Abstract
The ability of cells to repair DNA double-strand breaks (DSBs) is important for maintaining genome stability and eliminating oncogenic DNA lesions. Two distinct and complementary pathways, non-homologous end joining (NHEJ) and homologous recombination (HR), are employed by mammalian cells to repair DNA DSBs. Each pathway is tightly controlled in response to increased DSBs. The Ku heterodimer has been shown to play a regulatory role in NHEJ repair. Ku80 ubiquitination contributes to the selection of a DSB repair pathway by causing the removal of Ku heterodimers from DSB sites. However, whether Ku80 deubiquitination also plays a role in regulating DSB repair is unknown. To address this question, we performed a comprehensive study of the deubiquitinase specific for Ku80, and our study showed that the deubiquitinase OTUD5 serves as an important regulator of NHEJ repair by increasing the stability of Ku80. Further studies revealed that OTUD5 depletion impaired NHEJ repair, and hence reduced overall DSB repair. Furthermore, OTUD5-depleted cells displayed excess end resection; as a result, HR repair was facilitated by OTUD5 depletion during the S/G2 phase. In summary, our study demonstrates that OTUD5 is a specific deubiquitinase for Ku80 and establishes OTUD5 as an important and positive regulator of NHEJ repair.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DNA double-strand breaks (DSBs) are one of the most deleterious types of DNA damage resulting from exogenous agents, such as ionizing radiation (IR) and certain chemotherapeutic drugs, as well as from endogenous mechanical stress on chromosomes, such as chromosomal rearrangements during V(D)J recombination and immunoglobulin class-switch recombination [1]. DSB repair deficient cells often exhibit genomic instability, which is one of the key events that leads up to cancer, because the mammalian genome is at constant risk from genotoxic factors and accumulation of various mutations [2, 3]. Due to intrinsic cellular activities, two distinct and complementary pathways, non-homologous end joining (NHEJ) and homologous recombination (HR), are responsible for DNA DSB repair in mammalian cells [4]. NHEJ leaves “information scars” at most repair sites because resection of a few nucleotides and a random addition are necessary to bring the two DNA ends into a ligatable configuration [3]. In contrast, HR ensures error-free repair of a broken chromatid aided by the intact sister chromatid and is initiated by 5′–3′ nucleolytic degradation that generates single-stranded DNA (ssDNA) [5, 6].
The Ku heterodimer (Ku70/Ku80) forms a ring-shaped toroidal structure. Its abundance and high affinity for DNA ends are fundamental factors regulating the repair of DNA DSBs. During NHEJ, the Ku heterodimer is recruited to maintain the integrity of DSB end sites for blunt end ligation, independent sequence homology and the recruitment of the DNA-dependent protein kinase catalytic subunit (DNA–PKcs) [7, 8]. In contrast, during HR, the removal of the Ku heterodimer from DNA ends promotes the generation of ssDNA overhangs by the CtIP/Mre11-Rad50-Nbs1 (MRN) complex, which is critical for initiating HR [9].
Studies have shown that ubiquitination of Ku80 dislodges the Ku heterodimer from DNA by causing removal of Ku80 [10, 11]. In addition, ubiquitination-mediated degradation of Ku80 has been shown to be RNF8 dependent [12, 13]. RNF138 ubiquitinates Ku80 to displace Ku from DSB sites in the S and G2 phases of the cell cycle [12]. Furthermore, RNF126 has also been reported as a ubiquitin E3 ligase for Ku80 [14]. Recently, it was reported that ubiquitin carboxyl-terminal hydrolase L3 (UCHL3) deubiquitinated Ku80, and depleting UCHL3 resulted in reduced formation of Ku80 foci, moderately sensitizing cells to IR and decreasing NHEJ efficiency [15]. Extensive evidence has shown that Ku80 ubiquitination plays an important role in DSB repair through regulating the accumulation of Ku70/Ku80 at the DSB sites.
Although much of the study of Ku ubiquitination has been focused on the ubiquitinase of Ku80, little is known about the deubiquitinase of Ku80. The human genome encodes approximately 100 putative DUBs, which are divided into six DUB subfamilies based on their structures, including the ubiquitin-specific protease (USP) subfamily, ubiquitin C-terminal hydrolase (UCH) subfamily, Machado–Joseph disease (MJD) protein domain protease subfamily, ovarian tumor (OTU) protease subfamily, JAB1/MPN/Mov34 metalloenzyme (JAMM) motif protease subfamily and motif interacting with ub-containing novel DUB (MINDY) subfamily [16, 17]. In this study, we identified OTUD5 as a specific deubiquitinase for Ku80 by screening a deubiquitinase library, and we also demonstrated that OTUD5 served as an essential component in the regulation of DSB repair pathway selection through maintaining the stability of Ku80.
Results
Ku80 is ubiquitinated in response to DNA damage
To identify significant ubiquitinated factors in response to DNA damage, we designed a novel approach to distinguish DNA damage-dependent ubiquitinated lysine using heavy lysine (lys6) labeled H1299 cells, and SILAC-mass spectrometry (Fig. 1a). The results of the mass spectrometry experiments identified five lysine residues as ubiquitination sites in Ku80, and K481 experienced the largest increase in ubiquitination in response to IR (10 Gy) (Fig. 1b, Supplementary Table 1). To confirm that Ku80 is indeed ubiquitinated after DNA damage, we performed an immunoprecipitation (IP) assay with an antibody specific for ubiquitin without or with IR, and ubiquitinated Ku80 levels were significantly higher with IR than without IR (Fig. 1c). Additionally, co-expression of Ku80 with the ubiquitin E3 ligase RNF138 or RNF8 in 293T cells also increased the levels of ubiquitinated Ku80, consistent with previous studies (Supplementary Fig. 1A, B) [12, 13, 18].
Ku80 is ubiquitinated in response to DNA damage. a Strategy for identifying ubiquitinated factors in response to IR (10 Gy) by SILAC-MS/MS in H1299 cells. b Ku80 is ubiquitinated at lysine 481. c The levels of ubiquitinated Ku80 increased in response to DNA damage. H1299 cells were untreated or treated with ionizing radiation (10 Gy) and collected after 1 h. Ubiquitinated Ku80 was purified by immunoprecipitation using anti-ubiquitin antibodies and was analyzed by western blotting using an anti-Ku80 antibody
OTUD5 deubiquitinates Ku80 in vitro and in cells
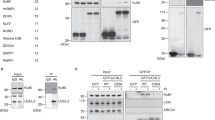
To identify a deubiquitinase specific for Ku80, we designed an in vitro assay using purified proteins (Fig. 2a). We purified poly-ubiquitinated Ku80 (Ku80-ubs) from 293T cells to use as the substrate for the in vitro deubiquitination assay. Ku80-ubs was then incubated with the 77 purified DUBs individually, followed by western blot analysis to reveal the levels of the remaining ubiquitin moiety on Ku80-ubs (Fig. 2b). We screened 16 DUBs that could reduce Ku80-ubs levels by at least 50% in vitro (Supplementary Table 2).
Identification of the specific deubiquitinase for Ku80 by complementary approaches using in vitro assays and cell-based assays. a Strategy for the screening of DUBs with defined substrates. b The in vitro deubiquitination assay using purified DUB enzymes and Ku80-ubs. The signal intensity of the Ku80-ubs was measured by Image Lab software (Supplementary Table 2). c Secondary screening of DUBs for Ku80 by using an shRNA knockdown in H1299 cells. d Confirmation of OTUD5 but not the other DUBs as the deubiquitinase of Ku80 by an siRNA knockdown assay. e Myc-Ku80 was stabilized when co-expressed with OTUD5, but not with USP7, OTUD6A or USP11
To confirm whether these DUBs indeed regulate Ku80 ubiquitination in vivo, we then used short hairpin RNAs (shRNAs) against 27 individual DUB candidates for Ku80 deubiquitination, which included 11 DUBs that were unable to be cloned previously and 16 DUBs identified by the in vitro deubiquitination assay. We performed IP assays using an anti-ubiquitin antibody and extracts from individual DUB-depleted H1299 cells. Knockdown of 4 of these 27 DUBs resulted in a significant increase in ubiquitinated Ku80 (Fig. 2c). Furthermore, the level of Ku80 was reduced significantly only when OTUD5 was depleted, suggesting that OTUD5 is the specific deubiquitinase for Ku80 (Fig. 2d). Supporting this finding, transfection with OTUD5 increased the levels of Ku80 in 293T cells (Fig. 2e). Taken together, these results demonstrated that OTUD5 is the specific deubiquitinase for Ku80.
OTUD5 is a unique deubiquitinase for Ku80
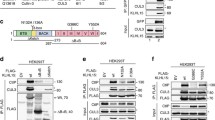
To further verify the role of OTUD5 in deubiquitination of Ku80, we performed an immunoprecipitation assay with H1299 cells in which OTUD5 was depleted by one of two different shRNAs against OTUD5 or one of two different siRNA sequences against OTUD5. Both types of RNAi-mediated OTUD5-depleted H1299 cells displayed increased levels of Ku80 ubiquitination (Fig. 3a, b). When we complemented OTUD5 levels by overexpression, ubiquitinated Ku80 was reduced. The protein level of Ku80 was rescued by wild-type OTUD5, but over-expression of catalytically inactive OTUD5 failed to rescue protein level of Ku80 (Fig. 3c). In addition, regardless of whether the cells in G1 or S/G2 phase, OTUD5 knockdown increased the level of Ku80-ubs (Supplementary Fig. 3E).
OTUD5 is a unique deubiquitinase for Ku80. a, b The levels of ubiquitinated Ku80 increased when OTUD5 was depleted by RNAi. a OTUD5 was knocked down by two different shRNAs, b OTUD5 was knocked down by two different siRNAs. c Complementation of OTUD5/WT reduced the level of ubiquitinated Ku80. OTUD5-depleted H1299 cells were transfected with vector plasmids or plasmids expressing wild-type OTUD5 or OTUD5/C224S. d, e OTUD5/C224S is inactive for deubiquitination of Ku80, and S177A decreased the capacity of OTUD5. For the in vitro assay, 293T cells were transfected with plasmid DNA expressing Flag-OTUD5/WT, Flag-OTUD5/C224S or Flag-OTUD5/S177A. The enzyme was then purified by M2-agarose beads under stringent conditions. All of the three types OTUD5 were incubated with Ku80-ubs for in vitro assay respectively. The assay was performed for three times. The signal intensity of Ku80-ubs was measured by Image Lab software. **p < 0.001. f Co-overexpressing WT OTUD5 but S177A/C224S reduced the level of ubiquitinated Ku80 induced by IR. MYC-Ku80 co-overexpressed with His-ub and plasmids encoding OTUD5/WT (lane 4), OTUD5/S177A (lane 5) or OTUD5/C224S (lane 6) respectively, treated with or without IR as indicated. g The levels of ubiquitinated Ku80 increased after OTUD5 was knocked down by siRNA in H1299 cells. H1299 cells were treated with IR (10 Gy, 1 h) (lanes 2–4) to induce Ku80-ubs. OTUD5 was knocked down by siRNA (lanes 3–4). Endogenous OTUD5 interacts with Ku80. Immunoprecipitation (IP) was conducted with antibodies specific for OTUD5 (h) or a Ku80 antibody (i). The IP-ed proteins and lysates were analyzed by western blotting
To firmly establish the role of OTUD5 in Ku80 deubiquitination, a catalytically inactive OTUD5 mutant protein (OTUD5/C224S) was purified, and its deubiquitination activity for Ku80 was determined. In addition, OTUD5 was shown to be activated by phosphorylation [19], and we also generated DNA to produce a mutant OTUD5 protein (OTUD5/S177A) to disable activation by phosphorylation. The results of the in vitro assay revealed that phosphorylated OTUD5 was more efficient than unphosphorylated OTUD5 in cleaving the ubiquitin chain of Ku80 (Fig. 3d, e, lane 2 vs lane 3), whereas the phosphorylation mutant OTUD5 (OTUD5/S177A) could only slightly cleave the ubiquitin chain (Fig. 3d, e, lane 4). Notably, the catalytically inactive mutant OTUD5 (OTUD5/C224S) lost its deubiquitination activity for Ku80, as indicated by unchanged levels of ubiquitinated Ku80 in the presence of OTUD5/C224S (Fig. 3d, e, lane 5).
To study the importance of Ku80 regulation by OTUD5 in vivo, we developed an assay using 293T cells co-transfected with plasmids expressing MYC-Ku80, His-ub and one of three types of OTUD5, OTUD5/WT, OTUD5/C224S and OTUD5/S177A. These cells were then treated with IR as indicated (Fig. 3f). The His-ub tagged proteins were purified from whole cell extracts using Ni-NTA bead purification. In the absence of OTUD5, Ku80 was ubiquitinated, and ubiquitinated Ku80 could be pulled down from whole cell extracts. In contrast, in the presence of OTUD5, ubiquitinated Ku80 was completely eliminated. This effect was dependent on OTUD5 activation by phosphorylation and OTUD5 activity, as co-expressing either OTUD5/S177A or OTUD5/C224S had little effect on the level of ubiquitinated Ku80 (Fig. 3f). In addition, the levels of Ku80 ubiquitination increased dramatically in OTUD5-knockdown cells after IR (Fig. 3g).
We then confirmed the interaction between endogenous OTUD5 and Ku80 in H1299 cells by an IP assay using either OTUD5 or a Ku80 antibody (Fig. 3h, i). Overall, these data suggest that OTUD5 was a specific deubiquitinase for Ku80 and played an important role in regulating the level of Ku80 ubiquitination in vivo.
OTUD5 stabilizes Ku80 and increases its accumulation at DSB sites
OTUD5 has been previously reported to be responsible for cleavage of ubiquitin chains formed through linkages at lysine48, and this linkage normally targets proteins for degradation by proteasome-mediated proteolytic destruction [20]. Ku80 was previously reported to be degraded by RNF8-mediated ubiquitination [13]. To study the role of OTUD5 in antagonizing the ubiquitination-mediated degradation of Ku80, H1299 cells were treated with MG132 or not, and Ku80 levels were analyzed by western blotting and immunofluorescence staining. We found that the protein level of Ku80 increased, accompanied by lengthening of the MG132 treatment (Supplementary Fig. 2B–D). We then co-expressed Ku80 with one of three types of OTUD5, OTUD5/WT, OTUD5/C224S or OTUD5/S177A, and MG132 treatment was added as a positive control. Indeed, the expression of OTUD5 markedly increased the protein levels of MYC-tagged Ku80, whereas OTUD5/S177A was not as potent as OTUD5/WT, and the expression of OTUD5/C224S had no effect on Ku80 protein levels (Fig. 4a). Furthermore, depletion of endogenous OTUD5 decreased the protein concentration of Ku80 (Fig. 4b), establishing the role of OTUD5 in maintaining Ku80 stability.
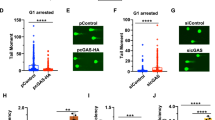
OTUD5 regulates the accumulation of Ku80 at DNA damage sites. a The effects of the expression of wild-type OTUD5 and mutant OTUD5 S177A and C224S on the stability of Ku80. MG132 (10 µM, 1 h) treatment was added as a positive control (lane 5). b Depletion of OTUD5 reduced the concentration of Ku80. H1299 cells were transfected with one of the two siRNAs (#1, #2) (lanes 2–3) or control siRNA (lane 1) for 48 h. Then, the lysate was analyzed by western blotting using the indicated antibodies. c OTUD5 knockdown impairs Ku80 focus formation at DNA damage sites. H1299 cells transfected with control siRNA or OTUD5 siRNA were exposed (or unexposed, top) to IR (5 Gy, 5 min). The Ku80 foci were visualized by immunofluorescence microscopy. d Quantitation of the numbers of foci in (c). Data are shown as the mean ± s. d.; n = 50 cells, data were obtained from three biological replicates. ***p < 0.0001. e ChIP assays to determine the recruitment of Ku80. The recruitment of Ku80 was reduced after OTUD5 knockdown. f Depletion of OTUD5 reduced the formation of XRCC4 foci after IR. H1299 cells transfected with control siRNA or OTUD5 siRNA were exposed (or unexposed, top) to IR (5 Gy, 30 min). The XRCC4 foci were visualized by immunofluorescence microscopy. g Quantitation of the numbers of foci in (f). Average XRCC4 foci per cell were counted and plotted as indicated. Data are shown as the mean ± s. d.; n = 50 cells, data were obtained from three biological replicates. ***p < 0.0001
Ku80 has been shown to regulate NHEJ repair through binding to DNA damage sites, and through recruiting factors involved in the DSB repair pathway. Recent studies also showed that Ku80 was ubiquitinated at DSB sites and that the initiation of HR depended on ubiquitination-mediated Ku80 removal from DNA [12,13,14]. These findings prompted us to investigate the physiological role of OTUD5 in regulating the accumulation of Ku80 at DSB sites and the recruitment of downstream NHEJ core factors. We utilized immunofluorescence staining in which the cells were extracted with CSK buffer combined with RNase A to obtain high-resolution images of Ku80 foci [21]. We introduced DSBs into the cells by IR to induce Ku80 accumulation at DSB sites. The Ku80 foci were visualized and increased dramatically during various cell cycle stages compared to the cells without IR treatment, suggesting that the binding of Ku80 to sites of DNA damage is not dependent on the cell cycle stage (Supplementary Fig. 3A, B). However, the formation of Ku80 foci was markedly reduced after depletion of OTUD5 by an OTUD5 siRNA (Fig. 4c, d).
These results were further supported by the I-SceI-induced DSB assay. The pCIN4-SceGFP-iGFP plasmid, which contains one recognition site for the I-SceI endonuclease [22], was integrated into the chromosomal DNA of H1299 cells as a substrate for I-SceI and HR repair. Cells harboring the recognition site for I-SceI induced DSB sites by transient transfection with the I-SceI expression vector pCDNA3.0–3 × NLS-I-SceI. After expression of I-SceI, the cells were fixed with formaldehyde and the chromatin was solubilized by sonication and purified. Immunoprecipitation was conducted with antibodies against Ku80. A significant portion of Ku80 recruitment was affected by Ku80 Ab, compared to the IgG control. This recruitment was reduced by 80% following OTUD5 depletion (Fig. 4e). These results demonstrated that Ku80 binding to DSB sites was regulated by OTUD5, as OTUD5-depleted cells displayed reduced Ku80 enrichment on DNA.
We then investigated the recruitment of downstream NHEJ core factors, using XRCC4 as an example. No XRCC4 foci were observed without IR, whereas XRCC4 foci increased dramatically after IR. Notably, XRCC4 foci decreased significantly after OTUD5 depletion. OTUD5 depletion indeed decreased the accumulation of the downstream NHEJ core factors (Fig. 4f, g).
Depletion of OTUD5 impairs NHEJ and overall DNA damage repair
Because of the central role of the Ku heterodimer in the recruitment of the nuclease, polymerases and ligases of NHEJ, we then explored the effect of OTUD5 on the efficiency of NHEJ through increasing the stability of Ku80.
To examine the specific impact of OTUD5 on NHEJ, the pCIN4-TK-EGFP plasmid, which contains two recognition sites for the I-SceI endonuclease in the reverse direction, was integrated into the chromosomal DNA of H1299 cells. Transient expression of I-SceI would introduce DNA DSB sites and increase the expression of EGFP resulting from NHEJ. Thus, the efficiency of NHEJ could be gauged by measuring the expression of GFP, and we confirmed the expression of I-SceI by co-transfecting dsRed plasmids [23]. OTUD5 depletion reduced the efficiency of NHEJ by approximately 50%, as measured by an I-SceI-mediated EGFP expression assay (Fig. 5a). We also studied 53BP1 focus formation in response to IR (5 Gy). After IR, 53BP1 foci increased dramatically. However, comparing the number of 53BP1 foci in cells synchronized in G1 phase with that in cells synchronized in S/G2 phases, we found no obvious changes following IR (Supplementary Fig. 3C, D). In contrast, ionizing radiation induced focus formation of 53BP1 decreased significantly due to the depletion of endogenous OTUD5 (Fig. 5c, d). Depletion of Ku80 had no effect on the formation of 53BP1 foci (Supplementary Fig. 5D). We then analyzed the cell extract and found that depletion of OTUD5 moderately reduced the protein level of 53BP1 (Supplementary Fig. 5C). These data further supported OTUD5 as a positive regulator of NHEJ.
OTUD5 promotes NHEJ repair. a OTUD5 depletion impairs the efficiency of NHEJ. Quantitation of the efficiency of NHEJ by counting EGFP positive cells using flow-cytometry. The H1299 cells were transfected with OTUD5, Ku80 or control siRNA, and plasmids expressing OTUD5/WT or OTUD5/C224S as indicated. Quantitative data represent the mean ± SEM from three independent experiments. ***p < 0.005. b Western blot analysis of the protein extracts from the cells used in (a). c OTUD5-depleted cells exhibited reduced 53BP1 foci. H1299 cells were transfected with different siRNAs as indicated. The cells were irradiated with ionizing radiation (5 Gy, 30 min), and the cells were fixed and stained. Images of 53BP1 foci were obtained by immunofluorescence microscopy. d Average numbers of 53BP1 foci per cell were counted from three independent experiments and plotted as indicated. Data are expressed as the mean ± S.D.; n = 50 cells, data were pooled across three experiments. ***p < 0.0001. e OTUD5 depletion reduced the viability of cells treated with increasing doses of IR. H1299 cells were transfected with control siRNA or siRNA against OTUD5 (OTUD5 #1, OTUD5 #2) before exposure to IR at the indicated dosages. Then, the cells were stained with methylene blue after 14 days of incubation. Error bars represent the s.e.m. from n = 3 biological experiments. f OTUD5 depletion delayed the clearance of γH2AX foci after IR. H1299 cells transfected with control siRNA or 2 different OTUD5 siRNAs were untreated or treated with IR (2 Gy) and subjected to immunofluorescence analysis of γH2AX at 0.5–12 h after irradiation. Nuclei were stained with DAPI. g The average numbers of γH2AX foci per cell were counted at 0.5, 1, 3, 6 and 12 h after irradiation from three independent experiments and plotted as indicated. Data represent the mean ± S.D.; n = 50 cells, data were pooled across three experiments. *p < 0.05, ***p < 0.0001
Reduced NHEJ repair increases cellular sensitivity to DNA DSBs and hence impairs cellular survival after DSB-inducing treatment. Indeed, OTUD5-depleted cells displayed reduced colony formation after IR (Fig. 5e). To determine the impact of OTUD5 on the overall repair of DSBs, γH2AX foci were used as a measure of overall DSB repair in the OTUD5-depleted cells after IR treatment. The results showed that depletion of OTUD5 delayed the disappearance of γH2AX foci in cells exposed to IR compared to the control cells (Fig. 5f, g), suggesting that OTUD5 plays a significant role in regulating overall DSB repair.
OTUD5 depletion promotes DNA end resection and HR repair during S/G2 phase
The switching of different DSB repair pathways involves direct competition between Ku70/Ku80 and MRN/CtIP for binding at DSBs, which affects the extent of DNA end resection and the balance between HR and NHEJ [24, 25]. The generation of short ssDNA overhangs by the Mre11 nuclease requires the removal of bound Ku at the DSB ends, thereby priming the ends for CtIP/Exo1-dependent DNA end resection. It is reasonable to believe that Ku plays a role in dictating the switching of DNA repair pathways by protecting DNA ends from end processing, which promoted us to examine the impact of OTUD5 on end resection and the extent of HR.
To obtain cells synchronized in different phases in culture, we used the thymidine double block system, which is based on inhibition of DNA synthesis by thymidine. A high concentration of thymidine interrupts the deoxynucleotide metabolism pathway, thereby arresting cells throughout early S phase [26]. After the thymidine double block procedure, the cells are synchronized at the G1/S boundary. The cells were synchronized in G1 phase during the initial 2 h, and S/G2 phase cells were obtained after 4–6 h of release (Supplementary Fig. 4A).
DNA ends that have already undergone resection will be coated by the RPA complex, composed of the subunits RPA1, RPA2 and RPA3. The complex is widely used as a proxy for end resection [25, 27]. For visualization of the RPA complex, H1299 cells expressing GFP-tagged RPA2 were extracted with CSK buffer combined with RNase A, before IR treatment. We hardly found any GFP-RPA2 foci in cells without synchronization because HR occurred during S/G2 phase (Supplementary Fig. 5A, B). Once the cells were arrested in the S/G2 phase, RPA2 focus-positive cells could be visualized easily. We found that the accumulation of RPA2 foci increased in OTUD5-depleted cells compared to that in control cells (Fig. 6a, b). There was no overlap between RPA2 foci and 53BP1 foci. In wild-type cells, the average number of 53BP1 foci was nearly fourfold higher than that of the RPA2 foci. In OTUD5-depleted cells, the ratio between the number of 53BP1 foci and the number of RPA2 foci decreased by twofold, indicating that NHEJ was suppressed and HR was stimulated by OTUD5 depletion during the S/G2 phase. As a complementary test for end resection, we compared 5-bromo-2-deoxyuridine (BrdU) focus formation in wild-type cells and OTUD5-depleted cells under non-denaturing conditions. As expected, there was no overlap between ssDNA (BrdU) foci and 53BP1 foci (Fig. 6c). The OTUD5-depleted cells exhibited increased formation of ssDNA (BrdU) foci in S/G2 phase (Fig. 6c, d). All of these results demonstrated that depletion of OTUD5 was required to prime DNA ends for HR repair. Unscheduled end resection would make the DNA ends unsuitable for NHEJ.
OTUD5 regulates switching of the DSB repair pathway. a OTUD5-depleted cells exhibited enhanced recruitment of RPA2 in cells synchronized in S/G2 phase. HeLa cells arrested in S/G2 phase were transfected with control siRNA or one of two different siRNAs against OTUD5 (OTUD5#1 and OTUD5#2) for 48 h. The cells were transfected with GFP-RPA2 before being irradiated with IR (5 Gy). The cells were stained as indicated. b The average numbers of RPA2 and 53BP1 foci per cell were counted from three independent experiments and plotted as indicated. Data represent the mean ± S.D.; n = 50 cells, data pooled across three experiments. c Depletion of OTUD5 increased the formation of ssDNA foci (BrdU signal) in cells synchronized in S/G2 phase. The effect of OTUD5 knockdown on ssDNA formation was tested using control siRNA and two different siRNAs against OTUD5 (OTUD5#1 and OTUD5#2). The cells were immune stained with antibodies against BrdU and 53BP1. d The average numbers of ssDNA foci and 53BP1 foci per cell were counted from three independent experiments and plotted as indicated. Data represent the mean ± S.D.; n = 50 cells, data were pooled across three experiments. e Depletion of OTUD5 enhanced the recruitment of RAD51 in response to IR in cells synchronized in S/G2 phase. HeLa cells were transfected with siRNA for 48 h before being treated with IR (5 Gy, 30 min). Immunostaining was performed with an anti-RAD51 antibody. f The average numbers of RAD51 foci per cell were counted from three independent experiments and plotted as indicated. Data represent the mean ± S.D.; n = 50 cells, data were pooled across three experiments. g The recruitment of proteins for HR was increased after OTUD5 knockdown, as determined by ChIP and real time PCR. The data represent the mean ± S.D. of three independent experiments; each sample was analyzed in triplicate. The fold enrichment was compared with the wild-type group. ***p < 0.0001. h OTUD5 knockdown increased HR efficiency. ***p < 0.0001. i Working model illustrating OTUD5 as a positive regulator of NHEJ
Rad51 is a key regulator of HR that helps to form a filament for strand exchange with a homologous DNA template. We examined the effect of OTUD5 depletion on Rad51 foci using immunofluorescence. RAD51 foci formed within two OTUD5-depleted cell lines were significantly more than the foci formed within the control cell line in response to IR (Fig. 6e, f). To further confirm these findings, we tested the accumulation of Mre11, RAD51 and Nbs1 at I-SceI-induced DSB sites. Cleavage of the I-SceI site resulted in enrichment of Mre11, RAD51 and NBS1 at the DSB sites in the S/G2 phase; however, depletion of OTUD5 significantly enhanced the enrichment of these core factors of HR, while recruitment of Ku80 was decreased (Fig. 6g). Consistent with this finding, OTUD5 depletion increased the efficiency of HR by approximately 70%, as measured by a chromosomal I-SceI-mediated gene conversion assay [28] (Fig. 6h).
We also checked whether OTUD5 knockdown had any effect on the cell cycle, and we found that there was no significant change in the population of cells in the S/G2 phase after OTUD5 knockdown (Supplementary Fig. 4B). In summary, these results suggest that OTUD5 is a negative regulator of HR repair (Fig. 6i).
Discussion
The Ku heterodimer, as a primary DNA end-binding factor that marks DNA ends for rejoining via NHEJ, has been studied extensively in past decades and is well understood [29]. The prevalent concept for DSB repair pathway switching holds that Ku should be removed from the DSB ends to initiate HR repair, as Ku trapped on DNA ends would preclude end resection [30, 31]. In addition, Ku80 is ubiquitinated at DNA damage sites, and ubiquitination regulates the removal of Ku80 from DSB sites [12,13,14, 32]. However, deubiquitination of Ku80 remains an active area of investigation. We studied the deubiquitination of Ku80 by multiple strategies (Fig. 2a) and demonstrated that OTUD5 was the specific deubiquitinase of Ku80. OTUD5 mutant proteins lost their Ku80 deubiquitination activity after we introduced C224S mutation. OTUD5 was previously reported to be responsible for cleaving ubiquitin chains formed through linkages at lysine48, and these modifications often lead to proteasome-mediated proteolytic destruction [20]. Consistent with this notion, we found that OTUD5 antagonized the ubiquitination-induced degradation of Ku80. We then explored the physiological roles of the Ku80 deubiquitination catalyzed by OTUD5. We studied the effects of OTUD5 depletion on the recruitment of Ku80 to DNA damage sites and demonstrated that depletion of OTUD5 impaired the accumulation of Ku80 at DNA damage sites. We verified this finding by testing the formation of foci of XRCC4, the downstream core factor of NHEJ. Additionally, knockdown of OTUD5 delayed the overall repair of DSBs. In the S/G2 phase, OTUD5 depletion results in excess end resection and increases the formation of foci of RAD51, the downstream core factor of HR, in response to DNA damage. These results demonstrate that OTUD5 regulates DSB repair pathway switching through promoting Ku80 deubiquitination. Given that recruitment of Ku80 is the initial step of NHEJ repair, our study reveals the unknown roles of OTUD5 in promoting NHEJ repair (Fig. 6i).
OTUD5 plays an important role in promoting genomic stability under conditions in which the dysfunction of NHEJ activates mutagenic alternative repair pathways. However, targeting Ku80 may not be the only mechanism whereby OTUD5 regulates the protein level of 53BP1. De et al. recently reported that OTUD5 localized to DNA double-strand breaks and regulated the accumulation of RPA complex at the DSB site [33], which is consistent with our results and further provides the possibility of OTUD5 playing an undiscovered role in DSB repair. OTUD5 may play a multiple role in DNA damage response. Some ubiquitin E3 ligases have been found being responsible for ubiquitination of K80, such as RNF8, RNF138 and RNF126. It is not surprising that Ku80-ubs is regulated by multiple DUBs to insure tight control of the levels of Ku80-ubs in cells, whereby ubiquitin carboxyl-terminal hydrolase L3 (UCHL3) was recently identified deubiquitinating Ku80 and participating in DNA damage response.
The Ku heterodimer is considered a caretaker because it regulates the processing of DNA DSB repair, otherwise leading to gross chromosomal rearrangements [34]. Ku80−/− mice with a loss of p53 or PARP-1 succumb to tumorigenesis, such that all ku80−/− p53−/− mice develop pro-B-cell lymphoma by 16 weeks [34, 35]. In addition, overexpression of Ku80 suppresses cellular proliferation and xenograft tumor growth in nude mice [36]. Ubiquitination-mediated removal of Ku80 from damage sites would be a reasonable mechanism to regulate the Ku heterodimer, and this regulation needs further investigation.
RNAi libraries were previously screened for the DUBs responsible for specific substrates, but this approach has limitations. It is usually difficult to achieve a high efficiency of uniform depletion for each DUB protein through screening because many DUBs are stable proteins in vivo with long half-lives. Additionally, DUB depletion may induce indirect effects, thus impacting the phenotypes obtained from RNAi screening [37]. To complement these results, we used an approach in which screening was performed using deubiquitinases and a defined substrate purified from mammalian cells in vitro, and the same approach could facilitate the identification of specific DUBs for many important cellular factors, similar to Ku80.
Materials and methods
Cell culture
HEK-293T, HeLa, and H1299 cells (obtained from the American Type Culture Collection) were maintained in DMEM, supplemented with 10% fetal bovine serum and penicillin/streptomycin in a humidified atmosphere with 5% CO2 at 37 °C.
Constructs
Each of the DUBs was cloned into the pCIN4-Flag expression vector, and its sequence was confirmed by DNA sequencing [37, 38]. Plasmids containing human OTUD5/C224S or OTUD5/S177A were obtained by mutagenesis according to the manufacturer’s protocol (Transgene). Ku80 was generated by PCR and cloned into the pRK5-MYC-Flag vector; RNF138 was generated by PCR and cloned into the pCIN4-HA vector. The RPA2 gene was cloned into the pcDNA3.1-GFP vector.
Immunoblots and antibodies
Western blotting was performed by the standard method. The primary antibodies used were specific for Ku80 (#2753; Cell Signaling Technology), OTUD5 (D8Y2U, Cat. No. 20087; Cell Signaling Technology), USP7 (SC-133204, Santa Cruz), USP11 (SC-365528, Santa Cruz), OTUD6A (HPA053304, Sigma) RAD51 (ab133534, Abcam), NBS1 (ab32074, Abcam), Mre11 (ab214, Abcam), RPA1 (2267S, Cell Signaling Technology), 53BP1 (4937S, Cell Signaling Technology), XRCC4 (ab97351, Abcam), c-MYC (9E10) (sc-40; Santa Cruz), HA (H9658, Sigma), Flag (F3165, sigma), ubiquitin (SC-8017, Santa Cruz), and β-actin (Sigma).
SILAC-MS/MS to identify significant ubiquitinated proteins in H1299 cells
H1299 cells were labeled with either “heavy” or “light” isotopic lysine using a SILAC Protein Quantitation Kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Briefly, two cell lines were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% dialyzed fetal bovine serum and with either the “heavy” (H) form of Lys6Arg10 or “light” (L) Lys0Arg0 for more than six passages before being used in the assay. Cells, either without or with IR treatment (10 Gy, 2 h), were harvested and washed twice with cold phosphate-buffered saline. The cells were lysed in RIPA buffer (40 mm Tris, pH 8.0, 200 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, and 1% SDS) on ice for 20 min. Equal amounts of protein from cells in the no-IR group and IR group were mixed. The proteins were digested with trypsin. To enrich di-GG-ubiquitinated peptides, the tryptic peptides were incubated with anti-di-GG agarose beads (PTM Biolabs Inc., Chicago, IL, USA) at 4 °C for 4 h with gentle shaking. The beads were washed four times and the bound peptides were eluted from the beads with 1% trifluoroacetic acid. HPLC/MS/MS analysis was performed. The ratio of H/L peptides was normalized by eliminating the impact of protein level change. The results are shown in Supplementary Table 1.
Transfection of cells
For the knockdown assay, cells were transfected with appropriate siRNAs against OTUD5 using Lipofectamine 3000, and scrambled siRNA was used as a control. After 48 h, the cells were harvested, and the efficiency of OTUD5 knockdown was verified by immunoblotting. For the overexpression assay, cells were transfected with the appropriate plasmid using Neofect DNA transfection reagent (Neofect Biotech) and harvested after 24–48 h.
Ni-NTA affinity purification and in vivo Ku80 ubiquitination
293T cells were transfected with plasmids expressing MYC-Ku80, His-ubiquitin and ubiquitin E3 ligase (HA-RNF138). Thirty-six hours post-transfection, one-tenth of the cells were lysed with RIPA buffer, and the extracts were used as input. The remaining cell extracts were mixed with Ni-NTA beads (Qiagen) in phosphate/guanidine buffer (6 M guanidine-HCl, 0.1 M Na2HPO4, 6.8 mM Na2H2PO4, 10 mM Tris–HCl pH 8.0, 0.2% Triton X-100, and freshly added 10 mM β-mercaptoethanol and 5 mM imidazole) to pull down his-tagged proteins overnight at 4 °C after sonication. The Ni-NTA resin-bound proteins were then washed once with wash buffer 1 (8 M urea, 0.1 M Na2HPO4, 6.8 mM Na2H2PO4, 10 mM Tris–HCl pH 8.0, 0.2% Triton X-100, and freshly added 10 mM β-mercaptoethanol and 5 mM imidazole) and washed three times with wash buffer 2 (8 M urea, 18 mM Na2HPO4, 80 mM Na2H2PO4, 10 mM Tris–HCl pH 6.3, 0.2% Triton X-100, and freshly added 10 mM β-mercaptoethanol and 5 mM imidazole). The bound proteins were eluted with loading buffer and resolved by SDS-PAGE.
Purification of DUBs and Ku80-ubs
The DUBs were purified from 293T cell lysates by immunoprecipitation using anti-Flag M2 beads [37, 38]. 293T cells were transfected with plasmids expressing each of the DUBs and harvested after 48 h. The cells were lysed in BC500 buffer (20 mM Tris–HCl pH 7.3, 500 mM NaCl, 20% glycerol, and 0.5% Triton X-100) with sonication before incubation with M2 beads overnight at 4 °C. After washing the beads three times with BC100 buffer (20 mM Tris–HCl pH 7.3, 100 mM NaCl, 20% glycerol, 0.1% Triton X-100), the DUBs were eluted with Flag peptide (Sigma). To purify ubiquitinated Ku80 (Ku80-ubs), 293T cells were transfected with plasmids expressing Flag-Ku80. After 48 h, the cells were treated with IR (10 Gy, recovery 1 h). The proteins were purified as described above.
In vitro deubiquitination
Ku80-ubs was incubated with each of the purified DUBs in deubiquitination buffer (50 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM EDTA, 10 mM DTT, and 5% glycerol) for 2 h at 37 °C. The reactions were stopped by adding loading buffer. The mixtures were resolved on SDS-PAGE for western blot analysis using an anti-ubiquitin antibody. The quantitative data of in vitro deubiquitination assay is shown in Supplementary Table 2.
Immunoprecipitation with the chromatin fraction
Cells were collected in PBS, and chromatin fractions were isolated in CSK buffer (20 mM HEPES pH 7.9, 50 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 0.5% Triton X-100). After sonication, the salt concentration was adjusted to 500 mM and the sample was further incubated for 30 min on ice. Lysates were clarified by centrifugation (20,800g, 15 min, 4 °C) and at least 1 mg of proteins was used per immunoprecipitation assay in IP buffer (50 mM Tris–HCl pH 7.3, 137 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X, 0.2% sarkosyl, 1 mM NaF, 1 mM Na3VO4, and 0.5 mM DTT). Target proteins were captured with an anti-ubiquitin antibody coupled to protein G magnetic Dynal beads. The complexes were extensively washed in IP buffer with the salt concentration adjusted to 500 mM NaCl. All extracts were pre-cleared using beads alone.
Immunofluorescence staining
For monitoring the formation of γH2AX, 53BP1 and RAD51 foci, the cells were fixed with 4% paraformaldehyde for 15 min and permeabilized by PBS containing 0.25% Triton X-100 for 10 min. After incubation with 5% bovine serum albumin (BSA) for 1 h at room temperature, the cells were incubated with the indicated primary antibodies overnight at 4 °C or for 2 h at room temperature. The cells were then washed with PBS and incubated with a fluorophore-conjugated secondary antibody. Finally, the cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei. The cells were imaged by a Zeiss 880 confocal laser-scanning microscope at 63 × magnification.
For visualization of Ku80 foci, RPA2 foci and XRCC4 foci, the cells were extracted with RNase A-containing CSK buffer before staining [21].
For non-denatured BrdU immunodetection, the cells were incubated with 20 mM BrdU for 24 h before DSB-induced treatment. The cells were extracted twice with CSK buffer for 5 min before being fixed, permeabilized and stained as described above.
RNA interference
The shRNA plasmids targeting the indicated DUBs were constructed in the pLVX-shRNA2 vector (Clontech). Lentiviral gene transduction was carried out using 293T packaging cells with the Lenti-X HTX Packaging System (Clontech). Medium containing virus was collected, supplemented with 8 μg/ml polybrene (Sigma) and incubated with target H1299 cells at 37 °C for 12 h. Infected H1299 cells were selected with 3 μg/ml puromycin for 3 days. Control cells were generated by lentivirus expressing scrambled shRNA. Sequences of all shRNAs used in this study are listed in Supplementary Table 3.
A stable OTUD5-depleted H1299 stable line was generated by shRNA. (Sequences of two shRNA against OTUD5: #1: 5′-tGGGCTGGGCCTGCCATCATTCttcaagagaGAATGAT GGCAGGCCCAGCCCttttttc-3′, #2: 5′-tGGGCCCTCATTCAGCAGATGTttcaagagaACATCTGCTGAATGAGGGCCCttttttc-3′).
H1299 cells were transfected with siRNAs (Gene Pharma) for OTUD5 knockdown. The sequences of the siRNA against OTUD5 are as follows: #1: 5′-GGGCUGGGCCUGCCAUCAUUC-3′, #2: 5′-GGGCCCUCAUUCAGCAGAUGU-3′. The sequences of the siRNAs against Ku80 are as follows: #1: 5′-AAUAUCCAGCUGACUUUUGCU-3′, #2: 5′-UCAAAUGGGGAUUCUAUACCA-3′.
ChIP assay
The effect of OTUD5 depletion on DNA repair protein recruitment to the defined DSB was determined by ChIP and PCR. Induction of a site-specific DSB was performed as described previously [39]. The forward primer was CCG ACA ACC ACT ACC T; the reverse primer was GCTGAACTTGTGGCCGTTTAC.
NHEJ assay
The pCIN4-TK-EGFP DNA was transfected into H1299 cells using the Lipofectamine 2000 reagent (Invitrogen), and the cells were subjected to puromycin screening. The retention of TK-EGFP DNA was confirmed by PCR. H1299 cells harboring two I-SceI sites were transiently transfected with the pCDNA3.0–3 × NLS-I-SceI plasmid after the depletion of OTUD5. Then the cells were subjected to trypsinization, washed with PBS, and analyzed with a flow cytometer (FACSCalibur, BD bioscience). The proportion of EGFP-positive cells was determined in 1 × 104 with the use of FACADiva software [23].
Tumor cell colony formation assays
Cells with OTUD5 depleted or not were seeded in triplicate onto 35 mm dishes, and incubated for 14 days after IR treatment. The resulting colonies were stained with 2% methylene blue/50% ethanol for 15 min. The stained cells were extracted with 1% SDS. To quantify relative cell number, the results were analyzed by a spectrophotometer, and the absorbance was detected at 600 nm. The results were normalized to the plating efficiency. The results are presented as the averages of data obtained from three independent experiments.
Synchronization of cells
Cells were synchronized by double thymidine block at the early S phase of the cell cycle. The cells grew to ~ 40% confluence, and were incubated with 2 mM thymidine for 14 h. The cells were released from thymidine block by washing with PBS five times and incubated with fresh medium for 6–8 h. After the second round of thymidine block and release, the cells were synchronized and allowed to progress into different phases in cell culture (Supplementary Fig. 4A).
Assay with synchronized cells
In the morning of the first day, the transfection assay was performed, and 8 h later the process of thymidine double block was started. On the third day, the synchronized cells were harvested at different time points for the subsequent analysis.
Statistical analysis
All results represent the average of experiments at least in triplicate, and all results are expressed as the mean ± standard derivation. The associations between categorical variables were assessed using the Chi-square test and Fisher’s exact test. Analysis of variance was performed to determine the statistical significance among groups. A value of p < 0.05 was considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001).
References
Khanna KK, Jackson SP (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27:247–254. https://doi.org/10.1038/85798
Streffer C (2010) Strong association between cancer and genomic instability. Radiat Environ Biophys 49:125–131. https://doi.org/10.1007/s00411-009-0258-4
Lieber MR (2008) The mechanism of human nonhomologous DNA end joining. J Biol Chem 283:1–5. https://doi.org/10.1074/jbc.R700039200
Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211. https://doi.org/10.1146/annurev.biochem.052308.093131
Mazon G, Mimitou EP, Symington LS (2010) SnapShot: homologous recombination in DNA double-strand break repair. Cell 142(646):e641. https://doi.org/10.1016/j.cell.2010.08.006
San Filippo J, Sung P, Klein H (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77:229–257. https://doi.org/10.1146/annurev.biochem.77.061306.125255
Chiruvella KK, Liang Z, Wilson TE (2013) Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol 5:a012757. https://doi.org/10.1101/cshperspect.a012757
Karanam K, Kafri R, Loewer A, Lahav G (2012) Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell 47:320–329. https://doi.org/10.1016/j.molcel.2012.05.052
Shim EY et al (2010) Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J 29:3370–3380. https://doi.org/10.1038/emboj.2010.219
Postow L (2011) Destroying the ring: freeing DNA from Ku with ubiquitin. FEBS Lett 585:2876–2882. https://doi.org/10.1016/j.febslet.2011.05.046
Postow L et al (2008) Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol 182:467–479. https://doi.org/10.1083/jcb.200802146
Ismail IH et al (2015) The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat Cell Biol 17:1446–1457. https://doi.org/10.1038/ncb3259
Feng L, Chen J (2012) The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 19:201–206. https://doi.org/10.1038/nsmb.2211
Ishida N et al (2017) Ubiquitylation of Ku80 by RNF126 promotes completion of nonhomologous end joining-mediated DNA repair. Mol Cell Biol 37:e00347-16. https://doi.org/10.1128/mcb.00347-16
Nishi R et al (2018) The deubiquitylating enzyme UCHL3 regulates Ku80 retention at sites of DNA damage. Sci Rep 8:17891. https://doi.org/10.1038/s41598-018-36235-0
Abdul Rehman SA et al (2016) MINDY-1 Is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell 63:146–155. https://doi.org/10.1016/j.molcel.2016.05.009
Nijman SM et al (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773–786. https://doi.org/10.1016/j.cell.2005.11.007
Bekker-Jensen S, Mailand N (2015) RNF138 joins the HR team. Nat Cell Biol 17:1375–1377. https://doi.org/10.1038/ncb3262
Huang OW et al (2012) Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat Struct Mol Biol 19:171–175. https://doi.org/10.1038/nsmb.2206
Mevissen TE et al (2013) OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154:169–184. https://doi.org/10.1016/j.cell.2013.05.046
Britton S, Coates J, Jackson SP (2013) A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol 202:579–595. https://doi.org/10.1083/jcb.201303073
Pierce AJ, Johnson RD, Thompson LH, Jasin M (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13:2633–2638
Ogiwara H et al (2011) Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 30:2135–2146. https://doi.org/10.1038/onc.2010.592
Chapman JR, Taylor MR, Boulton SJ (2012) Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47:497–510. https://doi.org/10.1016/j.molcel.2012.07.029
Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45:247–271. https://doi.org/10.1146/annurev-genet-110410-132435
Ma HT, Poon RY (2017) Synchronization of HeLa cells. Methods Mol Biol 1524:189–201. https://doi.org/10.1007/978-1-4939-6603-5_12
Gupta R et al (2018) DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 173:972–988. https://doi.org/10.1016/j.cell.2018.03.050
Pierce AJ, Hu P, Han M, Ellis N, Jasin M (2001) Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 15:3237–3242. https://doi.org/10.1101/gad.946401
Grob P et al (2012) Electron microscopy visualization of DNA-protein complexes formed by Ku and DNA ligase IV. DNA Repair (Amst) 11:74–81. https://doi.org/10.1016/j.dnarep.2011.10.023
Wu D, Topper LM, Wilson TE (2008) Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178:1237–1249. https://doi.org/10.1534/genetics.107.083535
Niu H, Raynard S, Sung P (2009) Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev 23:1481–1486. https://doi.org/10.1101/gad.1824209
Postow L, Funabiki H (2013) An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle 12:587–595. https://doi.org/10.4161/cc.23408
de Vivo A et al (2019) The OTUD5-UBR5 complex regulates FACT-mediated transcription at damaged chromatin. Nucleic Acids Res 47:729–746. https://doi.org/10.1093/nar/gky1219
Difilippantonio MJ et al (2000) DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404:510–514. https://doi.org/10.1038/35006670
Lim DS et al (2000) Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol Cell Biol 20:3772–3780
Wei S et al (2012) Ku80 functions as a tumor suppressor in hepatocellular carcinoma by inducing S-phase arrest through a p53-dependent pathway. Carcinogenesis 33:538–547. https://doi.org/10.1093/carcin/bgr319
Shan J, Zhao W, Gu W (2009) Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell 36:469–476. https://doi.org/10.1016/j.molcel.2009.10.018
Yu M et al (2016) USP11 is a negative regulator to gammaH2AX ubiquitylation by RNF8/RNF168. J Biol Chem 291:959–967. https://doi.org/10.1074/jbc.M114.624478
Rodrigue A et al (2006) Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J 25:222–231. https://doi.org/10.1038/sj.emboj.7600914
Acknowledgements
We are grateful to Dr. Jiadong Wang for his suggestion and help. We thank Qihua He for her suggestion and help, and Center of Medical and Health Analysis, Peking University, for confocal microscopy. We appreciate the ALENABIO (Xi’an, China) Company (http://www.alenabio.com) for the pathological micro-tissues (Cat. No. BC03119a). We deeply appreciate help from Ning Kon for editing our manuscript.
Funding
Wenhui Zhao was supported by the National Natural Science Foundation of China (Grant No. 85141044).
Author information
Authors and Affiliations
Contributions
FL and WZ, conceived and designed the study that led to the submission, acquired data, and interpreted the results; FL performed the experiment; QS, KL, HH, QH, ZC, YM, and FT participated in the revision of the manuscript; ZM and TT approved the final version; and WZ was the corresponding author. The authors declare that they have no conflicts of interest associated with the contents of this article.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Availability of data and material
All of the data and material in this paper are available upon request.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, F., Sun, Q., Liu, K. et al. The deubiquitinase OTUD5 regulates Ku80 stability and non-homologous end joining. Cell. Mol. Life Sci. 76, 3861–3873 (2019). https://doi.org/10.1007/s00018-019-03094-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03094-5