Abstract
The nuclear loss and cytoplasmic accumulation of TDP-43 (TAR DNA/RNA binding protein 43) are pathological hallmarks of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). Previously, we reported that the primate-specific cleavage of TDP-43 accounts for its cytoplasmic mislocalization in patients’ brains. This prompted us to investigate further whether and how the loss of nuclear TDP-43 mediates neuropathology in primate brain. In this study, we report that TDP-43 knockdown at the similar effectiveness, induces more damage to neuronal cells in the monkey brain than rodent mouse. Importantly, the loss of TDP-43 suppresses the E3 ubiquitin ligase PJA1 expression in the monkey brain at transcriptional level, but yields an opposite upregulation of PJA1 in the mouse brain. This distinct effect is due to the species-dependent binding of nuclear TDP-43 to the unique promoter sequences of the PJA1 genes. Further analyses reveal that the reduction of PJA1 accelerates neurotoxicity, whereas overexpressing PJA1 diminishes neuronal cell death by the TDP-43 knockdown in vivo. Our findings not only uncover a novel primate-specific neurotoxic contribution to the loss of function theory of TDP-43 proteinopathy, but also underscore a potential therapeutic approach of PJA1 to the loss of nuclear TDP-43.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
The nuclear protein TAR DNA/RNA binding protein 43 (TDP-43) regulates DNA transcription, RNA splicing, stability, and translation to mediate physiological functions [1,2,3,4,5]. TDP-43 pathology was first identified in postmortem spinal cord and cortex samples from patients with amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) [6], and later found in the brains of patients with other neurodegenerative diseases, including Alzheimer’s disease (AD), Lewy body dementia (LBD), and Huntington’s disease (HD) [7,8,9]. More than 50 mutations of TDP-43 associated with ALS/FTLD have been identified, further supporting its causal role [10, 11]. TDP-43 contains two conserved RNA recognition motifs, RRM1 and RRM2, which enables its nucleic acid-binding activity [12, 13] to regulate different functions of gene transcription or RNA processing [4, 12, 14, 15]. TDP-43 binds single-stranded RNA in the low nanomolar range with high specificity for UG-rich conserved sequences [16, 17], and is recruited to splice sites to inhibit exon recognition [12, 18] or mRNA instability [17, 19]. TDP-43 also interacts with (UG)n repeat elements, including VEGF (vascular endothelial growth factor), GRN (granulin precursor), NEFL (neurofilament light chain), and ADD2 (adducin 2), to mediate the degradation of mRNA, including its own mRNA, and suppress gene expression [16, 20,21,22,23,24].
Typical TDP-43 pathology is characterized by abnormal formation of cytosolic TDP-43 aggregates concurrent with loss of nuclear TDP-43. This has led to hypotheses that both gain- and loss-of-function of TDP-43 mutations can lead to pathogenic outcomes [2, 25,26,27]. In some studies, transgenic expression of wild-type or disease-associated TDP-43 mutants in mice recapitulated certain features of related diseases, supporting the gain-of-function theory. However, conditional TDP-43 knockout in excitatory neurons also caused neurodegeneration, arguing for a loss-of-function hypothesis [28]. Therefore, to better understand and treat these associated neurodegenerative diseases, research is urgently needed to resolve whether the gain or loss of TDP-43 function is the salient mechanism underlying neurodegeneration [2]. Although mouse models provided valuable information for important pathological events and mechanistic insights, species differences prevent small animals from recapitulating some important pathologic changes seen in patient brains. For example, most transgenic rodent models of ALS show the predominant nuclear localization of mutant TDP-43 [11, 29,30,31,32,33] or some minimal level of cytoplasmic TDP-43 [32, 34, 35],in contrast to ALS patient brains that display obvious cytoplasmic TDP-43 inclusions [6, 15], however, in a monkey brain expressing mutant TDP-43 [36], TDP-43 is localized in the cytoplasm to form aggregates consisting of highly ubiquitinated, phosphorylated, and cleaved TDP-43 forms, which are similar to those seen in patient brains [5, 37,38,39]. Additionally, human-specific TDP-43-missplicing was not found in mice [40,41,42].
To thoroughly investigate the impact of nuclear TDP-43 dysfunction in neuropathology, it is essential to utilize animal models that closely resemble humans. Given their proximity to humans and the presence of cytoplasmic localization of mutant TDP-43 in neuronal cytoplasm, we opted to employ cynomolgus monkeys for studying the role of TDP-43. In this study, we found that TDP-43 knockdown caused more neuronal damage and suppressed the expression of PJA1, an E3 ubiquitin ligase, in the monkey brain than rodent mice. These differential effects are due to the unique promoter sequences of the mouse and monkey PJA1 gene to which TDP-43 binds. Furthermore, we validated the critical function of PJA1 in TDP-43 deficiency-mediated toxicity through its rescuing effect via overexpression. This suggests that PJA1 can be a therapeutic target for neuropathology mediated by the nuclear loss of TDP-43.
Methods
Animals
The wild-type C57BL/6 mice at 8–12 months of age (n = 10 each group, 5 males and 5 females per group) and the WT cynomolgus monkeys aged 8–12 years (n = 6 each group, 3 males and 3 females per group) were used in the study.
Plasmids, viruses and antibodies
The shRNA designed for TDP-43 and PJA1 of different species was subcloned into the pLKO.1-GFP-puro vector (IGEbio Corp., Guangzhou) to generate lentivirus (LV)-sh-TDP-43 and LV-sh-PJA1 vectors. The control LV-GFP vector consisted of the same vector as LV-TDP-43 and contained the same ubiquitous expression promoter. The human PJA1 cDNA was subcloned into the pAAV9-EF1a vector (OBiO Corp., Shanghai) to generate the adeno-associated viruses (AAV)-human-PJA1 vector. The control AAV-GFP vector consists of the same vector as AAV-human-PJA1 and contains the same promoter. The titer of the LV vector genome is 1 × 109 vg/ml for LV-sh-TDP-43 and 1 × 109 vg/ml for LV-GFP. The titer of the AAV vector genome is 1 × 109 vg/ml for AAV-human-PJA1 and 1 × 1012 vg/ml for AAV-GFP.The primary antibodies used for immunoblot, immunofluorescence, and immunoprecipitation assays were as follows: rabbit anti-TDP-43 (Proteintech, 10,782–2-AP), rabbit anti-NeuN (Abcam, ab177487), mouse anti-Vinculin (Sigma, MAB3574), rabbit anti-PJA1 (Thermo, 72,845), chicken anti-MAP2 (Abcam, ab5392), rabbit anti-ChAT (Sigma, AB5042), rabbit anti-BOK (Abcam, ab186745), rabbit anti-BAK1 (CST, 12105S), rabbit anti-TUJ1 (Abcam, ab18207), rabbit anti-cleaved caspase-3 (CST, 9664S), rabbit anti-total caspase-3 (CST, 9662S), rabbit anti-DNM3 (BBI, D120571), rabbit anti-STAT1 (BBI, D120084), rabbit anti-SGK1 (BBI, D260927), and rabbit anti-PDGFRA (BBI, D151808). All secondary antibodies were purchased from Boster Bio and Thermo.
Stereotaxic injection
For injection of the mouse brain, C57BL/6 mice aged 8–12 months (n = 10 per group, 5 males and 5 females per group) were anesthetized by intraperitoneal injection of 2.5% Avertin. Their heads were placed in a Kopf stereotaxic frame (RWD Life Science) equipped with a digital manipulator, a UMP3-1 Ultra pump, and a 10 μl Hamilton microsyringe. A 33 G needle was inserted through a 1 mm drill hole in the scalp. Injections were performed at the following stereotaxic coordinates: 1.5 mm posterior to bregma, 1.0 mm lateral to the midline, 1.5 mm ventral to the dura, with bregma set at zero. The microinjections were carried out at a rate of 0.2 μl/min. The microsyringe was left in place for an additional 10 min before and after each injection. The LV-sh-TDP-43 or LV-sh-PJA1 viruses (1.5 μl) were stereotaxically injected into the right motor cortex of mice, and the control virus was injected into the left motor cortex.
Similarly, we injected LV-sh-TDP-43 or AAV-human-PJA1 into the right motor cortex of WT monkeys aged 8–12 years (n = 6 per group). AAV or LV-GFP was injected into the left motor cortex of the same monkey as a control. Each monkey was anesthetized by intraperitoneal injection of 0.3–0.5 mg atropine, followed by 10–12 mg ketamine and 15–20 mg pelltobarbitalumnatricum per kg body weight. Monkeys were secured in a stereotaxic apparatus (RWD Life Science). The precise location of the motor cortex for stereotaxic injection was determined by MRI before the injection. A 10 μl volume of the virus was injected into different locations in the right brain, and the needle insertion depth was calculated based on the MRI taken before surgery. After 8 weeks of injection, their brain tissues were isolated for immunohistochemical and Western blotting analysis.
Cell culture and transfection
The mouse neural crest-derived Neuro-2A cell line, human neuroblastoma SH-SY5Y cell line, and green monkey kidney fibroblast-like COS7 cell line were purchased from ATCC and cultured in DMEM/F12 medium containing 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin (Thermo Fisher Scientific), and 0.25 μg/ml amphotericin B (Thermo Fisher Scientific). The cells were maintained at 37 °C in 5% CO2 incubators. The medium was changed every 2 days. For transient transfection, cells were plated at 70–80% confluence and transfected with plasmid DNA using Lipofectamine 3000 (Invitrogen) for 24–72 h. Subsequently, cells were harvested for electrophoretic mobility shift assay (EMSA), luciferase reporter assay, and Western blotting.
Western blotting and Q-PCR
The cultured cells or brain tissues were homogenized in a soluble fraction buffer (10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.1% SDS, and 1% Triton X-100) with protease inhibitors (Sigma, P8340). The lysates were diluted in 1 × SDS sample buffer and sonicated for 15 s after incubation at 100 °C for 10 min. The total lysates (10–20 μg) were loaded onto a Tris–glycine gel and blotted onto a nitrocellulose membrane. The Western blotting was developed using the ECL (electrochemiluminescence) Prime Chemiluminescence kit (GE Healthcare Amersham).
For qPCR, reverse transcription reactions were performed with 1.5 µg of RNA isolated using the Superscript III First-Strand Synthesis System (Invitrogen). One microliter of cDNA was combined with 10 µl SYBR Select Master Mix (Applied Biosystems) and 1 µl of each primer in a 20 µl reaction. The reaction was performed in a real-time thermal cycler (Eppendorf, Realplex Mastercycler). The primer sequences for PJA1 cDNA in monkey and mouse are as follows: forward: TACACGGGCACGCGCAAACA and reverse: GGGACCAGACTGCAAAGGAGG. β-actin served as an internal control, with the following primer sequences: forward: ATCTGGCACCACACCTTCTACA and reverse: TACATGGCTGGGGTGTTGAAGGT. Relative expression levels were calculated using the 2−ΔΔCT method, with AAV-GFP or LV-GFP injection set as 1.
The primer sequences for the eight toxic substrates of PJA1 are as follows:
ARHGAP27: Forward: GGACGTGTACGTGCTGGTGGA.
ARHGAP27: Reverse: ACGTGCCACCAGTGCTCGGT.
MYCBP: Forward: AGGTCGCTGTCTTTGTAGT.
MYCBP: Reverse: GCACATCATCATCCTCGC.
PDGFRA: Forward: ACCCCATGTCTGAAGAAGA.
PDGFRA: Reverse: GATGTAAATGTGCCTGCCTT.
PML: Forward: TTGTGGTGATCAGCAGCT.
PML: Reverse: CATTGTCAATCTTGAGGTC.
DNM3: Forward: AGATGGGGAACCGGGAGAT.
DNM3: Reverse: AGAAAGTCCCTGCCCACGAA.
SGK1: Forward: GGGCAGTTTTGGAAAGGTTC.
SGK1: Reverse: AAGTGAAGGCCCACCAGGAAA.
SPHK1: Forward: GCACCCATGGGCCGCTGT.
SPHK1: Reverse: GCCAGGAAGAGGCGCAGC.
STAT1: Forward: AAAAGCAAGACTGGGAGCA.
STAT1: Reverse: AGATTACGCTTGCTTTTCC.
Immunofluorescence and immunohistochemistry
Mice or monkeys were anesthetized with 5% chloral hydrate and perfused with 0.9% NaCl followed by 4% paraformaldehyde (PFA). The brains were then removed and fixed in 4% PFA overnight at 4 °C. After fixation, the brains were transferred to 30% sucrose for 48 h and then cut into 20 or 30 μm sections using a cryostat at − 20 °C.
The sections were blocked in 4% donkey serum with 0.2% Triton X-100 and 3% BSA (bovine serum albumin, Roche Applied Science, BSAV-RO) in 1X PBS for 1 h. For immunofluorescent staining, 20 μm sections were incubated with primary antibodies in the same buffer at 4 °C overnight. After washing with 1X PBS, the sections were incubated with fluorescent secondary antibodies. Fluorescent images were acquired using a Zeiss microscope (Carl Zeiss Imaging, Axiovert 200 MOT) with either a 40X or 20X lens and a digital camera (Hamamatsu, Orca-100) with openlab software (Improvision).
For immunohistochemistry with DAB staining, after pre-blocking, 30 μm sections were incubated with antibodies for at least 24 h at 4 °C. A biotin/avidin immuno-assay kit (Vector Laboratories, PK-4001) and DAB kit (Thermo Scientific) were used for visualizing the staining. Images were acquired using a Zeiss microscope (Carl Zeiss Imaging, Imager A.2) with a 20X, 40X, or 63X lens and a digital camera (Carl Zeiss Imaging, AxioCam HRc) with AxioVision software.
Dual-luciferase reporter assay
For luciferase experiments, the mouse, monkey, or human PJA1 was subcloned into the pGL3-Basic vector using the double digestion enzymes Mlu (BioLabs, R3198S) and Kpn1 (BioLabs, R3142S).
For the mouse PJA1, the primers were as follows:
Mus-intron-1515 Forward: GTTCCTTCTCTCCCCTCCACCTACC.
Mus-intron-2967 Forward: AACCTGTGCTGCCTGTAATGAACGA.
Mus-pr/in-4257 Forward: TGCAAGCTCCAGCAACCTAGAAAGT.
Common Reverse: ATGCTTCTGTCAGTTTCAGGCCATCC.
Mus-prom-1 k Forward: TGCAAGCTCCAGCAACCTAGAAAGT.
Mus-prom-2 k Forward: GCATAGGTTAAATATGTTGACACTAGG.
Common Reverse: ATGCTTCTGTCAGTTTCAGGCCATCC.
For the monkey PJA1, the primers were as follows:
Mky-intron-1091 Forward: ATCTCTCTGCCACGTGTGTCAGACA.
Mky-intron-2251 Forward: CCAGCTGACTTCACAATCACTTCCG.
Mky-pr/in-3314 Forward: TTTCCCTCATTCCCTCCCTCCCTTC.
Common Reverse: ATTGAAAACCCTCCTAAGTCAGAGTG.
Mky-prom-1 k Forward: TTTCCCTCATTCCCTCCCTCCCTTC.
Mky-prom-2 k Forward: CTTGCCTTATTTCCTTTCTTCTTC.
Common Reverse: ATTGAAAACCCTCCTAAGTCAGAGTG.
For the human PJA1, the primers were as follows:
Hum-prom-1 k Forward: CTTCCCTCATTCCCTCTCTCCCTTT.
Hum-prom-2 k Forward: GCAACATGCTTTTTTCACTTAATATA.
Common Reverse: ATTGGAAACCCTCCTAAGTCAGAGTG.
Cells were plated in triplicate for each experiment and transfected with 500 ng of each firefly luciferase PJA1 construct (pGL3-T5-mouse/monkey/human-PJA1). The luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, E1910). The firefly luciferase activity of PJA1 constructs was normalized against the renilla luciferase output of the same pGL3-T5 vector.
Electrophoretic mobility shift assay (EMSA)
PJA1 DNA fragments were acquired using PCR. The PJA1 probe was designed with the following sequences: (Biotin) 5′-CACACACACGCACACACACACACACACACACACACACAGAAACCTCCTTTGCAGTCTGG-3′ (forward) and (Biotin) 5′-CCAGACTGCAAAGGAGGTTTCTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGTGTGTGTG-3′ (reverse). The PJA1 probe was incubated with 2 μg of nuclear protein in a 10 μl reaction solution containing 10 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, and 60 mM KCl at 26 °C for 30 min. The mixture was then electrophoresed through a 6% native polyacrylamide gel (PAGE) in 0.5X Tris–borate-EDTA buffer (TBE). The binding activity of TDP-43 to the biotin-labeled PJA1 probe was determined using a chemiluminescent EMSA kit (Beyotime, Shanghai), following the manufacturer’s protocol.
Chromatin immunoprecipitation assay (CHIP)
Chromatin immunoprecipitation (ChIP) was performed using the ChIP Assay kit (Millipore, 17-295) following the manufacturer’s instructions. Neuro-2A cells were collected after 48 h of incubation at 37 °C in DMEM/F12 medium containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were fixed with 1% formaldehyde for 10 min. Mouse brain tissue was ground and incubated with 1% formaldehyde for 15 min at 37 °C. The cross-linking reaction was quenched by adding glycine to a final concentration of 0.125 M for 15 min at room temperature.The cross-linked cells were harvested, washed with 1X PBS, and resuspended in 1 mL of SDS lysis buffer. The extract was sonicated on ice, and the soluble fraction, which predominantly contained DNA in the size range of 100 to 800 bp, was obtained by centrifugation. For each immunoprecipitation (IP), 60 µl of Protein G agarose was added to the pre-cleared chromatin and incubated for 1 h at 4 °C with rotation to remove proteins or DNA that might bind non-specifically to the Protein G agarose. Then, 100 µl of each IP sample was saved as input, and the remaining 900 µl was incubated with 5 µl of anti-TDP-43 antibody for 12 h at 4 °C with shaking. After that, the samples were incubated for an additional hour with the addition of 60 µl of Protein G Sepharose slurry. Immune complexes were collected by centrifugation and washed. The immunocomplexes were then collected and resuspended in 200 µl of elution buffer. Cross-linking was reversed by incubating for 4 h at 65 °C. Input and ChIP DNA samples were purified, resuspended in 20 µl TE, and analyzed by PCR using Thermo Scientific Maxima Hot Start Taq DNA Polymerase. The primer sequences for mouse PJA1 DNA were as follows: Forward: TACACGGGCACGCGCAAACA and Reverse: GGGACCAGACTGCAAAGGAGG. The primer sequences for monkey PJA1 cDNA were as follows: Forward: TACACGGGCACGCGCAAACA and Reverse: GGGACCAGACTGCAAAGGAGG.
Proteasome activity assay
All the samples were adjusted to a concentration of 0.5 mg/ml total protein by dilution with homogenization buffer and tested in triplicate. The chymotrypsin-like activity of 20S-beta-5 was determined using the substrate Suc-LLVY-aminomethyl coumarin (AMC) (40 μM; Bilmol), and the postglutamyl activity of 20S-beta-1 was determined using the substrate Z-LLE-AMC (400 μM; Bilmol).Equal amounts (10 μg) of the extracts were incubated with the corresponding substrates in 100 μl of proteasome activity assay buffer (0.05 M Tris–HCl, pH 8.0, 0.5 mM EDTA, 1 mM ATP, and 1 mM DTT) for 30–60 min at 37 °C. The reactions were stopped by adding 0.8 ml of cold water and placing the reaction mixtures on ice for at least 10 min. The free AMC fluorescence was quantified using a CytoFluor multi-well plate reader (FLUOstar; BMG Labtech) with excitation and emission wavelengths set at 380 nm and 460 nm, respectively. The fluorescence readings were then normalized by the protein concentrations to obtain the activity values in nmol/min/mg protein.
Statistical analysis
When comparing two groups, statistical significance was determined using the two-tailed Student’s t-test. One-way ANOVA was used to determine statistical significance when multiple groups were analyzed. Two-way ANOVA was used to analyze data for mice or monkeys that were repeatedly subjected to behavioral tests. Data are mean ± SEM. GraphPad Prism software was used for calculations. A P-value less than 0.05 was considered statistically significant.
Results
TDP-43 knockdown induces more neurotoxicity in the monkey brain than mouse
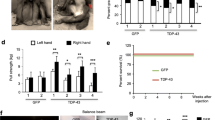
In a previous study, we demonstrated that overexpression of TDP-43(M337V) in the motor cortex of monkeys resulted in the distribution of cytoplasmic and cleaved TDP-43 in the primate brain, leading to neurotoxic gain-of-function [36]. One important question is whether the loss of TDP-43 can mediate neuropathology. Since species differences may prevent small animals from fully recapitulating important pathologic changes seen in patient brains, we first confirmed that endogenous TDP-43 is mainly located in the nucleus of both wild-type (WT) monkey and mouse cortex regions (Fig. S1). We then compared the effects of knocking down TDP-43 in the motor cortex of monkeys and mice, achieving similar effectiveness by injecting lentiviruses expressing TDP-43 shRNA (TDP-KD) or control GFP (Ctrl) (Fig. 1A and S2). Western blotting analysis using antibodies to NeuN and MAP2 revealed that the suppression of endogenous TDP-43 led to severe neuronal cell death in monkeys but milder neurotoxicity in mice (Fig. 1B and C). Immunohistochemical staining also indicated more neuronal loss in the injection area of the monkey cortex compared to the mouse brain (Fig. 1D and E).
TDP-43 knockdown induced more neurotoxicity in the monkey brain compared to mice. A Schematic representation of lentiviruses (LV) expressing TDP-43 shRNA (LV-TDP-43-shRNA) or scramble shRNA under the U6 promoter and GFP protein under the EF-1a promoter. The viruses were injected into the motor cortex of adult monkeys and mice (n = 6 animals per group; 8–12 years old for monkeys, 8–12 months old for mice). B Western blotting analysis indicated that the injection of LV-TDP-43-shRNA (KD) for two months suppressed approximately 50% of endogenous TDP-43 in both monkeys and mice, compared with non-targeted shRNA control (Ctrl). In the representative 4 animals, the reduced levels of neuronal proteins (NeuN and MAP2) were more pronounced in monkeys than mice. C Quantitative analysis of the ratios of TDP-43, NeuN and MAP2 to vinculin on western blots in panel (B). The data are mean ± SEM (n = 6 animals per group). *P < 0.05; ***P < 0.001; ns not significant. D Immunohistochemical staining with antibodies to C-terminal TDP-43 and NeuN showing that TDP-43 was knocked down by TDP-43 shRNA (KD) in the cortex of monkey (upper panel) and mouse (lower panel), but greater reduction of NeuN staining occurred in the monkey brain. Low magnification micrographs were shown in the left panel. Scale bar: 200 mm. E Quantitative analysis of the number of TDP-43 and NeuN positive cells in panel (D). Ten random fields (20×) in each slice from 3 animals in each group were used to generate data (mean ± SEM). *P < 0.05; ***P < 0.001
Transcriptome analysis showed that 8671 significantly differentially expressed genes (DEGs) were commonly present in both monkey and mouse cortex after TDP-43 knockdown, whereas 1023 and 394 DEGs were selectively changed in the monkey and mice, respectively (Fig. 2A and S3). The combinative and mean-value quantized heatmap displayed that some DEGs changed in the monkey were not significantly altered in the mouse cortex by TDP-43 knockdown (Fig. 2B). Gene networks of different functions via GO categorization indicated that the ‘ubiquitin-like protein transferase activity’-associated cluster was more extensive in the TDP-43 knockdown monkey brain (Figs. 2C and S4). The KEGG pathway analysis also showed enrichment of “ubiquitin-mediated proteolysis” related to the misfolded protein degradation process in the TDP-43 knockdown monkey (Figs. 2D and S5). Some genes that are important for the “UPS (ubiquitin–proteasome system)” functions showed opposite changes in the TDP-43 knockdown monkey and mouse brains, including the E3 ubiquitin ligase PJA1, which was significantly reduced in the TDP-43 knockdown monkey brain but increased in the mouse brain (Fig. 2E). By examining the expression of PJA1 mRNA and protein via Q-PCR (Fig. 2F) and western blotting (Fig. 2G and H, noting the same animals in Fig. 1B), we confirmed that the differential expression of PJA1 initially occurred at the transcriptional level. To address potential off-target effects, we designed two additional shRNAs targeting TDP-43 and confirmed that knocking down TDP-43 reduced PJA1 expression at the cellular level in monkey COS7 cells but upregulated PJA1 in the mouse Neuro2a cell line (Fig. S6).
Differential expression of PJA1 in the TDP-43 knocking down monkey and mouse. A Volcanic map for differentially expressed transcripts from the transcriptome analysis of the TDP-43 knockdown brain cortex in monkeys and mice. Colored points indicate statistically significant DEGs [false discovery rate (FDR) < 0.05]. Blue dots are downregulated genes [log2(fold change) < − 0.8], and red dots are upregulated genes [log2(fold change) > 0.8]. N = 3 biological replicates per genotype. There are 8671 differentially expressed genes that are commonly present in the brain cortex of both monkey and mouse; but 1023 and 394 were selectively found in the monkey and mouse, respectively. B Heatmap showing the combinative differentially expressing genes, with the mean-value (av. = average) quantization from 3 biological replicating animals, in the brain of monkey, but not mouse, after TDP-43 knockdown. C Gene networks of different functions via GO categorization between control and TDP-43 knockdown in the monkey and mouse brains. The red font highlight indicates “ubiquitin-like protein transferase activity”-associated cluster. D The important genes with “ubiquitin mediated proteolysis” by KEGG pathway enrichment, were also clustered after knocking down TDP-43 in the monkey and mouse brains. E The differentially expressing genes important for “ubiquitin-like protein transferase activity” and “ubiquitin mediated proteolysis” were also heat-mapped. Note that PJA1 was down-regulated in the monkey brain but up-regulated in the mouse brain after knocking down TDP-43. F Q-PCR analysis of the expression level of PJA1 mRNA, normalized to GAPDH, in TDP-43 knockdown tissues in mice and monkeys. The data are mean ± SEM (n = 6 animals per group). *P < 0.05; **P < 0.01. G Western blotting analysis of TDP-43 knockdown tissues (the same samples in Fig. 1B) in monkeys and mice, showed that the suppressing TDP-43 reduced PJA1 in the monkey brain, but increased PJA1 expression in the mouse brain. H Quantitative analysis of the intensity ratio of PJA1 levels to vinculin on western blotting in panel (G). The data are mean ± SEM (n = 6 animals per group). *P < 0.05; **P < 0.01
TDP-43 binds to the mouse, but not monkey, PJA1 promote to suppress its transcription
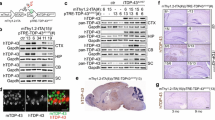
TDP-43 has been shown to interact with the UG repeats of the untranslated region to suppress the expression of target genes [4, 16, 20, 22, 24, 43,44,45,46]. In this study, we investigated whether TDP-43 binds differently to the promoters of the monkey and mouse PJA1 genes, leading to species-dependent transcriptional effects. We found that the TDP-43 binding region in the monkey and human PJA1 promoters is very similar. However, the mouse PJA1 gene promoter is followed by the intron 1 region, which contains a typical (TG)32 repeat before the translation initiation codon “ATG,” while this repeat region is not present in the monkey or human PJA1 introns (Fig. 3A). To determine the importance of these sequence differences for PJA1 transcription, we isolated different DNA regions of the PJA1 gene for luciferase assays in transfected mouse (Neuro-2a), monkey (COS7), and human (HEK293) cell lines. We found that different promoter regions produced varying reporter activities in the mouse and primate cell lines (Fig. 3B). Interestingly, the mouse PJA1 DNA region containing the TG repeat (mouse-intron 2967) promoted reporter activity, whereas the human intron 1 region without this repeat failed to activate the reporter (Fig. 3B). Following suppression of endogenous TDP-43 by siRNA (Fig. S7), the core promoter region (− 2000/ + 1) activity was decreased in the monkey and human cells, while the mouse core promoter (+ 1/ + 2967) activity was dramatically induced (Fig. 3C). Analysis of potential transcription factors (TFs) (https://jaspar.genereg.net/) that can bind to the PJA1 promoter revealed notable differences in TFs binding to the mouse-(+ 1/ + 2967) and monkey-(− 2000/ + 1) promoters. Only five TFs, including FOXO3, HOXA5, SOX10, HOXB5, and MYOG, were commonly present in both monkey and mouse sequences (Fig. 3D).
Species-dependent regulation of PJA1 by TDP-43 knockdown at the transcriptional level. A The putative DNA binding conserved sequences of (TG/GT)n motifs (32 units) were localized before the translation initiation codon “ATG” in the mouse PJA1 genes, but were not found in the human and monkey PJA1, with the sequences under the cartoon graphs. B Different promoter regions of the PJA1 gene were inserted into the pGL3-basic luciferase report vector for expression in cultured cells. The core promoter reporter of PJA1 was determined for transcription activity in transfected Neuro-2a, COS7 or HEK293 cells, and the results were obtained from three independent experiments. Note that the human and monkey PJA1 promoter region (– 2000/ + 1) had the highest reporter activity. However, the mouse PJA1 promoter region (– 2000/ + 1) had very low reporter activity, but the mouse (+ 1/ + 2967) region containing the 5'UTR and intron1 with the (TG/GT)n motifs showed the higher promote activity whereas the counterpart of the monkey or human promoter did not yield activity. C TDP-43 siRNA inhibited the transcription activity of the mouse PJA1 core promoter (+ 1/ + 2967) and the monkey promoter (– 2000/ + 1) activity, which was detected by the luciferase assays in three independent experiments. One-way ANOVA with Tukey’s test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Data are mean ± SEM. D The putative cis-elements in the PJA1 promoter of monkey and mouse were determined by https://jaspar.genereg.net/ program analyses. Note that only the limited five TFs including FOXO3, HOXA5, SOX10, HOXB5 and MYOG were commonly presented in the monkey and mouse sequences. E The chromatin immunoprecipitation (ChIP) assay was performed using mouse Neuro-2a cell lysates and wild-type (WT) mouse brain tissues. The endogenous TDP-43 was immunoprecipitated by the TDP-43 antibody and the associated DNAs were then detected by PCR. The results showed the binding of endogenous TDP-43 to (TG/GT)n motifs in the mouse PJA1 DNAs, which did not occur in monkey COS7 cells or brain tissues. IgG served as a negative control. F Q-PCR detection of PJA1 DNAs associated with TDP-43 that was immunoprecipitated by anti-TDP-43 in ChIP assay. The specific PJA1 DNA bound endogenous TDP-43 in mouse Neuro-2a cells and brain tissues. G The electrophoretic mobility shift assay (EMSA) was performed for the interaction of TDP-43 with (TG/GT)n motifs. The results showed that the probes targeting the (TG/GT)32 on the mouse PJA1 genes were shifted (arrows) by the TDP-43 antibody
TDP-43 is predominantly localized in the nucleus and likely binds to (UG/TG)n repeats with more than 6 units [47,48,49]. To confirm this, we obtained motif-containing (TG/GT)n repeats in the mouse and monkey brains and verified them through sequencing (Fig. S8). CHIP assay with the TDP-43 antibody precipitated endogenous TDP-43 in mouse and monkey cortex tissues. The DNAs associated with the precipitated nuclear TDP-43 in mouse Neuro-2a cells or WT mouse cortex were identified by PCR (Fig. 3E and F), revealing that the intron 1 segments of the PJA1 genes bound to endogenous TDP-43 and contained the (TG/GT)n repeats specifically in the mouse but not the monkey COS7 cell line or monkey cortex (Fig. 3E and F). Using the IGV genome browser to analyze the TDP-43-associated DNAs, we confirmed that the DNA segment located in front of the translation initiation codon “ATG” in the PJA1 gene was captured in mouse but not monkey brain tissues (Fig. S9). EMSA was also performed to examine the interaction of TDP-43 with (TG/GT)n motifs, which showed that the probes containing the (TG/GT)32 repeat on the mouse PJA1 gene were shifted by the TDP-43 antibody (Fig. 3G).
PJA1 protects neuronal cells against toxic substrates in monkey and mouse brain
PJA1 is a brain-expressed E3 ubiquitin ligase with an essential RING-H2 finger. It plays a role in ubiquitinating its targeted proteins, such as MAGED1/NRAGE/Dlxin1, leading to their subsequent degradation by the proteasome [50,51,52]. PJA1 is known as a common suppressor of protein aggregation associated with neurodegenerative diseases [53], It has suppressive effects on TDP-43 aggregate formation [54] and facilitates the degradation of ataxin-3 and huntingtin polyglutamine proteins, reducing polyglutamine-mediated toxicity [55]. Searching for PJA1 substrates using the database (http://ubibrowser.bio-it.cn/) (Fig. 4A) revealed several neuronal toxic proteins, including SPHK1 [56], STAT1 [57], SGK1 [58], DNM3 [59], PDGFRA [60], ARHGAP27 [61], PML [62], MYCBP2 [63] and others, which were induced at the transcript level by TDP-43 knockdown in both monkey and mouse cortex (Fig. 4B and C). However, Western blot analysis showed that the deficiency of TDP-43 led to the noticeable accumulation of these toxic substrates at the protein level in the monkey brain but not in mice (Fig. 4D and E). These results suggest that TDP-43 deficiency selectively affects PJA1-mediated degradation of targeted proteins in the monkey brain. Furthermore, the chymotrypsin-like and post-glutamyl activities were not significantly impaired by TDP-43 knockdown in both animals (Fig. S10). Considering the accumulation of toxic substrates in the monkey brain due to TDP-43 inhibition, we examined changes in apoptosis-related proteins, including cleaved caspase-3, BAK, and BOK1 (Fig. S11). The results showed increased levels of apoptotic proteins in the monkey brain when TDP-43 was knocked down. Consistently, there were the loss of motor neurons and activation of astrocytes in monkeys (Fig. S12). All these results suggest that apoptosis may be involved in neurodegeneration caused by the loss of TDP-43. Thus, in the primate brain, the suppression of PJA1 may contribute to severe neurotoxicity resulting from TDP-43 knockdown, whereas in the rodent brain, the elevated levels of PJA1 may directly remove the generated toxic substrates.
The neurotoxic substrates of PJA1 were increased by TDP-43 knockdown in monkey brain. A Prediction of the substrates of PJA1 by the database (http://ubibrowser.bio-it.cn/). The red dots indicate the known ESI (E3-substrate interaction), and the blue dots are the associated predicted ESI. B All the predicated ESI genes after TDP-43 knockdown in both monkey and mouse brains were heat-mapped (upper panel). Among these ESI of PJA1, SPHK1, STAT1, SGK1, DNM3, PDGFRA, ARHGAP27, PML and MYCBP2 with known neuronal toxicity were induced by TDP-43 knockdown in both the monkey and mouse cortex (lower panel). C Q-PCR analysis of the expression level of the known toxic genes of PJA1 in TDP-43 knockdown tissues in mice and monkeys, which was normalized to GAPDH. The data are mean ± SEM (n = 6 animals per group). *P < 0.05; **P < 0.01; ***P < 0.001. ns not significant. D Western blotting analysis of TDP-43 knockdown tissues in monkeys and mice showed that the accumulation of the neuronal toxic substrates of PJA1, including DNM3, STAT1, SGK1 and PDGFRA were detected in monkey brain, but not in TDP-43 knockdown mouse brain. E Quantitative analysis of the intensity ratio of TDP-43, DNM3, STAT1, SGK1 and PDGFRA to vinculin on western blotting in panel (D). The data are mean ± SEM (n = 6 animals per group). *P < 0.05; **P < 0.01; ***P < 0.001. ns Not significant
To confirm this, we generated a lentiviral vector expressing PJA1 shRNA and GFP under the EF1a promoter, along with its scramble control, and injected the viruses, along with LV-TDP-43 shRNA virus, into the motor cortex of mice (Fig. 5A). Western blot analysis revealed that suppressing PJA1 in the mouse cortex at 8–12 months of age significantly reduced the expression of neuronal proteins compared to the injection of the scramble control (Fig. 5B and C). Immunofluorescence double-staining also indicated that the inhibition of PJA1 induced pronounced neuronal cell death in the injected area, as evidenced by the loss of NeuN and MAP2 staining (Figs. 5D and E, and S13). These findings prompted us to investigate whether overexpression of PJA1 could enhance the clearance of toxic substrates in vivo and whether its downregulation could promote neurodegeneration. To test this, we generated an AAV9 vector expressing human PJA1 or GFP control under the same EF1a promoter (Fig. 5F). We then stereotaxically co-injected LV-TDP-43 shRNA and AAV-PJA1 or its AAV-GFP control into the motor cortex of monkeys aged 8–12 years. Western blot analysis showed that the overexpression of PJA1 in the monkey brain dramatically reduced TDP-43 knockdown-mediated neuronal cell loss, as evidenced by increased NeuN and MAP2 labeling (Fig. 5G and H). Double-staining also revealed that the restored PJA1 in the monkey cortex was associated with the clearance of toxic substrates in neuronal cells, indicating its protective effects (Figs. 5I and J, and S14). Immunohistochemical staining further confirmed that the restoration of PJA1 in the monkey cortex could rescue neuronal cell death caused by TDP-43 knockdown (Fig. S15).
Influence of PJA1 Alteration on Neurotoxicity in TDP-43 Knockdown Animal Brains. A Schematic representation of lentiviruses expressing PJA1 shRNA (LV-PJA1-shRNA) or scramble shRNA under the U6 promoter and GFP protein under the EF-1a promoter. These were stereotaxically co-injected with LV-TDP-43-shRNA into the motor cortex of mouse brains (n = 6 animals per group; 8–12 months old mice). B Western blot analysis revealed that PJA1-shRNA suppressed the expression of endogenous PJA1 and neuronal proteins (NeuN and MAP2) in representative mouse brains, two months after injection, compared to the scramble shRNA control. C Quantitative analysis of the intensity ratios of PJA1, NeuN, and MAP2 to vinculin on western blotting in panel (B). Data are presented as mean ± SEM (n = 6 animals per group). **P < 0.01; ***P < 0.001. D Immunofluorescence double-staining of PJA1, MAP2, and NeuN showed a pronounced reduction in MAP2 and NeuN levels by TDP-43 and PJA1 knockdown in the mouse cortex, compared to TDP-43 knockdown alone (n = 6 animals per group; 8–12 months old mice). Scale bar: 200 μm. E Quantitative analysis of the density of positive cells per image (40×). A total of 10 random fields in each section were examined, and the data (mean ± SEM) were obtained from three independent experiments (n = 6 for each group, 8–12 months old mice). ***P < 0.001. F Schematic representation of the adeno-associated virus expressing human PJA1 (pAAV-human PJA1) or GFP control (pAAV-GFP control) under the EF1a promoter, co-injected with LV-TDP-43-shRNA into the motor cortex of monkey brains (n = 5 animals per group; 8–12 years old monkeys). G Western blot analysis indicated that overexpression of PJA1 increased MAP2 and NeuN levels in representative TDP-43 knockdown monkey brains, two months after injection, compared to GFP control (pAAV-GFP control). H Quantitative analysis of the ratios of PJA1, NeuN, and MAP2 to vinculin on western blotting in panel (G). Data are presented as mean ± SEM (n = 5 animals per group). *P < 0.05; **P < 0.01. I Immunofluorescence double-staining of PJA1, MAP2, and NeuN showed that the reduction of MAP2 and NeuN was rescued by PJA1 overexpression in the monkey cortex (n = 5 animals per group; 8–12 years old monkeys). Scale bar: 200 μm. J Quantitative analysis of the density of positive cells per image (40X). A total of 10 random fields in each section were examined, and the data (mean ± SEM) were obtained from three independent experiments (n = 5 for each group, 8–12 years old monkeys). ***P < 0.001
Considering the pronounced nuclear loss of TDP-43 in primates, our studies suggest that the reduction of PJA1 mediated by TDP-43 deficiency could contribute to severe neurotoxicity in the monkey brain. However, due to the different binding of TDP-43 to the promoter of the mouse PJA1 gene, milder neurotoxicity was observed upon the loss of TDP-43 in the rodent brain. Therefore, PJA1-related neurotoxicity may be primate-specific and could serve as a potential therapeutic target for treating TDP-43-related neuropathology (see Fig. 6).
A proposed model for the nuclear loss of TDP-43-induced neurotoxicity. TDP-43 knockdown leads to heightened neurotoxicity in the monkey brain and decreases the expression of PJA1. This reduction in PJA1 expression can subsequently enhance the accumulation of toxic substrates. Conversely, in the rodent brain, the loss of TDP-43 results in increased PJA1 expression. This upregulation occurs due to the binding of TDP-43 to specific sequences within the mouse PJA1 promoter. These findings suggest that PJA1 plays a neuroprotective role in response to the nuclear loss of TDP-43 in the primate brain
Discussion
TDP-43 proteinopathy is characterized by the accumulation of cytoplasmic TDP-43 and aggregates in the brains and spinal cords of nearly all patients (~ 97%) with ALS and in ~ 45% of FTLD cases [1, 6, 64,65,66]. Furthermore, TDP-43 proteinopathies have been observed in 57% of Alzheimer’s disease cases and in some dementia patients with Lewy bodies [37,38,39, 67, 68]. However, only a small percentage (< 5%) of ALS patients carry mutations in the TDP-43 gene [5, 15, 69, 70]. Thus, factors other than TDP-43 mutations are primarily responsible for the cytoplasmic accumulation of TDP-43, and TDP-43 mutations may exacerbate this abnormal redistribution. The mislocalization of TDP-43 in the cytoplasm, leading to its loss from the nucleus, has been implicated in ALS, FTLD, [6, 64, 71] and other neurological disorders [5, 37,38,39, 67]. This combination of cytoplasmic inclusions, which can result in gain-of-function, and nuclear depletion of TDP-43, which can lead to loss-of-function, is thought to contribute significantly to TDP-43-associated neuropathology [72]. The cytoplasmic accumulation of TDP-43 appears to be independent of its mutations but is regulated by species-related factors. Transgenic rodent models overexpressing either normal or mutant TDP-43 predominantly show nuclear accumulation of TDP-43 [29,30,31,32, 73], while the expression of TDP-43 in the brains and spinal cords of monkeys through AAV vector injection leads to cytoplasmic distribution [36, 74]. Additionally, a transgenic TDP-43 pig model expresses mutant TDP-43 in the cytoplasm [75]. In previous studies using non-human primates, we discovered that caspase-4, which is selectively expressed in primate brains, cleaves TDP-43, removing N-terminal fragments carrying the nuclear import signal and resulting in the cytoplasmic accumulation of C-terminal fragments in primate brains [36]. Therefore, the loss of nuclear TDP-43 due to cytoplasmic distribution may be better replicated by knocking down TDP-43 in the primate brain, allowing for the identification of primate-specific effects of nuclear TDP-43 deficiency when compared to rodent models.
It is evident that TDP-43 is crucial for neuronal survival and development, as demonstrated by conditional knockout TDP-43 mouse models that exhibit neuronal loss [28, 76, 77]. The critical role of nuclear TDP-43 in gene transcription and RNA processing is well-documented [4, 14, 78]. Manipulation of TDP-43 levels in the nucleus, either through overexpression or depletion, significantly impacts gene expression and leads to corresponding phenotypes in mice [29, 79]. However, TDP-43 proteinopathy is characterized by the loss of nuclear TDP-43 and the accumulation of TDP-43 in the cytoplasm. When considering the neuropathology and phenotypes observed in humans with endogenous mutant TDP-43 expression, it is crucial to account for the loss of nuclear TDP-43. Therefore, investigating the neuropathology resulting from TDP-43 knockdown, rather than complete deletion, is essential. By comparing the phenotypes of monkey and mouse models, we discovered that TDP-43 knockdown induced more neurotoxicity in the monkey brain than in the mouse brain, highlighting the importance of using non-human primates to study TDP-43 proteinopathy.
Our findings suggest that PJA1 contributes to the primate-specific toxicity caused by the nuclear loss of TDP-43. Strong evidence supporting this idea is that the loss of TDP-43 selectively suppressed PJA1 expression in monkey cells but increased PJA1 expression in mouse cells. These differential effects are attributed to the distinct sequences of the PJA1 promoters in monkey and mouse cells, which result in different binding of nuclear TDP-43 and opposite effects on the regulation of PJA1 transcript expression in monkey and mouse. The differential binding of TDP-43 to the PJA1 gene in different species aligns with recent findings showing that TDP-43 binds to the human-specific UG-rich motif in the STMN2 intron, which is absent in the mouse STMN2 gene [80]. Additional evidence supporting the role of PJA1 in TDP-43 deficiency-mediated neuropathology includes the exacerbation of neurodegeneration upon PJA1 knockdown and the rescue effect achieved by overexpressing PJA1 in monkey brains with reduced TDP-43 levels. These findings are consistent with the critical role of PJA1 in eliminating misfolded or toxic proteins to prevent neurodegeneration [53,54,55] and suggest that PJA1 could be a potential therapeutic target for treating TDP-43 deficiency-associated neuropathology.
Data availability
The key data supporting the findings of this study are presented within the article and the Supplementary Information. The RNA sequencing reported in this paper can be found at https://doi.org/10.6084/m9.figshare.23684634.
References
Chen-Plotkin AS, Lee VM-Y, Trojanowski JQ (2010) TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol 6:211–220
Lee EB, Lee VM-Y, Trojanowski JQ (2012) Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci 13:38–50
Guo L, Shorter J (2017) Biology and pathobiology of TDP-43 and emergent therapeutic strategies. Cold Spring Harb Perspect Med 7:a024554
Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling S-C, Sun E, Wancewicz E, Mazur C (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14:459–468
Cohen TJ, Lee VM, Trojanowski JQ (2011) TDP-43 functions and pathogenic mechanisms implicated in TDP-43 proteinopathies. Trends Mol Med 17:659–667
Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133
Kwong LK, Neumann M, Sampathu DM, Lee VM-Y, Trojanowski JQ (2007) TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease. Acta Neuropathol 114:63–70
Neumann M, Kwong LK, Sampathu DM, Trojanowski JQ, Lee VM-Y (2007) TDP-43 proteinopathy in frontotemporal lobar degeneration and amyotrophic lateral sclerosis: protein misfolding diseases without amyloidosis. Arch Neurol 64:1388–1394
Kwong LK, Uryu K, Trojanowski JQ, Lee VM-Y (2008) TDP-43 proteinopathies: neurodegenerative protein misfolding diseases without amyloidosis. Neurosignals 16:41–51
Buratti E (2015) Functional significance of TDP-43 mutations in disease. Adv Genet 91:1–53
Watanabe S, Oiwa K, Murata Y, Komine O, Sobue A, Endo F, Takahashi E, Yamanaka K (2020) ALS-linked TDP-43 M337V knock-in mice exhibit splicing deregulation without neurodegeneration. Mol Brain 13:1–4
Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE (2005) TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem 280:37572–37584
Prakash A, Kumar V, Banerjee A, Lynn AM, Prasad R (2021) Structural heterogeneity in RNA recognition motif 2 (RRM2) of TAR DNA-binding protein 43 (TDP-43): clue to amyotrophic lateral sclerosis. J Biomol Struct Dyn 39:357–367
Freibaum BD, Chitta RK, High AA, Taylor JP (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res 9:1104–1120
Lagier-Tourenne C, Cleveland DW (2009) Rethinking als: the fus about tdp-43. Cell 136:1001–1004
Ayala YM, De Conti L, Avendaño-Vázquez SE, Dhir A, Romano M, D’ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E (2011) TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J 30:277–288
Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Župunski V (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14:452–458
Buratti E, Baralle FE (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator ofCFTR Exon 9. J Biol Chem 276:36337–36343
Tank EM, Figueroa-Romero C, Hinder LM, Bedi K, Archbold HC, Li X, Weskamp K, Safren N, Paez-Colasante X, Pacut C (2018) Abnormal RNA stability in amyotrophic lateral sclerosis. Nat Commun 9:2845
Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, Silani V, Ratti A (2012) TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem 287:15635–15647
Costessi L, Porro F, Iaconcig A, Muro AF (2014) TDP-43 regulates β-adducin (Add2) transcript stability. RNA Biol 11:1280–1290
Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C (2007) TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci 35:320–327
Volkening K, Leystra-Lantz C, Yang W, Jaffee H, Strong MJ (2009) Tar DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS). Brain Res 1305:168–182
Wang IF, Wu LS, Chang HY, Shen CKJ (2008) TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem 105:797–806
Kabashi E, Lin L, Tradewell ML, Dion PA, Bercier V, Bourgouin P, Rochefort D, Bel Hadj S, Durham HD, Velde CV (2010) Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum Mol Genet 19:671–683
Xu Z, Yang C (2014) TDP-43—the key to understanding amyotrophic lateral sclerosis. Rare Dis 2:e944443
Broeck LV, Callaerts P, Dermaut B (2014) TDP-43-mediated neurodegeneration: towards a loss-of-function hypothesis? Trends Mol Med 20:66–71
Wu L-S, Cheng W-C, Chen C-Y, Wu M-C, Wang Y-C, Tseng Y-H, Chuang T-J, Shen C-KJ (2019) Transcriptomopathies of pre-and post-symptomatic frontotemporal dementia-like mice with TDP-43 depletion in forebrain neurons. Acta Neuropathol Commun 7:1–31
Gendron TF, Rademakers R, Petrucelli L (2013) TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43. J Alzheimers Dis 33:S35–S45
Huang C, Tong J, Bi F, Zhou H, Xia X-G (2012) Mutant TDP-43 in motor neurons promotes the onset and progression of ALS in rats. J Clin Investig 122:107–118
Shan X, Chiang P-M, Price DL, Wong PC (2010) Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc Natl Acad Sci 107:16325–16330
Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH (2009) TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci 106:18809–18814
Yan S, Wang C-E, Wei W, Gaertig MA, Lai L, Li S, Li X-J (2014) TDP-43 causes differential pathology in neuronal versus glial cells in the mouse brain. Hum Mol Genet 23:2678–2693
Mitchell JC, Constable R, So E, Vance C, Scotter E, Glover L, Hortobagyi T, Arnold ES, Ling S-C, McAlonis M (2015) Wild type human TDP-43 potentiates ALS-linked mutant TDP-43 driven progressive motor and cortical neuron degeneration with pathological features of ALS. Acta Neuropathol Commun 3:1–16
Wils H, Kleinberger G, Janssens J, Pereson S, Joris G, Cuijt I, Smits V, Ceuterick-de Groote C, Van Broeckhoven C, Kumar-Singh S (2010) TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci 107:3858–3863
Yin P, Guo X, Yang W, Yan S, Yang S, Zhao T, Sun Q, Liu Y, Li S, Li X-J (2019) Caspase-4 mediates cytoplasmic accumulation of TDP-43 in the primate brains. Acta Neuropathol 137:919–937
Hasegawa M, Nonaka T, Masuda-Suzukake M (2017) Prion-like mechanisms and potential therapeutic targets in neurodegenerative disorders. Pharmacol Ther 172:22–33
Josephs KA, Whitwell JL, Weigand SD, Murray ME, Tosakulwong N, Liesinger AM, Petrucelli L, Senjem ML, Knopman DS, Boeve BF (2014) TDP-43 is a key player in the clinical features associated with Alzheimer’s disease. Acta Neuropathol 127:811–824
McAleese KE, Walker L, Erskine D, Thomas AJ, McKeith IG, Attems J (2017) TDP-43 pathology in Alzheimer’s disease, dementia with Lewy bodies and ageing. Brain Pathol 27:472–479
Ze M, López-Erauskin J, Baughn MW, Zhang O, Drenner K, Sun Y, Freyermuth F, McMahon MA, Beccari MS, Artates JW (2019) Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat Neurosci 22:180–190
Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, Mordes DA, Burberry A, Steinbaugh MJ, Gamage KK, Kirchner R (2019) ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci 22:167–179
Ling JP, Pletnikova O, Troncoso JC, Wong PC (2015) TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349:650–655
Buratti E, Dörk T, Zuccato E, Pagani F, Romano M, Baralle FE (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20:1774–1784
Kuo P-H, Chiang C-H, Wang Y-T, Doudeva LG, Yuan HS (2014) The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG-and TG-rich nucleic acids. Nucleic Acids Res 42:4712–4722
François-Moutal L, Felemban R, Scott DD, Sayegh MR, Miranda VG, Perez-Miller S, Khanna R, Gokhale V, Zarnescu DC, Khanna M (2019) Small molecule targeting TDP-43’s RNA recognition motifs reduces locomotor defects in a Drosophila model of amyotrophic lateral sclerosis (ALS). ACS Chem Biol 14:2006–2013
Yin P, Bai D, Deng F, Zhang C, Jia Q, Zhu L, Chen L, Li B, Guo X, Ye J (2022) SQSTM1-mediated clearance of cytoplasmic mutant TARDBP/TDP-43 in the monkey brain. Autophagy 18:1955–1968
Chang C-k, Wu T-H, Wu C-Y, Chiang M-h, Toh EK-W, Hsu Y-C, Lin K-F, Liao Y-h, Huang T-h, Huang JJ-T (2012) The N-terminus of TDP-43 promotes its oligomerization and enhances DNA binding affinity. Biochem Biophys Res Commun 425:219–224
Mompeán M, Romano V, Pantoja-Uceda D, Stuani C, Baralle FE, Buratti E, Laurents DV (2016) The TDP-43 N-terminal domain structure at high resolution. FEBS J 283:1242–1260
Qin H, Lim L-Z, Wei Y, Song J (2014) TDP-43 N terminus encodes a novel ubiquitin-like fold and its unfolded form in equilibrium that can be shifted by binding to ssDNA. Proc Natl Acad Sci 111:18619–18624
Mishra L, Tully R, Monga S, Yu P, Cai T, Makalowski W, Mezey E, Pavan W, Mishra B (1997) Praja1, a novel gene encoding a RING-H2 motif in mouse development. Oncogene 15:2361–2368
Sasaki A, Masuda Y, Iwai K, Ikeda K, Watanabe K (2002) A RING finger protein Praja1 regulates Dlx5-dependent transcription through its ubiquitin ligase activity for the Dlx/Msx-interacting MAGE/Necdin family protein, Dlxin-1. J Biol Chem 277:22541–22546
Zoabi M, Sadeh R, de Bie P, Ciechanover A (2011) PRAJA1 is a ubiquitin ligase for the polycomb repressive complex 2 proteins. Biochem Biophys Res Commun 408:393–398
Watabe K, Niida-Kawaguchi M, Tada M, Kato Y, Murata M, Tanji K, Wakabayashi K, Yamada M, Kakita A, Shibata N (2022) Praja1 RING-finger E3 ubiquitin ligase is a common suppressor of neurodegenerative disease-associated protein aggregation. Neuropathology 42:488–504
Watabe K, Kato Y, Sakuma M, Murata M, Niida-Kawaguchi M, Takemura T, Hanagata N, Tada M, Kakita A, Shibata N (2020) Praja1 RING-finger E3 ubiquitin ligase suppresses neuronal cytoplasmic TDP-43 aggregate formation. Neuropathology 40:570–586
Ghosh B, Karmakar S, Prasad M, Mandal AK (2021) Praja1 ubiquitin ligase facilitates degradation of polyglutamine proteins and suppresses polyglutamine-mediated toxicity. Mol Biol Cell 32:1579–1593
Lee JY, Han SH, Park MH, Baek B, Song I-S, Choi M-K, Takuwa Y, Ryu H, Kim SH, He X (2018) Neuronal SphK1 acetylates COX2 and contributes to pathogenesis in a model of Alzheimer’s disease. Nat Commun 9:1479
Morozko EL, Smith-Geater C, Monteys AM, Pradhan S, Lim RG, Langfelder P, Kachemov M, Kulkarni JA, Zaifman J, Hill A (2021) PIAS1 modulates striatal transcription, DNA damage repair, and SUMOylation with relevance to Huntington’s disease. Proc Natl Acad Sci 118:e2021836118
Elahi M, Motoi Y, Shimonaka S, Ishida Y, Hioki H, Takanashi M, Ishiguro K, Imai Y, Hattori N (2021) High-fat diet–induced activation of SGK1 promotes Alzheimer’s disease–associated tau pathology. Hum Mol Genet 30:1693–1710
Brown EE, Blauwendraat C, Trinh J, Rizig M, Nalls MA, Leveille E, Ruskey JA, Jonvik H, Tan MM, Bandres-Ciga S (2021) Analysis of DNM3 and VAMP4 as genetic modifiers of LRRK2 Parkinson’s disease. Neurobiol Aging 97:148.e117-148.e124
Sohn E-J, Nam Y-K, Park H-T (2021) Involvement of the miR-363-5p/P2RX4 axis in regulating Schwann cell phenotype after nerve injury. Int J Mol Sci 22:11601
Saeed M (2018) Genomic convergence of locus-based GWAS meta-analysis identifies AXIN1 as a novel Parkinson’s gene. Immunogenetics 70:563–570
Korb E, Finkbeiner S (2013) PML in the brain: from development to degeneration. Front Oncol 3:242
Coleman MP, Höke A (2020) Programmed axon degeneration: from mouse to mechanism to medicine. Nat Rev Neurosci 21:183–196
Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 351:602–611
Janssens J, Van Broeckhoven C (2013) Pathological mechanisms underlying TDP-43 driven neurodegeneration in FTLD–ALS spectrum disorders. Hum Mol Genet 22:R77–R87
Prpar Mihevc S, Darovic S, Kovanda A, Bajc Česnik A, Župunski V, Rogelj B (2017) Nuclear trafficking in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Brain 140:13–26
Colom-Cadena M, Grau-Rivera O, Planellas L, Cerquera C, Morenas E, Helgueta S, Muñoz L, Kulisevsky J, Martí MJ, Tolosa E (2017) Regional overlap of pathologies in Lewy body disorders. J Neuropathol Exp Neurol 76:216–224
Ticozzi N, LeClerc AL, Van Blitterswijk M, Keagle P, McKenna-Yasek DM, Sapp PC, Silani V, Wills A-M, Brown RH Jr, Landers JE (2011) Mutational analysis of TARDBP in neurodegenerative diseases. Neurobiol Aging 32:2096–2099
Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Velde CV, Bouchard J-P, Lacomblez L, Pochigaeva K, Salachas F (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 40:572–574
Renton A (2014) Chi∫ A, Traynor BJ State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 17:17–23
Geser F, Lee VMY, Trojanowski JQ (2010) Amyotrophic lateral sclerosis and frontotemporal lobar degeneration: a spectrum of TDP-43 proteinopathies. Neuropathology 30:103–112
Mackenzie IR, Neumann M, Baborie A, Sampathu DM, Du Plessis D, Jaros E, Perry RH, Trojanowski JQ, Mann DM, Lee VM (2011) A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol 122:111–113
Swarup V, Phaneuf D, Dupré N, Petri S, Strong M, Kriz J, Julien J-P (2011) Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor κB–mediated pathogenic pathways. J Exp Med 208:2429–2447
Uchida A, Sasaguri H, Kimura N, Tajiri M, Ohkubo T, Ono F, Sakaue F, Kanai K, Hirai T, Sano T (2012) Non-human primate model of amyotrophic lateral sclerosis with cytoplasmic mislocalization of TDP-43. Brain 135:833–846
Wang G, Yang H, Yan S, Wang C-E, Liu X, Zhao B, Ouyang Z, Yin P, Liu Z, Zhao Y (2015) Cytoplasmic mislocalization of RNA splicing factors and aberrant neuronal gene splicing in TDP-43 transgenic pig brain. Mol Neurodegener 10:1–20
Iguchi Y, Katsuno M, Niwa J-i, Takagi S, Ishigaki S, Ikenaka K, Kawai K, Watanabe H, Yamanaka K, Takahashi R (2013) Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain 136:1371–1382
Wu L-S, Cheng W-C, Shen C-KJ (2012) Targeted depletion of TDP-43 expression in the spinal cord motor neurons leads to the development of amyotrophic lateral sclerosis-like phenotypes in mice. J Biol Chem 287:27335–27344
Taylor JP, Brown RH Jr, Cleveland DW (2016) Decoding ALS: from genes to mechanism. Nature 539:197–206
Philips T, Rothstein JD (2015) Rodent models of amyotrophic lateral sclerosis. Curr Protoc Pharmacol. https://doi.org/10.1002/0471141755.ph0567s69
Baughn MW, Ze M, López-Erauskin J, Beccari MS, Ling K, Zuberi A, Presa M, Gonzalo-Gil E, Maimon R, Vazquez-Sanchez S (2023) Mechanism of STMN2 cryptic splice-polyadenylation and its correction for TDP-43 proteinopathies. Science 379:1140–1149
Acknowledgements
We thank Dr. Bang Li at Jinan University for animal care and experiment.
Funding
This work was supported by the National Natural Science Foundation of China (32270564, 81830032, 82071421, 82394422); the Guangdong Basic and Applied Basic Research (2023A1515010811, 2022A1515011205); the Department of Science and Technology of Guangdong Province (2021ZT09Y007, 2018B030337001); the Guangzhou Key Research Program on Brain Science (202007030008); and the Fundamental Research Funds for Central Universities (21622113).
Author information
Authors and Affiliations
Contributions
PY and X-JL designed research. LZ, FD and DB performed research. JH, QJ, CZ and KO analyzed data. SL provided technical guidance. PY and X-JL wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
All animals experimental procedures were approved by the local ethical committee of Jinan University for animal experiments and met governmental guidelines. Cynomolgus monkeys were bred at Guangdong Landao Biotechnology Co., Ltd., an institution accredited by the China Association for Assessment and Accreditation of Laboratory Animal Care. The company has dedicated breeders and veterinarians who closely monitor the water, temperature, and humidity in the cages where the monkeys are kept, as well as the monkeys’ health. Mice were bred in the animal facility of Jinan University. All animal-related protocols were approved in advance by the Animal Care and Use Committee of Guangdong Landao Biotechnology Co., Ltd. and Jinan University. This study strictly complied with the “Guidelines for the Care and Use of Laboratory Animals in the Institute of Laboratory Animal Science (2006 Edition)” and “The Use of Non-human Primates in the Institute of Laboratory Animal Science (2006 Edition)” “(est. 2006)” to ensure that personnel safety and animal welfare.
Consent to publish
All authors have approved the content of this manuscript and provided consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhu, L., Deng, F., Bai, D. et al. Loss of TDP-43 mediates severe neurotoxicity by suppressing PJA1 gene transcription in the monkey brain. Cell. Mol. Life Sci. 81, 16 (2024). https://doi.org/10.1007/s00018-023-05066-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-05066-2