Abstract
Understanding user emotion is essential for Human-AI Interaction (HAI). Thus far, many approaches have been studied to recognize emotion from signals of various physiological modalities such as cardiac activity and skin conductance. However, little attention has been paid to the fact that physiological signals are influenced by and reflect various factors that have little or no association with emotion. While emotion is a cross-modal factor that triggers responses across multiple physiological modalities, features used in existing approaches also reflect modality-specific factors that affect only a single modality and have little association with emotion. To address this, we propose an approach to extract features that exclusively reflect cross-modal factors from multimodal physiological signals. Our approach introduces a multilayer RNN with two types of layers: multiple Modality-Specific Layers (MSLs) for modeling physiological activity in individual modalities and a single Cross-Modal Layer (CML) for modeling the process by which emotion affects physiological activity. By having all MSLs update their hidden states using the CML hidden states, our RNN causes the CML to learn cross-modal factors. Using real physiological signals, we confirmed that the features extracted by our RNN reflected emotions to a significantly greater extent than the features of existing approaches.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Understanding user emotions is extremely important for various human-AI interaction (HAI) scenarios including goal and non-goal oriented dialogue [6, 8], user-adapted content creation [1], and content recommendation [2]. While most researchers collect ground truth of emotions by explicitly asking users what their emotions are, it is impractical to do so in real-world scenarios because doing so interferes with users and degrades the user experience. Therefore, there has been a great demand for recognizing user emotions from data that users generate.
Among various types of user-generated data, we focus on users’ physiological signals such as electroencephalogram (EEG), electrocardiogram (ECG), and galvanic skin response (GSR). Using wearable devices (e.g., watches, earphones), these signals can be collected in a less constrained context compared to other types of data such as texts, vocal tone, and facial expressions, which are available only when users write or say something or stay in front of a camera. In addition, unlike these data, physiological signals provide robust signs of emotion even when users exhibit their social masks to hide their true emotions [3].
Thus far, researchers have studied many approaches to recognize emotion from physiological signals and have confirmed their significant utility for emotion recognition [6]. However, little attention has been paid to the fact that physiological signals are influenced not only by emotion but also by various factors that have little or no association with emotion. Among them are factors that influence only a single physiological modality, i.e., a modality-specific factors. For example, heart muscle strength influences ECG signals, but has little influence on modalities other than cardiac activity such as brain activity and skin conductance. In contrast, emotion is a cross-modal factor, which triggers responses across multiple physiological modalities, e.g., anger increases heart rate and skin conductance level. Others are long-term factors such as body size and gender. These factors also influence physiological activity, but they are very different from emotion in a sense that they change very slowly or do not change, whereas emotion changes over short periods of time, i.e., a short-term factor.
As such, while emotion is a cross-modal and short-term factor, physiological signals are also influenced by and reflect factors that are modality-specific and/or long-term. Although they have little utility for emotion recognition, existing approaches extract and use features without distinguishing these factors, instead mixing them into the features. We posit this has degraded emotion recognition.
In light of the above, we propose an approach to extract features that exclusively reflect cross-modal and short-term factors. To achieve this, our approach distinguishes factors reflected in physiological signals along two axes: long- or short-term and modality-specific or cross-modal, and learns four types of factors that are distinct from each other. By adopting RNN, our approach separately models long- and short-term factors.
What is novel is that to model modality-specific and cross-modal factors, we introduce a multilayer RNN that consists of two types of layers: multiple Modality-Specific Layers (MSLs) that model physiological activity in individual modalities; and a single Cross-Modal Layer (CML) that learns cross-modal factors, among which is emotion. Our RNN takes sequences of multimodal physiological signals as input (e.g., ECG and GSR signals). Each MSL takes physiological signals of its corresponding modality (e.g., MSL1 takes ECG signals, MSL2 takes GSR signals) and reflects physiological states in its hidden state. When updating the hidden state, the MSL uses not only its own hidden state but also the CML’s hidden state. Since this is done in all the MSLs, it makes the CML’s hidden state affect physiological state in all the modalities the MSLs correspond to. In effect, therefore, this enables the CML to learn factors that affect physiological activities across multiple modalities, i.e., cross-modal factors.
To evaluate our approach, we recruited participants and measured their EEG, ECG, and GSR signals while presenting them with musical pieces and movie clips (i.e., stimuli). We trained our RNN by these signals and, using the CML’s hidden states, evaluated how accurately we could recognize emotions that the participants reported after each stimulus.
Our main contributions are as follows. 1) We propose a multilayer RNN that separates the RNN layer to learn factors that affect physiological activities across multiple modalities from the other layers designated to model modality-specific physiological activities. This enables our approach to extract features that exclusively reflect a cross-modal nature of emotion, which existing research has not focused on. 2) Using real physiological data, we demonstrate our RNN extracts features that reflect emotion to a greater extent than existing approaches.
2 Related Work
Similar to our approach, many existing approaches recognize emotion from multimodal physiological signals. Subramanian et al. [11] and Miranda et al. [7] used ECG, GSR, and EEG signals. Using feature extraction techniques that are widely used for each modality, they extracted features from each modality (physiological features; e.g., standard deviation of heartbeat intervals from ECG signals, mean skin conductance level from GSR signals). They then concatenated these physiological features and fed them into a classifier (i.e., early fusion). However, modality-specific factors reflected in the physiological features could not be removed by simple concatenation, thus limiting recognition accuracy. In addition, short- and long-term factors were not distinguished in the features. While they also tested late fusion, in which they combined recognition results in individual modalities to derive final results, the same issues remained because they used the same physiological features as in the early fusion, whose modality-specific factors hindered emotion recognition in each modality.
There are also multimodal approaches that adopt deep learning techniques. However, they have the same issues. Liu et al. [5] and Yin et al. [12] used deep autoencoders to learn shared representations of physiological features of multiple modalities (e.g., EEG and Electrooculogram) and recognized emotions by feeding the shared representations into classifiers. They trained the autoencoders so that the physiological features of each modality could be reproduced from the shared representations. This made the shared representations reflect not only cross-modal factors but also modality-specific factors. In addition, the use of the autoencoders did not help to distinguish between short- and long-term factors.
On the other hand, the approach proposed by Li et al. [4] can extract features that exclusively reflect short-term factors. Using the dataset built in [7], they fed time-series sequences of physiological features into LSTM, whose hidden states were then fed into an attention network. These steps enabled them to focus on emotionally salient parts of the sequences, from which they extracted the hidden states and fed them into a multilayer perceptron (MLP) to recognize emotions. However, they performed these steps in each physiological modality and derived final results by combining the results of individual modalities (i.e., late fusion). Therefore, as in [7, 11], emotion recognition in individual modalities was hindered by modality-specific factors, which also degraded the final recognition results.
3 Proposed Approach
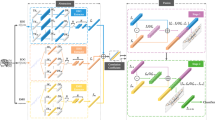
In contrast to the existing approaches, our RNN explicitly distinguishes the four types of factors that influence physiological activity. Figure 1 exemplifies our RNN (left) and shows how the four types, I−IV, are mapped to its variables (right). Modality-specific factors, I and II, are modeled by the MSLs. Each MSL corresponds to a single modality, e.g., MSL1 to EEG, MSL2 to ECG. It takes sequences of 1) physiological features of the corresponding modality, which are extracted in the same way as existing approaches (e.g., [7, 11]), and 2) one-hot vectors of user ID, by which a user representation (UR) is retrieved from the user matrix. Since the physiological features fed to the MSL are limited to the corresponding modality, its URs and hidden states reflect factors specific to this modality (I and II). In addition, while the hidden states are updated sequentially, the user matrix (set of URs) stays the same. This causes the MSL URs to reflect long-term factors (I) and its hidden states to reflect short-term factors (II).
On the other hand, cross-modal factors, III and IV, are modeled by the CML. As shown by link (A) in the figure, the CML sends its hidden states to the MSLs. The MSL cell uses these CML hidden states together with its input (the physiological features and URs) and its previous hidden states to update its hidden states. Because updated MSL hidden states are used to predict the physiological features at the next timeslot, it can be regarded as representing physiological state. Updating such MSL hidden states using the CML hidden states means that the CML hidden states affect physiological activity of individual modalities. Because all the MSLs update their hidden states in this way, the CML learns factors that affect physiological activity across multiple modalities, i.e., cross-modal factors (III and IV). As in the MSL, the CML also reflect short-term factors (III) in its hidden states and long-term factors (IV) in its URs, but the difference being they are cross-modal.
In addition to modeling I\(\sim \)IV, our RNN also models the process by which individual physiological differences moderate the relationship between emotion and physiological activity. For example, users with different heart muscle strength would have different ECG signals even when their emotions are the same. Our RNN models such moderating effect of individual differences by updating the MSL hidden states (reflecting physiological state) using both the CML hidden states (emotion) and the MSL URs (individual differences, e.g., heart muscle strength). This also differentiates our RNN from the existing approaches discussed in Section 2, all of which do not consider this moderating effect.
The next section describes in detail the hidden state updating in our RNN and its training process. See Table 1 for the notations and their descriptions.
3.1 Updating the Hidden States
Input to the CML and MSL are formatted as
where \(x^C_{a,t}=(\boldsymbol{i_u}, \boldsymbol{i_{s,t}})\) denotes user a’s t-th action (e.g., viewed t-th segment of a movie clip M); and \(x^n_{a,t}=(\boldsymbol{i_u}, \boldsymbol{p_t^n})\) denotes his physiological features extracted from signals during t-th action. Once \(x^C_{a,t}\) is input to the CML, it first retrieves a UR and stimulus segment (SS) representation (SR) from the user and SS matrices, i.e., \(\boldsymbol{e_u^C}=\boldsymbol{W_u^C}\boldsymbol{i_u}\) and \(\boldsymbol{e_{s, t}^C}=\boldsymbol{W_s^C}\boldsymbol{i_{s,t}}\), respectively. It then updates its hidden state \(\boldsymbol{h_t^C}\) as follows:
where \(f^C\) is a function implemented by LSTM. See the supplementary material at https://osf.io/mj3nr/ for detail.
After updating the hidden state, the CML sends it to all MSLs via link (A), which is done every time the CML updates its hidden state. When the MSL receives \(\boldsymbol{h_t^C}\), it retrieves a UR from its user matrix (\(\boldsymbol{e_u^n}=\boldsymbol{W_u^n}\boldsymbol{i_u}\)) and uses them together with input physiological features (\(\boldsymbol{p_{t-1}^n}\)) to update its hidden state \(\boldsymbol{h_t^n}\) as follows:
where \(f^n\) is a function implemented by LSTM (see the supplementary material).
3.2 Model Training
When \(\text {\textit{data}}^n_a\) is fed, each MSL predicts physiological features in each timeslot, e.g., if the input is \(\text {\textit{data}}^n_a=[x^n_{a, t}, x^n_{a,t+1},...,x^n_{a,T-1}]\), the output is \([\boldsymbol{\hat{p}^n_{t+1}}, \boldsymbol{\hat{p}^n_{t+2}}, ..., \boldsymbol{\hat{p}^n_{T}}]\). The predicted physiological features are compared with the actual features to calculate the loss that is used to learn the parameters of the MSL and CML cells and the user and SS matrices (\(\boldsymbol{W_u^n}\), \(\boldsymbol{W_u^C}\), and \(\boldsymbol{W_s^C}\)). Figure 2 shows how the prediction and loss calculation are performed. The MSL predicts physiological features using its hidden state as follows: \(\boldsymbol{\hat{p}^n_{t+1}}=f_{\text {MLP}}^n(\boldsymbol{h_{t+1}^n})\), where \(f^n_{\text {MLP}}\) is an MLP with ReLu activation. Then, the MSL calculates the residual sum of squares between actual and predicted physiological features as the loss.
4 Experiment
We built datasets and evaluated the extent to which the CML hidden states reflect emotion. We performed the following three steps: (1) Feature extraction - from the physiological features stored in our datasets, we extracted another set of features for emotion recognition (emotion features). In our RNN, the CML hidden states were used as the emotion features; (2) feature selection - we then performed LASSO regression to select the emotion features; and (3) linear regression - using the selected emotion features, we built models to predict emotions and evaluated their model fit and prediction accuracy.
We performed (1)–(3) for our RNN and three approaches to compare. The first approach, which was implemented following [7, 11], did not distinguish between the four types of factors at all when extracting the emotion features (hereafter “baseline”). The second one distinguished between long and short-term factors but not between cross-modal and modality-specific factors as in [4]; and the third one distinguished the four types of factors, but did not model the moderating effect of individual physiological differences. The last two were implemented by removing key features from our RNN (will be explained in 4.4 Ablation Study).
We built two different datasets and performed (1)-(3) for each dataset. In addition, because combinations of physiological modalities available in real-world scenario would be different depending on the devices users wear, we performed (1)-(3) for all possible modality combinations available in our datasets. That is, A) EEG+ECG+GSR, B) EEG+ECG, C) EEG+GSR, and D) ECG+GSR.
4.1 Dataset
Due to page limitations, only a brief summary of the datasets is described below. See the supplementary material (https://osf.io/mj3nr/) for detail. Although several datasets are publicly available today (e.g., [7, 11]), we built and used our own datasets. One reason is because the contacts of these datasets did not respond to our requests. The other is because they used only videos as stimuli when collecting physiological signals. Because music is another popular type of stimulus that would be played more often especially while working, studying, etc., we considered evaluation should be done for both music and video.
We built Music and Movie datasets by conducting data collection experiments, in which 54 and 52 (out of 54) subjects participated, respectively. They were presented with multiple stimuli, each of which was 60 s long, while their EEG, ECG, and GSR signals were measured. In total, 2,336 and 2,119 trials were performed for the music and movie datasets, respectively (one trial denotes one subject listening to/viewing one stimulus). After listening to/viewing each stimulus, they reported emotions according to the six dimensions whose scores ranged 0–15, (a) sad-happy, (b) nervous-relaxed, (c) fear-relieved, (d) lethargic-excited, (e) depressed-delighted, and (f) angry-serene. Although Russel’s circumplex model [10] has been widely used to determine emotion, we did not use it because it is not easy for lay participants to report “arousal” and “valence” defined in the model. We selected the six dimensions so that the participants can easily report their emotions and the dimensions cover the Russel’s circumplex as much as possible.
After collecting the physiological signals, we extracted the physiological features from the raw signals by feature extraction techniques that are widely used for each modality as in [7, 11]. We extracted two types of features: window and stimulus features, which are summarized in Table 2. For the window features, we applied sliding window to the raw signals measured during one stimulus and extracted features from each window. We set the window size to ten seconds and used two different slide sizes, three and five seconds. That is, we had 17 and 11 windows for each stimulus, respectively. The stimulus features were extracted from entire signals measured during a stimulus. We stored the physiological features in the datasets after performing z-standardization for each dimension.
4.2 Step1 - Extraction of Emotion Features
Proposed Approach. Of the two types of the physiological features, we used the window features as the input to our RNN. That is, an input sequence to the CML and MSL n (\(\text {\textit{data}}^C_a\) and \(\text {\textit{data}}^n_a\)) corresponds to a trial. An element of \(\text {\textit{data}}^C_a\) (i.e., \(\boldsymbol{i_{s,t}}\)) and \(\text {\textit{data}}^n_a\) (i.e., \(\boldsymbol{p^n_t}\)) correspond to t-th window of a stimulus and the physiological features extracted from the raw signals in t-th window of the stimulus, respectively. The total number of input sequences was equal to the number of trials, out of which 80% were used for training and 20% for validation. We did not use the stimulus features because \(\boldsymbol{i_{s,t}}\) corresponds to a stimulus if we do so and thus the number of input sequences, which is equal to the number of participants, was too small for training our RNN.
The hyper parameters were as follows: slide size of the sliding window \(=[3 \text {sec}, 5 \text {sec}]\), learning rate \(= [5\times 10^{-4},1\times 10^{-3}]\), dimension of hidden layers of the MSL’s MLP (i.e., \(f_{\text {MLP}}^n\)) \(= [(16,8),(32,16)]\) (from input to output layer), batch size \(= [16, 32]\), and dimension of UR, SR, and hidden state of the CML and MSL \(= [8, 16]\). For all possible combinations of the hyper parameters, we conducted training and validation for 100 epochs and extracted the CML hidden states of the validation samples when we observed the minimum validation loss. We repeated this changing training and validation samples so that we could obtain the CML hidden states for all trials. Because the prediction target is emotion after each trial, we used the last CML hidden state of each trial as the emotion features, i.e., if the last element of \(\text {\textit{data}}^C_a\) was \(\boldsymbol{i_{s,T}}\), we used \(\boldsymbol{h^C_T}\).
Baseline. Following [7, 11], we first concatenated the physiological features across modalities. This was done for both the stimulus and window features. For example, if the modality combination was A) EEG+ECG+GSR, we built 137 (\(116+10+11\)) dimension features from the stimulus features and 748 (\((29+7+8)\times 17\)) dimension features from the window features (if there are 17 windows in a stimulus) for each trial. We then reduced their dimension by performing PCA and extracted top n features in terms of their contribution ratio so that their cumulative contribution ratio is maximum below a threshold. We used these features as the emotion features. We set three different thresholds, 0.85, 0.90, and 0.95. In the following, S and W denote the emotion features extracted from the stimulus and window features, respectively. Because Miranda et al. [7] reported that recognition by unimodal features outperformed multimodal features, we also extracted S and W for each physiological modality.
4.3 Step 2 and 3 - Feature Selection and Linear Regression
After extracting the emotion features, we performed feature selection and linear regression. These were done for each of the six emotion dimensions.
We performed the feature selection because dimension of the emotion features of the baseline was large relative to the sample size (i.e., the number of trials). For fair comparison, this was done for both the baseline and our approach. We first finetuned the LASSO parameter \(\lambda \), which controls the strength of the imposed regularization based on the number of selected features. Over a set of \(\lambda \) values, we sought the value that output the most accurate prediction (i.e., minimum mean squared error between the actual and predicted emotion scores) performing five-fold cross-validation multiple times. Second, we conducted the LASSO regression again using the value of \(\lambda \) determined in the previous step and selected features for which the regression coefficients were not zero.
After the feature selection, we performed two types of linear regression. One is model fit evaluation using all samples. The other is prediction evaluation by performing five-fold cross validation.
4.4 Ablation Study
To determine the effectiveness of the key features of our RNN, we evaluated its variants without the key features, which are shown in Fig. 3. One is a single layer RNN (AB1) that takes concatenated multimodal physiological features (\(\boldsymbol{p_{s,t}}\) in the figure) as input and the other is a multilayer RNN without the MSL URs (AB2). We extracted their hidden states (the CML hidden states in AB2) as the emotion features and evaluated them in the same way as our RNN.
Similar to [4], while AB1 can extract emotion features that exclusively reflect short-term factors, it cannot distinguish between modality-specific and cross-modal factors, mixing both into the features. While the emotion features of AB2 would exclusively reflect short-term and cross-modal factors, the MSL in AB2 cannot model the moderating effect of individual physiological differences due to lack of the MSL URs.
5 Results and Discussion
Table 3 shows the results. Due to page limitations, the table shows only the Akaike Information Criterion (AIC; model fit metric; the lower the better) and the Root Mean Square Error (RMSE; prediction accuracy metric). See the supplementary material (https://osf.io/mj3nr/) for the results of other metrics. As the table shows, the regression models of our RNN (ours) outperformed the baseline models (BL, U-BL), AB1, and AB2 in most conditions not limited to specific stimulus types, emotion dimensions, or modality combinations.
Compared to the baseline models, which do not distinguish between the four types of factors at all, ours outperformed them in all conditions of both datasets with only one exception (RMSE of (b)-C in the Movie dataset). The differences are significant according to the relative likelihood (RL) that are calculated from their AICs; \(\text {RL}=\exp ((\text {AIC}(\text {ours})-\text {AIC}(\text {BL or U-BL}))/2)\), where \(\text {AIC}(M)\) denotes the AIC of regression model M. In all conditions, the RLs between ours and the best baseline models are less than 0.05 (see the supplementary material), which means that the likelihood of the best baseline models being closer to the true model than ours is less than 0.05. This indicates that the features extracted by our RNN reflect emotions to a significantly greater extent than the baseline.
The same is true between our RNN and its variants, AB1 and AB2. Out of 24 conditions, ours outperformed them in 23 conditions in the Music dataset and 20 conditions in the Movie dataset for both AIC and RMSE. The RLs between ours and the better of AB1 and AB2 were less than 0.05 in all 23 conditions in the Music dataset and 15 out of 20 conditions in the Movie dataset. These results indicate that the following key features of our RNN, which were not implemented in AB1 and AB2, significantly contributed to causing its emotion features to reflect emotion. That is, the multilayer structure for distinguishing cross-modal and modality-specific factors and the MSL URs for modeling the moderating effect of individual physiological differences.
What is notable in our RNN is that using more modalities does not necessarily make the emotion features (i.e., the CML hidden states) reflect emotion more. As shown in the table, using all three modalities (i.e., A) performed best in only three out of 12 cases (six emotion dimensions \(\times \) two datasets). This accords with the existing studies [7, 11]. For example, in [11], ECG+GSR outperformed EEG+ECG+GSR for recognizing arousal. The authors considered this would be because EEG did not reflect arousal as well as the other two modalities and would be noise for the recognition.
Although our RNN differs from them in the feature extraction, we consider this is also true for our approach. In our RNN, the CML learns latent common factors that affect all input physiological modalities. While this prevents the CML from learning modality-specific factors, it would be also possible that the CML fails to learn factors that are common to only a subset of input modalities and useful for emotion recognition but do not affect the remaining input modalities. For example, in the Music dataset, C) EEG+GSR outperformed A) EEG+ECG+GSR to recognize c) fear-relieved. We consider using ECG as input would have prevented the CML from learning factors that are common only to EEG and GSR and useful for recognizing this emotion dimension.
In light of the above, as in the existing approaches, it is necessary to compare possible modality combinations to identify the best combination in our approach. Since the best modality combinations are different between emotion dimensions and stimulus types (music and movie), the comparison of modality combinations should be done for each emotion dimension and stimulus type.
6 Conclusions, Limitations and Future Direction
In this paper, we proposed a multilayer RNN to extract features from multimodal physiological signals for emotion recognition. Using a multilayer structure, our RNN models the process by which emotion affects physiological activities across multiple modalities. This enables our RNN to extract features that are cross-modal, which is one of the characteristics of emotion but has been overlooked in existing studies. The experiments conducted on EEG, ECG, and GSR signals showed that the features extracted by our RNN reflected the participants’ emotions to a significantly greater extent than existing approaches.
One limitation is that our RNN only models unidirectional relationship between emotion and physiological activity, i.e., the former affects the latter. According to Roberts et al [9], perception of internal physiological state (known as interoception) would also affect emotion. Modeling this inverse relationship in our RNN would make the features reflect emotion more. This possibility should be explored. Another limitation is that we only examined physiological signals collected while the participants stayed still. In real-world scenarios, however, physiological signals would contain noise caused by body movements. Further studies are warranted to investigate how our RNN performs with such signals.
References
Aranha, R.V., Corrêa, C.G., Nunes, F.L.: Adapting software with affective computing: a systematic review. IEEE Trans. Affect. Comput. 12(4), 883–899 (2019)
Deng, J.J., Leung, C.H., Milani, A., Chen, L.: Emotional states associated with music: Classification, prediction of changes, and consideration in recommendation. ACM Trans. Interact. Intell. Syst. (TiiS) 5(1), 1–36 (2015)
Kim, J., André, E.: Emotion recognition based on physiological changes in music listening. IEEE Trans. Pattern Anal. Mach. Intell. 30(12), 2067–2083 (2008)
Li, C., Bao, Z., Li, L., Zhao, Z.: Exploring temporal representations by leveraging attention-based bidirectional LSTM-RNNs for multi-modal emotion recognition. Inf. Process. Manag. 57(3), 102185 (2020)
Liu, W., Zheng, W.-L., Lu, B.-L.: Emotion recognition using multimodal deep learning. In: Hirose, A., Ozawa, S., Doya, K., Ikeda, K., Lee, M., Liu, D. (eds.) ICONIP 2016. LNCS, vol. 9948, pp. 521–529. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46672-9_58
Ma, Y., Nguyen, K.L., Xing, F.Z., Cambria, E.: A survey on empathetic dialogue systems. Inf. Fusion 64, 50–70 (2020)
Miranda-Correa, J.A., Abadi, M.K., Sebe, N., Patras, I.: Amigos: a dataset for affect, personality and mood research on individuals and groups. IEEE Trans. Affect. Comput. 12(2), 479–493 (2018)
Peng, W., Hu, Y., Xing, L., Xie, Y., Sun, Y.: Do you know my emotion? emotion-aware strategy recognition towards a persuasive dialogue system. In: ECML PKDD 2022, Proceedings, Part II. pp. 724–739. Springer (2023). https://doi.org/10.1007/978-3-031-26390-3_42
Roberts, T.A., Pennebaker, J.W.: Gender differences in perceiving internal state: Toward a his-and-hers model of perceptual cue use. In: Advances in Experimental Social Psychology, vol. 27, pp. 143–175. Elsevier (1995)
Russell, J.A.: A circumplex model of affect. J. Pers. Soc. Psychol. 39(6), 1161 (1980)
Subramanian, R., Wache, J., Abadi, M.K., Vieriu, R.L., Winkler, S., Sebe, N.: Ascertain: emotion and personality recognition using commercial sensors. IEEE Trans. Affect. Comput. 9(2), 147–160 (2016)
Yin, Z., Zhao, M., Wang, Y., Yang, J., Zhang, J.: Recognition of emotions using multimodal physiological signals and an ensemble deep learning model. Comput. Methods Programs Biomed. 140, 93–110 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Ishikawa, Y. et al. (2024). Learning Cross-Modal Factors from Multimodal Physiological Signals for Emotion Recognition. In: Liu, F., Sadanandan, A.A., Pham, D.N., Mursanto, P., Lukose, D. (eds) PRICAI 2023: Trends in Artificial Intelligence. PRICAI 2023. Lecture Notes in Computer Science(), vol 14325. Springer, Singapore. https://doi.org/10.1007/978-981-99-7019-3_40
Download citation
DOI: https://doi.org/10.1007/978-981-99-7019-3_40
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-7018-6
Online ISBN: 978-981-99-7019-3
eBook Packages: Computer ScienceComputer Science (R0)