, each sample avoided light and added PI mixing and then incubated with light-avoidance strips at room temperature for 30 min. The cell cycle was detected by flow cytometry after transferring the assayed samples to 5 mL flow tubes, and the cell cycle analysis was performed using Flowjo or modifit software.
Cell scratching assay
Cells in logarithmic growth phase were taken and inoculated into 12-well plates with 4 × 105 cells per well. After the cells were attached to the wall, the cells were scribed with 200 μL of yellow lance tip, and the cells were washed with PBS buffer for 2 to 3 times to remove the scribed cells, so that the gaps left behind would be clearly visible to the naked eye, and then the medium was replaced with 2% low serum. The cells were incubated in a 37 °C, 5% CO2 incubator. Then they were removed at appropriate time points, observed through a microscope and photographed. The results of the experiments were analyzed using Image J, and data statistics were performed using GraphPad Prism 8.0.
Western blot assay
A549 cells were treated with different concentrations of compounds in 6 cm dishes for 12 h, followed by centrifugation at 20,000 g for 10 min after treatment with 0 °C lysate for 0.5 h. The supernatant was then analyzed for protein concentration by bicinchoninine acid (BCA). The protein concentration in the supernatant was determined by bicinchoninine acid (BCA). Lysed proteins were treated with 12% sodium, sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE), and electrophoretically transferred to a PVDF membrane (amcBiobindNT-200). After blotting, the membranes were closed in 5% blocking solution for 1 h and incubated with specific primary antibodies at 4 °C overnight, followed by enzyme-labeled anti-mouse or anti-rabbit secondary antibodies, and the proteins were visualized with the Super Signal West Pico Chemiluminescent Substrate Kit (Pierce).
Molecular docking study
Molecular docking was performed with compound 8 u using the protein structure of HDAC2 retrieved from the LeDock software website. The docked compound was prepared using Chem.3D ultra 12.0 software. The energy of molecular docking was minimized by removing water molecules, adding polar hydrogen atoms, and applying a Charmm force field to the protein structure. The results were manifested by Discovery Studio Visualizer.
Experimental section
General procedure
All reagents and solvents were purchased from commercial sources and used without further purification. The reaction process was monitored by thin-layer chromatography at 254 nm on a UV fluorescence analyzer, and the corresponding groups were verified by ninhydrin, ferric chloride, and iodine colorimetry. 1H-NMR results were measured by an AVANCE NEO 400 M or 600 M NMR spectrometer. ESI-MS was detected by an Agilent Trap XCT mass spectrometer. The structures of the target compounds were determined by NMR hydrogen spectroscopy and mass spectrometry, and the purity was determined by selecting a GH0525010C18AB column (250 × 10 mm I.D.) with a mobile phase of methanol: water = 35%: 65% and a detection wavelength of 254 nm.
General procedure for the synthesis of compounds 1–5, 6a–6w, 7a–7w
Compounds 1–3 were synthesized according to published literature and the characterization data are consistent with literatures.
Methyl 4-(bromomethyl) benzoate (1.55 g, 1 eq, 6.7 mmol) was dissolved with appropriate amount of acetonitrile, anhydrous K2CO3 (1.77 g, 2 eq, 13.6 mmol) and tert-butyl pi-perazine-1-carboxylate (1.4 g, 1.2 eq, 7.5 mmol) were added, the reaction was carried out in an ice bath for half an hour and then transferred to room temperature for 16 h. The filtrate was filtered through filter paper and the cake was removed, and the filtrate was isolated and purified to obtain compound 4.
tert-butyl 4-(4-(methoxycarbonyl) benzyl) piperazine-1-carboxylate (4)
Faint white solid, yield: 90.5%, 1H NMR (400 MHz, DMSO-d6) δ 7.93 (d, J = 8.2 Hz, 2H), 7.46 (d, J = 8.2 Hz, 2H), 3.85 (s, 3H), 3.55 (s, 2H), 3.32 (brs, 4H), 2.31 −2.33 (m, 4H), 1.39 (s, 9H).
Compound 4 After drying, the compound was added with HCl-ethyl acetate stirring overnight, filtration and leave the filter cake to obtain compound 5. compound 5 and different chloroyl chloride or sulfonyl chloride with appropriate amount of anhydrous DCM dissolved in TEA reaction to obtain compounds 6a–6w.
methyl 4-((4-(4-methoxybenzoyl) piperazin-1-yl) methyl) benzoate (6a)
Faint white solid, yield: 72.8%, 1H NMR (400 MHz, DMSO-d6) δ 7.93 (d, J = 8.2 Hz, 2H), 7.47 (d, J = 8.1 Hz, 2H), 7.35 (d, J = 8.7 Hz, 2H), 6.97 (d, J = 8.7 Hz, 2H), 3.85 (s, 3H), 3.79 (s, 3H), 3.58 (s, 2H), 3.50 (brs, 4H), 2.39 (brs, 4H).
methyl 4-((4-(4-(trifluoromethyl) benzoyl) piperazin-1-yl) methyl) benzoate (6b)
Faint white solid, yield: 73.5%,1H NMR (400 MHz, DMSO-d6) δ 7.93 (d, J = 8.2 Hz, 2H), 7.81 (d, J = 8.1 Hz, 2H), 7.61 (d, J = 8.1 Hz, 2H), 7.47 (d, J = 8.2 Hz, 21H), 3.85 (s, 3H), 3.66 (brs, 2H), 3.60 (s, 2H), 3.30 (brs, 2H), 2.46 (brs, 2H), 2.36 (brs, 2H).
methyl 4-((4-(3-chloro-4-fluorobenzoyl) piperazin-1-yl) methyl) benzoate (6c)
Faint white solid, yield: 75.8%,1H NMR (400 MHz, CDCl3) δ 8.00 (d, J = 8.2 Hz, 2H), 7.48 (dd, J = 7.0, 2.0 Hz, 1H), 7.41 (d, J = 8.1 Hz, 2H), 7.30 (m, 1H), 7.17 (t, J = 8.6 Hz, 1H), 3.91 (s, 3H), 3.77 (brs, 2H), 3.61 (s, 2H), 3.46 (brs, 2H), 2.51 (brs, 4H).
methyl 4-((4-(3-chlorobenzoyl) piperazin-1-yl) methyl) benzoate (6d)
Faint yellow solid, yield: 74.3%,1H NMR (400 MHz, Chloroform-d) δ 8.01 (d, J = 8.0 Hz, 1H), 7.47–7.37 (m, 2H), 7.37–7.23 (m, 1H), 3.92 (s, 2H), 3.82 (s, 1H), 3.66 (s, 1H), 3.48 (s, 1H), 2.59 (s, 1H), 2.47 (s, 1H), 1.26 (s, 2H), 0.96–0.81 (m, 1H), 0.86 (s, 2H).
methyl 4-((4-(3,5-dimethylbenzoyl) piperazin-1-yl) methyl) benzoate (6e)
Faint white solid, yield: 76.5%, 1H NMR (400 MHz, CDCl3) δ 8.00 (d, J = 8.2 Hz, 2H), 7.41 (d, J = 8.1 Hz, 2H), 7.02 (s, 1H), 6.98 (s, 2H), 3.91 (s, 3H), 3.80 (brs, 2H), 3.60 (s, 2H), 3.45 (brs, 2H), 2.54 (brs, 1H), 2.39 (brs, 1H), 2.32 (s, 6H).
methyl 4-((4-(3-(trifluoromethyl) benzoyl) piperazin-1-yl) methyl) benzoate (6 f)
Faint white solid, yield: 76.5%,1H NMR (400 MHz, CDCl3) δ 8.00 (d, J = 8.2 Hz, 2H), 7.68 (d, J = 6.2 Hz, 2H), 7.56 (dt, J = 15.7, 7.7 Hz, 2H), 7.42 (d, J = 8.0 Hz, 2H), 3.91 (s, 3H), 3.83 (brs, 2H), 3.62 (s, 2H), 3.43 (brs, 2H), 2.57 (brs, 2H), 2.42 (brs, 2H).
methyl 4-((4-(1-naphthoyl) piperazin-1-yl) methyl) benzoate (6 g)
Faint white solid, yield: 75.5%,1H NMR (400 MHz, CDCl3) δ 7.98 (d, J = 8.2 Hz, 2H), 7.85 (dd, J = 12.9, 7.7 Hz, 3H), 7.51-7.54 (m, 2H), 7.46–7.49 (m, 1H), 7.39–7.41 (m, 3H), 3.97 (brs, 1H), 3.90 (brs, 1H), 3.60 (s, 2H), 3.23 (s, 2H), 2.64 (s, 2H), 2.31 (s, 2H).
methyl 4-((4-(4-(dimethylamino) benzoyl) piperazin-1-yl) methyl) benzoate (6 h)
Faint white solid, yield: 73.5%, 1H NMR (400 MHz, CDCl3) δ 8.00 (d, J = 8.2 Hz, 2H), 7.41 (d, J = 8.2 Hz, 2H), 7.35 (d, J = 8.8 Hz, 2H), 6.66 (d, J = 8.8 Hz, 2H), 3.91 (s, 3H), 3.67 (s, 4H), 3.60 (s, 2H), 2.98 (s, 6H), 2.48 (s, 4H).
methyl 4-((4-([1,1’-biphenyl]-3-carbonyl) piperazin-1-yl) methyl) benzoate (6i)
Faint white solid, yield: 79.6%,1H NMR (400 MHz, DMSO-d6) δ 7.93 (d, J = 8.2 Hz, 2H), 7.74 (d, J = 7.9 Hz, 1H), 7.69 (d, J = 7.5 Hz, 2H), 7.63 (s, 1H), 7.46-7.55 (m, 5H), 7.39 (dd, J = 12.8, 7.4 Hz, 2H), 3.84 (s, 3H), 3.66 (brs, 2H), 3.59 (s, 2H), 3.40 (brs, 2H), 2.45 (brs, 2H), 2.38 (brs, 2H).
methyl 4-((4-(3-methoxybenzoyl) piperazin-1-yl) methyl) benzoate (6j)
Faint white solid, yield: 76.5%,1H NMR (400 MHz, DMSO-d6) 1H NMR (400 MHz, DMSO) δ 7.93 (d, J = 8.2 Hz, 2H), 7.47 (d, J = 8.2 Hz, 2H), 7.35 (t, J = 7.8 Hz, 1H), 7.01 (dd, J = 8.2, 2.0 Hz, 1H), 6.93 (d, J = 8.3 Hz, 2H), 3.85 (s, 2H), 3.78 (s, 3H), 3.63 (brs, 2H), 3.58 (s, 2H), 3.37 (brs, 2H), 2.43 (brs, 2H), 2.36 (brs, 2H).
methyl 4-((4-(2-naphthoyl) piperazin-1-yl) methyl) benzoate (6k)
Faint white solid, yield: 78.3%,1H NMR (400 MHz, CDCl3) δ 8.00 (d, J = 8.2 Hz, 2H), 7.90 (s, 1H), 7.86 (t, J = 7.2 Hz, 3H), 7.52–7.54 (m, 2H), 7.48 (dd, J = 8.5, 1.4 Hz, 1H), 7.42 (d, J = 7.8 Hz, 2H), 3.91 (s, 3H), 3.85 (brs, 2H), 3.61 (s, 2H), 3.51 (brs, 2H), 2.57 (brs, 2H), 2.42 (brs, 2H).
methyl 4-((4-(3-phenylpropanoyl) piperazin-1-yl) methyl) benzoate (6 l)
Faint white solid, yield: 72.5%,1H NMR (400 MHz, DMSO-d6) δ 7.93 (d, J = 8.2 Hz, 2H), 7.46 (d, J = 8.2 Hz, 2H), 7.15–7.28 (m, 5H), 3.84 (s, 3H), 3.54 (s, 2H), 3.46 (t, J = 4.4 Hz, 2H), 3.41 (t, J = 4.4 Hz, 2H), 2.80 (t, J = 7.7 Hz, 2H), 2.59 (t, J = 7.7 Hz, 2H), 2.29 (d, J = 4.6 Hz, 4H).
methyl 4-((4-(cyclopentanecarbonyl) piperazin-1-yl) methyl) benzoate (6 m)
Faint yellow solid, yield: 74.5%,1H NMR (400 MHz, DMSO-d6) δ 7.93 (d, J = 8.1 Hz, 2H), 7.47 (d, J = 8.1 Hz, 2H), 3.85 (s, 3H), 3.56 (s, 2H), 3.47–3.49 (m, 4H), 2.90–2.98 (m, 1H), 2.33 (dd, J = 13.5, 4.3 Hz, 4H), 1.48–1.77 (m, 8H).
methyl 4-((4-(cyclobutanecarbonyl) piperazin-1-yl) methyl) benzoate (6n)
Faint white solid, yield: 72.5%,1H NMR (400 MHz, DMSO-d6) δ 7.93 (d, J = 8.2 Hz, 2H), 7.46 (d, J = 8.2 Hz, 2H), 3.85 (s, 3H), 3.55 (s, 2H), 3.44 (t, J = 3.2 Hz, 2H), 3.28-3.33 (m, 3H), 2.31 (d, J = 3.2 Hz, 4H), 2.01-2.09 (m, 4H), 1.83–1.90 (m, 1H), 1.70–1.75 (m, 1H).
methyl 4-((4-((3-bromophenyl) sulfonyl) piperazin-1-yl) methyl) benzoate (6o)
Faint white solid, yield: 76.5%, 1H NMR (400 MHz, CDCl3) δ 7.96 (d, J = 8.1 Hz, 2H), 7.89 (d, J = 1.6 Hz, 1H), 7.74 (d, J = 8.0 Hz, 1H), 7.68 (d, J = 8.1 Hz, 1H), 7.42 (t, J = 7.9 Hz, 1H), 7.33 (d, J = 8.0 Hz, 1H), 3.90 (s, 1H), 3.54 (s, 1H), 3.06 (s, 2H), 2.54 (m, 2H).
methyl 4-((4-((4-fluorophenyl) sulfonyl) piperazin-1-yl) methyl) benzoate (6p)
Faint white solid, yield: 76.5%,1H NMR (400 MHz, DMSO-d6) δ 7.88 (d, J = 8.2 Hz, 2H), 7.74 (td, J = 8.0, 5.6 Hz, 1H), 7.61 (ddd, J = 18.4, 9.6, 2.1 Hz, 2H), 7.39 (d, J = 8.2 Hz, 1H), 3.83 (s, 3H), 3.55 (s, 2H), 2.95 (brs, 4H), 2.44 (t, J = 4.5 Hz, 4H).
methyl 4-((4-((3-fluorophenyl) sulfonyl) piperazin-1-yl) methyl) benzoate (6q)
Faint yellow solid, yield: 72.8%,1H NMR (400 MHz, CDCl3) δ 7.96 (d, J = 8.1 Hz, 2H), 7.75–7.79 (m, 2H), 7.32 (d, J = 7.7 Hz, 2H), 7.22 (t, J = 8.6 Hz, 2H), 3.90 (s, 3H), 3.54 (s, 2H), 3.04 (brs, 4H), 2.53 (brs, 4H).
methyl 4-((4-((4-(trifluoromethyl) phenyl) sulfonyl) piperazin-1-yl) methyl) benzoate (6r)
Faint white solid, yield: 78.5%,1H NMR (400 MHz, DMSO-d6) δ 8.05 (d, J = 8.2 Hz, 2H), 7.95 (d, J = 8.1 Hz, 2H), 7.88 (d, J = 8.0 Hz, 2H), 7.39 (d, J = 8.1 Hz, 2H), 3.83 (s, 3H), 3.55 (s, 2H), 2.97 (brs, 4H), 2.45 (brs, 4H).
methyl 4-((4-((4-chlorophenyl) sulfonyl) piperazin-1-yl) methyl) benzoate (6 s)
Faint white solid, yield: 74.5%, 1H NMR (400 MHz, DMSO-d6) δ 7.88 (d, J = 8.2 Hz, 2H), 7.74 (s, 4H), 7.39 (d, J = 8.2 Hz, 2H), 3.83 (s, 3H), 3.55 (s, 2H), 2.92 (brs, 4H), 2.44 (brs, 4H).
methyl 4-((4-tosylpiperazin-1-yl) methyl) benzoate (6t)
Faint white solid, yield: 81.6%, 1H NMR (400 MHz, CDCl3) δ 7.95 (d, J = 8.2 Hz, 2H), 7.63 (d, J = 8.2 Hz, 2H), 7.31–7.34 (m, 4H), 3.90 (s, 3H), 3.53 (s, 2H), 3.02 (brs, 4H), 2.52 (brs, 4H), 2.44 (s, 3H).
methyl 4-((4-(naphthalen-1-ylsulfonyl) piperazin-1-yl) methyl) benzoate (6 u)
Faint white solid, yield: 68.4%, 1H NMR (400 MHz, CDCl3) δ 8.76 (d, J = 8.1 Hz, 1H), 8.19 (dd, J = 7.4, 0.9 Hz, 1H), 8.08 (d, J = 8.2 Hz, 1H), 7.93–7.95 (m, 3H), 7.53–7.64 (m, 3H), 7.32 (d, J = 6.7 Hz, 2H), 3.89 (s, 3H), 3.54 (s, 2H), 3.23 (s, 4H), 2.51 (s, 4H).
methyl 4-((4-(phenylcarbamoyl) piperazin-1-yl) methyl) benzoate (6 v)
Faint yellow solid, yield: 71.5%, 1H NMR (400 MHz, DMSO-d6) δ 8.49 (s, 1H), 7.94 (d, J = 8.2 Hz, 2H), 7.46 (dd, J = 16.9, 8.0 Hz, 4H), 7.22 (t, J = 7.9 Hz, 2H), 6.92 (t, J = 7.3 Hz, 1H), 3.85 (s, 3H), 3.59 (s, 2H), 3.44-3.47 (m, 4H), 2.38-2.40 (m, 4H).
methyl 4-((4-(1H-indole-2-carbonyl) piperazin-1-yl) methyl) benzoate (6w)
Faint white solid, yield: 76.5%, 1H NMR (400 MHz, CDCl3) δ 9.43 (s, 1H), 8.02 (d, J = 8.2 Hz, 2H), 7.63 (d, J = 8.0 Hz, 1H), 7.43 (t, J = 7.7 Hz, 3H), 7.27–7.29 (m, 1H), 7.13 (t, J = 7.5 Hz, 1H), 6.76 (d, J = 1.2 Hz, 1H), 3.96 (brs, 4H), 3.92 (s, 3H), 3.62 (s, 2H), 2.55 (brs, 4H).
Compound 6a–6w hydrolyzed with methanol: 2 mol/L NaOH = 8:1, evaporated to dryness and adjusted with 1 mol/L HCl PH to 5–6, evaporated to dryness to obtain 7a–7w.
General procedure for the synthesis of compounds 8a–8w
A 100 mL reaction flask was charged with compounds 7a–7w (0.8 g, 1.0 eq, 0.02 mmol), dissolved in dry DMF, O-Pheny-lenediami (0.73 g, 3.0 eq, 0.06 mmol), 1-Hydroxybenzotriazole (0.45 g, 1.5 eq, 0.003 mmol), 1-(3-Dimethylaminopropyl)-3-(3-Di-methylaminopropyl) ethylcarbodiimide hydrochloride (0.65 g, 1.5 eq, 0.003 mmol) and N-Eth-yldiisopropylamine (0.98 ml, 2.5 eq, 0.007 mol/L), stirred at room temperature for 12 h. Adding water and EA extraction, column chromatography separation and purification, TCL detection, methanol ultrasonic precipitation, filtration to obtain the target compounds 8a–8w.
N-(2-aminophenyl)-4-((4-(4-methoxybenzoyl) piperazin-1-yl) methyl) benzamide (8a)
Faint white solid, yield: 71.5%, 1H NMR (400 MHz, DMSO-d6) δ 9.64 (s, 1H), 7.95 (d, J = 7.8 Hz, 2H), 7.45 (d, J = 7.8 Hz, 2H), 7.36 (d, J = 8.4 Hz, 2H), 7.16 (d, J = 7.8 Hz, 1H), 6.98 (d, J = 8.0 Hz, 3H), 6.78 (d, J = 7.9 Hz, 1H), 6.60 (t, J = 7.5 Hz, 1H), 4.89 (s, 2H), 3.79 (s, 3H), 3.59 (s, 7H), 2.41 (s, 4H).13C NMR (151 MHz, DMSO-d6) δ 169.46, 165.69, 160.75, 143.72, 142.01, 134.01, 129.61, 129.56, 129.27, 128.36, 127.25, 127.04, 123.93, 116.83, 116.70, 114.21, 61.95, 55.72, 53.09, 40.22. HRMS (ESI) calcd for C26H28N4O3 [M + H] +: 445.2234, found: 445.2233.
N-(2-aminophenyl)-4-((4-(4-(trifluoromethyl) benzoyl) piperazin-1-yl) methyl) benzamide (8b)
Faint white solid, yield: 68.3%, 1H NMR (400 MHz, DMSO-d6) δ 9.65 (s, 1H), 7.96 (d, J = 7.9 Hz, 2H), 7.82 (d, J = 8.1 Hz, 2H), 7.61 (d, J = 7.9 Hz, 2H), 7.45 (d, J = 8.0 Hz, 2H), 7.20–7.13 (m, 1H), 6.97 (td, J = 7.6, 1.6 Hz, 1H), 6.78 (dd, J = 8.0, 1.5 Hz, 1H), 6.60 (td, J = 7.5, 1.5 Hz, 1H), 4.89 (s, 2H), 3.66 (s, 2H), 3.60 (s, 2H), 2.47 (s, 2H), 2.38 (s, 2H). 13C NMR (151 MHz, DMSO-d6) δ 168.11, 165.66, 143.69, 141.96, 140.55, 133.99, 130.29, 130.08, 129.22, 128.35, 128.29, 127.22, 126.07, 125.38, 123.92, 123.57, 116.81, 61.87, 53.25, 52.63, 47.60. HRMS (ESI) calcd for C25H25F3N4O2 [M + H] +: 483.2002, found: 483.1999.
N-(2-aminophenyl)-4-((4-(3-chloro-4-fluorobenzoyl) piperazin-1-yl) methyl) benzamide (8c)
Faint white solid, yield: 72.5%, 1H NMR (400 MHz, DMSO-d6) δ 9.65 (s, 1H), 7.96 (d, J = 8.0 Hz, 2H), 7.65 (dd, J = 7.2, 1.9 Hz, 1H), 7.55–7.34 (m, 4H), 7.17 (d, J = 7.5 Hz, 1H), 7.04–6.92 (m, 1H), 6.83–6.75 (m, 1H), 6.60 (t, J = 7.1 Hz, 1H), 4.90 (s, 2H), 3.59 (s, 4H), 2.41 (s, 4H). 13C NMR (151 MHz, DMSO-d6) δ 167.18, 165.66, 159.02, 157.37, 141.99, 134.21, 130.01, 129.22, 128.57, 128.34, 127.23, 123.92, 120.40, 120.28, 117.69, 116.69, 61.87, 53.18, 47.75. HRMS (ESI) calcd for C25H24ClFN4O2 [M + H] +: 467.1645, found: 467.1643.
N-(2-aminophenyl)-4-((4-(3-chlorobenzoyl) piperazin-1-yl) methyl) benzamide (8d)
Faint yellow solid, yield: 70.8%, 1H NMR (400 MHz, DMSO-d6) δ 9.65 (s, 1H), 7.96 (d, J = 7.9 Hz, 2H), 7.56–7.41 (m, 5H), 7.35 (dt, J = 7.4, 1.5 Hz, 1H), 7.17 (dd, J = 7.8, 1.5 Hz, 1H), 6.97 (td, J = 7.6, 1.5 Hz, 1H), 6.78 (dd, J = 8.1, 1.4 Hz, 1H), 6.60 (td, J = 7.5, 1.5 Hz, 1H), 4.90 (s, 2H), 3.61 (d, J = 18.4 Hz, 4H), 2.41 (d, J = 25.0 Hz, 4H). 13C NMR (151 MHz, DMSO-d6) δ 167.87, 165.67, 143.69, 141.99, 138.56, 133.99, 131.01, 129.98, 128.35, 127.28, 127.23, 126.04, 123.94, 116.82, 116.71, 61.88, 53.22 47.66. HRMS (ESI) calcd for C25H25ClN4O2 [M + H] +: 449.1739, found: 449.1735.
N-(2-aminophenyl)-4-((4-(3,5-dimethylbenzoyl) piperazin-1-yl) methyl) benzamide (8e)
Faint white solid, yield: 72.5%, 1H NMR (400 MHz, DMSO-d6) δ 9.64 (s, 1H), 7.95 (d, J = 7.9 Hz, 2H), 7.44 (d, J = 8.0 Hz, 2H), 7.16 (dd, J = 7.8, 1.5 Hz, 1H), 7.07 (s, 1H), 7.02–6.91 (m, 3H), 6.78 (dd, J = 8.0, 1.4 Hz, 1H), 6.60 (td, J = 7.5, 1.5 Hz, 1H), 4.89 (s, 2H), 3.59 (s, 4H), 2.43 (d, J = 30.9 Hz, 4H), 2.29 (s, 6H). 13C NMR (151 MHz, DMSO-d6) δ 169.70, 165.66, 143.68, 141.94, 138.15, 136.46, 133.99, 131.31, 129.24, 128.34, 127.22, 127.16, 124.99, 123.94, 116.82, 116.71, 61.90, 47.64, 40.06, 21.34. HRMS (ESI) calcd for C27H30N4O2 [M + H] +: 443.2442, found: 443.2440.
N-(2-aminophenyl)-4-((4-(3-(trifluoromethyl) benzoyl) piperazin-1-yl) methyl) benzamide (8f)
Faint white solid, yield: 69.5%, 1H NMR (400 MHz, DMSO-d6) δ 9.64 (s, 1H), 7.95 (d, J = 7.9 Hz, 2H), 7.87–7.77 (m, 1H), 7.76–7.65 (m, 3H), 7.45 (d, J = 8.0 Hz, 2H), 7.16 (dd, J = 7.8, 1.5 Hz, 1H), 6.97 (td, J = 7.6, 1.5 Hz, 1H), 6.78 (dd, J = 8.0, 1.4 Hz, 1H), 6.60 (td, J = 7.5, 1.5 Hz, 1H), 4.89 (s, 2H), 3.60 (s, 4H), 2.44 (d, J = 35.1 Hz, 4H). 13C NMR (151 MHz, DMSO-d6) δ 167.94, 165.67, 143.70, 142.01, 137.55, 133.99, 131.48, 129.97, 129.21, 128.35, 127.15, 127.01, 126.72, 123.93, 123.53, 116.82, 116.70, 61.86, 53.18, 47.69. HRMS (ESI) calcd for C26H25F3N4O2 [M + H] +: 483.2002, found: 483.2000.
4-((4-(1-naphthoyl) piperazin-1-yl) methyl)-N-(2-aminophenyl) benzamide (8g)
Faint white solid, yield: 68.6%, 1H NMR (400 MHz, DMSO-d6) δ 9.64 (s, 1H), 8.05–7.89 (m, 4H), 7.81–7.71 (m, 1H), 7.68–7.51 (m, 3H), 7.49–7.39 (m, 3H), 7.16 (dd, J = 7.9, 1.5 Hz, 1H), 6.97 (td, J = 7.6, 1.6 Hz, 1H), 6.78 (dd, J = 8.0, 1.5 Hz, 1H), 6.59 (td, J = 7.5, 1.5 Hz, 1H), 4.90 (s, 2H), 3.87 (s, 1H), 3.74 (s, 1H), 3.58 (s, 2H), 3.22–3.01 (m, 2H), 2.54 (t, J = 4.6 Hz, 2H), 2.41–2.11 (m, 2H). 13C NMR (151 MHz, DMSO-d6) δ 168.41, 165.66, 143.68, 134.76, 134.03, 133.53, 129.60, 129.33, 128.96, 128.36, 127.55, 127.23, 125.93, 125.19, 124.23, 123.93, 116.83, 116.71, 61.80, 52.81, 47.13. HRMS (ESI) calcd for C29H28N4O2 [M + H] +: 465.2285, found: 465.2284.
N-(2-aminophenyl)-4-((4-(4-(dimethylamino) benzoyl) piperazin-1-yl) methyl) benzamide (8h)
Faint white solid, yield: 71.3%, 1H NMR (400 MHz, DMSO-d6) δ 9.64 (s, 1H), 7.95 (d, J = 7.9 Hz, 2H), 7.45 (d, J = 7.9 Hz, 2H), 7.32–7.21 (m, 2H), 7.16 (dd, J = 7.9, 1.6 Hz, 1H), 7.02–6.92 (m, 1H), 6.78 (dd, J = 8.0, 1.5 Hz, 1H), 6.75–6.65 (m, 2H), 6.60 (td, J = 7.5, 1.5 Hz, 1H), 4.89 (s, 2H), 3.59 (s, 2H), 3.52 (t, J = 5.2 Hz, 4H), 2.94 (s, 6H), 2.40 (t, J = 4.8 Hz, 4H). 13C NMR (151 MHz, DMSO-d6) δ 170.22, 165.69, 151.73, 143.69, 142.01, 133.99, 129.59, 129.27, 128.35, 127.22, 127.01, 123.94, 122.58, 116.83, 116.71, 111.56, 61.98, 53.19, 40.60, 40.20. HRMS (ESI) calcd for C27H31N5O2 [M + H] +: 458.2551, found: 458.2549.
4-((4-([1,1’-biphenyl]-3-carbonyl) piperazin-1-yl) methyl)-N-(2-aminophenyl) benzamide (8i)
Faint white solid, yield: 65.5%, 1H NMR (400 MHz, DMSO-d6) δ 9.64 (s, 1H), 7.95 (d, J = 7.9 Hz, 2H), 7.75 (dt, J = 7.9, 1.4 Hz, 1H), 7.71–7.66 (m, 2H), 7.64 (t, J = 1.8 Hz, 1H), 7.58–7.34 (m, 7H), 7.16 (dd, J = 8.0, 1.5 Hz, 1H), 6.97 (td, J = 7.6, 1.5 Hz, 1H), 6.78 (dd, J = 8.0, 1.4 Hz, 1H), 6.60 (td, J = 7.5, 1.4 Hz, 1H), 4.89 (s, 2H), 3.64 (d, J = 27.6 Hz, 4H), 3.52–3.35 (m, 2H), 2.44 (d, J = 26.2 Hz, 4H). 13C NMR (151 MHz, DMSO-d6) δ 169.37, 165.62, 143.67, 140.91, 139.91, 137.03, 134.30, 129.71, 129.59, 129.48, 128.42, 127.38, 127.26, 127.06, 126.50, 123.91, 116.86, 116.74, 61.47, 52.52 47.32. HRMS (ESI) calcd for C31H30N4O2 [M + H] +: 491.2442, found: 491.2440.
N-(2-aminophenyl)-4-((4-(3-methoxybenzoyl) piperazin-1-yl) methyl) benzamide (8j)
Faint white solid, yield: 67.4%, 1H NMR (600 MHz, DMSO-d6) δ 9.64 (s, 1H), 7.96 (d, J = 7.7 Hz, 2H), 7.46 (d, J = 7.8 Hz, 2H), 7.36 (dd, J = 8.3, 7.5 Hz, 1H), 7.17 (dd, J = 7.9, 1.5 Hz, 1H), 7.02 (ddd, J = 8.4, 2.6, 0.9 Hz, 1H), 6.98 (td, J = 7.6, 1.6 Hz, 1H), 6.95–6.88 (m, 2H), 6.79 (dd, J = 8.0, 1.4 Hz, 1H), 6.61 (td, J = 7.5, 1.5 Hz, 1H), 4.90 (s, 2H), 3.78 (s, 3H), 3.66 (s, 4H), 2.41 (d, J = 33.0 Hz, 4H). 13C NMR (151 MHz, DMSO-d6) δ 169.17, 165.66, 159.66, 143.68, 137.93, 134.08, 130.21, 129.35, 128.37, 127.24, 127.03, 123.92, 119.41, 116.84, 116.71, 115.68, 112.82, 61.78, 55.76, 52.69, 47.51. HRMS (ESI) calcd for C26H28N4O3 [M + H] +: 445.2234, found: 445.2233.
4-((4-(2-naphthoyl) piperazin-1-yl) methyl)-N-(2-aminophenyl) benzamide (8k)
Faint white solid, yield: 70.6%, 1H NMR (600 MHz, DMSO-d6) δ 9.64 (s, 1H), 8.04–7.99 (m, 1H), 8.01–7.95 (m, 4H), 7.96 (d, J = 5.4 Hz, 1H), 7.63–7.55 (m, 2H), 7.51 (dd, J = 8.3, 1.7 Hz, 1H), 7.47 (d, J = 7.9 Hz, 2H), 7.17 (d, J = 7.8 Hz, 1H), 6.97 (td, J = 7.6, 1.6 Hz, 1H), 6.79 (dd, J = 8.0, 1.4 Hz, 1H), 6.60 (td, J = 7.5, 1.5 Hz, 1H), 4.89 (s, 2H), 3.43 (s, 2H), 3.33 (d, J = 1.4 Hz, 4H), 2.43 (s, 4H). 13C NMR (151 MHz, DMSO-d6) δ 169.53, 165.71, 143.72, 142.05, 134.02, 133.84, 133.66, 132.83, 129.26, 128.93, 128.54, 128.25, 127.67, 127.31, 127.25, 125.03, 123.96, 116.73, 61.95, 52.87, 47.86. HRMS (ESI) calcd for C29H28N4O2 [M + H] +: 465.2285, found: 465.2284.
N-(2-aminophenyl)-4-((4-(3-phenylpropanoyl) piperazin-1-yl) methyl) benzamide (8l)
Faint white solid, yield:67.5%, 1H NMR (600 MHz, DMSO-d6) δ 9.63 (s, 1H), 7.96 (d, J = 7.9 Hz, 2H), 7.44 (d, J = 7.9 Hz, 2H), 7.32–7.20 (m, 4H), 7.20–7.13 (m, 2H), 6.98 (td, J = 7.6, 1.5 Hz, 1H), 6.79 (dd, J = 8.1, 1.5 Hz, 1H), 6.61 (td, J = 7.5, 1.5 Hz, 1H), 4.89 (s, 2H), 3.55 (s, 2H), 3.45 (dt, J = 29.3, 5.1 Hz, 4H), 2.81 (dd, J = 8.6, 6.9 Hz, 2H), 2.61 (dd, J = 8.6, 6.9 Hz, 2H), 2.31 (dt, J = 7.7, 4.9 Hz, 4H). 13C NMR (151 MHz, DMSO-d6) δ 170.39, 165.71, 143.70, 142.08, 141.95, 133.99, 129.23, 129.00, 128.80, 128.65, 127.23, 127.03, 126.42, 123.96, 116.85, 116.73, 61.97, 52.88, 40.22, 34.51, 31.39. HRMS (ESI) calcd for C27H30N4O2 [M + H] +: 443.2442, found: 443.2440.
N-(2-aminophenyl)-4-((4-(cyclopentanecarbonyl) piperazin-1-yl) methyl) benzamide (8m)
Faint yellow solid, yield: 63.2%, 1H NMR (600 MHz, DMSO-d6) δ 9.64 (s, 1H), 7.96 (d, J = 7.9 Hz, 2H), 7.45 (d, J = 8.1 Hz, 2H), 7.17 (dd, J = 7.9, 1.5 Hz, 1H), 6.98 (dd, J = 8.0, 7.3, 1.6 Hz, 1H), 6.79 (dd, J = 8.0, 1.5 Hz, 1H), 6.61 (td, J = 7.5, 1.5 Hz, 1H), 4.89 (s, 2H), 3.58 (s, 2H), 3.49 (dt, J = 20.3, 5.4 Hz, 4H), 3.00–2.92 (m, 1H), 2.40–2.36 (m, 2H), 2.33 (t, J = 5.2 Hz, 2H), 1.74 (s, 1H), 1.78–1.70 (m, 1H), 1.64 (dd, J = 14.8, 10.9, 7.1, 1.8 Hz, 2H), 1.60 (s, 1H), 1.61–1.55 (m, 1H), 1.51 (qt, J = 5.9, 3.8 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 173.86, 165.68, 143.69, 142.11, 133.96, 129.21, 128.33, 127.21, 127.01, 123.92, 116.81, 116.69, 61.97, 53.60, 53.01, 45.45, 30.14, 26.21. HRMS (ESI) calcd for C24H30N4O2 [M + H] +: 407.2442, found: 407.2440.
N-(2-aminophenyl)-4-((4-(cyclobutanecarbonyl) piperazin-1-yl) methyl) benzamide (8n)
Faint white solid, yield: 59.6%, 1H NMR (600 MHz, DMSO-d6) δ 9.63 (s, 1H), 7.95 (d, J = 7.8 Hz, 2H), 7.44 (d, J = 8.1 Hz, 2H), 7.17 (dd, J = 7.8, 1.5 Hz, 1H), 6.98 (td, J = 7.6, 1.6 Hz, 1H), 6.79 (dd, J = 8.0, 1.4 Hz, 1H), 6.61 (td, J = 7.5, 1.4 Hz, 1H), 4.89 (s, 2H), 3.56 (s, 2H), 3.45 (t, J = 5.1 Hz, 2H), 3.34–3.27 (m, 2H), 2.33 (dt, J = 9.9, 4.9 Hz, 4H), 2.19–2.10 (m, 2H), 2.13–2.03 (m, 2H), 1.88 (dp, J = 10.8, 8.9 Hz, 1H), 1.78–1.69 (m, 1H). 13C NMR (151 MHz, DMSO-d6) δ 172.49, 165.69, 143.67, 142.10, 133.97, 129.20, 128.34, 127.19, 127.0, 123.98, 116.84, 116.74, 62.01, 53.48, 44.97, 36.77, 25.09, 17.97. HRMS (ESI) calcd for C23H28N4O2 [M + H] +: 393.2285, found: 393.2284.
N-(2-aminophenyl)-4-((4-((3-bromophenyl) sulfonyl) piperazin-1-yl) methyl) benzamide (8o)
Faint white solid, yield: 70.8%, 1H NMR (600 MHz, DMSO-d6) δ 9.62 (s, 1H), 7.98 (dd, J = 8.0, 2.0, 1.0 Hz, 1H), 7.92 (d, J = 7.8 Hz, 2H), 7.87 (t, J = 1.8 Hz, 1H), 7.76 (dt, J = 7.9, 1.3 Hz, 1H), 7.64 (t, J = 7.9 Hz, 1H), 7.38 (d, J = 7.9 Hz, 2H), 7.16 (d, J = 7.8 Hz, 1H), 6.97 (td, J = 7.6, 1.6 Hz, 1H), 6.78 (dd, J = 8.0, 1.4 Hz, 1H), 6.60 (td, J = 7.5, 1.5 Hz, 1H), 4.88 (s, 2H), 3.57 (s, 2H), 2.97 (s, 4H), 2.47 (s, 4H), 0.89–0.80 (m, 0H). 13C NMR (151 MHz, DMSO-d6) δ 165.66, 143.69, 141.85, 137.57, 136.77, 133.99, 132.30, 130.27, 129.11, 128.33, 127.21, 123.93, 123.06, 117.87, 116.84, 115.12, 61.45, 51.96, 46.54. HRMS (ESI) calcd for C24H25BrN4O3S [M + H] +: 529.0904, found: 529.0903.
N-(2-aminophenyl)-4-((4-((3-fluorophenyl) sulfonyl) piperazin-1-yl) methyl) benzamide (8p)
Faint white solid, yield: 68.5%, 1H NMR (400 MHz, DMSO-d6) δ 9.67 (s, 1H), 7.94 (d, J = 7.9 Hz, 2H), 7.74 (td, J = 8.1, 5.7 Hz, 1H), 7.68–7.53 (m, 3H), 7.43 (d, J = 7.0 Hz, 2H), 7.16 (d, J = 7.6 Hz, 1H), 7.04–6.88 (m, 1H), 6.79 (dd, J = 8.0, 1.1 Hz, 1H), 6.61 (t, J = 7.5 Hz, 1H), 3.72 (s, 2H), 3.02 (s, 4H), 2.79–2.51 (m, 4H). 13C NMR (151 MHz, DMSO-d6) δ 165.63, 163.27, 161.63, 143.66, 137.59, 134.03, 132.45, 129.17, 128.34, 127.24, 124.39, 123.91, 121.15, 116.84, 115.08, 61.35, 51.92, 46.42. HRMS (ESI) calcd for C24H25FN4O3S [M + H] +: 469.1704, found: 469.1703.
N-(2-aminophenyl)-4-((4-((4-fluorophenyl) sulfonyl) piperazin-1-yl) methyl) benzamide (8q)
Faint yellow solid, yield: 73.6%, 1H NMR (400 MHz, DMSO-d6) δ 9.62 (s, 1H), 7.91 (d, J = 8.0 Hz, 2H), 7.81 (dd, J = 8.8, 5.1 Hz, 2H), 7.51 (t, J = 8.8 Hz, 2H), 7.37 (d, J = 8.1 Hz, 2H), 7.15 (d, J = 7.7 Hz, 1H), 7.01–6.91 (m, 1H), 6.78 (d, J = 8.0 Hz, 1H), 6.59 (t, J = 7.5 Hz, 1H), 4.88 (s, 2H), 3.55 (s, 2H), 2.92 (s, 4H), 2.46 (d, J = 4.9 Hz, 4H). 13C NMR (151 MHz, DMSO-d6) δ 166.11, 165.66, 164.44, 143.69, 141.88, 133.99, 131.80, 131.27, 129.36, 128.34, 127.24, 127.04, 123.94, 117.30, 116.85, 61.46, 51.98, 46.50. HRMS (ESI) calcd for C24H25FN4O3S [M + H] +: 469.1704, found: 469.1702.
N-(2-aminophenyl)-4-((4-((4-(trifluoromethyl) phenyl) sulfonyl) piperazin-1-yl) methyl) benzamide (8r)
Faint white solid, yield: 68.5%, 1H NMR (400 MHz, DMSO-d6) δ 9.62 (s, 1H), 8.05 (d, J = 8.3 Hz, 2H), 7.96 (d, J = 8.2 Hz, 2H), 7.91 (d, J = 8.1 Hz, 2H), 7.37 (d, J = 8.1 Hz, 2H), 7.15 (d, J = 7.5 Hz, 1H), 7.02–6.92 (m, 1H), 6.78 (dd, J = 8.0, 1.1 Hz, 1H), 6.59 (t, J = 7.5 Hz, 1H), 4.88 (s, 2H), 3.55 (s, 2H), 2.98 (s, 4H), 2.46 (s, 4H). 13C NMR (151 MHz, DMSO-d6) δ 165.65, 143.71, 141.86, 139.55, 134.00, 133.57, 133.36, 129.11, 128.34, 127.23, 127.04, 124.93, 123.93, 123.13, 116.84, 61.45, 52.02, 46.47. HRMS (ESI) calcd for C25H25F3N4O3S [M + H] +: 519.1672, found: 519.1670.
N-(2-aminophenyl)-4-((4-((4-chlorophenyl) sulfonyl) piperazin-1-yl) methyl) benzamide (8s)
Faint white solid, yield: 70.2%, 1H NMR (400 MHz, DMSO-d6) δ 9.62 (s, 1H), 7.91 (d, J = 7.9 Hz, 2H), 7.75 (s, 5H), 7.37 (d, J = 8.0 Hz, 2H), 7.15 (dd, J = 7.5, 1.5 Hz, 1H), 6.97 (td, J = 7.6, 1.6 Hz, 1H), 6.78 (dd, J = 8.0, 1.5 Hz, 1H), 6.59 (td, J = 7.5, 1.4 Hz, 1H), 4.88 (s, 2H), 3.55 (s, 2H), 2.93 (s, 4H), 2.45 (t, J = 4.9 Hz, 4H). 13C NMR (151 MHz, DMSO-d6) δ 165.64, 143.67, 141.86, 138.87, 134.29, 133.97, 130.03, 129.32, 128.32, 127.21, 127.02, 123.93, 116.84, 116.72, 61.43, 51.97, 46.46. HRMS (ESI) calcd for C24H25ClN4O3S [M + H] +: 485.1409, found: 485.1407.
N-(2-aminophenyl)-4-((4-tosylpiperazin-1-yl) methyl) benzamide (8t)
Faint white solid, yield: 65.5%, 1H NMR (400 MHz, DMSO-d6) δ 9.62 (s, 1H), 7.90 (d, J = 7.8 Hz, 2H), 7.62 (d, J = 8.1 Hz, 2H), 7.47 (d, J = 8.0 Hz, 2H), 7.36 (d, J = 7.9 Hz, 2H), 7.15 (d, J = 7.8 Hz, 1H), 6.97 (td, J = 7.6, 1.6 Hz, 1H), 6.78 (dd, J = 8.0, 1.5 Hz, 1H), 6.59 (td, J = 7.6, 1.5 Hz, 1H), 4.88 (s, 2H), 3.54 (s, 2H), 2.88 (s, 4H), 2.43 (d, J = 7.7 Hz, 7H). 13C NMR (151 MHz, DMSO-d6) δ 165.66, 144.27, 143.68, 141.94, 133.97, 132.34, 130.42, 129.08, 128.33, 128.21, 127.22, 127.03, 123.94, 116.85, 116.73, 61.48, 52.02, 46.51, 21.60. HRMS (ESI) calcd for C25H28N4O3S [M + H] +: 465.1955, found: 465.1952.
N-(2-aminophenyl)-4-((4-(naphthalen-1-ylsulfonyl) piperazin-1-yl) methyl) benzamide (8u)
Faint white solid, yield: 68.5%, 1H NMR (400 MHz, DMSO-d6) δ 9.60 (s, 1H), 8.68 (d, J = 8.4 Hz, 1H), 8.32 (d, J = 8.2 Hz, 1H), 8.15 (t, J = 7.8 Hz, 2H), 7.89 (d, J = 8.0 Hz, 2H), 7.73 (ddd, J = 12.3, 8.6, 6.5 Hz, 3H), 7.35 (d, J = 8.1 Hz, 2H), 7.28 (q, J = 8.2 Hz, 1H), 7.15 (d, J = 7.8 Hz, 1H), 6.97 (t, J = 7.6 Hz, 1H), 6.78 (d, J = 8.0 Hz, 1H), 6.60 (t, J = 7.5 Hz, 1H), 4.88 (s, 2H), 3.52 (s, 2H), 3.11 (d, J = 5.3 Hz, 4H), 2.92 (d, J = 31.7 Hz, 1H), 2.40 (t, J = 5.0 Hz, 4H). 13C NMR (151 MHz, DMSO-d6) δ 165.66, 143.67, 141.83, 135.20, 134.54, 133.95, 132.49, 130.86, 129.66, 128.81, 128.76, 128.30, 127.56, 127.21, 127.01, 125.25, 123.93, 116.84, 61.49, 52.27, 46.04. HRMS (ESI) calcd for C28H28N4O3S [M + H] +: 501.1955, found: 501.1953.
4-(4-((2-aminophenyl) carbamoyl) benzyl)-N-phenylpiperazine-1-carboxamide (8v)
Faint yellow solid, yield: 59.8%,1H NMR (400 MHz, DMSO-d6) δ 9.65 (s, 1H), 8.50 (s, 1H), 7.97 (d, J = 7.9 Hz, 2H), 7.46 (dd, J = 8.0, 6.0 Hz, 4H), 7.32–7.12 (m, 3H), 7.04–6.87 (m, 2H), 6.79 (dd, J = 8.1, 1.4 Hz, 1H), 6.61 (td, J = 7.5, 1.5 Hz, 1H), 4.90 (s, 2H). 13C NMR (151 MHz, DMSO-d6) δ 165.77, 155.55, 143.71, 142.16, 141.09, 133.97, 129.26, 128.85, 128.36, 127.26, 123.98, 122.28, 120.22, 116.88, 116.75, 62.10, 53.07, 44.34. HRMS (ESI) calcd for C25H27N5O2 [M + H] +:430.2238, found: 430.2236.
4-((4-(1H-indole-2-carbonyl) piperazin-1-yl) methyl)-N-(2-aminophenyl) benzamide (8w)
Faint white solid, yield: 63.3%, 1H NMR (400 MHz, DMSO-d6) δ 11.58 (d, J = 2.3 Hz, 1H), 9.65 (s, 1H), 7.98 (d, J = 7.9 Hz, 2H), 7.61 (d, J = 8.0 Hz, 1H), 7.48 (d, J = 8.0 Hz, 2H), 7.43 (d, J = 8.3 Hz, 1H), 7.19 (t, J = 7.2 Hz, 2H), 7.05 (t, J = 7.5 Hz, 1H), 6.98 (td, J = 7.6, 1.5 Hz, 1H), 6.85–6.73 (m, 2H), 6.61 (td, J = 7.5, 1.5 Hz, 1H), 4.91 (s, 2H), 3.63 (s, 2H), 2.48 (d, J = 5.2 Hz, 3H). 13C NMR (151 MHz, DMSO-d6) δ 165.72, 162.51, 143.71, 142.03, 136.47, 134.01, 130.42, 129.27, 128.36, 127.54, 127.36, 127.03, 123.95, 123.76, 121.90, 120.29, 116.84, 116.71, 112.60, 104.56, 61.95, 55.47, 53.24. HRMS (ESI) calcd for C27H27N5O2 [M + H] +: 454.2238, found: 454.2235.
References
Ansari J, Shackelford RE, El-Osta H. Epigenetics in non-small cell lung cancer: from basics to therapeutics. Transl Lung Cancer Res. 2016;5:155–71. https://doi.org/10.21037/tlcr.2016.02.02
Bondarev AD, Attwood MM, Jonsson J, Chub-arev VN, Tarasov VV, Schiöth HB. Recent developments of HDAC inhibitors: emerging indications and novel molecules. Br J Clin Pharmacol. 2021;87:4577–97. https://doi.org/10.1111/bcp.14889
Ramaiah MJ, Tangutur AD, Manyam RR. Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sci. 2021;277:119504–22. https://doi.org/10.1016/j.lfs.2021.119504
Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed and Biotechnol. 2011;2011:1–10. https://doi.org/10.1155/2011/371832
Cheng B, Pan W, Xiao Y, Ding Z, Zhou Y, Fei X, et al. HDAC-targeting epigenetic modulators for cancer immunotherapy. Eur J Med Chem. 2024;265:116129–45. https://doi.org/10.1016/j.ejmech.2024.116129
Botrugno OA, Santoro F, Minucci S. Histone deacetylase inhibitors as a new weapon in the arsenal of differentiation therapies of cancer. Cancer Lett. 2009;280:134–44. https://doi.org/10.1016/j.canlet.2009.02.027
Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone deacetylase inhibitors as anticancer D-rugs. Int J Mol Sci. 2017;18:1414–38. https://doi.org/10.3390/ijms18071414
Shetty MG, Pai P, Deaver RE, Satyamoorthy K, Babitha KS. Histone deacetylase 2 selective inhibitors: a versatile therapeutic strategy as next generation drug target in cancer therapy. Pharmacol Res. 2021;170:105695–7. https://doi.org/10.1016/j.phrs.2021.105695
Sangwan R, Rajan R, Mandal PK. HDAC as onco target: Reviewing the synthetic approaches with SAR study of their inhibitors. Eur J Med Chem. 2018;158:620–706. https://doi.org/10.1016/j.ejmech.2018.08.073
Yuan H, Marmorstein R. Structural basis for sirtuin activity and inhibition. J Biolog Chem. 2012;287:42428–35. https://doi.org/10.1074/jbc.R112.372300
Smalley JP, Baker IM, Pytel WA, Lin L-Y, Bowman KJ, Schwabe JWR, et al. Optimization of class I histone deacetylase PROTACs reveals that HDAC1/2 degradation is critical to induce apoptosis and cell arrest in cancer cells. J Med Chem. 2022;65:5642–59. https://doi.org/10.1021/acs.jmedchem.1c02179
Schübeler D, Hess L, Moos V, Lauber AA, Reiter W, Schuster M, et al. A toolbox for class I HDACs reveals isoform specific roles in gene regulation and protein acetylation. PLOS Genet. 2022;18:1010376–92. https://doi.org/10.1371/journal.pgen.1010376
Li J, Lu L, Liu L, Ren X, Chen J, Yin X, et al. HDAC1/2/3 are major histone desuccinylases critical for promoter desuccinylation. Cell Discov. 2023;9:85–101. https://doi.org/10.1038/s41421-023-00573-9
Federico M, Bagella L. Histone deacetylase inhibitors in the treatment of hematological malignancies and solid tumors. J Biomed Biotechnol. 2011;2011:1–12. https://doi.org/10.1155/2011/475641
Guerrant W, Mwakwari SC, Chen PC, Khan SI, Tekwani BL, Oyelere AKJC. A structure–activity relationship study of the antimalarial and antileishmanial activities of nonpeptide macrocyclic histone deacetylase inhibitors. 2010;5:1232-5. https://doi.org/10.1002/cmdc.201000087.
Cheshmazar N, Hamzeh-Mivehroud M, Nozad Charoudeh H, Hemmati S, Melesina J, Dastmal-chi S. Current trends in development of HDAC-based chemotherapeutics. Life Sci. 2022;308. https://doi.org/10.1016/j.lfs.2022.120946.
Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. https://doi.org/10.1038/nbt1272
Sawas A, Radeski D, O’Connor OA. Belinostat in patients with refractory or relapsed peripheral T-cell lymphoma: a perspective review. Ther Adv Hematol. 2015;6:202–8. https://doi.org/10.1177/2040620715592567
Sharma S, Bailey H, Stenehjem D. Panobinostat for the treatment of multiple myeloma: the evidence to date. J Blood Med. 2015;6:269–76. https://doi.org/10.2147/jbm.S69140
Reddy SA. Romidepsin for the treatment of relapsed/refractory cutaneous T-cell lymphoma (mycosis fungoides/Sézary syndrome): Use in a community setting. Crit Rev Oncol Hematol. 2016;106:99–107. https://doi.org/10.1016/j.critrevonc.2016.07.001
Ning ZQ, Li ZB, Newman MJ, Shan S, Wang XH, Pan DS, et al. Chidamide (CS055/HBI-8000): a new histone deacetylase inhibitor of the benzamide class with antitumor activity and the ability to enhance immune cell-mediated tumor cell cytotoxicity. Cancer Chemother Pharmacol. 2011;69:901–9. https://doi.org/10.1007/s00280-011-1766-x
Han H, Feng X, He T, Wu Y, He T, Yue Z, et al. Discussion on structure classification and regulation function of histone deacetylase and their inhibitor. Chem Biol Drug Des. 2023;103:14366–84. https://doi.org/10.1111/cbdd.14366
Yu F, Ran J, Zhou J. Ciliopathies: does HDAC6 represent a new therapeutic target? Trends Pharmacol Sci. 2016;37:114–9. https://doi.org/10.1016/j.tips.2015.11.002
San JE, Gimenez C, Agirre X, Prosper F. HDAC inhibitors in acute myeloid leukemia. Cancers. 2019;11:1794–17. https://doi.org/10.3390/cancers11111794
Thomas M, Clarhaut J, Tranoy-Opalinski I, Gesson JP, Roche J, Papot S. Synthesis and biological evaluation of glucuronide prodrugs of the histone deacetylase inhibitor CI-994 for application in selective cancer chemotherapy. Bioorg Med Chem. 2008;16:8109–16. https://doi.org/10.1016/j.bmc.2008.07.048
Jespersen H, Olofsson Bagge R, Ullenhag G, Carneiro A, Helgadottir H, Ljuslinder I, et al. Phase II multicenter open label study of pembrolizumab and entinostat in adult patients with metastatic uveal melanoma (PEMDAC study). Ann Oncol. 2019;19:415–21. https://doi.org/10.1093/annonc/mdz394.068
Huang R, Zhang X, Min Z, Shadia AS, Yang S, Liu X. MGCD0103 induces apoptosis and simultaneously increases the expression of NF‑κB and PD‑L1 in classical Hodgkin’s lymphoma. Exp Ther Med. 2018;16:3827–34. https://doi.org/10.3892/etm.2018.6677
Nakagawa-Saito Y, Saitoh S, Mitobe Y, Sugai A, Togashi K, Suzuki S, et al. HDAC Class I inhibitor domatinostat preferentially targets glioma stem cells over their differentiated progeny. Int J Mol Sci. 2022;23:8084–95. https://doi.org/10.3390/ijms23158084
Shinke G, Yamada D, Eguchi H, Iwagami Y, Asaoka T, Noda T, et al. Role of histone deacetylase 1 in distant metastasis of pancreatic ductal cancer. Cancer Sci. 2018;109:2520–31. https://doi.org/10.1111/cas.13700
Zhang RH, Guo HY, Deng H, Li J, Quan ZS. Piperazine skeleton in the structural modification of natural products: a review. J Enzyme Inhib Med Chem. 2021;36:1165–97. https://doi.org/10.1080/14756366.2021.1931861
Hatnapure GD, Keche AP, Rodge AH, Birajd-ar SS, Tale RH, Kamble VM. Synthesis and biological evaluation of novel piperazine derivatives of flavone as potent anti-inflammatory and antimicrobial agent. Bioorg Med Chem Lett. 2012;22:6385–90. https://doi.org/10.1016/j.bmcl.2012.08.071
Li R, Wu J, He Y, Hai L, Wu Y. Synthesis and in vitro evaluation of 12-(substituted aminomethyl) berberrubine derivatives as anti-diabetics. Bioorg Med Chem Lett. 2014;24:1762–5. https://doi.org/10.1016/j.bmcl.2014.02.032
Cao F, Zwinderman MRH, van Merkerk R, Ettema PE, Quax WJ, Dekker FJ. Inhibitory selectivity among class I HDACs has a major impact on inflammatory gene expression in macrophages. Eur J Med Chem. 2019;177:457–66. https://doi.org/10.1016/j.ejmech.2019.05.038
Ruzic D, Ellinger B, Djokovic N, Santibanez JF, Gul S, Beljkas M, et al. Discovery of 1-Benzhydryl-Piperazine-Based HDAC inhibitors with anti-breast cancer activity: synthesis, molecular modeling, in vitro and in vivo biological evaluation. Pharmaceutics. 2022;14:2600–22. https://doi.org/10.3390/pharmaceutics14122600
Trivedi P, Adhikari N, Amin SA, Bobde Y, Ganesh R, Jha T, et al. Design, synthesis, biological evaluation and molecular docking study of arylcarboxamido piperidine and piperazine-based hydroxamates as potential HDAC8 inhibitors with promising anticancer activity. Eur J Pharm Sci. 2019;138:105046–60. https://doi.org/10.1016/j.ejps.2019.105046
Li L, Mei DT, Zeng Y. HDAC2 promotes the migration and invasion of non-small cell lung cancer cells via upregulation of fibronectin. Biomed Pharmacother. 2016;84:284–90. https://doi.org/10.1016/j.biopha.2016.09.030
Acknowledgements
This work was supported by the National Science Foundation for Young Scientists of China to Yepeng Luan (NSFC No.81602947).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cai, C., Luan, Y. Design, synthesis and antitumor activity evaluation of novel benzamide HDAC inhibitors. Med Chem Res 33, 748–763 (2024). https://doi.org/10.1007/s00044-024-03210-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-024-03210-6