Abstract
Aromatase inhibitors are the most important targets in treatment of estrogen-dependent cancers. In order to search for potent non-steroidal aromatase inhibitors (NSAIs) with lower side effects and overcome cellular resistance, Genetic Algorithm with Linear Assignment of Hypermolecular Alignment of Database was used to derive 3D pharmacophore models. The obtained best pharmacophore model contains one acceptor atom, one donor atom, and two hydrophobes, which was used in effective alignment of dataset. In succession, comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA) were performed on 84 structurally diverse NSAIs to build 3D-QSAR models based on both pharmacophore and docking alignments. The CoMFA and CoMSIA models based on the pharmacophore alignment show better statistical results (CoMFA: q 2 = 0.634, r 2ncv = 0.986, r 2pred = 0.737; CoMSIA: q 2 = 0.668, r 2ncv = 0.926, r 2pred = 0.708). This 3D-QSAR approach provides significant insights that can be used to develop novel and potent NSAIs. In addition, the best pharmacophore model was used as a 3D query for virtual screening against NCI2000 database. The hit compounds were further filtered by docking, and their biological activities were predicted by the CoMFA and CoMSIA models, and six structurally diverse compounds with good predicted pIC50 values were obtained, which are expected to design novel NSAIs with new skeletons.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aromatase is a cytochrome P-450 dependent enzyme that catalyzes the aromatization of androgens to estrogens. Aromatase inhibitors (AIs) reduce the synthesis of estrogens and offer a therapeutic alternative for the treatment of estrogen-dependent cancers such as breast cancer (Winer et al., 2005; Perez, 2006; Jordan and Brodie, 2007). There are two classes of AIs, steroidal and non-steroidal compounds, which cause potent estrogen suppression (Brueggemeier et al., 2005). The non-steroidal aromatase inhibitors (NSAIs) are mostly azole type compounds such as the clinically used anastrozole and letrozole, which compete with the substrate for binding to the enzyme active site (Recanatini et al., 2002). Among steroidal aromatase inhibitors (SAIs), formestane was widely used during the early 1990s, but it is not used nowadays because of the need to administer it by intramuscular injection. Therefore, the orally active exemestane is the main steroidal inhibitor (Seralini and Moslemi, 2001). These SAIs mimic the natural substrate androstenedione and are converted by the enzyme to reactive intermediates, which bind irreversibly to the enzyme active site, resulting in inactivation of aromatase (Hong et al., 2011). Despite the success of the third-generation NSAIs (anastrozole and letrozole) and SAIs (exemestane), they still have some major side effects, such as increase of bone loss, joint pain, and heart problems (Dutta and Pant, 2008). In addition, after some years of usage they can develop cellular resistance. For these reasons, it is important to search for other potent and specific molecules with lower side effects and which can overcome the resistance phenomena.
Pharmacophore searches are the best option to find a range of chemical structures with viable features. A pharmacophore model can be considered as the ensemble of steric and electrostatic features of different compounds which are necessary to ensure optimal supramolecular interactions with a specific biological target structure and to trigger or to block its biological response. Thus, pharmacophore modeling is the method of choice for the first round of compound selection. This ability of a pharmacophore model is used to find new classes of inhibitors when one class is known. This is known as ‘scaffold hopping’ (Bhatt and Patel, 2012).
Three-dimensional quantitative structure–activity relationship (3D-QSAR) methods have been successfully employed to assist the design of new small molecule drug candidates (Honorio et al., 2007; Salum et al., 2007). Comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA) are two of the most widely used 3D-QSAR methodologies. CoMFA calculates the energies of steric and electrostatic interactions between the compound and the probe atom at various intersections of a regular 3D lattice according to Lennard-Jones and Coulomb potentials. The resulting energies derived from these two potential functions can be contoured to offer a quantitative spatial description of the molecular properties (Cramer et al., 1988). CoMSIA introduces the Gaussian function for the distance dependence between the molecular atoms and the probe atom in order to avoid some inherent deficiencies arising from the Lennard-Jones and Coulomb potential functional forms. CoMSIA is applied to gain an insight into how steric fields, electrostatic fields, hydrophobic fields, hydrogen bond donor (HBD), and hydrogen bond acceptor (HBA) influence the activity of inhibitors (Klebe et al., 1994).
Several structurally diverse NSAIs such as resveratrol analogs, isoflavanone derivatives, and tetrahydropyrroloquinolinone type compounds (Table 1), have been reported in the recent literatures (Sun et al., 2010; Bonfield et al., 2012; Hu et al., 2012; Yin et al., 2013). To understand the structural basis for inhibitory activity and design more potent agents, pharmacophore models were created and 3D-QSAR studies were performed for the fist time for these structurally diverse NSAIs using CoMFA and CoMSIA based on both pharmacophore and docking alignments. In addition, the obtained best pharmacophore model was used as a 3D query for virtual screening against NCI2000 database. The hit compounds were further filtered by docking, and their biological activities were predicted using CoMFA and CoMSIA models.
Materials and methods
Dataset
Compounds 1–26 (resveratrol analogs) (Sun et al., 2010), compounds 27–45, (isoflavanone derivatives) (Bonfield et al., 2012), compounds 46–61 (Hu et al., 2012), and compounds 62–84 (tetrahydropyrroloquinolinone type) (Yin et al., 2013) were used for this analysis, and their structures and bioactivity values are presented in Table 1. The pIC50 (−log IC50) values were used to derive 3D-QSAR models. The whole dataset of 84 compounds was divided into two groups in the ratio of 4:1:a training set with 63 compounds, a test set with 21 compounds (Table 1). The selection of the training and test sets was done manually such that low, moderate, and high activity compounds were present in roughly equal proportions in both sets. The training set was used to build predictive models, while the test set was used to validate the predictive ability of the models.
Computational approach
The 3D-QSAR modeling analyses, calculations, and visualizations were performed using the SYBYL 7.3 molecular modeling package from Tripos Inc., St. Louis, Mo, USA, installed on Red Hat Linux workstations. All the structures were built, and energy was minimized with the following steps: (i) optimization by Steepest Descent with initial optimization of 200 simplex iterations using Tripos force field and Gasteiger–Marsili charges; (ii) optimization by conjugate gradient; and (iii) optimization by BFGS (Hu et al., 2009).
Pharmacophore hypothesis
The pharmacophore hypothesis was generated using Genetic Algorithm with Linear Assignment of Hypermolecular Alignment of Database (GALAHAD) module of SYBYL, which operates in two main stages: the ligands are aligned to each other in internal coordinate space, and then the conformations produced are aligned in Cartesian space. The feature considered in developing the pharmacophore model includes HBD atoms, HBA atoms, and hydrophobic and charged centers (Richmond et al., 2006; Shepphird and Clark, 2006; Andrade et al., 2008). In our study, twelve compounds shown in Table 1 were selected to carry out the pharmacophore hypothesis, and the genetic algorithm was used to create conformers for all molecules. The compounds selected to generate the pharmacophore hypothesis are highly active and structurally diverse.
Molecular docking
The crystal structure of aromatase complexed with NSAIs in present study has not yet been reported. Therefore, the crystal structure of aromatase complexed with ASD (PDB code: 3S79) from the RCSB Protein Data Bank was used (Ghosh et al., 2012). The molecular docking was performed using the Surflex-Dock module of SYBYL. All parameters were set with default values in the whole process. Before docking, the ligand was extracted, all the water molecules were removed, and hydrogen atoms were added to the receptor. The protomol was produced using docking-based method: ligand location in the same coordinate space in the receptor. In our study, each conformer of all 84 inhibitors was docked into the binding site ten times, and the docking score values were used to evaluate the docking analysis. The top ranked conformations for each molecule were extracted and aligned together for the subsequent CoMFA and CoMSIA modeling (Wang et al., 2012). The Surflex-Dock was also used to filter the hit compounds in the virtual screening.
Molecular alignment
The 3D structural alignment is a crucial component in 3D-QSA studies, and affects the outcome of the CoMFA and CoMSIA statistical analysis. There are three main different procedures proposed for aligning molecules for 3D-QSAR: maximum common substructures overlap, pharmacophore overlap and docking-based alignment (Hu et al., 2009). Both of pharmacophore- and docking-based alignment procedures were performed in the present study because of structural diversity of the studied compounds. Pharmacophore-based alignment was done using GALAHAD and docking-based alignment was done using Surlflex-Dock.
CoMFA and CoMSIA models
In CoMFA, the steric fields were calculated using a Lennard-Jones potential, while the electrostatic fields were calculated using a Coulombic potential. To calculate the CoMFA fields, a 3D-cubic lattice with grid spacing of 2.0 Å in X, Y, and Z directions was created automatically by SYBYL. The grid pattern extended 4.0 Å units in all directions beyond the dimensions of each molecule. The steric and electrostatic probe-ligand interaction energies were calculated using a sp3 carbon probe atom and a +1.0 charge with a distance-dependent dielectric function at each lattice point. The cut-off for energies was set to ±30 kcal/mol, and the electrostatic contributions were ignored at lattice points with maximal steric interactions (Cramer et al., 1988). In CoMSIA, five different similarity fields (steric, electrostatic, hydrophobic, HBD, and HBA) were calculated. CoMSIA models were also derived with the same lattice box, and all five fields were calculated using a probe of charge +1, a radius of 1, hydrophobicity and hydrogen bonding properties of +1, and an attenuation factor of 0.3 for the Gaussian distance-dependent function (Klebe et al., 1994).
Statistical analysis
In order to derive 3D-QSAR models, CoMFA and CoMSIA descriptors were used as independent variables and the pIC50 values as the dependent variables. PLS method with cross-validation (leave-one-out) was used in SYBYL to determine the optimal numbers of components using cross-validated coefficient q 2 (r 2cv ). After obtaining the optimal numbers of components, a PLS analysis was performed with no validation and column filtering 2.0 to generate the final model with the training set. The obtained final non-cross-validated correlation coefficient (r 2ncv ) is a measure of the quality of the model. The predictive capability of the 3D-QSAR models was determined from the predictive correlation (r 2pred ). The predicted activities for the test set were obtained from the model produced by the training set.
Results and discussion
Pharmacophore generation
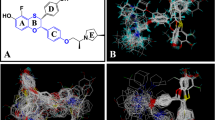
Twenty pharmacophore models were generated with default parameters after GALAHAD run. Each of the obtained models represents a different tradeoff among the conflicting demands of maximizing steric consensus, maximizing pharmacophore consensus, and minimizing energy. All the twenty models had Pareto rank 0, which means no one model is superior to any other one. Some models had very high energy, which is recognized that high energy values are due to steric clashes (Dorfman et al., 2008). Small value of energy and high values of N_hits, Sterics, and Mol_Qry are desired for the best model (Caballero, 2010), so six models were chosen for the analysis, and their statistical values are shown in Table 2. All the selected models were used for the molecular alignment to produce CoMFA and CoMSIA models. Model_08 was considered to be the best model as the best CoMFA and CoMSIA results were obtained when Model_08 was used to align the dataset. This model contains one acceptor atom, one donor atom, and two hydrophobes, which is shown in Fig. 1. The acceptor atom and donor atom are very close to each other. In addition, this model was further converted into a UNITY query for virtual screening studies.
CoMFA and CoMSIA statistical results
The structural alignment of compounds plays a very important role in the development of successful 3D-QSAR models, so all compounds of the dataset were aligned according to both pharmacophore and docking to derive the CoMFA and CoMSIA models in the present study. Figure 2a, b show pharmacophore-based and docking-based alignments of training set molecules used in 3D-QSAR models, respectively.
To obtain an effective 3D-QSAR model, a number of statistical parameters, q 2(r 2cv ), r 2ncv , standard error estimate (SEE), and F-statistic values (F) were computed as defined in SYBYL. The CoMFA and CoMSIA results obtained from both pharmacophore- and docking-based alignments are shown in Table 3, which show that the pharmacophore-based CoMFA and CoMSIA models exhibit better statistical results than the docking-based CoMFA and CoMSIA models, where the pharmacophore-based modeling yielded q 2(r 2cv ) = 0.634, r 2ncv = 0.986 for CoMFA model and q 2(r 2cv ) = 0.668, r 2ncv = 0.962 for CoMSIA model, while the docking-based modeling gave q 2(r 2cv ) = 0.563, r 2ncv = 0.824 for CoMFA model, q 2(r 2cv ) = 0.469, r 2ncv = 0.761 for CoMSIA model, respectively.
We mainly focus on the CoMFA and CoMSIA obtained from pharmacophore-based alignment due to its more satisfactory statistical results. As shown in Table 3, the CoMFA model has a high q 2 (r 2cv ) of 0.634 with seven optimal components. This CoMFA model has r 2ncv of 0.986, SEE of 0.131 and F value of 564.951. The corresponding field contributions of steric and electrostatic are 0.354 and 0.646, respectively, which means the electrostatic field gives more contribution to activity than the steric field. The CoMSIA model (q 2 = 0.668, r 2ncv = 0.962, F = 236.690, SEE = 0.215) was obtained using the combination of steric, electrostatic, hydrophobic, HBD, and HBA fields with six optimal components. The corresponding field contributions are 0.130, 0.319, 0.200, 0.128, and 0.224, respectively. Both CoMFA and CoMSIA models obtained from pharmacophore-based alignment are fairly good as their q 2 values are more than 0.6.
In order to validate the 3D-QSAR models, r 2pred was used to determine the predictive abilities of the CoMFA and CoMSIA models from the 21 compounds (test set) which were not included in the generation of the models. The obtained r 2pred of the test set is 0.737 and 0.708 for the CoMFA and CoMSIA model, respectively, which indicates that both models have high predictive ability. The observed and predicted pIC50 by the CoMFA and CoMSIA model of the training and test sets are given in Table 4, and the correlations between the observed and predicted pIC50 of training and test sets are depicted in Fig. 3 for CoMFA model, Fig. 4 for CoMSIA model, respectively.
CoMFA contour maps
The steric contour maps for the CoMFA model with two most active inhibitor compounds 55 and 79 are shown in Fig. 5a, b, respectively. In these figures, the green contours represent regions of high steric tolerance (80 % contribution), while the yellow contours represent regions of low steric bulk tolerance (20 % contribution). Figure 5a combined with compound 55 shows that a large green contour near the –OBz group attached to the 2-position of benzene ring indicates that a bulky group in this position is favorable to bioactivity. It is confirmed by the fact that compounds 52–55 and 60 with bulky substitution in that position have higher bioactivity while compounds 56–59 with no substitution in that position have lower bioactivity. A large green contour near the 3-ClPh group can also be observed in Fig. 5b combined with compound 79, which suggests that a bulky group in this region will increase inhibitory activity. This is supported by the higher activity of compounds 74–82, which have large substituents in that position, compared with the lower activity of compounds 68–70, which have small substituents in that position.
CoMFA contour maps (standard deviation × coefficient) in combination with compounds 55 and 79. a, b Steric contour maps: green contours (80 % contribution) refer to sterically favored regions; yellow contours (20 % contribution) indicate sterically disfavored regions. c, d Electrostatic contour maps: blue contours (80 % contribution) refer to regions where positively charged substituents are favored; red contours (20 % contribution) indicate regions where negatively charged substituents are favored (Color figure online)
The CoMFA electrostatic contour maps in combination with compounds 55 and 79 are shown in Fig. 5c, d, respectively. The red areas are the regions where a negative potential is favorable to activity, while a negative potential is unfavorable in the blue areas. Figure 5c combined with compound 55 displays a red contour near the nitrogen atom of the pyridine ring, which indicates that the bioactivity can be enhanced if an electronegative atom is present. It is clear that most of the compounds 46–60 with a nitrogen atom in that position have high inhibitory activity. Similarly, Fig. 5d combined with compound 79 also shows a red contour near the nitrogen atom of the pyridine ring, which means that an electronegative atom in this position can increase the bioactivity. This can be confirmed by the higher activity of compounds 72, 74, and79, compared with the lower activity of compounds 63, 64 and 65, respectively.
CoMSIA contour maps
CoMSIA not only calculates steric and electrostatic fields as in CoMFA, but also additionally computes hydrophobic, HBD, and HBA fields. The steric and electrostatic contour maps of CoMSIA are consistent with those of CoMFA. The contour maps of CoMSIA hydrophobic, HBD, and HBA fields are presented in Fig. 6. For each field, the favorable and disfavored contours represent 80 and 20 % level contributions, respectively. The hydrophobic contour maps in combination with compounds 73 and 79 are shown in Fig. 6a, b, respectively, in which yellow contours indicate regions where hydrophobic groups are favorable to bioactivity while white contours represent areas where hydrophilic groups are favorable. Figure 6a combined with compound 73 shows that Ph group is near the white region and –OH group is near the yellow region, while Fig. 6b combined with compound 79 shows that –ClPh group is near the yellow region. This is supported by the case that the activity of compound 73 is much lower than that of compound 79.
CoMSIA contour maps (standard deviation × coefficient). a, b Hydrophobic contour maps in combination with compounds 73 and 79: white contours refer to regions where hydrophilic substituents are favored; yellow contours indicate regions where hydrophobic substituents are favored. c HBD contour map in combination with compound 1: cyan contours indicate HBD substituents in this region are favorable to activity; purple contours represent that HBD groups in this area are unfavorable. d, e HBA contour maps in combination with compounds 55 and 79: magenta contours show regions where HBA substituents are expected; red contours refer to areas where HBA substituents are unexpected (Color figure online)
The HBD contour map in combination with compound 1 is present in Fig. 6c, in which cyan contours indicate that HBD substituents in this region are favorable to activity while purple contours represent that HBD groups in this area are unfavorable. Two cyan contours are shown near the –NH2 group of the compound 1, which indicates the necessity of the hydrogen atoms at this position for high bioactivity. It is confirmed by the fact that the compounds 2, 8, and 12 with –NH2 group at 6-position have higher bioactivity compared with the compounds 3, 7, and 11 with –NH2 group at 5-position have lower bioactivity, respectively.
Figure 6d, e show the HBA contour maps in combination with compounds 55 and 79, respectively, where magenta and red contours represent areas where HBA substituents are favored and disfavored, respectively. Figure 6d combined with compound 55 displays that one magenta contour is near the carbonyl group, and the other magenta contour is near the nitrogen atom of the pyridine ring, which indicates that the activity can be enhanced if the electronegative atoms are in these two positions. It is supported by the case that most of the compounds 46–60, which have nitrogen and oxygen atoms in these two positions, show high bioactivity. Similarly, Fig. 6e combined with compound 79 also shows that one magenta contour is near the carbonyl group, and the other magenta contour is near the nitrogen atom of the pyridine ring, which represents that the electronegative atoms in these two positions can increase the bioactivity. This is confirmed by the higher activity of compounds 72, 74, and 79 compared with the lower activity of compounds 63, 64, and 65, respectively.
Docking analysis
In order to validate the docking model in the virtual screening, the C_score values of all 84 inhibitors were tested using Surflex-Dock, and the correlations between the C_score values and pIC50 values are depicted in Fig. 7. It can be seen that 75 of 84 inhibitors have positive C_score values, so the hit compounds should have positive C_score values in the virtual screening.
Virtual screening
The obtained best GALAHAD model (Fig. 1) was converted into a UNITY query, which was screened against NCI2000 database. The “flexible database search” option was implemented to perform virtual screening. Primary filters such as Lipinski’s rule of five, Van der Waals bumps, and QFIT (pharmacophoric match between query and the hit compound) were applied to reduce the dataset (Kothandan et al., 2013). The screening of the pharmacophore query yielded 336 hits that met the specific requirements. The 336 hit compounds were further subjected to molecular docking using the Sulflex-Dock module of SYBYL. 174 compounds were selected based on the docking score values (C_score value > 0). The pIC50 values of the selected 174 compounds were predicted using CoMFA and CoMSIA models generated on the basis of pharmacophore alignment. Finally, six structurally diverse compounds with good pIC50 values predicted by both CoMFA and CoMSIA models (both pIC50 > 6.40) are listed in Table 5. Thus, these potential hit compounds are expected to design novel NSAIs with new skeleton.
Conclusion
Aromatase inhibitors have proven to be the most important targets for treatment of estrogen-dependent cancers. In order to search for more potent NSAIs with lower side effects and overcome the drug resistance, pharmacophore modeling, virtual screening, and 3D-QSAR studies were performed. Pharmacophore model was derived from twelve highly active and structurally diverse compounds using GALAHAD. The best pharmacophore model includes one acceptor atom, one donor atom, and two hydrophobes, which was used as a query to search NCI2000 database. 336 hit compounds were obtained after the screening of the pharmacophore query, and 90 compounds were selected by molecular docking. Finally, six structurally diverse compounds with good predicted pIC50 values were obtained, which are expected to design novel NSAIs with new skeletons. The 3D-QSAR techniques based on both pharmacophore and docking alignments, CoMFA and CoMSIA, were applied to 84 NSAIs with different skeletons. The CoMFA and CoMSIA models obtained from pharmacophore-based alignment shows better statistical results, q 2 = 0.634, r 2ncv = 0.986, r 2pred = 0.737 for CoMFA and q 2 = 0.668, r 2ncv = 0.962, r 2pred = 0.708 for CoMSIA, which indicate that these structurally diverse NSAIs must bind to the same active site of aromatase, and it is feasible to drive CoMFA and CoMSIA models based on structurally diverse compounds. The present pharmacophore modeling, virtual screening, and 3D-QSAR approach provides useful information to design and synthesize novel NSAIs.
References
Andrade CH, Salum LB, Pasqualoto KFM, Ferreira EI, Andricopulo AD (2008) Three-dimensional quantitative structure-activity relationships for a large series of potent antitubercular agents. Lett Drug Des Discov 5:377–387
Bhatt HG, Patel PK (2012) Pharmacophore modeling, virtual screening and 3D-QSAR studies of 5-tetrahydroquinolinylidine aminoguanidine derivatives as sodium hydrogen exchanger inhibitors. Bioorg Med Chem Lett 22:3758–3765
Bonfield K, Amato E, Bankemper T, Agard H, Steller J, Keeler JM, Roy D, McCallum A, Paula S, Ma L (2012) Development of a new class of aromatase inhibitors: Design, synthesis and inhibitory activity of 3-phenylchroman-4-one (isoflavanone) derivatives. Bioorg Med Chem 20:2603–2613
Brueggemeier RW, Hackett JC, Diaz-Cruz ES (2005) Aromatase inhibitors in the treatment of breast cancer. Endocr Rev 26:331–345
Caballero J (2010) 3D-QSAR (CoMFA and CoMSIA) and pharmacophore (GALAHAD) studies on the differential inhibition of aldose reductase by flavonoid compounds. J Mol Graph Model 29:363–371
Cramer RD, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967
Dorfman RJ, Smith KM, Masek BB, Clark RD (2008) A knowledge-based approach to generating diverse but energetically representative ensembles of ligand conformers. J Comput Aided Mol Des 22:681–691
Dutta U, Pant K (2008) Aromatase inhibitors: past, present and future in breast cancer therapy. Med Oncol 25:113–124
Ghosh D, Lo J, Morton D, Valette D, Xi J, Griswold J, Hubbell S, Egbuta C, Jiang W, An J, Davies HM (2012) Novel Aromatase Inhibitors by Structure-Guided Design. J Med Chem 55:8464–8476
Hong Y, Rashid R, Chen S (2011) Binding features of steroidal and nonsteroidal inhibitors. Steroids 76:802–806
Honorio KM, Garratt RC, Polikatpov I, Andricopulo AD (2007) 3D QSAR comparative molecular field analysis on nonsteroidal farnesoid X receptor activators. J Mol Graph Model 25:921–927
Hu RJ, Barbault F, Delamar M, Zhang RS (2009) Receptor- and ligand-based 3D-QSAR study for a series of non-nucleoside HIV-1 reverse transcriptase inhibitors. Bioorg Med Chem 17:2400–2409
Hu QZ, Yin L, Hartmann RW (2012) Selective dual inhibitors of CYP19 and CYP11B2: targeting cardiovascular diseases hiding in the shadow of breast cancer. J Med Chem 55:7080–7089
Jordan VC, Brodie AMH (2007) Development and evolution of therapies targeted to the estrogen receptor for the treatment and prevention of breast cancer. Steroids 72:7–25
Klebe G, Abraham U, Mietzner T (1994) Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem 37:4130–4146
Kothandan G, Madhavan T, Gadhe CG, Cho SJ (2013) A combined 3D QSAR and pharmacophore-based virtual screening for the identification of potent p38 MAP kinase inhibitors: an in silico approach. Med Chem Res 22:1773–1787
Perez EA (2006) Appraising adjuvant aromatase inhibitor therapy. Oncologist 11:1058–1069
Recanatini M, Cavalli A, Valenti P (2002) Nonsteroidal aromatase inhibitors: recent advances. Med Res Rev 22:282–304
Richmond NJ, Abrams CA, Wolohan PRN, Abrahamian E, Willett P, Clark RD (2006) GALAHAD: 1. Pharmacophore identification by hypermolecular alignment of ligands in 3D. J Comput Aided Mol Des 20:567–587
Salum LD, Polikarpov I, Andricopulo AD (2007) Structural and chemical basis for enhanced affinity and potency for a large series of estrogen receptor ligands: 2D and 3D QSAR studies. J Mol Graph Model 26:434–442
Seralini GE, Moslemi S (2001) Aromatase inhibitors: past, present and future. Mol Cell Endocrinol 178:117–131
Shepphird JK, Clark RD (2006) A marriage made in torsional space: using GALAHAD models to drive pharmacophore multiplet searches. J Comput Aided Mol Des 20:763–771
Sun B, Hoshino J, Jermihov K, Marler L, Pezzuto JM, Mesecar AD, Cushman M (2010) Design, synthesis, and biological evaluation of resveratrol analogues as aromatase and quinone reductase 2 inhibitors for chemoprevention of cancer. Bioorg Med Chem 18:5352–5366
Wang JN, Wang FF, Xiao ZT, Sheng GW, Li Y, Wang YH (2012) Molecular simulation of a series of benzothiazole PI3 K alpha inhibitors: probing the relationship between structural features, anti-tumor potency and selectivity. J Mol Model 18:2943–2958
Winer EP, Hudis C, Burstein HJ et al (2005) American society of clinical oncology technology assessment on the use of aromatase inhibitors as adjuvant therapy for postmenopausal women with hormone receptor-positive breast cancer: status report 2004. J Clin Oncol 23:619–629
Yin L, Hu QZ, Hartmann RW (2013) Tetrahydropyrroloquinolinone type dual inhibitors of aromatase/aldosterone synthase as a novel strategy for breast cancer patients with elevated cardiovascular risks. J Med Chem 56:460–470
Acknowledgments
This work was financially supported by the Science and Technology Planning Project of Yunnan Province (No. 2011FZ096).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Xie, H., Qiu, K. & Xie, X. Pharmacophore modeling, virtual screening, and 3D-QSAR studies on a series of non-steroidal aromatase inhibitors. Med Chem Res 24, 1901–1915 (2015). https://doi.org/10.1007/s00044-014-1257-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-014-1257-9