Abstract
Calvarial bone is one of the most complex sequences of developmental events in embryology, featuring a uniquely transient, pluripotent stem cell-like population known as the cranial neural crest (CNC). The skull is formed through intramembranous ossification with distinct tissue lineages (e.g. neural crest derived frontal bone and mesoderm derived parietal bone). Due to CNC’s vast cell fate potential, in response to a series of inductive secreted cues including BMP/TGF-β, Wnt, FGF, Notch, Hedgehog, Hippo and PDGF signaling, CNC enables generations of a diverse spectrum of differentiated cell types in vivo such as osteoblasts and chondrocytes at the craniofacial level. In recent years, since the studies from a genetic mouse model and single-cell sequencing, new discoveries are uncovered upon CNC patterning, differentiation, and the contribution to the development of cranial bones. In this review, we summarized the differences upon the potential gene regulatory network to regulate CNC derived osteogenic potential in mouse and human, and highlighted specific functions of genetic molecules from multiple signaling pathways and the crosstalk, transcription factors and epigenetic factors in orchestrating CNC commitment and differentiation into osteogenic mesenchyme and bone formation. Disorders in gene regulatory network in CNC patterning indicate highly close relevance to clinical birth defects and diseases, providing valuable transgenic mouse models for subsequent discoveries in delineating the underlying molecular mechanisms. We also emphasized the potential regenerative alternative through scientific discoveries from CNC patterning and genetic molecules in interfering with or alleviating clinical disorders or diseases, which will be beneficial for the molecular targets to be integrated for novel therapeutic strategies in the clinic.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Cranial neural crest cell and its fate labeling
Cranial neural crest cell (CNC) is an embryonic structure with developmental potential in vertebrates. Morphologically, pre-migratory cranial neural crest cells (CNCs) are initiated in the dorsal folds of the neural tube during neurulation (Fig. 1), pre-migratory CNCs undergo an epithelial-to-mesenchyme transition, and then delaminate from the neural tube to become migratory CNCs, which will further migrate into differently destined sites in early embryos. The migratory CNCs can give rise to diverse types of cells including peripheral neurons and glia, melanocytes and the mesectodermal derivatives, which include osteoblasts and chondrocytes at the craniofacial level, as well as the smooth muscle cells in cardiovascular structures.
The cell fate of the neural crest can be traced using transgenesis and genome editing technologies in mice. The Cre-loxP system is a frequently used tool, in which expression of a Cre-recombinase in CNCs or their derivatives genetically enables the expression of a Cre-reporter allele, thus permanently tracing CNC-derived cells [1]. Wnt1-Cre [2,3,4] and Pax3-Cre [5] have been used to genetically trace pre-migratory CNCs and their derivatives. Multiple Cre transgenic mouse lines have been generated to label migratory CNCs and their derivatives, e.g., Dhh-Cre [6, 7], HtPA-Cre [8], Sox10-Cre [9], Sox10ER(T2)-Cre [10], Mef2c-F10N-Cre [11], and P0-Cre [12,13,14] (Fig. 1).
However, Wnt1-Cre transgene is observed to induce ectopic activation of Wnt signaling which results in defective midbrain development [15]. Using Cre immunosignals, we revealed the differences in P0-Cre/R26-lacZ and P0-Cre/R26-RFP in E8.0–9.5 (4–19 somites) embryos in labeling CNC [16]. P0-Cre labels migrating CNC cells and are more extensive in the forebrain and hindbrain but not apparent in the midbrain. Wnt1-Cre labels extensive in the midbrain. The difference between P0-Cre and Wnt1-Cre in labeling CNC suggests a better explanation of the differential distributions of CNC derivatives and the phenotypes caused by Cre-driven genetic modifications.
Neural crest and mesoderm lineage in vertebrate-specific structure
Using a transgenic mouse with a permanent neural crest cell lineage marker, Wnt1-Cre/R26R showed that the frontal bones are neural crest-derived and the parietal bones mesodermal [4], providing solid perspectives on skull evolution. Cranial neural crest and mesodermal lineages contribute to the development of most craniofacial structures. CNC-derived two frontal bones located anteriorly in the skull vault, and two mesoderm lineage-derived parietal bones located in the middle posterior part of the skull vault [4, 17, 18]. Interparietal bone is located in the posterior part of the skull containing mixed lineages from both neural crest and mesoderm [19]. Between the cranial bones, different sutures connect each other. Sagittal suture is from the neural crest, separating two mesoderm-derived parietal bones. Coronal suture is a mesoderm origin, separating the frontal and parietal bone [4].
Calvarial bones are essential to support and protect brain growth and expansion. The neural crest and mesoderm lineages endow a regional difference in vertebrate-specific structures during the development [20]. What is more, two different lineages in the cranial bones were found conserved in different species, such as in humans, chickens, and rats. The craniofacial structures from different tissue lineage exhibited evolutional features, providing novel insights for the comparative analysis on the development and evolution of vertebrates and vertebrate-specific structures [21].
Gene regulatory network in CNCs and mesoderm-derived osteoblasts and calvarial bone
The differences in the intrinsic osteogenic potential are highly associated with tissue lineage in cranial bone. CNC-derived osteoblasts grow faster with less differentiation compared to mesoderm-derived osteoblasts [22]. When CNC-derived osteoblasts were cultured into mesodermal osteoblasts, CNC-derived osteoblasts are capable to nucleate ossification centers [23]. CNC-derived osteoblasts demonstrate to be low apoptosis and higher osteogenic capacity [24] (Fig. 2).
A shared gene regulatory network in cranial bones in mouse and human. CNC-derived frontal bone (in green) and osteoblasts show higher proliferation and osteogenesis and strong activities of BMP, FGF and WNT signaling. Mesoderm derived parietal bone (in gray) and osteoblasts display a higher level of differentiation, apoptosis and TGF signaling
Higher levels of FGF ligands and receptors were observed in frontal bone [25], suggesting that FGF signaling mediates osteogenic potential difference [26]. Later evidence showed that FGF1 is a positive regulator of Runx2 and functions as a unique molecule in CNC-derived osteoblast differentiation [27]. Canonical Wnt signaling and BMP signaling is active in CNC-derived frontal bone [20, 28]. Using Axin2 knockout model to activate the Wnt signaling, parietal bone is capable to reach a higher level of osteogenic potential similar to that in frontal bone [29]. TGF-β signaling is a main positive regulator of apoptosis in cranial osteoblasts [24, 30] (Fig. 2). A shared gene regulatory network in the canonical WNT, TGF-β, BMP, and FGF pathways was verified in the mouse and human to govern the regional differences upon osteogenic potential within the cranial bone [31], indicating that the findings from mouse models are suitable for the translational potential to human (Fig. 2).
High throughput sequencing from mouse and human frontal and parietal compartments showed a broad spectrum of differently expressed genes covering the cell matrix, transcriptional factors, cytokines and receptors [32, 33]. At embryonic development, a large number of differently expressed genes were found at different axial levels of CNC-derived tissues using transgenic mouse lines, indicating again the potential regulatory networks in controlling the formation of specific skeletal elements and promoting migration through different molecular pathways [34].
Gene regulatory network from CNC patterning into osteoblasts
We previously revealed some genes to be functional during the development of the skull [18], and proposed a gene regulatory network to maintain CNC-derived frontal bone characteristic [35]. A diverse set of interacting signals, transcription factors, and downstream effectors have been indicated to endow CNC cell features to influence fate decision, migration and differentiation [36]. Therefore, we summarized recent discoveries from conserved signaling pathways including BMP, TGF, FGF, Wnt, Hippo, PGDF, Notch, Hedgehog signaling, and integrated them together to be a gene regulatory network within CNCs to orchestrate spatial orientation, developmental stability and plasticity, which are hallmarks of osteoblast differentiation and calvarial bone development.
BMP signaling
BMP signaling is transduced through the binding of BMP ligands to BMP receptor (BMPR) type I and type II (BMPRI and BMPRII), which further activate the intracellular Smads (Smad1, Smad5, and Smad8) proteins to form a complex with Smad4, which can translocate into the nucleus and trigger target gene expressions [20]. Bmp2 deletion in mice lacks both branchial arches and detectable migratory CNCs [37] and plays a crucial role in craniofacial bone development [38]. CNC inactivation of Bmp2, Bmp4 and Bmp7 leads to multiple loss of CNC-derived skeletal elements [39]. Double loss of Bmp5 and Bmp7 in mice exhibited underdeveloped branchial arches due to defective proliferation of migrating CNCs [40]. Bmp4 exposure to mouse embryonic stem cells in vitro can induce neural ectodermal differentiation [41]. Bmp4-treated CNCs were capable to differentiate into osteocytes [41]. Noggin and chordin, the secreted Bmp antagonists, were used as an early inducer to CNC induction and regulated the emigration of CNC from the neural tube [42]. Gremlin 1, a secreted Bmp antagonist, is expressed in CNC and required for neural crest development [43]. During the differentiation from iPSC to CNC in vitro, precise regulation of BMP activity is critically needed for efficient differentiation [44].
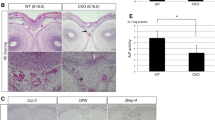
At the receptor level, BMPR1A is a major type 1 BMP receptor for BMP-Smad signaling during skull development [45]. Enhanced BMPR1A in CNC can cause premature suture fusion in mice [46] and result in midline craniosynostosis via mTOR activation [47]. Mice lacking a BMP type I receptor called activin A receptor type 1 (ACVR1) in CNCs display an alteration of cell fate from odontoblasts to osteoblasts [48]. The constitutively activated Acvr1(ca-Acvr1) mouse line was generated to investigate the functions of BMP-dependent signaling [49], and found that constitutively active ACVR1 in CNCs caused CNC fate switch to a chondrogenic fate in mice [50]. Suture mesenchymal stem cells (SuSC) are controversial for the tissue origins due to different transgenic mouse models that have provided the tracing properties in SuSC [20]. BMPR1A deletion in SuSC-specific cells triggered precocious differentiation, resulting in craniosynostosis in mice [51]. Therefore, BMP signaling is vitally important for CNC migration, fate commitment and shaping the development of craniofacial structures in mice (Fig. 3).
An integrated gene regulatory network from CNC patterning into osteoblast lineage. CNC undergoes induction, delamination and migration at an early stage. The migratory CNCs follow a gene regulatory network pathway to differentiate into osteoblast to shape the development of craniofacial structures. Multiple signaling pathways (e.g. BMP, TGF, Wnt, Hippo, PGDF, Hedgehog) can integrate together to converge into specific transcription factors to control CNC spatial orientation, developmental stability and plasticity during calvarial bone development
TGF-β signaling
Transforming growth factor-β (TGF-β) superfamily members signal through a heteromeric receptor (TGF-β type II receptor, Tgfbr2; TGF-β type I receptor, Alk5) complex to induce intracellular Smads (Smad2/3) in response to regulate CNC patterning during craniofacial development. Tgfbr2 loss in CNCs showed craniofacial skeletal malformations [52] and skull defects [53]. Alk5 controls CNC survival and regulates the fate of CNCs [54]. TGF-β signaling removal in CNC leads to alterations in proliferation and differentiation of CNC-derived osteogenic cells [55].
At a cellular level, Smad4 mutants display the underdevelopment of craniofacial structures [56]. For mesoderm-derived bone, TGF-β signaling can influence the fate of mesoderm-derived cells via a TGF-β/Msx2 cascade to regulate the skull development [57]. Modulation of TGF-β signaling in CNC is found useful for the prevention of congenital craniofacial birth defects [58]. What is more, transferrin receptor (Tfrc) deletion in CNC can cause craniofacial malformation by working as a facilitator to TGF-β/BMP signaling pathways [59]. For Smad-independent TGF-β signaling, TGF-β-activated kinase 1 (Tak1) provides a critical interaction between the canonical and noncanonical TGF-β signaling. Tak1 deficiency in CNC displays the round skull, hypoplastic maxilla and mandible [60]. Splicing factor Rbfox2 is expressed in CNC, and functions through Rbfox2-TGF-β-Tak1 interaction to control CNC patterning. Rbfox2 deletion in CNC leads to defective craniofacial bone development [61] (Fig. 3). Therefore, TGF-β signaling plays a critical role in instructing CNCs to form the craniofacial skeleton.
Wnt signaling
Most Wnt genes will result in distinct phenotypes once eliminated from the genome. Through the binding of Wnts ligand to Frizzleds (FZDs) receptors, it will lead to elevated levels of β-catenin [62]. Wnt signaling is an early signal for CNC specification, induction, delamination and migration during craniofacial development [63]. Temporal control by Wnt/β-catenin is an important factor for CNC fate decisions [64]. Wnt ligands secretion and Wnt/β-catenin signaling in cranial mesenchyme are dispensable for specification and proliferation of early meningeal progenitors, which are derived from CNCs [65]. Knockout of β-catenin in CNC causes increased apoptosis in pre-migratory CNCs [66]. Enhanced activation of Wnt signaling using Axin2 knockout mice can increase the osteogenic potential and osteogenesis in mesoderm derived cranial bones [29] (Fig. 3).
Tcf7l1, a transcriptional repressor of Wnt target genes, is expressed in the anterior neural fold region during neurulation and is required for forebrain development [67]. Conditional inactivation of Tcf7l1 using AP2α-Cre leads to CNC fate conversion and aberrant activation of Wnt/β-catenin signaling [67]. The other Wnt antagonist Dkk1, which is secreting by the prechordal mesoderm, can inhibit CNC formation and prevent the formation of neural fold in mouse [68]. Protein arginine methyltransferase 1 (Prmt1) can negatively regulate canonical Wnt signaling. Disruption of Prmt1 in CNC causes craniofacial defects [69]. CNC-secreted Bmp4 genetically interacts with Msx1, which further represses Osr2-dependent expression of Wnt antagonists Dkk2 and Sfrp2 in mouse to result in craniofacial malformation [70]. In short, the gradient of Wnt/β-catenin signaling is crucial for CNC-derived osteoblasts differentiation at a cellular and molecular level.
Hippo signaling
Hippo is a fundamentally conserved signaling pathway in regulating cell proliferation, survival, and differentiation for normal CNC development. YAP/TAZ are transcriptional factors responding to MST1/2 and LTS1/2 activities to induce target gene expressions [71]. Nf2 acts as upstream of Hippo signaling, transgenic mice carrying a 2.4-kb Nf2 promoter to drive β-galactosidase (β-gal) with a nuclear localization signal were generated to visualize Nf2 expression pattern. Strong Nf2 promoter activity was observed in the developing brain and migrating neural cells, suggesting a specific function of Nf2 in CNC migration [72].
The knockout of the Hippo pathway gene Nf2, Mst1/2, or Lats2 leads to embryonic lethality in the mouse germline. Yap and Taz deletion in CNC using Wnt1-Cre and Wnt1-Cre2SOR resulted in reduced proliferation in branchial arch mesenchyme, and transcriptional factor Foxc1 was found involved in regulating YAP/TAZ activity [73]. In addition, BAF complex works through interaction with Hippo-Yap signaling to modulate gene regulatory networks for neural crest development [74]. Therefore, CNC-specific deletion of BAF155/BAF170 leads to a wide range of craniofacial defects [74]. The transcription factor FoxO6 in CNC was found to be an activator of Hippo signaling. FoxO6 is specifically expressed in craniofacial tissues. FoxO6 knockout mice lead to the expanded face and skull. Mechanically, FoxO6 activates Lats1 expression, which further increases the level of Yap phosphorylation to activate Hippo signaling. Accordingly, FoxO6 knockout mice result a decrease in Lats1 expression, which significantly reduced Shh and Runx2 activities, suggesting that Shh and Runx2 are interplayed with Hippo signaling [75]. Interestingly, PITX2 is able to activate FoxO6 expression, suggesting that PITX2-FoxO6-Hippo interaction is capable to coordinate the osteogenic differentiation and skull growth during CNC patterning [75] (Fig. 3).
FGF signaling
Fibroblast growth factor (FGF) signaling consists of 22 ligands that interact with 4 receptors (Fgfrs), and the intracellular signaling is mediated by multiple pathways including PI3K-AKT, PLCγ, STAT and MAPK [76]. FGF is essential in CNC-derived skeletogenic differentiation [76]. Fgf8 is a negative regulator to control osteogenic fate and is sufficient to switch CNC-derived mesenchyme into cartilage [77]. Fgfr1 loss in CNC induced multiple malformations including heterotopic chondrogenesis and osteogenesis at the interface of the anterior portions of frontal bones [78], defective cleft palate and severe craniofacial pattern [79]. Conditional expression of the Fgfr2-S252W mutation in CNC results in severe craniofacial phenotype [80], and simultaneous expression of Fgfr2 (S252W) is sufficient to induce craniosynostosis [81]. Ectopic expression of Fgfr2 has been implicated in the development of craniosynostosis in mice and humans [82] through the induction of Runx2-dependent osteogenic program [82].
Interaction with FGF signaling pathway is crucial for the CNC-derived craniofacial bone development. Bcl11b, a transcriptional factor, expresses in osteogenic and sutural mesenchyme [82], Bcl11b acts as a regulator to Fgfr2. Bcl11b knockout in CNCs exhibits increased osteoprogenitors, premature osteoblast differentiation, and enhanced cranial mineralization [82]. Homeoprotein engrailed 1 (EN1) interacts with Fgfr2 during osteogenic differentiation [83]. Robo1 deficiency in CNC showed defective cranial frontal and parietal bones [84] in part through the interaction with FGF signaling [84]. Additionally, Dex, a common used drug in treating benign and malignant conditions, can disrupt CNC development via the inhibition of FGF signaling, which in turn causes defective cranial bones [85]. Recent evidence using Fgfr1 and Fgfr2 allelic knock-in mouse strains showed that Fgfr1 and Fgfr2 play combinatorial roles in craniofacial development, uncoupling a novel Fgfr kinase-dependent cell adhesion property in CNCs [86].
Hedgehog signaling and primary cilium
The Hedgehog signalling (Hh) pathway consists of sonic hedgehog (Shh), desert hedgehog (Dhh), and Indian hedgehog (Ihh). In the presence of Hh, Hh binds to its receptor Ptc, so that the Ptc inhibition to Smo is released, which leads to activation of Gli transcriptional factors [87]. Ihh deletion in CNC displays the gradual dwarfism in mice [88]. Shh sends signals to multipotent CNCs to control normal craniofacial development [89]. Disruption of Hh signals leads to abnormal CNC development and malformed skull base [90]. Hh-responsiveness removal in CNC results in the absence of CNC-derived skeletal components [91], suggesting Hh signaling is essential to establish intrinsic and extrinsic patterning cues for the craniofacial skeleton [92]. Fox mediates the action of Shh in regulating facial development [91], so Shh-Fox interactions are crucial for CNC proliferation. Inactivation of suppressor of Fused (Sufu) in CNC or mesoderm, which is a critical repressor of Hedgehog signaling, results in abnormal osteogenic differentiation [93].
The primary cilium is a microtubule-based organelle, where intraflagellar transport (IFT) plays a pivotal role in assembling primary cilia. The cilia are also the main center for Hh signaling transduction. IFT20 mutants in CNC did not secrete procollagen and results in skeletal dysplasia via the dysregulation of intracellular collagen trafficking [94]. Mechanically, IFT20 disruption in CNC down-regulates PDGFRα production, which further causes the suppression of PDGF-Akt signaling, resulting in decreased osteogenic proliferation and increased apoptosis [94]. IFT88 deletion in CNC results in a decreased rate of cell proliferation at early stages [95]. Motor proteins within primary cilia are also essential for the signaling and development of the skull, such as Kif3a is a motor protein. Kif3a loss in CNC causes a dramatic defect in intramembranous ossification, resulting in the missing of Shh signaling in Kif3a-deficient CNC-derived mesenchyme [96]. Polycystin 2 (Pkd2) is localized in primary cilia, and Pkd2 deletion in CNC exhibits malformed skull structures [97] (Fig. 3).
Ciliary proteins EVC and EVC2 are positive regulators of Hedgehog signaling and express in craniofacial tissues [98]. EVC2 removal in CNC does not cause obvious skull defects [99] but shows distinct defects in the skull base [100]. However, malfunction of EVC2 in mice and human shows striking phenotype, which is paralleling to human-chimpanzee craniofacial differences, suggesting that a regulatory divergence of Hedgehog signaling may contribute to the unique craniofacial morphology in human [101]. Besides, Fuz, serving as a crucial regulator of ciliogenesis, Fuz removal in CNCs results in expanded frontal bone mainly via the excessive Fgf8 expression [102].
Notch signaling
Notch signaling consists of delta-like (DLL1, DLL3, DLL4) and the Jagged (JAG1, JAG2) families, which serve as ligands. Ligand-receptor binding leads to a structural change in the Notch receptor, and the intracellular domain (NICD) can translocate into the nucleus for the association with transcription factors. Conditional gain or loss of Notch signaling in CNC results in craniofacial abnormalities, suggesting an autonomous role for gradient Notch signaling in CNC migration, proliferation, and differentiation [103].
As a membrane-bound Notch ligand JAG1, deletion of Jagged1 in CNC exhibited underdeveloped mesenchyme and aberrant growth of craniofacial structure [104]. JAG1 stimulates osteoblast-specific genes expressions in CNCs, contributing to the differentiation and mineralization of osteoblasts [105]. Exogenous JAG1 exposure to CNCs can induce osteoblast differentiation and maturation along with target genes expressions of Notch signaling such as Hes1 and Hey1 [105]. Using synthetic hydrogels to deliver exogenous JAG1 in CNC cell line is sufficient to induce in vivo osteogenesis [106].
PDGF signaling
The platelet-derived growth factor (PDGF) family has four ligands (PDGF-A, PDGF-B, PDGF-C, PDGF-D) and two tyrosine kinase receptors (PDGFRα and PDGFRβ). PDGFRα and PDGFRβ are expressed in the craniofacial mesenchyme [107]. PDGFRα is expressed in the migratory progenitors including the CNCs. PDGFRα disruption in CNCs resulted in defective craniofacial development [108]. Conditionally expressing an auto-activated PDGFRα in CNC enhances osteoprogenitor proliferation and osteoblasts ossification [109]. The defective cell survival and patterning in PDGFRα deleted CNC is a cause of malformation of early embryogenesis [110] (Fig. 3). PDGF-responsive SRF-driven transcriptional program has been identified [111]. SRF and PDGFRα mutants genetically interact in regulating CNC proliferation and migration during craniofacial bone development.
PDGFRβ contributed to CNC-derived craniofacial bone development [112].PDGFRs genetically interact with each other but play distinct mechanisms to regulate CNC activity and subsequent craniofacial development, such as PDGFRα plays a predominant role in CNC migration whereas PDGFRβ primarily contributes to the proliferation of mesenchyme [113].
Signaling crosstalk and transcriptional factors
Signaling crosstalk
Integrated signaling pathways and factors are converged into specific transcriptional factors to regulate CNC plasticity and patterning. FGF, BMP and Shh mediated signaling pathways are required for cranial suture morphogenesis and calvarial bone development [138]. SMAD4 as the common mediator of TGF-β/BMP signaling, Smad4 loss results in increased Wnt/β-catenin activity partially through the downregulation of Dkk1 and Sfrp1, indicating the crosstalk between TGF-β/BMP and WNT signaling to ensure the proper CNC cell fate decision during organogenesis [139]. Fhl3, a scaffold LIM domain protein, modulates BMP gradient interpretation during CNC induction. Differential Fhl3 expression in underlying paraxial mesoderm cells can ensure a finely tuned coordination of BMP and WNT signaling at several stages of CNC patterning [140].
BMP/Wnt signaling, mTORC and autophagy interaction have been found to be functional in CNC fate determination. Using a constitutively active ACVR1 mouse model, the increased BMP signaling can induce a higher level of Sox9 activity in CNC, which further driving CNC to adopt a chondrogenic fate and results in the ectopic formation of craniofacial cartilage [50]. Additionally, autophagy plays a central role in degrading Wnt/β-catenin activities, which can be activated by BMP-mediated mTORC1 activities, suggesting that coordinated signaling pathways are crucial for CNC cell fate selection and performance in vivo [50] (Fig. 3).
Sox (SRY-related HMG-box) transcriptional factor
Sox genes play diverse functions in CNC patterning. Sox9 is a crucial factor for the chondrogenic lineage commitment in CNCs. Sox10 is one of the earliest CNC-specifying genes and plays an autonomous role in driving CNC delamination and directly regulates numerous downstream effectors [114]. SoxE contains cis-regulatory sequences which can direct its expression in migrating CNCs [115]. Murine Sox2 plays an essential role in controlling progenitor behavior. Sox2 mutant embryos deregulate CNC progenitors, resulting in aberrant Sox10+ CNC migration and exacerbated branchial arches [116]. Med23 binds to the promoter region of Sox9 to repress Sox9 expression in vitro. Med23 mutants in CNC showed the enhanced interaction of Sox9 binds to beta-catenin, which in turn resulted in altered skeletal differentiation [117] (Fig. 3).
Transcription factor AP2 and Alx3
AP2 mutant mice died with severe malformed face and skull [118]. AP2 can specify CNC commitment as an early cell-autonomous factor, but induce osteogenesis as a non-autonomous factor at a later stage [119]. AP2α is prominently expressed in migrating CNC, and later expresses in the regulatory regions during craniofacial development [120]. AP2α-IRES-Cre mice displayed perturbed cranial bones [121]. AP-2β is expressed in CNC and its derived tissue. AP-2β deletion in CNCs results in post-natal ocular defects [122]. Our previous findings show that CNC-derived frontal bone has a high level of AP-2β at the embryonic stage, suggesting that AP-2β is an important factor in CNC-derived frontal bone [33]. The stage-dependent function of AP2 was due to its ability to target distinct genomic regions, suggesting that AP2 can be an evolutionary strategy to diversify the regulators to control embryonic development.
Single-cell RNA sequencing analysis shows that Alx transcription factor is enriched in the frontonasal population of neural crest cells. Alx3 regulates the timing of distinct differentiation and cellular morphologies among frontonasal CNC subpopulations. Alx3/Alx4 double mutant newborn mice show malformed CNC-derived skull elements, which are anatomically manifest at embryonic 10.5, suggesting that the Alx factor is crucial to control developmental timing to shape craniofacial skeletal elements [123] (Fig. 3).
Msx1/2, Twist1/2, Dlx and Runx2
Msx1 and Msx2 are early identified genes associated with craniofacial formation. Msx2 is an important factor to drive CNC differentiation and establish a balance between CNC survival and apoptosis for proper craniofacial morphogenesis [124]. Msx2 deletion in CNC shows defective skeletogenic mesenchyme and frontal bone [125]. Double deletion of Msx1 and Msx2 display defective CNC patterning and cranial bones development [126]. Msx2 can interact with Twist to coordinate proliferation and differentiation of skeletogenic mesenchyme and calvarial bone formation. Double mutants of Twist1 and Msx2 reduce the population of cranial mesenchyme [127]. Special deletion of Twist1 in the mesoderm-derived parietal bone leads to defective cranial bones and extracellular matrix production and cell–matrix interaction [128]. Of note, Twist1, Msx2 and EphA4 can form a complex to regulate coronal synostosis [129]. EphA4 works as a Twist1 effector which further regulates the development of frontal and parietal bones [130]. Twist1 can trigger a selective downregulation of Fgf23 on mesoderm-derived osteoblasts to interfere with the osteogenic activity in mesoderm-derived parietal bone [131] (Fig. 3).
Dlx2 controls CNC-derived craniofacial skeleton. Dlx2 and Dlx3 deletion leads to defective calvaria morphogenesis [132]. Dlx5 and Dlx6 coordinate together in shaping craniofacial development. MEF2C is required for the expressions of the Dlx5 and Dlx6 within the pharyngeal [133]. Besides, Dlx5 expression requires Msx1, so the Msx1/Dlx5 interaction is crucial for osteogenic induction during frontal bone development [134]. Msx2 is reported to be a direct downstream target of BMP signaling and Twist1 is found to be a downstream target of the WNT signaling pathway in craniofacial bone development [135] (Fig. 3).
Runx2 is a master transcriptional factor to control osteoblast differentiation. Msx genes are critical for the Runx2 activity in CNC-derived osteogenic lineage. Runx2 is heterogeneously expressed in Prx1-GFP+ cells, which are located at the intracutural mesenchyme in the calvaria. Runx2 activity starts at the Prx1+Sca1+ mesenchymal stem cell stage and ends at the Osx+Prx1−Sca1− osteoblast precursor stage [136]. Runx2 deficiency in CNC resulted in defective craniofacial bones. Both mesoderm-derived cells and CNC-derived cells require Runx2 activity to differentiate during intramembranous ossification, suggesting that distinct dependency upon Runx2 for proper ossification in the calvaria [137].
Epigenetic factors
Epigenetic factors pose precise timing in controlling CNC differentiation during craniofacial development. Polycomb domains provide a chromatin template to regulate CNC positional identity in vivo [141]. Deletion of histone H3 lysine 9 methyltransferase (G9A) in CNC results in incomplete ossification. G9A and H3K9me2 physically interacted to regulate Twist activity to control its temporal and tissue-specific expression [142]. G9A can enhance the transcriptional activity of Runx2. Lacking G9A expression in Sox9-positive CNC-derived cells displayed severe hypo-mineralization in cranial vault bone [143]. Ezh2 catalyzes trimethylation of lysine 27 in histone 3 (H3K27me3), Ezh2 removal in CNC leads to abnormal formation of craniofacial bones [144].
A highly conserved acetyltransferase Gcn5 (or KAT2A) is required for murine craniofacial development. Gcn5 mutation in CNC demonstrated defective craniofacial skeleton and abnormal activity of histone 3 lysine 9 (H3K9) acetylation [145]. Gcn5 acts as an epigenetic regulator of H3K9 acetylation, and the underlying pathway of Gcn5 is via direct activation of mTORC1 [146]. Ankyrin repeat domain 11 (ANKRD11) is another chromatin regulator in CNC cell fate modulation. Ankrd11 deletion in CNC leads to a defective reduction in intramembranous ossification [147].
Histone deacetylase (Hdac) activity is essential to guide CNC patterning. Hdac8 mutation in CNC develops skull instability [148]. Hdac3 knockout in CNC exhibits penetrant craniofacial abnormalities in part through the upregulations of Msx1, Msx2 and BMP4 in the CNC-derived mesenchyme [149]. Ubiquitin proteasomal pathway is involved in epigenetic regulation. Wwp2 E3 ubiquitin ligase can work with paired-like homeobox transcription factors during craniofacial development. Conditionally deletion APC (Cdh1) E3 ubiquitin ligase in CNC displays bone malformation, similar defective phenotypes were found compared to that in Wwp2-deficient mice such as a domed skull, a short snout and a twisted nasal bone [150]. Nedd4 works as an E3 ubiquitin ligase. Ablation of Nedd4 in CNC or osteoblasts showed profound craniofacial defects with a marked reduction in cranial bones [151].
CNC patterning dysregulation and associated human diseases
Birth defects are the most common craniofacial anomalies, frequently involving defective CNC migration, proliferation, and fate determination. Defects in post-migratory CNC can result in similar phenotypes of developing craniofacial skeleton and craniosynostosis in the clinic, such as premature fusion of cranial bones/cranial sutures. Activated mutations of Fgfr1-3 or inactivation of Twist1 in CNC are the most common causes of the occurrence of craniosynostosis [81, 130]. Treacher Collins syndrome is an autosomal dominant congenital disorder with a characterization of craniofacial deformities [152]. TCOF1 encodes a serine/alanine-rich nucleolar phosphoprotein protein called TREACLE, which plays a role in ribosome biogenesis in CNC. TCOF1 deficiency in CNCs in mice contributes to a high similarity to the clinical phenotype of Treacher Collins syndrome [153].
Excessive intake of vitamin A can lead to decreased Shh signaling and elevated CNC apoptosis during early pregnancy, resulting in an increased incidence of cleft palate in offspring in humans and animal models [154], which is a congenital craniofacial anomaly in humans. Constitutively active ACVR1 in CNCs can result in ectopic craniofacial cartilage [50], and this defect is similar to the phenotype found in fibrodysplasia ossificans progressiva (FOP) patients. Richieri-Costa-Pereira syndrome (RCPS) is an autosomal recessive condition mainly characterized by craniofacial and limb malformations. EIF4A3 mutation in CNCs causes multiple defects in mice, which looks like the phenotype in RCPS patients, suggesting a valuable mouse model to study RCPS disorders [155]. Age-related hearing loss is a progressive pathophysiological process. A disfunction of Kir4.1 in CNC was found to be an important contributing factor in the aged human cochlea [156].
Transcriptional factor ALX1 has been associated with frontonasal dysplasia (FND) pathogenic. ALX1(L165F/L165F) mutants in CNC lead to more sensitivity to apoptosis and migration [157]. Kabuki syndrome (KS) is a congenital craniofacial disorder. KMT2D in CNC knockout mice demonstrates hypoplasia with reductions in frontonasal bone [158]. Ankrd11 inactivation can cause a rare autosomal dominant congenital disorder. Ankrd11 deletion in CNC leads to reduced ossification in midfacial bones, suggesting transgenic Ankrd11 can serve as pre-clinical models in humans [147]. The phenotype of the Pierre Robin Sequence (PRS) consists of cleft palate, glossoptosis and micrognathia. Transferrin receptor (Tfrc) deletion in CNC demonstrated multiple disorders, which are highly resemble human PRS. Tfrc deletion dramatically suppressed TGF/BMP signaling in CNC-derived mandibular tissues [59]. Besides, the phenotypes of Bmp2 deletion [38] and Mycn ablation [159] in CNC were also similar to that of PRS in humans, suggesting different genes may involve in the regulation of PRS pathology.
Alagille syndrome included biliary, cardiac and craniofacial anomalies. Deleted Jagged1 in CNC leads to a similar phenotype of Alagille syndrome, such as reduced cellular proliferation and aberrant craniofacial growth [104]. Anterior segment dysgenesis (ASD) encompasses a group of developmental disorders and 50% of patients develop glaucoma. The phenotype of AP-2β deletion in CNC resulted in post-natal ocular defects typified by opacity, suggesting that AP-2β in CNC knockout mice can serve as a new and exciting model to study the pathology of ASD and glaucoma in human [122] (Table 1).
CNC translational potential in regenerative medicine
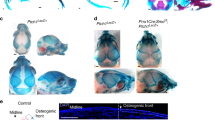
CNC-derived MSC/progenitor cells are a promising source for tissue regeneration, especially due to CNC’s distinct cell-autonomous and paracrine properties [160]. CNC-derived tissues exhibited superior properties for optimal translation in regenerative medicine [161], such as CNC-derived chondrocyte exhibits particular therapy in cartilage repair [162]. Scaffolds containing CNC-derived stem cell demonstrated superior bone formation in mouse calvarial bone injury model [163]. Using a biodegradable material to deliver suture stem cell is sufficient to regenerate normal cranial suture to restore skull deformity [164]. CNCs from the differentiation of induced pluripotent stem (iPS) cells represent alternative sources [165] for translational potential in the clinic, optimizing the crucial parameters of CNC differentiation will be valuable in tissue homeostasis and endogenous regeneration [166] (Fig. 4).
The strategies used for CNC translational potential in medicine. Differences in Gene regulatory network lead to distinct osteogenic potential and bone regeneration in frontal and parietal bones. CNC transplantation from iPS, or CNC transplantation with scaffold, or suitable molecular target delivery with scaffold are working strategies to enhance endogenous bone regeneration in cranial bones injury model (indicated in a dotted circle in red)
Suitable targets from scientific discoveries are capable to enhance CNC’s translational potential. JAG1 can induce CNC-derived osteoblast commitment during craniofacial intramembranous ossification. Exogenous JAG1 delivery using synthetic hydrogels containing CNCs into critical-sized calvarial defects can promote robust bone regeneration in mice, demonstrating exogenous JAG1 delivery is a potential bone-regenerative pathway [106]. Fgf2, Fgf9 and Fgf18 treated parietal bone exhibits superior bone regeneration both in juvenile and adult mice [28]. Active canonical Wnt signaling contributed to the superior intrinsic osteogenic potential and tissue regeneration in CNC-derived frontal bone [28]. Enhanced activation of Wnt signaling is capable to improve the capacity of bone regeneration in mesoderm-derived parietal bone similar to that in frontal bone [29] (Fig. 4).
Summary
Cranial neural crest patterning is a vital developmental process to coordinate cell proliferation, migration and differentiation at cellular and molecular levels. Most cranial bones are derived from CNCs, and CNC-derived cranial bones are endowed superior osteogenic potential and regeneration in vivo and in vitro. The underlying differences from conserved signaling pathways have been demonstrated functional in shaping the morphology of skull development in mice and human. Through the genetic mouse models, we bring forward integrated signaling pathways as gene regulatory network to better understand CNC spatial orientation and developmental stability and plasticity during cranial bone development.
What is more, the dysregulation of CNC patterning is highly relevant to birth defects in the clinic. However, less information or models are available to guide the basic research into clinical practice, which will be a severe hindrance to understand the molecular pathology of craniofacial birth defects or diseases in the clinic. Advancing the scientific discoveries from transgenic mouse models will be essential to observe potential links to the phenotypes found in clinical diseases, which in turn will be very important to understand the causes of the diseases or birth defects, and in the long term, it will be possible to timely diagnose, prevent or alleviate disease occurrence in the clinic. Taken together, the gene regulatory network underlying CNC patterning in mice models provides novel insight into the interplays from different signaling pathways, transcriptional factors, downstream effectors and epigenetic factors, which will be beneficial to the identification of new targets to be considered for the translational potential to treat human diseases or disorders in clinic.
Availability of data and materials
Not applicable.
Abbreviations
- CNC:
-
Cranial neural crest cells
- BMP:
-
Bone morphogenetic proteins
- FGF:
-
Fibroblast growth factors
- Fb:
-
Frontal bone
- Pb:
-
Parietal bone
- PDGF:
-
Platelet-derived growth factor
- TGF-β:
-
Transforming growth factor-β
- Hh:
-
Hedgehog signaling
References
Debbache J, Parfejevs V, Sommer L (2018) Cre-driver lines used for genetic fate mapping of neural crest cells in the mouse: an overview. Genesis 56:e23105. https://doi.org/10.1002/dvg.23105
Echelard Y, Vassileva G, McMahon AP (1994) Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development 120:2213–2224
McMahon AP, Joyner AL, Bradley A, McMahon JA (1992) The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 69:581–595
Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM (2002) Tissue origins and interactions in the mammalian skull vault. Dev Biol 241:106–116. https://doi.org/10.1006/dbio.2001.0487
Jarad G, Miner JH (2009) The Pax3-Cre transgene exhibits a rostrocaudal gradient of expression in the skeletal muscle lineage. Genesis 47:1–6. https://doi.org/10.1002/dvg.20447
Gershon TR et al (2009) Enteric neural crest differentiation in ganglioneuromas implicates Hedgehog signaling in peripheral neuroblastic tumor pathogenesis. PLoS ONE 4:e7491. https://doi.org/10.1371/journal.pone.0007491
Wang Q, Kumar S, Mitsios N, Slevin M, Kumar P (2007) Investigation of downstream target genes of PAX3c, PAX3e and PAX3g isoforms in melanocytes by microarray analysis. Int J Cancer 120:1223–1231. https://doi.org/10.1002/ijc.22316
Lee RT et al (2013) Cell delamination in the mesencephalic neural fold and its implication for the origin of ectomesenchyme. Development 140:4890–4902. https://doi.org/10.1242/dev.094680
Simon C, Lickert H, Gotz M, Dimou L (2012) Sox10-iCreERT2: a mouse line to inducibly trace the neural crest and oligodendrocyte lineage. Genesis 50:506–515. https://doi.org/10.1002/dvg.22003
He F, Soriano P (2015) Sox10ER(T2) CreER(T2) mice enable tracing of distinct neural crest cell populations. Dev Dyn 244:1394–1403. https://doi.org/10.1002/dvdy.24320
Aoto K et al (2015) Mef2c-F10N enhancer driven beta-galactosidase (LacZ) and Cre recombinase mice facilitate analyses of gene function and lineage fate in neural crest cells. Dev Biol 402:3–16. https://doi.org/10.1016/j.ydbio.2015.02.022
Zhang SM, Marsh R, Ratner N, Brackenbury R (1995) Myelin glycoprotein P0 is expressed at early stages of chicken and rat embryogenesis. J Neurosci Res 40:241–250. https://doi.org/10.1002/jnr.490400213
Yamauchi Y et al (1999) A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev Biol 212:191–203. https://doi.org/10.1006/dbio.1999.9323
Wang SK, Komatsu Y, Mishina Y (2011) Potential contribution of neural crest cells to dental enamel formation. Biochem Biophys Res Commun 415:114–119. https://doi.org/10.1016/j.bbrc.2011.10.026
Lewis AE, Vasudevan HN, O’Neill AK, Soriano P, Bush JO (2013) The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev Biol 379:229–234. https://doi.org/10.1016/j.ydbio.2013.04.026
Chen G et al (2017) Specific and spatial labeling of P0-Cre versus Wnt1-Cre in cranial neural crest in early mouse embryos. Genesis. https://doi.org/10.1002/dvg.23034
Kuratani S (2005) Cephalic neural crest cells and the evolution of craniofacial structures in vertebrates: morphological and embryological significance of the premandibular-mandibular boundary. Zoology 108:13–25. https://doi.org/10.1016/j.zool.2004.12.001
Wu T, Chen G, Tian F, Liu HX (2017) Contribution of cranial neural crest cells to mouse skull development. Int J Dev Biol 61:495–503. https://doi.org/10.1387/ijdb.170051gc
Koyabu D, Maier W, Sanchez-Villagra MR (2012) Paleontological and developmental evidence resolve the homology and dual embryonic origin of a mammalian skull bone, the interparietal. Proc Natl Acad Sci U S A 109:14075–14080. https://doi.org/10.1073/pnas.1208693109
Chen G et al (2020) BMP signaling in the development and regeneration of cranium bones and maintenance of calvarial stem cells. Front Cell Dev Biol 8:135. https://doi.org/10.3389/fcell.2020.00135
Depew MJ, Bertocchini F (1976) Avenues for investigating the neural crest and its derivatives in non-model (unconventional) vertebrates: a craniofacial skeleton perspective. Methods Mol Biol 207–221:2019. https://doi.org/10.1007/978-1-4939-9412-0_16
Xu Y, Malladi P, Zhou D, Longaker MT (2007) Molecular and cellular characterization of mouse calvarial osteoblasts derived from neural crest and paraxial mesoderm. Plast Reconstr Surg 120:1783–1795. https://doi.org/10.1097/01.prs.0000279491.48283.51
Doro D, Liu A, Grigoriadis AE, Liu KJ (2019) The osteogenic potential of the neural crest lineage may contribute to craniosynostosis. Mol Syndromol 10:48–57. https://doi.org/10.1159/000493106
Senarath-Yapa K, Li S, Meyer NP, Longaker MT, Quarto N (2013) Integration of multiple signaling pathways determines differences in the osteogenic potential and tissue regeneration of neural crest-derived and mesoderm-derived calvarial bones. Int J Mol Sci 14:5978–5997. https://doi.org/10.3390/ijms14035978
Quarto N, Behr B, Li S, Longaker MT (2009) Differential FGF ligands and FGF receptors expression pattern in frontal and parietal calvarial bones. Cells Tissues Organs 190:158–169. https://doi.org/10.1159/000202789
Li S, Quarto N, Longaker MT (2010) Activation of FGF signaling mediates proliferative and osteogenic differences between neural crest derived frontal and mesoderm parietal derived bone. PLoS ONE 5:e14033. https://doi.org/10.1371/journal.pone.0014033
Kidwai F et al (2020) Lineage-specific differentiation of osteogenic progenitors from pluripotent stem cells reveals the FGF1-RUNX2 association in neural crest-derived osteoprogenitors. Stem Cells 38:1107–1123. https://doi.org/10.1002/stem.3206
Quarto N et al (2010) Origin matters: differences in embryonic tissue origin and Wnt signaling determine the osteogenic potential and healing capacity of frontal and parietal calvarial bones. J Bone Miner Res 25:1680–1694. https://doi.org/10.1359/jbmr.091116
Li S et al (2015) Enhanced activation of canonical wnt signaling confers mesoderm-derived parietal bone with similar osteogenic and skeletal healing capacity to neural crest-derived frontal bone. PLoS ONE 10:e0138059. https://doi.org/10.1371/journal.pone.0138059
Li S, Meyer NP, Quarto N, Longaker MT (2013) Integration of multiple signaling regulates through apoptosis the differential osteogenic potential of neural crest-derived and mesoderm-derived Osteoblasts. PLoS ONE 8:e58610. https://doi.org/10.1371/journal.pone.0058610
Menon S, Huber J, Duldulao C, Longaker MT, Quarto N (2021) An evolutionary conserved signaling network between mouse and human underlies the differential osteoskeletal potential of frontal and parietal calvarial bones. Front Physiol 12:747091. https://doi.org/10.3389/fphys.2021.747091
Homayounfar N et al (2015) Transcriptional analysis of human cranial compartments with different embryonic origins. Arch Oral Biol 60:1450–1460. https://doi.org/10.1016/j.archoralbio.2015.06.008
Hu B et al (2017) Physiological signatures of dual embryonic origins in mouse skull vault. Cell Physiol Biochem 43:2525–2534. https://doi.org/10.1159/000484496
Lumb R, Buckberry S, Secker G, Lawrence D, Schwarz Q (2017) Transcriptome profiling reveals expression signatures of cranial neural crest cells arising from different axial levels. BMC Dev Biol 17:5. https://doi.org/10.1186/s12861-017-0147-z
Roth DM, Bayona F, Baddam P, Graf D (2021) Craniofacial development: neural crest in molecular embryology. Head Neck Pathol 15:1–15. https://doi.org/10.1007/s12105-021-01301-z
Meulemans D, Bronner-Fraser M (2004) Gene-regulatory interactions in neural crest evolution and development. Dev Cell 7:291–299. https://doi.org/10.1016/j.devcel.2004.08.007
Kanzler B, Foreman RK, Labosky PA, Mallo M (2000) BMP signaling is essential for development of skeletogenic and neurogenic cranial neural crest. Development 127:1095–1104
Chen Y, Wang Z, Chen Y, Zhang Y (2019) Conditional deletion of Bmp2 in cranial neural crest cells recapitulates Pierre Robin sequence in mice. Cell Tissue Res 376:199–210. https://doi.org/10.1007/s00441-018-2944-5
Bonilla-Claudio M et al (2012) Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development 139:709–719. https://doi.org/10.1242/dev.073197
Solloway MJ, Robertson EJ (1999) Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development 126:1753–1768
Mimura S et al (2016) Bone morphogenetic protein 4 promotes craniofacial neural crest induction from human pluripotent stem cells. Int J Dev Biol 60:21–28. https://doi.org/10.1387/ijdb.160040mk
Anderson RM, Stottmann RW, Choi M, Klingensmith J (2006) Endogenous bone morphogenetic protein antagonists regulate mammalian neural crest generation and survival. Dev Dyn 235:2507–2520. https://doi.org/10.1002/dvdy.20891
Pegge J, Tatsinkam AJ, Rider CC, Bell E (2020) Heparan sulfate proteoglycans regulate BMP signalling during neural crest induction. Dev Biol 460:108–114. https://doi.org/10.1016/j.ydbio.2019.12.015
Hackland JOS et al (2017) Top-down inhibition of BMP signaling enables robust induction of hpscs into neural crest in fully defined, Xeno-free Conditions. Stem Cell Reports 9:1043–1052. https://doi.org/10.1016/j.stemcr.2017.08.008
Pan H et al (2017) BmpR1A is a major type 1 BMP receptor for BMP-Smad signaling during skull development. Dev Biol 429:260–270. https://doi.org/10.1016/j.ydbio.2017.06.020
Komatsu Y et al (2013) Augmentation of Smad-dependent BMP signaling in neural crest cells causes craniosynostosis in mice. J Bone Miner Res 28:1422–1433. https://doi.org/10.1002/jbmr.1857
Kramer K et al (2018) Rapamycin rescues BMP mediated midline craniosynostosis phenotype through reduction of mTOR signaling in a mouse model. Genesis 56:e23220. https://doi.org/10.1002/dvg.23220
Zhang X et al (2019) Distinctive role of ACVR1 in dentin formation: requirement for dentin thickness in molars and prevention of osteodentin formation in incisors of mice. J Mol Histol 50:43–61. https://doi.org/10.1007/s10735-018-9806-z
Yang J et al (2021) Generation of a new mouse line with conditionally activated signaling through the BMP receptor, ACVR1: a tool to characterize pleiotropic roles of BMP functions. Genesis 59:e23419. https://doi.org/10.1002/dvg.23419
Yang J et al (2021) Augmented BMP signaling commits cranial neural crest cells to a chondrogenic fate by suppressing autophagic beta-catenin degradation. Sci Signal. https://doi.org/10.1126/scisignal.aaz9368
Maruyama T et al (2021) BMPR1A maintains skeletal stem cell properties in craniofacial development and craniosynostosis. Sci Transl Med. https://doi.org/10.1126/scitranslmed.abb4416
Ho TV et al (2015) Integration of comprehensive 3D microCT and signaling analysis reveals differential regulatory mechanisms of craniofacial bone development. Dev Biol 400:180–190. https://doi.org/10.1016/j.ydbio.2015.02.010
Ito Y et al (2003) Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development 130:5269–5280. https://doi.org/10.1242/dev.00708
Zhao H, Oka K, Bringas P, Kaartinen V, Chai Y (2008) TGF-beta type I receptor Alk5 regulates tooth initiation and mandible patterning in a type II receptor-independent manner. Dev Biol 320:19–29. https://doi.org/10.1016/j.ydbio.2008.03.045
Iwata J et al (2010) Transforming growth factor-beta regulates basal transcriptional regulatory machinery to control cell proliferation and differentiation in cranial neural crest-derived osteoprogenitor cells. J Biol Chem 285:4975–4982. https://doi.org/10.1074/jbc.M109.035105
Ko SO et al (2007) Smad4 is required to regulate the fate of cranial neural crest cells. Dev Biol 312:435–447. https://doi.org/10.1016/j.ydbio.2007.09.050
Hosokawa R et al (2007) TGF-beta mediated Msx2 expression controls occipital somites-derived caudal region of skull development. Dev Biol 310:140–153. https://doi.org/10.1016/j.ydbio.2007.07.038
Iwata J et al (2012) Modulation of noncanonical TGF-beta signaling prevents cleft palate in Tgfbr2 mutant mice. J Clin Invest 122:873–885. https://doi.org/10.1172/JCI61498
Lei R et al (2016) Transferrin receptor facilitates TGF-beta and BMP signaling activation to control craniofacial morphogenesis. Cell Death Dis 7:e2282. https://doi.org/10.1038/cddis.2016.170
Yumoto K et al (2013) TGF-beta-activated kinase 1 (Tak1) mediates agonist-induced Smad activation and linker region phosphorylation in embryonic craniofacial neural crest-derived cells. J Biol Chem 288:13467–13480. https://doi.org/10.1074/jbc.M112.431775
Cibi DM et al (2019) Neural crest-specific deletion of Rbfox2 in mice leads to craniofacial abnormalities including cleft palate. Elife. https://doi.org/10.7554/eLife.45418
Nusse R, Clevers H (2017) Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 169:985–999. https://doi.org/10.1016/j.cell.2017.05.016
Ji Y, Hao H, Reynolds K, McMahon M, Zhou CJ (2019) Wnt signaling in neural crest ontogenesis and oncogenesis. Cells. https://doi.org/10.3390/cells8101173
Hari L et al (2012) Temporal control of neural crest lineage generation by Wnt/beta-catenin signaling. Development 139:2107–2117. https://doi.org/10.1242/dev.073064
DiNuoscio G, Atit RP (2019) Wnt/beta-catenin signaling in the mouse embryonic cranial mesenchyme is required to sustain the emerging differentiated meningeal layers. Genesis 57:e23279. https://doi.org/10.1002/dvg.23279
Brault V et al (2001) Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128:1253–1264
Masek J, Machon O, Korinek V, Taketo MM, Kozmik Z (2016) Tcf7l1 protects the anterior neural fold from adopting the neural crest fate. Development 143:2206–2216. https://doi.org/10.1242/dev.132357
Carmona-Fontaine C, Acuna G, Ellwanger K, Niehrs C, Mayor R (2007) Neural crests are actively precluded from the anterior neural fold by a novel inhibitory mechanism dependent on Dickkopf1 secreted by the prechordal mesoderm. Dev Biol 309:208–221. https://doi.org/10.1016/j.ydbio.2007.07.006
Bikkavilli RK et al (2012) Dishevelled3 is a novel arginine methyl transferase substrate. Sci Rep 2:805. https://doi.org/10.1038/srep00805
Jia S et al (2016) Bmp4-Msx1 signaling and Osr2 control tooth organogenesis through antagonistic regulation of secreted Wnt antagonists. Dev Biol 420:110–119. https://doi.org/10.1016/j.ydbio.2016.10.001
Zhao X, Le TP, Erhardt S, Findley TO, Wang J (2021) Hippo-yap pathway orchestrates neural crest ontogenesis. Front Cell Dev Biol 9:706623. https://doi.org/10.3389/fcell.2021.706623
Akhmametyeva EM et al (2006) Regulation of the neurofibromatosis 2 gene promoter expression during embryonic development. Dev Dyn 235:2771–2785. https://doi.org/10.1002/dvdy.20883
Wang J et al (2016) Yap and Taz play a crucial role in neural crest-derived craniofacial development. Development 143:504–515. https://doi.org/10.1242/dev.126920
Bi-Lin KW et al (2021) Critical role of the BAF chromatin remodeling complex during murine neural crest development. PLoS Genet 17:e1009446. https://doi.org/10.1371/journal.pgen.1009446
Sun Z et al (2018) FoxO6 regulates Hippo signaling and growth of the craniofacial complex. PLoS Genet 14:e1007675. https://doi.org/10.1371/journal.pgen.1007675
Sarkar S, Petiot A, Copp A, Ferretti P, Thorogood P (2001) FGF2 promotes skeletogenic differentiation of cranial neural crest cells. Development 128:2143–2152
Xu J et al (2018) FGF8 signaling alters the osteogenic cell fate in the hard palate. J Dent Res 97:589–596. https://doi.org/10.1177/0022034517750141
Kawai M et al (2019) Fgfr1 conditional-knockout in neural crest cells induces heterotopic chondrogenesis and osteogenesis in mouse frontal bones. Med Mol Morphol 52:156–163. https://doi.org/10.1007/s00795-018-0213-z
Wang C et al (2013) Type 1 fibroblast growth factor receptor in cranial neural crest cell-derived mesenchyme is required for palatogenesis. J Biol Chem 288:22174–22183. https://doi.org/10.1074/jbc.M113.463620
Heuze Y et al (2014) Morphological comparison of the craniofacial phenotypes of mouse models expressing the Apert FGFR2 S252W mutation in neural crest- or mesoderm-derived tissues. Bone 63:101–109. https://doi.org/10.1016/j.bone.2014.03.003
Holmes G, Basilico C (2012) Mesodermal expression of Fgfr2S252W is necessary and sufficient to induce craniosynostosis in a mouse model of Apert syndrome. Dev Biol 368:283–293. https://doi.org/10.1016/j.ydbio.2012.05.026
Kyrylkova K, Iwaniec UT, Philbrick KA, Leid M (2016) BCL11B regulates sutural patency in the mouse craniofacial skeleton. Dev Biol 415:251–260. https://doi.org/10.1016/j.ydbio.2015.10.010
Deckelbaum RA et al (2012) Regulation of cranial morphogenesis and cell fate at the neural crest-mesoderm boundary by engrailed 1. Development 139:1346–1358. https://doi.org/10.1242/dev.076729
Li Y et al (2017) Robo signaling regulates the production of cranial neural crest cells. Exp Cell Res 361:73–84. https://doi.org/10.1016/j.yexcr.2017.10.002
Cheng X et al (2017) From the cover: usage of dexamethasone increases the risk of cranial neural crest dysplasia in the chick embryo. Toxicol Sci 158:36–47. https://doi.org/10.1093/toxsci/kfx073
Ray AT et al (2020) FGF signaling regulates development by processes beyond canonical pathways. Genes Dev 34:1735–1752. https://doi.org/10.1101/gad.342956.120
Yang J, Andre P, Ye L, Yang YZ (2015) The Hedgehog signalling pathway in bone formation. Int J Oral Sci 7:73–79. https://doi.org/10.1038/ijos.2015.14
Amano K, Okuzaki D, Aikawa T, Kogo M (2020) Indian hedgehog in craniofacial neural crest cells links to skeletal malocclusion by regulating associated cartilage formation and gene expression. FASEB J 34:6791–6807. https://doi.org/10.1096/fj.201903269R
Sun MR et al (2020) Sonic hedgehog signaling in cranial neural crest cells regulates microvascular morphogenesis in facial development. Front Cell Dev Biol 8:590539. https://doi.org/10.3389/fcell.2020.590539
Berio A, Piazzi A (2007) Serious cerebral malformations (corpus callosum aplasia, prosencephalic cyst), internal carotid canal and facial malformations due to neural crest abnormalities, associated with choleosteatoma. Minerva Pediatr 59:403–408
Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP (2004) Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev 18:937–951. https://doi.org/10.1101/gad.1190304
Swartz ME, Nguyen V, McCarthy NQ, Eberhart JK (2012) Hh signaling regulates patterning and morphogenesis of the pharyngeal arch-derived skeleton. Dev Biol 369:65–75. https://doi.org/10.1016/j.ydbio.2012.05.032
Li J et al (2017) Suppressor of Fused restraint of Hedgehog activity level is critical for osteogenic proliferation and differentiation during calvarial bone development. J Biol Chem 292:15814–15825. https://doi.org/10.1074/jbc.M117.777532
Noda K, Kitami M, Kitami K, Kaku M, Komatsu Y (2016) Canonical and noncanonical intraflagellar transport regulates craniofacial skeletal development. Proc Natl Acad Sci U S A 113:E2589-2597. https://doi.org/10.1073/pnas.1519458113
Tian H et al (2017) Intraflagellar transport 88 (IFT88) is crucial for craniofacial development in mice and is a candidate gene for human cleft lip and palate. Hum Mol Genet. https://doi.org/10.1093/hmg/ddx002
Kolpakova-Hart E, Jinnin M, Hou B, Fukai N, Olsen BR (2007) Kinesin-2 controls development and patterning of the vertebrate skeleton by Hedgehog- and Gli3-dependent mechanisms. Dev Biol 309:273–284. https://doi.org/10.1016/j.ydbio.2007.07.018
Khonsari RH et al (2013) Multiple postnatal craniofacial anomalies are characterized by conditional loss of polycystic kidney disease 2 (Pkd2). Hum Mol Genet 22:1873–1885. https://doi.org/10.1093/hmg/ddt041
Badri MK et al (2016) Expression of Evc2 in craniofacial tissues and craniofacial bone defects in Evc2 knockout mouse. Arch Oral Biol 68:142–152. https://doi.org/10.1016/j.archoralbio.2016.05.002
Zhang H et al (2015) Generation of Evc2/Limbin global and conditional KO mice and its roles during mineralized tissue formation. Genesis. https://doi.org/10.1002/dvg.22879
Kulkarni AK et al (2018) A ciliary protein EVC2/LIMBIN plays a critical role in the skull base for mid-facial development. Front Physiol 9:1484. https://doi.org/10.3389/fphys.2018.01484
Gokhman D et al (2021) Human-chimpanzee fused cells reveal cis-regulatory divergence underlying skeletal evolution. Nat Genet 53:467–476. https://doi.org/10.1038/s41588-021-00804-3
Tabler JM, Rice CP, Liu KJ, Wallingford JB (2016) A novel ciliopathic skull defect arising from excess neural crest. Dev Biol 417:4–10. https://doi.org/10.1016/j.ydbio.2016.07.001
Mead TJ, Yutzey KE (2012) Notch pathway regulation of neural crest cell development in vivo. Dev Dyn 241:376–389. https://doi.org/10.1002/dvdy.23717
Humphreys R et al (2012) Cranial neural crest ablation of Jagged1 recapitulates the craniofacial phenotype of Alagille syndrome patients. Hum Mol Genet 21:1374–1383. https://doi.org/10.1093/hmg/ddr575
Kamalakar A et al (2019) A non-canonical JAGGED1 signal to JAK2 mediates osteoblast commitment in cranial neural crest cells. Cell Signal 54:130–138. https://doi.org/10.1016/j.cellsig.2018.12.002
Kamalakar A et al (2021) JAGGED1 stimulates cranial neural crest cell osteoblast commitment pathways and bone regeneration independent of canonical NOTCH signaling. Bone 143:115657. https://doi.org/10.1016/j.bone.2020.115657
Fantauzzo KA, Soriano P (2016) PDGFRbeta regulates craniofacial development through homodimers and functional heterodimers with PDGFRalpha. Genes Dev 30:2443–2458. https://doi.org/10.1101/gad.288746.116
Richarte AM, Mead HB, Tallquist MD (2007) Cooperation between the PDGF receptors in cardiac neural crest cell migration. Dev Biol 306:785–796. https://doi.org/10.1016/j.ydbio.2007.04.023
Moenning A et al (2009) Sustained platelet-derived growth factor receptor alpha signaling in osteoblasts results in craniosynostosis by overactivating the phospholipase C-gamma pathway. Mol Cell Biol 29:881–891. https://doi.org/10.1128/MCB.00885-08
Soriano P (1997) The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development 124:2691–2700
Vasudevan HN, Soriano P (2014) SRF regulates craniofacial development through selective recruitment of MRTF cofactors by PDGF signaling. Dev Cell 31:332–344. https://doi.org/10.1016/j.devcel.2014.10.005
Corsinovi D, Giannetti K, Cericola A, Naef V, Ori M (2019) PDGF-B: the missing piece in the mosaic of PDGF family role in craniofacial development. Dev Dyn 248:603–612. https://doi.org/10.1002/dvdy.47
Mo J, Long R, Fantauzzo KA (2020) Pdgfra and Pdgfrb genetically interact in the murine neural crest cell lineage to regulate migration and proliferation. Front Physiol 11:588901. https://doi.org/10.3389/fphys.2020.588901
Betancur P, Bronner-Fraser M, Sauka-Spengler T (2010) Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc Natl Acad Sci U S A 107:3570–3575. https://doi.org/10.1073/pnas.0906596107
Jandzik D et al (2015) Evolution of the new vertebrate head by co-option of an ancient chordate skeletal tissue. Nature 518:534–537. https://doi.org/10.1038/nature14000
Mandalos N et al (2014) Sox2 acts as a rheostat of epithelial to mesenchymal transition during neural crest development. Front Physiol 5:345. https://doi.org/10.3389/fphys.2014.00345
Dash S, Bhatt S, Falcon KT, Sandell LL, Trainor PA (2021) Med23 regulates Sox9 expression during craniofacial development. J Dent Res 100:406–414. https://doi.org/10.1177/0022034520969109
Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ (1996) Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381:235–238. https://doi.org/10.1038/381235a0
Knight RD, Javidan Y, Zhang T, Nelson S, Schilling TF (2005) AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development 132:3127–3138. https://doi.org/10.1242/dev.01879
Enkhmandakh B, Bayarsaihan D (2015) Genome-wide chromatin mapping defines AP2alpha in the etiology of craniofacial disorders. Cleft Palate Craniofac J 52:135–142. https://doi.org/10.1597/13-151
Machon O, Masek J, Machonova O, Krauss S, Kozmik Z (2015) Meis2 is essential for cranial and cardiac neural crest development. BMC Dev Biol 15:40. https://doi.org/10.1186/s12861-015-0093-6
Martino VB et al (2016) Conditional deletion of AP-2beta in mouse cranial neural crest results in anterior segment dysgenesis and early-onset glaucoma. Dis Model Mech 9:849–861. https://doi.org/10.1242/dmm.025262
Mitchell JM et al (2021) The alx3 gene shapes the zebrafish neurocranium by regulating frontonasal neural crest cell differentiation timing. Development. https://doi.org/10.1242/dev.197483
Winograd J et al (1997) Perinatal lethality and multiple craniofacial malformations in MSX2 transgenic mice. Hum Mol Genet 6:369–379. https://doi.org/10.1093/hmg/6.3.369
Roybal PG et al (2010) Inactivation of Msx1 and Msx2 in neural crest reveals an unexpected role in suppressing heterotopic bone formation in the head. Dev Biol 343:28–39. https://doi.org/10.1016/j.ydbio.2010.04.007
Ishii M et al (2005) Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development 132:4937–4950. https://doi.org/10.1242/dev.02072
Bildsoe H et al (2009) Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Dev Biol 331:176–188. https://doi.org/10.1016/j.ydbio.2009.04.034
Bildsoe H et al (2016) Transcriptional targets of TWIST1 in the cranial mesoderm regulate cell-matrix interactions and mesenchyme maintenance. Dev Biol 418:189–203. https://doi.org/10.1016/j.ydbio.2016.08.016
Merrill AE et al (2006) Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum Mol Genet 15:1319–1328. https://doi.org/10.1093/hmg/ddl052
Ting MC et al (2009) EphA4 as an effector of Twist1 in the guidance of osteogenic precursor cells during calvarial bone growth and in craniosynostosis. Development 136:855–864. https://doi.org/10.1242/dev.028605
Quarto N et al (2018) Twist1-haploinsufficiency selectively enhances the osteoskeletal capacity of mesoderm-derived parietal bone through downregulation of Fgf23. Front Physiol 9:1426. https://doi.org/10.3389/fphys.2018.01426
McKeown SJ, Newgreen DF, Farlie PG (2005) Dlx2 over-expression regulates cell adhesion and mesenchymal condensation in ectomesenchyme. Dev Biol 281:22–37. https://doi.org/10.1016/j.ydbio.2005.02.004
Ruest LB et al (2003) dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev Biol 257:263–277. https://doi.org/10.1016/s0012-1606(03)00068-x
Chung IH, Han J, Iwata J, Chai Y (2010) Msx1 and Dlx5 function synergistically to regulate frontal bone development. Genesis 48:645–655. https://doi.org/10.1002/dvg.20671
Mishina Y, Snider TN (2014) Neural crest cell signaling pathways critical to cranial bone development and pathology. Exp Cell Res 325:138–147. https://doi.org/10.1016/j.yexcr.2014.01.019
Takarada T et al (2016) Genetic analysis of Runx2 function during intramembranous ossification. Development 143:211–218. https://doi.org/10.1242/dev.128793
Shirai Y et al (2019) Runx2 function in cells of neural crest origin during intramembranous ossification. Biochem Biophys Res Commun 509:1028–1033. https://doi.org/10.1016/j.bbrc.2019.01.059
Kim HJ, Rice DP, Kettunen PJ, Thesleff I (1998) FGF-, BMP- and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development 125:1241–1251
Li J et al (2011) SMAD4-mediated WNT signaling controls the fate of cranial neural crest cells during tooth morphogenesis. Development 138:1977–1989. https://doi.org/10.1242/dev.061341
Alkobtawi M, Pla P, Monsoro-Burq AH (2021) BMP signaling is enhanced intracellularly by FHL3 controlling WNT-dependent spatiotemporal emergence of the neural crest. Cell Rep 35:109289. https://doi.org/10.1016/j.celrep.2021.109289
Minoux M et al (2017) Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science. https://doi.org/10.1126/science.aal2913
Higashihori N et al (2017) Methyltransferase G9A regulates osteogenesis via twist gene repression. J Dent Res 96:1136–1144. https://doi.org/10.1177/0022034517716438
Ideno H et al (2020) G9a is involved in the regulation of cranial bone formation through activation of Runx2 function during development. Bone 137:115332. https://doi.org/10.1016/j.bone.2020.115332
Schwarz D et al (2014) Ezh2 is required for neural crest-derived cartilage and bone formation. Development 141:867–877. https://doi.org/10.1242/dev.094342
Sen R et al (2018) Kat2a and Kat2b acetyltransferase activity regulates craniofacial cartilage and bone differentiation in zebrafish and mice. J Dev Biol. https://doi.org/10.3390/jdb6040027
Pezoa SA, Artinger KB, Niswander LA (2020) GCN5 acetylation is required for craniofacial chondrocyte maturation. Dev Biol 464:24–34. https://doi.org/10.1016/j.ydbio.2020.05.006
Roth DM et al (2021) The chromatin regulator Ankrd11 controls palate and cranial bone development. Front Cell Dev Biol 9:645386. https://doi.org/10.3389/fcell.2021.645386
Haberland M, Mokalled MH, Montgomery RL, Olson EN (2009) Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev 23:1625–1630. https://doi.org/10.1101/gad.1809209
Singh N et al (2013) Murine craniofacial development requires Hdac3-mediated repression of Msx gene expression. Dev Biol 377:333–344. https://doi.org/10.1016/j.ydbio.2013.03.008
Shao R et al (2016) Cdh1 regulates craniofacial development via APC-dependent ubiquitination and activation of Goosecoid. Cell Res 26:699–712. https://doi.org/10.1038/cr.2016.51
Wiszniak S, Harvey N, Schwarz Q (2016) Cell autonomous roles of Nedd4 in craniofacial bone formation. Dev Biol 410:98–107. https://doi.org/10.1016/j.ydbio.2015.12.001
Trainor PA (2010) Craniofacial birth defects: the role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A 152A:2984–2994. https://doi.org/10.1002/ajmg.a.33454
van Gijn DR, Tucker AS, Cobourne MT (2013) Craniofacial development: current concepts in the molecular basis of Treacher Collins syndrome. Br J Oral Maxillofac Surg 51:384–388. https://doi.org/10.1016/j.bjoms.2012.09.008
Wang Q et al (2019) Perturbed development of cranial neural crest cells in association with reduced sonic hedgehog signaling underlies the pathogenesis of retinoic-acid-induced cleft palate. Dis Model Mech. https://doi.org/10.1242/dmm.040279
Miller EE et al (2017) EIF4A3 deficient human iPSCs and mouse models demonstrate neural crest defects that underlie Richieri-Costa-Pereira syndrome. Hum Mol Genet 26:2177–2191. https://doi.org/10.1093/hmg/ddx078
Liu T et al (2019) Age-dependent alterations of Kir4.1 expression in neural crest-derived cells of the mouse and human cochlea. Neurobiol Aging 80:210–222. https://doi.org/10.1016/j.neurobiolaging.2019.04.009
Pini J et al (2020) ALX1-related frontonasal dysplasia results from defective neural crest cell development and migration. EMBO Mol Med 12:e12013. https://doi.org/10.15252/emmm.202012013
Shpargel KB, Mangini CL, Xie G, Ge K, Magnuson T (2020) The KMT2D Kabuki syndrome histone methylase controls neural crest cell differentiation and facial morphology. Development. https://doi.org/10.1242/dev.187997
Yang R et al (2021) Mycn deficiency underlies the development of orofacial clefts in mice and humans. Hum Mol Genet. https://doi.org/10.1093/hmg/ddab288
Zhang M et al (2020) Investigate the odontogenic differentiation and dentin-pulp tissue regeneration potential of neural crest cells. Front Bioeng Biotechnol 8:475. https://doi.org/10.3389/fbioe.2020.00475
Kim HJ et al (2021) Nasal turbinate mesenchymal stromal cells preserve characteristics of their neural crest origin and exert distinct paracrine activity. J Clin Med. https://doi.org/10.3390/jcm10081792
Taihi I, Nassif A, Isaac J, Fournier BP, Ferre F (2019) Head to knee: cranial neural crest-derived cells as promising candidates for human cartilage repair. Stem Cells Int 2019:9310318. https://doi.org/10.1155/2019/9310318
Yoshida H et al (2021) Neural crest-derived cells in nasal conchae of adult mice contribute to bone regeneration. Biochem Biophys Res Commun 554:173–178. https://doi.org/10.1016/j.bbrc.2021.03.079
Yu M et al (2021) Cranial suture regeneration mitigates skull and neurocognitive defects in craniosynostosis. Cell 184:243-256.e18. https://doi.org/10.1016/j.cell.2020.11.037
Kawano E et al (2017) Induction of neural crest cells from human dental pulp-derived induced pluripotent stem cells. Biomed Res 38:135–147. https://doi.org/10.2220/biomedres.38.135
Hoving AL et al (2021) Between fate choice and self-renewal-heterogeneity of adult neural crest-derived stem cells. Front Cell Dev Biol 9:662754. https://doi.org/10.3389/fcell.2021.662754
Author information
Authors and Affiliations
Contributions
JL, YH, QW, SC, CZ, DW, ZL, XZ: data analysis, literature formation. GC, MW: supervision and grant holder.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All the authors agree the current state of the manuscript to be submitted to the journal.
Funding
This work was supported by grants by the Zhejiang Qianjiang Talent Program (21040040-E), a startup grant from Zhejiang SCI-TECH University (18042290-Y; 2021Q031), Department of Sci-Tech of Zhejiang Province (LGF19H140002), National Natural Science Foundation of China (81400489) and Jiaxing Science Technology Foundation (2020AY10001).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liao, J., Huang, Y., Wang, Q. et al. Gene regulatory network from cranial neural crest cells to osteoblast differentiation and calvarial bone development. Cell. Mol. Life Sci. 79, 158 (2022). https://doi.org/10.1007/s00018-022-04208-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04208-2