Abstract
Imerubrine, a biologically active natural product, is one of the initial members of tropoloisoquinolines and biosynthetically related to the more common azafluoranthene alkaloids. We perform a comprehensive quantum chemical analysis on imerubrine using density functional theory at B3PW91/6-311 + G(d,p) level. The equilibrium molecular structure of imerubrine has been obtained. The weak intra-molecular C–H⋯O interactions are recognized, characterized and quantified by quantum theory of atoms in molecule and relaxed force constants. The chemical reactivity of imerubrine is explained and discussed with the help of highest occupied molecular orbital, lowest unoccupied molecular orbital and molecular electro static potential surfaces as well as a number of reactivity descriptors. The infrared spectrum of imerubrine has been calculated and the vibrational modes have been assigned on the basis of the potential energy distribution with the highest possible accuracy. The nuclear magnetic resonance spectra of imerubrine have been calculated, analyzed and compared with available experimental data. A good agreement between experimental and calculated values has been observed. The molecular docking of imerubrine into B1 bradykinin receptor (PDB ID: 1HZ6) shows that it is capable to bind with the receptor and hence, it can act as an effective bradykinin receptor agonist.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Imerubrine is one of the initial members of a rare class of naturally occurring tropoloisoquinolines (Buck, 1984), which is mainly extracted from Abuta imene of the Menispermacae family. It is biosynthetically related to the more common azafluoranthene alkaloids (Zhao and Snieckus, 1984; Boger and Brotherton, 1984). The azafluoranthene alkaloids show various properties of biological and technological interest. They have been patented as constituents of wound-healing agents (Lewis et al., 1992) and have been reported to possess antidepressant activity (Schwan, 1976). Furthermore, Scherowsky et al. (1997) synthesized the azafluoranthene derivative and determined its crystal structure, confirming the suitability of the tetracycle for the formation of discotic phases, and attempted to develop new approaches to discotic liquid crystals, forming tilted columnar phases with ferroelectric properties. The natural products of the azafluoranthene family and their unnatural analogs can be synthesized by direct arylation (Ponnala and Harding, 2013) and electrocyclization (Silveira et al., 2009). The total syntheses of imerubrine and related molecules based on cycloaddition have been reported by Boger and Takahashi (1995), as well as Lee and Cha (2001).
Recently, a comparative quantum chemical investigation on two azafluoranthene natural products, triclisine and rufescine, has been carried out (Srivastava et al., 2015). Imerubrine is a close analog of rufescine in which additional oxygen is substituted at a six membered ring. It seems, therefore, interesting to compare the molecular properties of imerubrine with those of rufescine. Vibrational spectroscopy provides immensely invaluable information about the structure and properties of molecules if used in synergy with quantum chemical calculations. Prediction of vibrational frequency of molecules by quantum chemical computation has become very popular (Srivastava et al., 2014b; Srivastava et al., 2014c; Kumar et al., 2015; Srivastava et al., 2014a) because of its accurate and therefore consistent description of the experimental data. Inter-molecular and intra-molecular bonding have important consequences on the structure and activity of the molecule (Jeffrey and Saenger, 1991). Particularly, C–H⋯O interactions are important in crystal engineering (Aakeroy and Seddon, 1993) due to their influence on packing motifs (Desiraju et al., 1993). Biologically, their occurrence in carbohydrate (Steiner and Saenger, 1992) and nucleosides (Saenger, 1984) can never be ignored. Using quantum theory of atoms-in-molecule (QTAIM) (Bader, 1990) method in the present study, we establish imerubrine as a C–H⋯O interactions rich system. The detection, characterization and estimation of C–H⋯O interactions have been efficiently performed within the QTAIM. In addition, the strength of these interactions is also described by relaxed force constant (RFC) which has been conceived as a “chemically more meaningful bond strength parameter than the regular force constant” (Swanson, 1976).
Methodology
The initial geometry of imerubrine molecule was modeled by standard geometrical parameters as implemented in Gauss View 5.0 program (Dennington et al., 2005) and then, it was optimized without any symmetry constraint in the potential energy surface (PES) using a hybrid functional B3PW91 (Becke, 1993; Perdew and Wang, 1992) in combination with 6-311 + G(d,p) basis set. The vibrational IR frequency calculations were performed using optimized geometry at the same level of theory. All frequencies were found to be positive ensuring the geometry corresponding to true minimum in the PES. The calculated frequencies were scaled by the factor of 0.9648 (Merrick et al., 2007) in order to account for anharmonicity of vibrations and other basis set deficiencies. All calculations were performed using Gaussian 09 program package (Frisch et al., 2010). The wavefunction output of imerubrine generated by Gaussian 09 was employed for QTAIM analysis which is performed with AIMAll program (Keith, 2012). The computations were performed with the help of a computer cluster using 8 processors.
Results and discussion
Molecular structure and intra-molecular interaction
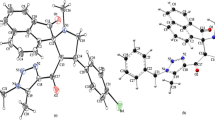
The optimized structure of imerubrine has been displayed in Fig. 1a. Imerubrine consists of four planar ring systems: three six-membered (R1, R2, and R4) and one five-membered (R3). The upper ring R1 has nitrogen atom in the core thus making it a heterocycle. The lower rings R2 and R4 are substituted with three –OCH3 groups and one –OCH3 as well as =O groups, respectively. The structure of imerubrine appears to be a close analog of rufescine with a positional change in –OCH3 group and substitution of =O in the ring R4. The optimized parameters (bond lengths and angles) of imerubrine can be found in supplementary Table S1.
QTAIM describes the chemical bonding and structure of the chemical system based on the topology of the electron density (ρ). In addition to bonding, QTAIM allows the calculation of certain physical properties on a per atom basis, by dividing space up into atomic volumes containing exactly one nucleus which acts as a local attractor of the electron density. The bonding is characterized by the presence of a bond critical point (BCP) between the atomic pairs. Molecular graph of imerubrine is shown in Fig. 1b which clearly reveals three intra-molecular interactions (by dotted lines). Topological parameters associated with these C–H⋯O interactions are given in Table 1. According to Rozas et al. (Rozas et al., 2000), the nature of C–H⋯O bonds are characterized as weak and electrostatic in nature due to Laplacian, ∇2 ρ > 0 and total electron energy density, H > 0. Espinosa et al. (Espinosa et al., 1998) have proposed proportionality between hydrogen bond energy (ΔE) and potential energy density (V) at BCP as:
According to this equation, the energy of O20⋯H32, O20⋯H28 and O25⋯H19 interactions are calculated to be 2.94, 2.84 and 1.35 kcal/mol, respectively. Evidently, all these interactions are too weak to provide any significant stabilization within the molecule. However, these interactions are relatively larger in strength as compared to those in rufescine (Srivastava et al., 2015).
In order to get further insights into strength of these C–H⋯O interactions, we have also evaluated their RFC values. Use of the compliance constant, which is inverse of the force constant matrix elements, over the regular force constant, has been addressed by many workers (Brandhorst and Grunenberg, 2008; Jones and Swanson, 1976; Madhav and Manogaran, 2009; Majumder and Manogaran, 2013). The reciprocal of the diagonal compliance matrix element is known as the RFC which was successfully applied to the covalent bonds and also extended to non-covalent interactions like hydrogen bonding (Brandhorst and Grunenberg, 2008). Nevertheless, it was brought up that the RFCs of many non-bonded pairs have close values as bonded pairs in several molecules (Baker and Pulay, 2006; Baker, 2006). The compliance constants are calculated directly by using:
where R kk is the deviation in O⋯H bond distance for partial optimization and ΔE is the difference in energies of partial and full optimization of the molecule. Therefore, RFC values are calculated as:
The calculated RFC values further suggest the order of strength of C–H⋯O interactions, O20⋯H32 > O20⋯H28 > O25⋯H19, which are in accordance with the QTAIM calculations.
Vibrational spectroscopic analysis
Imerubrine (C20H17NO5) contains 43 atoms and therefore, 123 normal modes (3N-6) of vibration. For the sake of simplicity of discussion, we have assigned all vibrational modes up to 400 cm−1. These assignments have made on the basis of potential energy distribution (PED) calculated by VEDA 4 program (Jamroz, 2004; 2013). The internal coordinates are optimized repeatedly to maximize the PED contributions and then, the modes with PED < 15 % have been neglected. Fig. 2 plots calculated IR spectrum for a visual indication. We discuss the significant mode of vibrations in the subsections below.
C–H vibrations
Aromatic compounds commonly exhibit multiple weak bands in the region 3100–3000 cm−1 (George, 2001), which is characteristic region for the identification of C–H stretching vibration. Hence, the infrared bands appearing in the region 3108–3062 cm−1 have been assigned to C–H stretching vibrations associated with ring systems. The C–H modes associated with –OCH3 groups are found between 3047 and 2914 cm−1.
The bands due to C–H in-plane bending vibrations are observed in the region 1300–1000 cm−1. In imerubrine, these modes are calculated at 1270, 1265, and 1221 cm−1. The C–H out-of-plane bending vibrations are strongly coupled vibrations, occurring in the region 1000–750 cm−1 (Sundaraganesan et al., 2007). The C–H out-of-plane bending vibrations are observed in the region 866–682 cm−1 in imerubrine.
C–C vibrations
The bands ranging 1650–1200 cm−1 in the aromatic ring compounds are assigned to C–C stretching modes. These modes are sensitive to the substitution. However, the actual positions are determined not so much by the nature of the substitution but by their positions (Arjunan et al., 2013). The bands calculated in the range 1602–1473 cm−1 have been assigned to C–C stretching vibrations. The C–C in-plane and out-of-plane bending vibrations are identified at lower frequencies and listed in Table 2.
C–N vibrations
In aromatic compounds the C–N stretching vibrations usually lies in the region 1400–1200 cm−1. The identification of C–N stretching frequency is a difficult task due to the mixing of vibrations in this region (Arivazhagan and Jevavijayan, 2011; Krishnakumar and Balachandran, 2005). The infrared band appearing at 1374 cm−1 has been assigned to C–N stretching vibration. The in-plane and out-of-plane bending C–N vibrations have also been identified and assigned in Table 2.
C–O and C=O vibrations
In imerubrine, the strong absorption band at 1519 cm−1 is assigned to the stretching of C=O group. The C–O stretching vibrations are obtained in the region 1356–884 cm−1, i.e., in a lower frequency region in the present case due to the delocalization of lone pair of electrons. These observations are in good agreement with the literature value (Arjunan et al., 2004; Mukherjee and Mishra, 1996). The other mode of vibrations are also assigned within the characteristic region and presented in Table 2.
Nuclear magnetic resonance (NMR) spectroscopic analysis
NMR provides the detailed information for the structural prediction of large bio-molecules (Schlick, 2010). The geometry of the studied compound, together with that of tetramethylsilane (TMS) is fully optimized at the same level of theory. 1H and 13C-NMR chemical shifts are calculated with the “gauge-independent atomic orbital” approach at B3PW91/6–311 + G(d,p) method. The chemical shift of any “x” proton (δ X) is equal to the difference between isotropic magnetic shielding (IMS) of TMS and proton (x) and also reported in parts per million (ppm) relative to TMS for 1H and 13C-NMR spectra. It is defined by the equation: δ X = IMSTMS−IMSX. The IMS values of C and H atoms of imerubrine are displayed in Fig. 3. The calculated values for 1H and 13C-NMR chemical shifts are listed in Table 3 and compared with corresponding experimental values reported by Boger and Takahashi, 1995.
The chemical shift of non-equivalent protons have different chemical shifts and also because of the powerful deshielding due to the non-bonding electrons of carbon atom. So the chemical shift observed for the 7H, 11H, 18H, and 19H are 8.02, 8.94, 7.37, and 7.64 respectively and are in good agreement with the experimental adta. The methyl group hydrogen atom of imerubrine absorbs far up field and the chemical shift of 23H, 24H and 27H are found to be 4.14, 3.97 and 4.23 ppm respectively. Its corresponding experimental values are 4.04, 4.00, and 4.12 ppm, which are also in good agreement with experimental data. These unusual shifts are due to diamagnetic anisotropy.
Aromatic carbon atom gives peaks in the range of 100–150 ppm in overlapped areas of the spectrum. The NMR peak of carbon atom 5C, 13C, and 15C are calculated at the 128.3, 145.3, and 114.0 ppm, which agree well with corresponding experimental values. Nitrogen atom increases the electron density of carbon atoms 9C and 10C hence corresponding 13C-NMR peaks are obtained at the 163.0, and 152.0 ppm, respectively. For the methyl group carbon of imerubrine chemical shift are observed at 62.2 and 57.6 ppm. All the theoretical data of 13C-NMR spectra are also in good agreement with experimental data, which are shown in Table 4.
Chemical reactivity
The chemical reactivity reflects the susceptibility of a molecule towards a specific chemical reaction. It plays a key role in the design of new molecules as well as in the understanding of biological systems and materials science. The chemical reactivity of imerubrine is described by various reactivity surfaces and reactivity descriptors as below.
Highest occupied molecular orbital (HOMO), lowest unoccupied molecular orbital (LUMO), and molecular electro static potential (MESP) analyses
The frontier molecular orbitals, namely, the highest occupied (HOMO) and lowest unoccupied (LUMO) are important due to the fact that they explore the way, a molecule interacts with other species. The energy difference between HOMO and LUMO, i.e., HOMO–LUMO gap is recognized as a stability index of the molecular system. A large HOMO–LUMO gap can be associated with high-kinetic stability because it is energetically unfavorable to add electrons to a high-lying LUMO or to extract electrons from a low-lying HOMO and so to form the activated complexes of any potential reaction (Manolopoulos et al., 1991). The HOMO and LUMO surfaces of imerubrine are plotted in Fig. 4. One can see that the HOMO is delocalized over rings R2, R3, and R4 excluding upper ring (R1). This is in contrast to rufescine in which HOMO is delocalized over whole molecule (Srivastava et al., 2015). On the contrary, the LUMO of imerubrine is delocalized over whole rings including R1. Therefore, the transition from HOMO→LUMO indicates the charge transfer to upper ring (R1). This can be expected due to the fact that the upper ring contains electronegative nitrogen atom. The HOMO–LUMO gap of 2.93 eV measures the strength of this charge transfer interaction. This is smaller than that of rufescine (3.56 eV), suggesting more chemically reactive nature of imerubrine.
The MESP is related to the electron density and is very useful in understanding the sites for electrophilic (electronegative region) and nucleophilic (electropositive region) reactions (Luqul et al., 2000). MESP is also well suited for analyzing process based on the “recognition” of one molecule by another, as in drug receptor binding and enzyme-substrate interactions, because it is through their potentials that the two species first “see” each other (Scrocco andTomasi, 1973). The MESP surface of imerubrine is also displayed in Fig. 4 in color coding scheme. The color code in the title molecule ranges between −0.07022 a.u. for deepest red and +0.07022 a.u. for deepest blue, where red and blue indicate the most electronegative, i.e., electron rich region and electropositive, i.e., electron poor region, respectively. From the MESP surface, it is evident that the most electronegative region is located over oxygen substituted at R4 ring system which effectively acts as an easy target for electrophilic attack in the molecule. On the contrary, the most electronegative region is located over the nitrogen substituted at upper ring (R1), which effectively acts as electron donor in rufescine (Srivastava et al., 2015).
Reactivity descriptors
To further describe the chemical reactivity of imerubrine, we have calculated various reactivity descriptors viz. ionization potentials (I), electron affinity (A), absolute electronegativity (χ) and chemical hardness (η) etc. I and A are calculated as the negative of energy eigen values of HOMO and LUMO, respectively. χ and η can be calculated by using finite-difference approximations (Parr and Yang, 1989) as χ = ½ (I + A) and η = ½ (I-A). These parameters are listed in Table 4. One can note that I and χ values of imerubrine are smaller than those of rufescine (Srivastava et al., 2015), whereas A and η values are larger. Therefore, imerubrine is chemically less hard, i.e., more reactive than rufescine due to the presence of O atom. Molecular dipole moment (μ) provides a signature of the geometry and charge distribution within the molecular system. The dipole moment of imerubrine is 9.54 Debye, directed from R3 to R2 along C5-C6 moiety. This value is fairly large to establish that imerubrine is a highly polar molecule and hence, can be easily soluble into polar solvents.
Molecular docking
The molecular docking explores the way in which two molecules, such as ligand and receptor fit together and dock to each other well. The molecular docking studies have been performed with SwissDock web server (Grosdidier et al., 2011). In this process all the possible conformers of the molecule (ligand) and their corresponding all the energy values are calculated and finally the best binding modes are ranked according to the full fitness (FF) score. In order to avoid sampling bias the whole docking process performed by SwissDock as blind by covering the entire protein and not defining any specific region of the protein as bonding pocket. The resulting output clusters obtained after each run and the result shows that cluster having the best FF score. The highest negative FF score indicates a more favorable binding site between ligand and receptor. The suitable targets (receptors) have been predicted using the SwissTargetPrediction (Gfeller et al., 2013). This prediction result suggests that the suitable target for imerubrine is B1 bradykinin receptor (PDB ID: 1HZ6) (http://www.rcsb.org/pdb/explore.do?structureId=1hz6), which is a G-protein coupled receptor having principal ligand is the bradykinin. It is one of two G-protein coupled receptors, which binds with bradykinin and mediate responses to pathophysiologic conditions such as inflammation, trauma, burns, shock, and allergy.
The FF score and binding affinity obtained for protein targets clearly shows that the molecule effectively bonded with 1HZ6 target with one hydrogen bond 2.099 Å (FF score = −348.03 kcal/mol, binding affinity ∆G = −6.76 kcal/mol). The docking picture has been obtained from the UCSF chimera software, displayed in Fig. 5, which clearly indicates valine (val) as the active binding sites. Thus, imerubrine is capable to be an effective 1HZ6 receptor agonist, meaning thereby, it can activate the 1HZ6 receptor for bind up with bradykinin.
Conclusions
We have performed a detailed quantum chemical studies on imerubrine using density functional theory at B3PW91/6-311 + G(d,p) level. QTAIM analysis reveals three intra-molecular C–H⋯O interactions and characterizes them as weak, which is further supported by their RFC values. Vibrational spectroscopic analysis has been performed and all modes up to 400 cm−1 are assigned with their PED values. The NMR chemical shifts have been calculated for C and H atoms, which are found to be in good agreement with the available experimental data. The HOMO→LUMO transition in imerubrine corresponds to the charge transfer to nitrogen containing ring and HOMO–LUMO gap, as well as other reactivity descriptors suggests its more reactive nature than rufescine. The molecular docking of imerubrine into 1HZ6 receptor suggests that it can bind and activate the receptor and therefore, it can act as an effective 1HZ6 receptor agonist. These finding may stimulate further observations on the biological activity of imerubrine and related natural products.
References
Aakeroy CB, Seddon KR (1993) The hydrogen bond and crystal engineering. Chem Soc Rev 22:397–407
Arivazhagan M, Jevavijayan S (2011) Vibrational spectroscopic, first-order hyperpolarizability and HOMO, LUMO studies of 1,2-dichloro-4-nitrobenzene based on Hartree–Fock, and DFT calculations. Spectrochim Acta A 79:376–38
Arjunan V, Sakiladevi S, Marchewka MK, Mohan S (2013) FT-IR, FT-Raman, FT-NMR and quantum chemical investigations of 3-acetylcoumarin. Spectrochim Acta A 109:79
Arjunan V, Subramaniam S, Mohan S (2004) Synthesis, Fourier transform infrared and Raman spectra, assignments and analysis of N-(phenyl)- and N-(chloro substituted phenyl)-2,2-dichloroacetamides. Spectrochim Acta A 60:1141–1159
Bader RFW (1990) Atoms in Molecules. A Quantum Theory, 2nd ed. Oxford, New York
Baker J (2006) A critical assessment of the use of compliance constants as bond strength descriptors for weak interatomic interactions. J Chem Phys 125:014103
Baker J, Pulay P (2006) The interpretation of compliance constants and their suitability for characterizing hydrogen bonds and other weak interactions. J Am Chem Soc 128:11324–11325
Becke AD (1993) Density functional thermochemistry.III. The role of exact exchange. J Chem Phys 98:5648
Boger DL, Brotherton CE (1984) Plementary use of 3-Methoxycarbonyl-2-pyrones. J Org Chem 49:4050
Boger DL, Takahashi K (1995) Total Synthesis of granditropone, grandirubrine, imerubrine and isoimerubrine. J Am Chem Soc 117:12452–12459
Brandhorst K, Grunenberg J (2008) How strong is it? The interpretation of force and compliance constants as bond strength descriptors. Chem Soc Rev 37:1558–1567
Buck KT (1984) The Alkaloids: Chemistry and Pharmacology 23:301–325
Dennington R, Keith T and Millam J (2009) GaussView Version 5.0. Semichem Inc. Shawnee Mission, KS, USA
Desiraju GR, Kashino S, Coombs MM, Glusker JP (1993) C-H--O packing motifs in some cyclopenta[a]phenanthrenes. Acta Cryst B 49:880–892
Espinosa E, Molins E, Lecomte C (1998) Hydrogen bond strength revealed by topological analyses of experimentally observed electron densities. Chem Phys Lett 285:170
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zheng G, Petersson GA, Voth GA, Scalmani G, Nakatsuji H, Nakai H, Hratchian HP, Brothers E, Ogliaro F, Bloino J, Cioslowski J, Foresman JB, Burant JC, Cross JB, Jaramillo J, Knox JE, Montgomery JA, Millam JM, Normand J, Peralta JE, Heyd JJ, Dannenberg JJ, Sonnenberg JL, Hasegawa J, Tomasi J, Ortiz JV, Ochterski JW, Salvador P, Morokuma K, Raghavachari K, Toyota K, Kudin KN, Bearpark M, Caricato M, Cossi M, Ehara M, Hada M, Ishida M, Klene M, Rega N, Farkas O, Kitao O, Yazyev O, Cammi R, Fukuda R, Gomperts R, Kobayashi R, Stratmann RE, Martin RL, Iyengar SS, Dapprich S, Keith T, Nakajima T, Vreven T, Barone V, Bakken V, Rendell V, Staroverov VN, Zakrzewski VG, Li X, Honda Y, Daniels AD, Izmaylov AF, Austin AJ, Mennucci B, Adamo C, Pomelli C, Fox DJ (2010) Gaussian 09 Revision B.01. Gaussian Inc., Wallingford CT
George S (2001) Infrared and Raman characteristic group frequencies-Tables and Charts, 3rd ed. Wiley, Chichester
Gfeller D, Michielin O and Zoete V (2013) Bioinformatics 29: 3073
Grosdidier A, Zoete V and Michielin O (2011) Nucleic Acids Res. 39: 270
Jamroz MH (2004) Vibrational energy distribution analysis, VEDA 4 Program. Warsaw, Poland
Jamroz MH (2013) Vibrational Energy Distribution Analysis (VEDA): Scopes and limitations. Spectrochim Acta A 114:220–230
Jeffrey GA, Saenger W (1991) Hydrogen Bonding in Biological Structures. Springer–Verlag, Berlin Heidelberg
Jones LH, Swanson BI (1976) Bonding in Metal Carbonyls. Acc Chem Res 9:128
Keith TA (2012) AIMAll Version 12.09.23 TK Gristmill Software Overland Park KS USA
Krishnakumar V, Balachandran V (2005) Analysis of vibrational spectra of 5-fluoro, 5-chloro and 5-bromo-cytosines based on density functional theory calculations. Spectrochim Acta A 61:1001–1006
Kumar A, Srivastava AK, Mondal A, Brahmachari G, Misra N, Gangwar S (2015) Combined experimental (FT-IR, UV-visible spectra, NMR) and theoretical studies on the molecular structure, vibrational spectra, HOMO, LUMO, MESP surfaces, reactivity descriptor and molecular docking of Phomarin. J Mol Struct 1096:94–101
Lee JC, Cha JK (2001) Total synthesis of tropoloisoquinolines: imerubrine, isoimerubrine, and grandirubrine. J Am Chem Soc 123:3243–3246
Lewis WH, Stonard RJ, Porras-Reyes B and Mustoe TA (1992) (US Patent No. 5,156,847)
Luqul FJ, Lopez JM, Orozco M (2000) Perspective on electrostatic interactions of a solute with a continuum, a direct utilization of ab initio molecular potentials for the prevision of solvent effects. Theor Chem Acc 103:343
Madhav MV, Manogaran S (2009) A relook at the compliance constants in redundant internal coordinates and some new insights. J Chem Phys 131:174112–174116
Majumder M, Manogaran S (2013) Redundant internal coordinates, compliance constants and non-bonded interactions-Some new insights. J Chem Sci 125:9–15
Manolopoulos DE, May JC, Down SE (1991) Systematic relationships between fullerenes without spirals. Chem Phys Lett 181:105
Merrick JP, Moran D, Radom L (2007) An evaluation of harmonic vibration frequency scale factors. J Phys Chem A 111:11683–11700
Mukherjee KM, Mishra TN (1996) Surface-enhanced raman spectra of 2- and 3-hydroxypyridines in silver sol. J Raman Spectrosc 27:595–600
Parr RG, Yang W (1989) Density Functional Theory of Atoms and Molecules. Oxford University Press and Clarendon Press, New York and Oxford
Perdew JP, Wang Y (1992) Accurate and simple analytic representation of the electron-gas correlation energy. Phys Rev B 45:13244
Ponnala S, Harding WW (2013) A route to azafluoranthene natural products through direct arylation. Eur J Org Chem 6:1107
Rozas I, Alkorta I, Elguero J (2000) Behavior of ylides containing N, O and C atoms as hydrogen bond acceptors. J Am Chem Soc 122:11154
Saenger W (1984) Principles of Nucleic Acid Structure. Springer, Berlin
Scherowsky G, Frackowiak E, Adam D (1997) 4,5,6,7,8,9-Hexamethoxyindeno[1,2,3-ij] isoquinoline. Acta Cryst C 53:5
Schlick T (2010) Molecular modelling and simulation: An Interdisciplinary Guide Vol. 21, 2nd ed. New York
Schwan TJ (1976) (US Patent No. 3,971,788)
Scrocco E, Tomasi J (1973) The electrostatic molecular potential as a tool for the interpretation of molecular properties. Top Curr Chem 42:95–170
Silveira CC, Larghi EL, Mendes SR, Bracca ABJ, Rinaldi F, Kaufman TS (2009) Electrocyclization-mediated approach to 2-methyltriclisine, an unnatural analog of the azafluoranthene alkaloid triclisine. Eur J Org Chem 2:4637
Srivastava AK, Baboo V, Narayana B, Sarojini BK, Misra N (2014a) Comparative DFT study on reactivity, acidity and vibrational spectra of halogen substituted phenylacetic acids. Indian J Pure Appl Phys 52:507–519
Srivastava AK, Narayana B, Sarojini BK, Misra N (2014b) Vibrational, structural and hydrogen bonding analysis of N’-[(E)-4-hydroxybenzylidene]-2-(naphthalene-2-yloxy) acetohydrazide: combined density functional and atoms-in-molecule based theoretical studies. Indian J Phys 88:547–556
Srivastava AK, Pandey AK, Gangwar SK, Misra N (2014c) Structural, vibrational and electronic properties of cis and trans conformers of 4-hydroxy-l-proline: a density functional approach. J At Mol Sci 5:279–288
Srivastava AK, Pandey AK, Misra N, Jain S (2015) FT-IR spectroscopy, intra-molecular C-H--O interactions, HOMO, LUMO, MESP analysis and biological activity of two natural products, triclisine and rufescine: DFT and QTAIM approaches. Spectrochim Acta A 136:682–689
Steiner T, Saenger W (1992) Geometry of C–H---O Hydrogen bonds in Carbohydrate crystal structures, Analysis of neutron diffraction data. J Am Chem Soc 114:10146–10154
Sundaraganesan N, Ilakiamani S, Joshua BD (2007) FT-Raman and FT-IR spectra, ab initio and density functional studies of 2-amino-4,5-difluorobenzoic acid. Spectrochim Acta A 67:287–297
Swanson BI (1976) Minimum energy coordinates. A relation between molecular vibrations and reaction coordinates. J Am Chem Soc 98:3067–3071
Zhao B, Snieckus V (1984) Integrated aromatic metalation—cross coupling methodologies. A concise synthesis of the azafluoranthene alkaloid imeluteine. Tetrahedron Lett 32:5277–5278
Acknowledgments
A.K. Srivastava is thankful to Council of Scientific and Industrial Research (CSIR), New Delhi, India for providing a research fellowship via grant no. 09/107(0359)/2012-EMR-I. The Central Facility for Computational Research (CFCR), Department of Chemistry, University of Lucknow is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Srivastava, A.K., Kumar, A., Pandey, S.K. et al. Spectroscopic analyses, intra-molecular interaction, chemical reactivity and molecular docking of imerubrine into bradykinin receptor. Med Chem Res 25, 2832–2841 (2016). https://doi.org/10.1007/s00044-016-1710-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-016-1710-z