Abstract
Valosine containing protein (VCP/p97), which involves in several important cellular functions and plays an important role in ubiquitin-mediated misfolding protein degradation, has been found to be a novel target for the treatment of a range of cancers, such as lung cancer and breast cancer. In this study, the atom-based three-dimensional quantitative structure–activity relationship (3D-QSAR) and docking models were developed using PHASE and GLIDE modules of Schrödinger software, respectively. The theoretical models were generated from 38 N-benzylpyrimidin-4-amine inhibitors of p97. An AADRRR model consisting of two hydrogen bond acceptors (A), one hydrogen bond donor (D), and three aromatic rings (R) was obtained. Thirty eight derivatives were divided into a training set with 27 molecules to generate 3D-QSAR models and a test set with 11 molecules to validate 3D-QSAR model. A robust QSAR model with good prediction in internal and external verification was constructed, where R2, Q2, and Pearson-R were 0.924, 0.701, and 0.8783, respectively. QSAR model showed the hydrogen bond donor, electron-withdrawing group, and hydrophobic characteristics affecting the p97 activity. Molecular docking studies indicated that the H-bond and hydrophobic interactions existed between the inhibitors and p97, which was consistent with the results of 3D-QSAR. These results provided some useful information for designing new and effective p97 inhibitors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Valosine containing protein (VCP, also called p97 or Cdc48 in yeast), a member of the AAA ATPase family, plays a crucial part in the processes of cellular activities, including proteostasis, endoplasmic reticulum-associated degradation (ERAD), golgi reassembly, and autophagosome maturation (Ding et al. 2019; Bursavich et al. 2010; Alverez et al. 2015; Song et al. 2003; Wang et al. 2004; Smith et al. 2011). The cellular activities of p97 protein mainly depend on its binding to different protein cofactors. For example, the ubiquitin–proteasome system was regulated as p97 binds to Npl4/Ufd1, whereas the lysosomal function was affected when p97 binds to UBXD1 (Qiu et al. 2007). As a homehexamer, p97 is composed of three major regions: one N-terminal domain and two AAA ATPs enzyme domains (D1 and D2; Ding et al. 2019; Zhang et al. 2017; Alverez et al. 2015; Meyer, Weihl 2014). The flexible N-terminal domain is responsible for various cofactors that interact with various substrate proteins (Zhou et al. 2015). The D1 domain is the basal ATPase activity, partly due to ADP’s low dissociation rate (Meyer et al. 2012), while the D2 domain is believed to the major ATPase activity of p97 under physiological conditions due to its higher ATP Km and faster ATP hydrolysis to ADP (Wang et al. 2019; Song et al. 2003). Numerous studies reported that VCP/p97 ATPase played a key role in the degradation of misfolded membranes and secreted proteins by removing misfolded poly-ubiquitinated proteins from the ER into the cytoplasm and then transporting them to the proteasome for degradation (Segura-Cabrera et al. 2017; Chou et al. 2011). Therefore, p97 is an attractive target for diseases involving excessive ERAD, such as cancer and cystic fibrosis (Chapman et al. 2011; Cabrera et al. 2017).
Related studies have shown that the high expression of p97 in lung cancer, prostate cancer, pancreatic cancer, and breast cancer was associated with poor prognosis of the tumor (Ding et al. 2019). In many physiological and pathological processes, p97 mainly controls the protein quality by eliminating abnormal proteins in cells and maintains the survival of normal cells and tumor cells (Smith et al. 2011; Chou et al. 2011). VCP/p97 has been an attractive target for cancer due to VCP/p97-induced protein cytotoxic ER stress and cell death in cancer cells (Cabrera et al. 2017), thus several papers about p97 inhibitors have been published in recent years (Chapman et al. 2011; Chou and Deshaies 2011; Chapman et al. 2015). The most famous reversible p97 inhibitor is CB-5083, which selectively acts on the D2 domain, competitively inhibits ATPase activity and has good oral bioavailability (Le Moigne et al. 2017; Wang et al. 2019; Tang et al. 2019). Although CB-5083 is of excellent antitumor activity and has entered phase I clinical trials, the toxicity of CB-5083 impeded further clinical study (Zhou et al. 2015; Chapman et al. 2015). Therefore, it is necessary to develop new methods to illustrate the mechanism between molecules and p97, and provide inspiration for designing and discovering novel p97 inhibitors.
The computer-aided drug design technology can predict the biological activity of a compound by its association with its structural properties, and can also be used to design new active molecules (van Vlijmen et al. 2017). Especially quantitative structure–activity relationship (QSAR) studies have been successfully applied to the bioactivity modeling of natural and synthetic chemicals. In this manuscript, we construct atom-based three-dimensional (3D)-QSAR models studies using 38 N-benzylpyrimidin-4-amine derivatives to obtain key groups of p97 inhibitors, which was performed in the PHASE (PHASE 2015) module of Schrödinger suite. 3D-QSAR model suggested that the hydrogen bond donor, electron-withdrawing group, and hydrophobic characteristics of inhibitors affected the p97 activity. To verify the conclusion of 3D-QSAR model, the molecular docking was carried out by Glide (GLIDE 2015) module of Schrödinger suite, which was consistent with the results of 3D-QSAR.
Materials and methods
Data set
Thirty eight N-benzylpyrimidin-4-amine derivatives of p97 inhibitors (Table 1) were selected from the published work and used in our study (Zhou et al. 2015; Wang et al. 2019). Inhibitory potencies of the compounds were reported as IC50 values varying from 0.015 to 21.865 μM and then were converted to pIC50 using the formula pIC50 = −lg IC50 (Evans et al. 2007). The original data set was randomly divided into training and test set comprising 27 and 11 molecules, respectively. The training set was used to generate pharmacophore models and the test set was used to validate the expected models.
Pharmacophore modeling
Based on a set of active molecules, the development of pharmacophore model is the primary mean of generating a hypothetical phase of the pharmacophore, which is then used to construct the 3D-QSAR model. In this 3D-QSAR study, PHASE version 4.3 implemented in the Schrödinger software package (Maestro 2015) was used.
Ligand preparation
The two-dimensional (2D) structure of 38 molecules were drawn using Maestro version 10.2 (Maestro 2015). These 2D structures were converted into the 3D structures using LigPrep module (LigPrep 2015) incorporated in PHASE, and then these compounds were subjected to energy minimization and geometry optimization using OPLS-2005 as force field, with distance-dependent dielectric model (Dixon et al. 2006). The rapid twist angle method “ConfGen” was used to generate their conformers for each ligand and to eliminate structures with high estimated energies. For each structure, a maximum of 1000 conformers were generated using preprocess minimization of 100 steps and postprocess minimization of 50 steps. Each minimized conformer was filtered through a relative energy window of 10 Kcal/mol and a minimum atom deviation of 1.00 Å. This value (10 Kcal/mol) sets the energy threshold relative to the lowest energy conformer. Conformers having higher energy than the threshold were discarded. The root mean square deviation (RMSD) of all pairs of corresponding heavy atoms must be below this cutoff for two conformers to be considered identical. This criterion is applied only after the energy difference threshold and only if two conformers are within 1 Kcal/mol.
Creation of pharmacophoric sites
During the creation of pharmacophoric sites, the pharmacophore features were used to define the model or add new features, and then pharmacophore sites were created for the features in each conformer of ligand. The six built-in pharmacophore features, hydrogen bond acceptor (A), hydrogen bond donor (D), hydrophobic group (H), aromatic ring (R), positively charged group (P), and negatively charged group (N) were used to create pharmacophore sites. The compounds were defined as active (pIC50 > 6.5) and inactive (pIC50 < 6.0) ligands by setting the activity thresholds.
Finding common pharmacophore and scoring hypotheses
The pharmacophore features were produced by a set of variants that were tree-based partitioning technique to define pharmacophore characteristics. The final box size is 1.0 Å and its size determines the size of the matching tolerance. The smaller the box is, the tighter it matches. The appropriate pharmacophore hypothesis was generated by setting the number of sites and matched active molecules. In this study, the number of sites was varied from six to four until at least one hypothesis was identified, and all active molecules must be matched to the pharmacophore model. The scoring function was used to score the common pharmacophore hypotheses, with RMSD values <1.2 Å, vector scores values at 0.5 and the weight of the term to the default value. After analyzing the scores and alignments of the active ligands, the optimal pharmacophore hypothesis was selected for further study.
Construction of 3D-QSAR model
The QSAR model divides the space occupied by the ligand into cubic grids. Regression was completed by constructing a model of partial least squares (PLS) factor. The actual and predicted activity values of 27 training set molecules were analyzed and compared, and an atom-based 3D-QSAR model was constructed. In the process of constructing 3D-QSAR, the PLS factor was no >1/5 of the number of molecules in the training set and the grid spacing is 1 Å. All models were validated by predicting the activity values of the test set.
Validation of pharmacophore model
External verification is a very important step in the design of 3D-QSAR models, which is the basis for judging whether the constructed model has strong stability and excellent prediction. Validation was performed by fitting statistics of training sets and test sets, including R regression values (R2), regression standard deviations (SD), variance ratios (F), significance level of variance ratio (P), root mean square error of the test set (RMSE), value of Q2 for the predicted activities of the test set (Q2) and Pearson-R. In this study, the model was validated by the ability of the model generated by the training set molecules to predict the molecular activity of the test set, and the correlation between the experimental values of the test set and the predicted values was compared.
Molecular docking
To investigate the interaction between N-benzylpyrimidin-4-amine derivatives and key parts of the VCP/P97, molecular docking study was performed with the Glide. The p97 X-ray crystal structure (PDB ID: 6MCK) complexed with CB-5083 at the resolution of 3.77 Å was obtained from the RCSB protein data bank (Tang et al. 2019). Before molecular docking, the Protein Preparation Wizard in the Maestro 2015 program of Schrödinger software was successively used to add hydrogens, remove water molecules, and minimize energy at the force field of OPLS-2005. Then the receptor grid was generated in the Receptor Grid Generation panel of Glide and a 20 × 20 × 20 Å cube box was set to define the docking sites for the ligand. Subsequently, the ligands were docked into the receptor based on the grid using extra-precision (XP) mode, and the most active compound and the least active compound were used for further analysis. Finally, the ligands were scored primarily based on the Glide score, the H-bond length, and the number of hydrogen bonds formed.
Results and discussion
Pharmacophore and 3D-QSAR models
Pharmacophore model and 3D-QSAR studies were performed successfully on a series of N-benzylpyrimidin-4-amine derivatives to understand the important pharmacophoric characteristics for p97 inhibitors. The activity values of these 38 molecules ranged from 4.66 to 7.83, spanning three orders of magnitude. The activity threshold was divided into active and inactive molecules by >6.5 (containing 20 active molecules) and <6.0 (containing ten inactive molecules). The pharmacophore model was generated using active molecules. The 20 active molecules were matched using a tree-based partitioning algorithm, while four hypotheses based on six-pharmacophore characteristics were generated in the variant list. Screening a common pharmacophore from these four variants, we finally determined that there were six-pharmacophore features (AADRRR) consisting of hydrogen bond acceptor (A), hydrogen bond donor (D), and aromatic rings (R). The pharmacophore hypothesis AADRRR was aligned with all active molecules, as shown in Fig. 1. Twenty molecules in the active group matched the hypothetical AADRRR. The hypothesis of the group feature (Fig. 2a) was followed by a scoring function and 3D-QSAR study. The summary of the result was shown in Supplementary Table 1.
The results of the six-pharmacophore feature hypotheses with PLS of 3, labeled AADRRR.576, AADRRR.551, and AADRRR.552, were presented in Table 2. These hypotheses were aligned with the training set and finally the test set were used to verify the predictive activity of these three hypotheses.
Analysis of PHASE 3D-QSAR validation
The pharmacophore models were evaluated by statistical parameters R2, Q2, SD, F, P, RMSD, and Pearson-R, an optimal model is the one with higher R2 and F, and lower SD, P, and RMSD (Suryanarayanan et al. 2012). From Table 2, we can conclude that the hypothesis AADDHR.576 yielded a statistically significant 3D-QSAR model with PLS factors (R2 = 0.9238, SD = 0.2389, F = 92.8, and P = 5.402e−0.13) for training set of 27 compounds and PLS factors (Q2 = 0.7012, RMSE = 0.3635, and Pearson-R = 0.8783) for test set of 11 compounds. In addition, another way to verify the optimal model is comparing the magnitude of the value of Q2. Compared with other two hypotheses, AADRRR.576 showed good external prediction ability for its high Q2 value (0.7012). Therefore, as the best model among these three hypotheses, AADRRR.576 would be explored for further 3D-QSAR study. The hypothesis matched the ligand fitness scoring 3, as shown in Fig. 2b. In terms of hypothesis AADRRR.576, the distance and angle data between different pharmacophore feature points were shown in Supplementary Tables 2 and 3, respectively.
In the validated 3D-QSAR model, the test set was used to evaluate the validity and predictability of the hypothesis AADRRR.576. In this study, the activity of the 11 molecules in test set were predicted using hypothesis AADRRR.576, then the values of predicted and actual activity were listed in Table 1. According to Alexander Torpsha’s research (Tropsha 2010), when the value of R2 is >0.6 and Q2 is >0.5, the model could be considered to have strong robustness and excellent predictive ability. The correlation value Q2 between the predicting activity and actual activity of test set is 0.701 (Fig. 3), indicating that AADRRR.576 could rationally predict p97 inhibitory activity.
Analysis of atom-based PHASE 3D-QSAR model
According to the characteristics of the model, the relevance of the activity in the hypothesis are explained and the QSAR visual analysis of the model is described. The QSAR visualization is performed in the form of a 3D contour map associated with the model of the atom and structural features, and the spatial distribution contributed to the model can be viewed. Figure 4 shows the effect of electron withdrawing, hydrogen bond donor and hydrophobicity of the most active compound 3 and the least active compound 27 on the inhibitory activity, respectively. For the selected pharmacophore, the blue cubes indicated favorable regions, whereas red cubes indicated unfavorable regions for p97 inhibitory activity. Increasing the number of blue cubes certainly would enhance the inhibition of p97, while the inhibition of p97 alleviates as the number of red cubes increases.
Figure 4a shows the hydrogen bond donor characteristics for the selected hypothesis. The blue region seen at the NH2 of amide group on the most active compound 3 was favorable for inhibitory activity, and the addition of a hydrogen bond donor at the amino site would result in an increase in the activity of the p97 inhibitor. Adding a hydrogen bond donor to the nitrogen atom on the six-membered ring of molecule 27 would result in a decrease in inhibitor activity. Figure 4b illustrates the electron-withdrawing characteristics of the selected hypothesis. The visual analysis of Fig. 4b shows that the blue cubic cluster at the amide bond of the indole ring represents the positive potential of the electron-withdrawing feature of the molecule and is essential for inhibition activity. Therefore, it can be considered that adding a suitable electron-withdrawing group to the amino group and the imino group would increase the inhibitory effect of compound 3. However, the electron-withdrawing group at C2 position of the indole ring resulted in a decrease in receptor binding, which decrease the inhibitory activity of the compound. It was also found by visual analysis that the electron-withdrawing group at the C4 position increased the activity of 3, while the electron-withdrawing group at the C6 position decreased the activity. For example, compound 30 (IC50 = 0.354 μM) with a cyano group at the C4 is more active than compound 27 (IC50 = 21.865 μM) with a cyano group at C6. Similarly, Fig. 4c illustrates the significant favorable regions and unfavorable hydrophobic interactions that arise when the QSAR model is applied to the most active compound 3 and most inactive compound 27. The red region on the six-membered ring indicates that a hydrophobic group on this position reduces the inhibitory activity. Although there are few blue cubes in six-membered ring, the inhibition of p97 still slightly increases. The blue regions around C4 and C6 on the anthracene ring indicate that hydrophobic group at these positions can enhance the inhibition of p97.
Molecular docking analysis
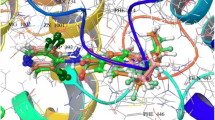
Docking studies are performed on Glide module to explore the interaction mechanism between inhibitors and the receptor. GLIDE conducts a complete systematic search of the conformation, orientation, and positional space of the docking ligand, and then uses a series of graded filters to search for possible positions of the ligand in the active site of the receptor (Gao et al. 2011). Firstly, in order to verify the reliability of the docking algorithm, the co-crystallized ligand CB-5083 has been redocked into the active site of p97. Figure 5 showed that the redocking conformation of CB-5083 overlapped with the conformation in the crystal structure and the RMSD value is 0.897 Å. The result demonstrates that the docking methods are reliable, as well as can restore the conformation in the crystal structure. Then, all ligands were docked into the active sites of p97 to investigate the interaction between p97 inhibitors and the key amino acid residues of p97. The docking scores ranged from −2.043 to −10.311 Kcal/mol of all ligands have been presented in Supplementary Table 4. Hydrogen bond interaction between the backbone carbonyl group of Asp478 and the NH of all inhibitors was conserved. Some inhibitors have also shown the additional hydrogen bond interactions with Thr688, Arg662, and Ala685. Hence, after analyzing all the collected molecules, it is evident that hydrogen bonding with negatively charged amino acid (Asp478) and polar amino acid (Thr688) play an essential role in the binding of ligands. For further understanding the effect of different scaffolds on their activity, the most active ligand 3 and least active ligand 27 were studied. Figure 6a illustrated the binding mode of compound 3 (most active) with p97 with a docking score of −7.82 Kcal/mol. The benzylamino moiety of compound 3 penetrates deep into the pocket, occupying a hydrophobic niche, in direct contact with the enzyme residues of Asp 478 and Thr688. Moreover, the benzylamino moiety groups trigger hydrophobic interactions with some residues in the pocket (Val 474, ILE 479, TRP 476, VAL 485, and LEU 482). The carbonyl group of Asp478 forms a hydrogen bond with the NH of compound 3 with a length of 2.52 Å and the -OH of Thr688 forms a hydrogen bond interaction with the O atom in the amide group of the molecule with a length of 2.13 Å. The docking score of compound 27 (least active) is −6.15 Kcal/mol (Fig. 6b) and it binds with amino acids Asp478 and Gly523 with the docking energy of −43.894 Kcal/mol. The difference lied in H-bond lengths that was more in compound 27 between polar hydrogen of nitrogen and carbonyl oxygen of Asp478 (2.73 Å), and polar hydrogen of -NH2 on Gly523 with cyano nitrogen (2.21 Å). This, in turn, resulted into low binding affinity of compound 27 toward p97. This observation complemented the results obtained from the 3D-QSAR model.
Conclusion
In this manuscript, we carried out a molecular modeling study on 38 N-benzylpyrimidin-4-amine derivatives as p97 inhibitors using 3D-QSAR and molecular docking. Generation of a reliable pharmacophore model AADRRR.576, 3D-QSAR model (PLS = 3) and molecular docking analysis demonstrated the involvement of H-bond donor, hydrophobic, and charged regions as crucial factors for ligand–receptor interaction. The hypothesis demonstrates pharmacophoric or functional groups and their spatial arrangement were suitable for the development of specific p97 inhibitors. With the 3D-QSAR model visualization, presence of hydrogen donor groups in the amino will favor the inhibitory activity. Substitution of electron-withdrawing groups around C4′ which is more active than the group at the C6′ favors p97 inhibitory activity. Hydrophobic interaction of the molecule with the p97 can be enhanced by incorporating hydrophobic amino acid into benzyl groups. Molecular docking not only demonstrates the way ligands interact with p97, but also further validates the contour plots generated by the 3D-QSAR model. The study about potential binding mode of the compounds from a data set of 38 p97 inhibitors with its receptor is also conducted. Analysis of the most active compound 3 and the least active compound 27, the inhibitors form a strong hydrogen bond with the key amino acid residues THR688, ASP478, and ARG662 in the protein. The hydrophobic group on the benzyl group corresponds to the hydrophobic interaction of the group in the active pocket, shown by the docking result. The results obtained from the 3D-QSAR studies and docking simulation could be used for designing new and potent p97 inhibitors.
References
Alverez C, Bulfer SL, Chakrasali R, Chimenti MS, Deshaies RJ, Green N, Kelly M, LaPorte MG, Lewis TS, Liang M, Moore WJ, Neitz RJ, Peshkov VA, Walters MA, Zhang F, Arkin MR, Wipf P, Huryn DM (2015) Allosteric indole amide inhibitors of p97: Identification of a novel probe of the ubiquitin pathway. ACS Med Chem Lett 7:182–187
Bursavich MG, Parker DP, Willardsen JA, Gao ZH, Davis T, Ostanin K, Robinson R, Peterson A, Cimbora DM, Zhu JF, Richards B (2010) 2-Anilino-4-aryl-1,3-thiazole inhibitors of valosin-containing protein (VCP or p97). Bioorg Med Chem Lett 20:1677–1679
Chapman E, Fry AN, Kang MJ (2011) The complexities of p97 function in health and disease. Mol Biosyst 7:700–710
Chapman E, Maksim N, de la Cruz F, La Clair JJ (2015) Inhibitors of the AAA+ chaperone p97. Molecules 20:3027–3049
Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, Patricelli MP, Hodder P, Rosen H, Deshaies RJ (2011) Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc Natl Acad Sci USA 108:4834–4839
Chou TF, Deshaies RJ (2011) Development of p97 AAA ATPase inhibitors. Autophagy 7:1091–1092
Ding R, Zhang T, Wilson DJ, Xie JS, Williams J, Xu Y, Ye YH, Chen LQ (2019) Discovery of irreversible p97 inhibitors. J Med Chem 62:2814–2829
Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE, Friesner RA(2006) PHASE: a new engine for pharmacophore perception 3D QSAR model Dev, 3D database Screen: 1 Methodol preliminary results. J Comput Aid Mol Des 20:647–671
Evans DA, Doman TN, Thorner DA, Bodkin MJ (2007) 3D QSAR methods: phase and catalyst compared. J Chem Inf Model 47:1248–1257
Gao L, Zu M, Wu S, Liu AL, Du GH (2011) 3D QSAR and docking study of flavone derivatives as potent inhibitors of influenza H1N1 virus neuraminidase. Bioorg Med Chem Lett 21:5964–5970
GLIDE, version 6.7 (2015) Schrödinger, LLC, New York
Le Moigne R, Aftab BT, Djakovic S, Dhimolea E, Valle E, Murnane M, King EM, Soriano F, Menon MK, Wu ZY, Wong ST, Lee GJ, Yao B, Wiita AP, Lam C, Rice J, Wang J, Chesi M, Bergsagel PL, Kraus M, Driessen C, Kiss von Soly S, Yakes FM, Wustrow D, Shawver L, Zhou HJ, Martin TG, Wolf JL, Mitsiades CS, Anderson DJ, Rolfe M (2017) The p97 Inhibitor CB-5083 is a unique disrupter of protein homeostasis in models of multiple myeloma. Mol Cancer Ther 16:2375–2386
LigPrep, version 3.4 (2015) Schrödinger, LLC, New York
Maestro, version 10.2 (2015) Schrödinger, LLC, New York
Meyer H, Bug M, Bremer S (2012) Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14:117–123
Meyer H, Weihl CC (2014) The VCP/p97 system at a Slance: connecting cellular function to disease pathogenesis. J Cell Sci 127:3877–3883
PHASE, version 4.3 (2015) Schrödinger, LLC, New York
Qiu Y, Tomita Y, Zhang B, Nakamichi I, Morii E, Aozasa K (2007) Pre-B-cell leukemia transcription factor 1 regulates expression of valosin-containing protein, a gene involved in cancer growth. Am J Pathol 170:152–159
Segura-Cabrera A, Tripathi R, Zhang XY, Gui L, Chou TF, Komurov K (2017) A structure- and chemical genomics-based approach for repositioning of drugs against VCP/p97 ATPase. Sci Rep 7:1–10
Smith MH, Ploegh HL, Weissman JS (2011) Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334:1086–1090
Song CC, Wang Q, Li CC (2003) ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J Biol Chem 278:3648–3655
Suryanarayanan V, Sudha A, Rajamanikandan S, Vanajothi R, Srinivasan P (2012) Atom-based 3D QSAR studies on novel N-β-d-xylosylindole derivatives as SGLT2 inhibitors. Med Chem Res 22:615–624
Tang WK, Odzorig T, Jin W, Xia D (2019) Structural basis of p97 inhibition by the site-selective anticancer compound CB-5083. Mol Pharm 95:286–293
Tropsha A (2010) Best practices for QSAR model development, validation, and exploitation. Mol. Inform 29:476–488
van Vlijmen H, Desjarlais RL, Mirzadegan TJ (2017) Computational chemistry at Janssen. J Comput Aid Mol Des 31:267–273
Wang Q, Song CC, Li CC (2004) Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J Struct Biol 146:44–57
Wang XY, Bai EH, Zhou H, Sha SJ, Miao H, Qin YR, Liu ZG, Wang J, Zhang HY, Lei M, Liu J, Hai O, Zhu YQ (2019) Discovery of a new class of valosine containing protein (VCP/P97) inhibitors for the treatment of non-small cell lung cancer. Bioorg Med Chem 27:533–544
Zhang T, Midshra P, Hay BA, Chan D, Guo M (2017) Valosin-containing protein (VCP/p97) inhibitors relieve Mitofusin-dependent mitochondrial defects due to VCP disease mutants. eLife 6:1–28
Zhou HJ, Wang JH, Yao B, Wong S, Djakovic S, Kumar B, Rice J, Valle E, Soriano F, Menon MK, Madriaga A, Kiss von Soly S, Kumar A, Parlati F, Yakes FM, Shawver L, Moigne RL, Anderson DJ, Rolfe M, Wustrow D (2015) Discovery of a first-in-class, potent, selective, and orally bioavailable inhibitor of the p97 AAA ATPase (CB-5083). J Med Chem 58:9480–9497
Acknowledgements
This research work was supported by the Technology Support Program of Science and Technology Department of Jiangsu Province (BE2015703) and the Major Program for the Natural Science Fundamental Research of the Higher Education Institutions of Jiangsu Province (15KJA350002). We thank Yadong Chen’s group (College of Science, China Pharmaceutical University) for providing the computational facilities, Schröedinger, for this research work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Gu, C., Zhang, Y., Xie, X. et al. Pharmacophore modeling, atom-based 3D-QSAR and molecular docking studies on N-benzylpyrimidin-4-amine derivatives as VCP/p97 inhibitors. Med Chem Res 29, 727–737 (2020). https://doi.org/10.1007/s00044-020-02517-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-020-02517-4