Abstract
The implementation of photosynthetic organisms has received tremendous attention in the last two decades in order to achieve the products of high industrial value at the minimal cost of environmental carbon dioxide, water and nutrients. The advancement of molecular biology tools including the availability of completely sequenced genome of a variety of photoautotrophs has made their genetic modification possible for it. In the past decade, there was an increase in the discovery of novel biosynthetic pathways in photosynthetic organisms, but there are several challenges that need to be reported and solved for developing an improved engineered strain with desired traits. Genetic engineering tools are required not only to introduce novel pathways in the photosynthetic organisms but also to modify host metabolism. For solar biofuels, most of the metabolic engineering attempts have been applied on cyanobacteria and microalgae, mainly focused in this chapter. To modify cyanobacteria for production of biofuel, the efficiency of photon conversation should be targeted which in turn may allow effective utilization of solar energy. A combinatorial approach for developments of these strains, their selection, genetic engineering, optimization of bioreactors and processing technology may pave the way for the production of biofuels that can ensure future energy security in a sustainable manner. In a larger perspective, efficient photosynthetic machinery provides a solution for an efficient and large-scale biofuel production which holds the promise of replacing harmful non-renewable fossil fuels, which may eventually delay a shift in global climate change.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
10.1 Background
Humans have adversely changed the living conditions on earth with their ever-growing demand for energy. To meet such a large sum of energy, a great amount of fossil fuel has been combusting every day that eventually increased the global carbon dioxide level and as a result earth’s climate started to react more severely. In the year 2011 alone, 82% of global primary energy usage was based on fossil fuels (EIA 2012). It is clear that the overuse of petroleum, coal and natural gas resulted in major global environmental issues (Goncalves et al. 2016). This climate change is negatively influencing the life on our planet (Kerr 2007), which, if not checked sooner, may cause a permanent threat to the ecosystem and environment on our planet. These issues pose two important concerns for mankind: (1) to find a sustainable and clean energy source soon and (2) to find an alternative source of energy that can be used if the non-renewable source will be exhausted or deplete to a critical level. These are the most challenging problems humankind is going to possibly face in the future since they are directly associated with our economic development, global stability and strategic planning (Mata et al. 2010).
To valiantly deal with these circumstances, a growing popular option among scientists, industrialists and ecologists is “biofuel”, which is produced by means of biological processes. Biofuels are carbon-neutral (minimal CO2 output) fuels which can be reproduced relatively in a short period of time and do not contribute to the rise in global atmospheric CO2 levels upon combustion (Zeman and Keith 2008). Gasoline and diesel can be replaced by relatively less harmful biofuels such as bioethanol and biodiesel, respectively (Mata et al. 2010). Biofuel in form of biodiesel is synthesized from transesterification of fats/oils, and the total content of solid particles and polycyclic aromatic hydrocarbons are significantly lower in respect to typical diesel fuel that is derived from fossils (Zajac 2008). Today the primary sources of biodiesel are vegetable oils. On earth’s surface, approximately 200 W/m2 solar energy is available per year, out of which a large proportion (29.2%) is harvested by photosynthesis (Lan and Liao 2011). This is a significant amount of renewable and sustainable energy source, which could be used for the synthesis of biofuels. With recent advancement in photosynthesis research, photosynthetic microbes such as microalgae and cyanobacteria have emerged as one of the most promising host organisms for biofuel production (Savakis and Hellingwerf 2015).
Microalgae has the impeccable ability to grow whenever they receive a sufficient amount of light and water source; however, for the production of biofuels at a commercial scale, a larger sum of biomass is needed which is not possible through natural means. Therefore, an artificial cultivation system should be required for growth in order to improve the overall process. The conditions that should be considered for this might be CO2, H2O, light, salts and temperature (Chisti 2013). According to the reports, approximately 183 tons of CO2 consumed by algae leads to the production of 100 tons of biomass (Chisti 2013). Despite the fundamental constituents such as carbon and hydrogen, the inorganic macronutrients such as nitrogen, phosphorous and iron are the important components of algal biomass (Grobbelaar 2003), which is represented by the formulae CO0.48 H1.83 N0.11 P0.01. For commercial growth of microalgae at the optimum conditions, the main cultivation systems extensively used are closed photobioreactors and open ponds.
10.2 Photosynthetic Factories for Biofuel
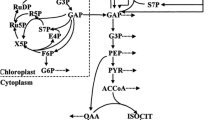
Cyanobacteria are prokaryotic microorganisms that capture their energy from photosynthesis via utilizing CO2 and light and in return produce oxygen as a by-product and certain useful products (Fig. 10.1) (Machado and Atsumi 2012). Microalgae are slightly complex as they are eukaryotes and similar to cyanobacteria having the capability of performing photosynthesis (Sydney et al. 2010). Both play a crucial role for primary production in aquatic ecosystems and participate in the utilization of global CO2 (Patel et al. 2019). On the other hand, heterotrophic bacteria utilize organic carbon sources (originally derived from photosynthesis), which increases the overall number of steps involved in metabolic reactions to generate the resultant fuel compound. In the timeline of recent metabolic engineering research, two heterotrophic model organisms Escherichia coli and Saccharomyces cerevisiae (Budding yeast) have been the main sources of microbial fuels. However, there are some drawbacks associated with them such as the cost of their organic feedstock, fuel extraction recovery cost and the cost for additional chemical modifications (Rude and Schirmer 2009). In comparison to heterotrophic microbes, plants represent a much attractive model for biofuel production as they can synthesize fuels directly from CO2 and do not need expensive feedstocks. However, the use of a food source for fuel, need for scalability, the requirement of freshwater resources and the need of land for their growth are some convincing negative aspects of plants as biofuel production hosts (Al-Haj 2016). Among these options, photosynthetic microbes (microalgae and cyanobacteria) seem to be the best alternatives (Ducat et al. 2011; Dismukes et al. 2008). As a proof, if we compare sugarcane (for ethanol), palm oil (for biodiesel), algae (for biodiesel) and cyanobacteria (for ethanol), the current yield (ha−1 year−1) is 6000 litre, 5500 litre, 58,700 litre (Maximal theoretical yield) and 50,000 litre, respectively (Anemaet et al. 2010).
Figure 10.2 represents a flow chart that shows the basic steps involved in the production of cyanobacterial and microalgal biofuels. Microalgae do not require a large land area and irrigation for growth. Since microalgae are resilient to environmental stress and cosmopolitan, several species of microalgae can live and are able to thrive in diverse and adverse environmental conditions, which gives them an advantage over other systems for the production of biofuel that needs tedious optimization efforts (Jagadevan et al. 2018). Furthermore, in past few years, several compatible genetic manipulation tools have been developed for many photosynthetic microbes. In addition, their growth conditions are continuously being optimized for similar and other algal species to broaden their applications (Nasir et al. 2019). In addition, phototrophic microbes are an efficient transducer of solar energy (3%) to biofuels as compared to higher plants (0.5%) (Melis 2009). Moreover, the photosynthetic ability of algae is approximately three times higher than terrestrial plants, by virtue of their simple cellular structure (Shimizu 1996).
One more benefit of phototrophic microbes is that they can grow within an enclosed photo-bioreactor (Angermayr et al. 2009) and are able to withstand higher CO2 content in gas streams (Zhou and Li 2010). Designing photobioreactors is a very important factor to take into consideration. There are two types of photobioreactors: open and closed. They must be chosen depending on the final use of the biomass. In the case of high-quality biomass as a cosmetic or nutraceutical use, close photobioreactors are recommended. In the case of biofertilizers or aquaculture feed, open reactors should be used. That is because open reactors are more vulnerable to environmental contamination than the closed ones. In summary, the important features that make a phototrophic microbes as a good alternative for biofuel production is their fast growth rate, simplicity of cultivation and the availability of facile genetic manipulation techniques to alter the fundamental aspects of their cellular metabolism (Surzycki et al. 2009). Yet, the most advantageous feature enabling ongoing progress in the application of algal biotechnology is the availability of robust and well-established transformation systems for these cells (Purton et al. 2013). A beneficial feature of microalgae making it an easy host for biofuel research is that many of them are considered as GRAS (“generally recognized as safe”) organisms, attributed to the purification of the bio-available products which can be substantially reduced or eliminated altogether (Matos 2017).
Among microalgae, Chlorella sorokiniana and Chlamydomonas reinhardtii have been studied as a potential candidate for biofuel production (Illman et al. 2000; Gouveia and Oliveira 2009), and genetic manipulation techniques are widely available for these species. Further, between cyanobacteria and microalgae, cyanobacteria have even faster growth rates and are easily amenable to genetic manipulation (Berla et al. 2013). Two model cyanobacterial strains, Synechocystis sp. PCC 6803 and Synechococcus sp. PCC 7002, represent the most studied system for biofuel production to date. Interestingly, Synechocystis (glucose-tolerant strains) can grow heterotrophically (in the absence of photosynthesis) (Vermaas 1998), which gives it an added advantage as a model strain to study the role of essential photosynthetic genes. The availability of complete genomic sequence has facilitated the biotechnological research on these organisms as their genomics, transcriptomics and proteomics information can be easily extracted (Ikeuchi and Tabata 2001) from web-resources (e.g. CyanoEXpress: http://cyanoexpress.sysbiolab.eu/). This huge amount of information has allowed the appropriate genetic engineering of these strains by pathway modification (Wu and Vermaas 1995), which allowed the detection and analysis of bottlenecks for improved metabolic production of bioenergy compounds such as hydrogen and ethanol (Knoop et al. 2010). A good example of metabolic engineering success in cyanobacteria is an improved strain of Synechocystis PCC 6803, known as the iSyn731 strain, that integrates all the functions and combines them to an improved metabolic capability (Berla et al. 2013). This strain contains nearly 322 unique reactions particularly in lipid biosynthetic pathways (Saha et al. 2012).
10.3 Biosynthetic Pathways for Biofuel Production: Promises and Challenges
In order to achieve the fundamental goals of biofuel production, researchers have been able to introduce novel pathways utilizing the existing molecular biology resources and knowledge. Although these efforts have rapidly increased the number of synthetic and engineered pathways in cyanobacteria, many bigger problems are still needed to be solved. For instance, less is known about how to modify the genetic makeup of the host cells/transportation system to improve the excretion of the intracellular compound from the cytoplasm during the growth of cyanobacteria (Niederholtmeyer et al. 2010). Another problem with cyanobacteria is that many biofuel-producing pathways in the cells are using anaerobic fermentative metabolism for production of reduced metabolites which involves oxygen-sensitive enzymes. Since cyanobacteria are dependent on oxygenic photosynthesis for their survival, spatial compartmentalization of the heterologous enzymes is a crucial aspect that should not be overlooked while considering metabolic engineering (Agapakis et al. 2012). Further, the toxicity caused by end product in higher titres can impair the cell growth as well. Thus, there is also a need to study the tolerance to biofuel compounds in phototrophic microbes (Anfelt et al. 2013). In light of that, it was reported that overexpression of a sigma factor (a transcription regulator protein) gene, SigB in Synechocystis PCC 6803, increased butanol tolerance, which demonstrated that genetic manipulation of transcriptional regulators can be a useful tool to improve metabolic pathways in the direction of biofuel production and consequent tolerance against biofuel-associated metabolites (Kaczmarzyk et al. 2014).
In order to produce fuel compounds in cyanobacteria, basically, the photon conversion efficiency needs to be improved, which would allow more effective utilization of sunlight flux (Badger and Price 2003; Ducat et al. 2012). Furthermore, pathways are needed to be optimized for host metabolism for increased flux. An example of this approach is the adjustment of heterologous biofuel pathways from Clostridia, by excluding oxygen-sensitive bioconversion and introducing oxygen-tolerant enzymes (Lan et al. 2013). Alternatively, the separation of oxygen-sensitive biological processes could also take place spatially, by targeting nitrogen-fixing heterocyst (Ihara et al. 2013; Savakis and Hellingwerf 2015).
In cyanobacteria, if energy storage metabolism associated with glycogen and polyhydroxybutyrate (PHB) are inactivated, the extra carbon can be directed towards the formation of products (Panda et al. 2006). Therefore, there is not only a need for the introduction of novel target pathways but also a radical modification of host metabolism (Savakis and Hellingwerf 2015; Wang et al. 2013). It has also been reported that in cyanobacteria when decarboxylation and phosphodiester bond cleavage reactions are modified, it can cause a diversion in the pathway for improved product synthesis (Savakis and Hellingwerf 2015; Oliver et al. 2013). Similarly, the overexpression of alcohol dehydrogenase and pyruvate decarboxylase leads to enhanced production of ethanol in Synechococcus elongatus PCC 7942 (Deng and Coleman 1999). In Synechococcus sp. PCC 7002, carbon flux was directed towards D-mannitol production using mannitol-1-phosphate dehydrogenase and mannitol-1-phosphatase (Jacobsen and Frigaard 2014).
Apart from scientific efforts for rewiring native metabolic pathways, there are also interesting examples available for construction of new pathways in the microorganism. One such example is the enhanced production of farnesene in E. coli by Wang et al. (2011) via the reconstruction of the yeast mevalonate pathway. Farnesene is categorized as sesquiterpenes which are important materials in the pharmaceutical industry. Further, in a recent successful attempt, the reactions and enzymes involved in chlorophyll biosynthesis have been reconstituted in E. coli expressing the full pathway accumulating chlorophyll (Chen et al. 2018). However, they are not capable of showing any photosynthetic activity as chlorophyll is just one of the main components needed for photosynthesis. Nonetheless, this report is optimistic news for cyanobacterial synthetic biology for future genetic manipulation of biofuel pathway engineering.
It is also useful to consider that while optimizing metabolic pathways, it is a necessity to maintain a balanced redox state in the engineered microbes (Mukhopadhyay et al. 2008), to avoid the accumulation of toxic by-products that will ultimately reduce the desired product formation. Furthermore, low activity of pathway enzymes can also limit the production potentials; therefore, it is an important factor that needs to be addressed while considering regulation in pathway engineering.
10.4 Tools for Manipulating the Photosynthetic Pathways
Genomic sequence information along with genetic engineering tools and methods are being developed and getting advanced rapidly for the modification of cyanobacterial and microalgal species in the last two decades. Molecular biology techniques employed to engineered cyanobacterial and microalgal species enlisted in Table 10.1. Cyanobacteria can obtain exogenous DNA by natural transformation (Porter 1986) or can intake plasmid DNA by triparental conjugation using E. coli strains (Elhai and Wolk 1988); however, natural transformation is still the primary way of mobilizing the foreign DNA into unicellular cyanobacteria (Golden et al. 1987). Conjugation method involves the transfer of foreign DNA from a donor strain to a recipient strain via cell-to-cell contact (Stucken et al. 2012). For the strains that are not naturally transformable, one common strategy is electroporation, where the cells are exposed with an electrical field leading to DNA uptake by cells due to temporary loss of semi-permeability of cell membranes (Tsong 1989). Regarding microalgae, which are a bit complicated than cyanobacteria as they require two separate transformation location, either nuclear or chloroplast (Gong et al. 2011), the most suitable and efficient method for the delivery of foreign DNA is microparticle (either tungsten or gold coated with DNA) bombardment using the biolistic gun method (Heiser 1992). This method is particularly effective for the delivery of DNA into the chloroplast as the DNA must pass through multiple membranes. Once inside the organelle, the foreign DNA integrates into the genome of chloroplast via homologous recombination. Other than this, one popular, simple and efficient method for the nuclear transformation of C. reinhardtii is agitation of cell/DNA suspension with ~0.4 mm glass beads (Kindle 1998). In this series of methods, agitation with silicon carbide whiskers (Dunahay 1993), sonication (Jarvis and Brown 1991) or co-cultivation with Agrobacterium tumefaciens (Kumar and Rajam 2007) have also proved to be suitable for nuclear transformation of microalgae.
Except for transformation, other tools such as “Bio-Brick” method are quite important for successful genetic manipulation of desired genes in organisms. Bio-brick method involves the expression of desired genes in cells by the addition of recombinant plasmids that are constructed to contain genetic “parts” such as promoters, ribosome-binding sites, genes, terminators and other regulatory elements in microorganisms (Berla et al. 2013; Huang and Lindblad 2013). This modern tool is now a valuable technology to design a programmed cell with desired traits, where the assembly of the biological parts occurs in a hierarchal manner (Andrianantoandro et al. 2006).
Further, gene modification is also a key factor for successful development of engineered algal strains. The most common approach in algal synthetic biology is cis gene modification (through chromosomal editing) (Berla et al. 2013), although trans gene modification (through foreign plasmid addition) is also occasionally used. If we look at further improvement, modernization of DNA synthesis technologies has allowed the design of coding sequences and the use of codon optimized trans genes to improve the heterologous protein expression in cyanobacterial cells (Gustafsson et al. 2004).
The functional genomics toolbox of cyanobacteria consists of two basic strategies to abolish gene function: (a) gene knockout and (b) gene knockdown. Gene knockout strategy is a tool of reverse genetics, which is used to determine the functional role of the target gene by knocking out that gene either by gene deletion or by gene inactivation leaving all other genes unaffected. Gene knockout through insertional inactivation and in-frame deletion has been routinely employed to block the function of the target genes in cyanobacteria, but in some cases, the mutants are not viable and/or cannot segregate completely due to essential life function encoded or controlled by them. Therefore, to study such essential genes, instead of making the cells completely devoid of their gene product, their transcript levels are targeted by antisense RNA (asRNA) for their down-regulation (Blanco et al. 2011; Lin et al. 2013). In contrast, Antisense RNA approach is not only promising for the study of essential genes, but it can also provide an advantage to tune-up or tune-down the expression of genes when controlled under a regulatable promoter (e.g. copper-responsive PpetE or nickel inducible PprsB). Promoters are important constituent for controlled gene expression and crucial for asRNA-mediated approach in a cyanobacterium. Native promoters from the genes related to photosynthesis (PrbcL, Pcmp, Psbt, PpsaA, PpsaD, PpsbA1, PpsbA2 and Pcpc) have been investigated for their potential in gene expression in cyanobacteria. Many studies have shown that higher concentration of antisense RNA is important for efficient suppression of gene expression, which could easily be achieved by using strong promoters (Robert et al. 1990; Cannon et al. 1990; Van der Meer et al. 1992). Besides native promoters, heterologous promoters can also be utilized in cyanobacteria. Notably, some strong E. coli promoters (e.g. Ptac/Ptrc) have shown to express the target genes in higher levels in cyanobacteria, whereas others (e.g. Plac, Ptet, and λ PR) display little to undetectable activity in Synechocystis (Wang et al. 2012). To find out the role of a unique sigma factor, SigJ in Anabaena PCC 7120, Srivastava et al. (2017) employed antisense RNA-mediated knockdown technique, achieving an approximately threefold down-regulation of sigJ mRNA in the knockdown mutant eventually increasing its carotenoid (myxoxanthophylls) content and improved photo-protection, supporting the feasibility of this technique in both basic and applied research.
Further, among all the tools used in metabolic engineering, the choice of plasmid vectors is elementary while considering transformation in cyanobacteria. They can be categorized into two groups based on their mode of replication inside the host (Koksharova and Wolk 2002): (1) integrative plasmids, which cannot replicate independently and would be lost eventually in further cell divisions (Wang et al. 2012), and (2) replicative plasmids, which are capable of maintaining themselves stably in the cells and are able to replicate in both cyanobacteria and E. coli (Deng and Coleman 1999, Koksharova and Wolk 2002; Wang et al. 2012). A number of replicative plasmids harbouring diverse selectable markers and RSF1010-derived plasmids have been generated for cyanobacteria (Berla et al. 2013; Golden et al. 1987; Wang et al. 2012). However, to express the biofuel-related genes, only a few expression plasmids are available for cyanobacteria. Generally, the native plasmids are stable inside the cells and hence a good candidate to modify for genetic engineering applications. Several native high copy number plasmids were already reported in cyanobacteria, which could be good candidates for the construction of cloning/expression vectors as they provide the possibility of isolation, restriction and ligation in a similar way to E. coli (Miyak et al. 1999).
10.5 CRISPR/Cas Gene Editing Tool as an Emerging Approach for Engineering Biofuel Production
Many bacteria and archaea have an adaptive immune system called CRISPR-Cas9 system (Clustered regularly interspaced short palindromic repeats/CRISPR associated proteins), which has been widely employed for gene inactivation and expression modification in several organisms (Peters et al. 2015; Hsu et al. 2014). The main advantage of this technique is multiplexing and marker less modification of genes. This allows a faster construction of strains with more than one modification in a relatively shorter time (Behler et al. 2018). In principle, the prokaryote intakes a piece of invading virus/plasmid DNA (the protospacer) and incorporates in a form of array in the genome. Thus, with each intake, a new piece is incorporated in the array and the host immunity further grows (Horvath and Barrangou 2010). Adjacent to the CRISPR array, often Cas9 protein is present that eventually carries out the immune response (Horvath and Barrangou 2010; Sander and Joung 2014). The main function of the Cas9 protein is to cut DNA and make a double-strand break. Scientists have been using it for gene editing using the CRISPR/Cas9 system by taking advantage of the host’s own immunity and DNA repair strategy. Besides that this system is also used for modifying the gene expression (La Russa and Qi 2015). Further, this technique was also further improvised to down-regulate the genes by mutating the catalytic active site of Cas9 making Cas9 dead protein (dCas9) which stably binds and interferes with transcription of the target genes but does not cleave (Qi et al. 2013; Zhang et al. 2017). Gene editing in Synechococcus elongatus PCC 7942 and Synechococcus elongatus UTEX 2973 has been already carried out using Streptococcus pyogenes-derived CRISPR-Cas9 system; however, the high expression of Cas9 effector proved to be toxic for the cells (Li et al. 2016; Wendt et al. 2016). Further, this technique has been tried on Synechocystis, Anabaena PCC 7120, Phaeodactylum tricornutum, Nannochloropsis spp. and Chlamydomonas reinhardtii (Ungerer and Pakrasi 2016; Daboussi et al. 2014; Weyman et al. 2015; Wang et al. 2016; Huang et al. 2016; Baek et al. 2016), which gave promising results. Therefore, this genome editing tool is providing a new insight for microalgae-based biofuel applications.
10.6 Future Prospects
A photoautotrophic microorganism is well-equipped with sophisticated biosynthetic pathways and thriving on least of nutrient supplements with a relatively faster growth rate. They are gaining much attention for their ability to produce biotechnologically important products in an environmentally sustainable manner. Until now, it is not considered a very cheap technology which must be improved to have a competitive price. Thus, recycling of nutrients and water by using wastewater as an input should be taken into consideration to make this technology competitive into the market. However, higher productivity and genetic makeup for production of complex molecules make them a promising candidate for future biofuel production. Therefore, it is seemingly desired to design cyanobacterial and microalgal cell factories utilizing available synthetic biology resources and metabolic engineering approaches. However, it is imperative to gain an in-depth knowledge of metabolic regulation and mechanism of the biosynthetic pathway to improve the strain for desired traits. A collective effort of multi-omics approach is needed to improve the production and tolerance (in the host) of biofuel-related product.
References
Agapakis CM, Boyle PM, Silver PA (2012) Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat Chem Biol 8(6):527
Al-Haj L, Lui Y, Abed R, Gomaa M, Purton S (2016) Cyanobacteria as chassis for industrial biotechnology: progress and prospects. Life 6(4):42
Andrianantoandro E, Basu S, Karig DK, Weiss R (2006) Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol 2(1):2006-0028
Anemaet IG, Bekker M, Hellingwerf KJ (2010) Algal photosynthesis as the primary driver for a sustainable development in energy, feed, and food production. Mar Biotechnol 12(6):619–629
Anfelt J, Hallström B, Nielsen J, Uhlén M, Hudson EP (2013) Using transcriptomics to improve butanol tolerance of Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 79(23):7419–7427
Angermayr SA, Hellingwerf KJ, Lindblad P, de Mattos MJT (2009) Energy biotechnology with cyanobacteria. Curr Opin Biotechnol 20(3):257–263
Badger MR, Price GD (2003) CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot 54(383):609–622
Baek K, Kim DH, Jeong J, Sim SJ, Melis A, Kim JS, Jin E, Bae S (2016) DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci Rep 6:30620
Behler J, Vijay D, Hess WR, Akhtar MK (2018) CRISPR-based technologies for metabolic engineering in cyanobacteria. Trends Biotechnol 36(10):996–1010
Berla BM, Saha R, Immethun CM, Maranas CD, Moon TS, Pakrasi H (2013) Synthetic biology of cyanobacteria: unique challenges and opportunities. Front Microbiol 4:246
Blanco NE, Ceccoli RD, Segretin ME, Poli HO, Voss I, Melzer M, Bravo-Almonacid FF, Scheibe R, Hajirezaei MR, Carrillo N (2011) Cyanobacterial flavodoxin complements ferredoxin deficiency in knocked-down transgenic tobacco plants. Plant J 65(6):922–935
Cannon M, Platz J, O’Leary M, Sookdeo C, Cannon F (1990) Organ-specific modulation of gene expression in transgenic plants using antisene RNA. Plant Mol Biol 15(1):39–47
Chen GE, Canniffe DP, Barnett SF, Hollingshead S, Brindley AA, Vasilev C, Bryant DA, Hunter CN (2018) Complete enzyme set for chlorophyll biosynthesis in Escherichia coli. Sci Adv 4(1):eaaq1407
Chisti Y (2013) Constraints to commercialization of algal fuels. J Biotechnol 167(3):201–214
Chung YS, Lee JW, Chung CH (2017) Molecular challenges in microalgae towards cost-effective production of quality biodiesel. Renew Sust Energ Rev 74:139–144
Daboussi F, Leduc S, Maréchal A, Dubois G, Guyot V, Perez-Michaut C, Amato A, Falciatore A, Juillerat A, Beurdeley M, Voytas DF (2014) Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat Commun 5:3831
Deng MD, Coleman JR (1999) Ethanol synthesis by genetic engineering in cyanobacteria. Appl Environ Microbiol 65(2):523–528
Dismukes GC, Carrieri D, Bennette N, Ananyev GM, Posewitz MC (2008) Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr Opin Biotechnol 19(3):235–240
Ducat DC, Way JC, Silver PA (2011) Engineering cyanobacteria to generate high-value products. Trends Biotechnol 29(2):95–103
Ducat DC, Avelar-Rivas JA, Way JC, Silver PA (2012) Rerouting carbon flux to enhance photosynthetic productivity. Appl Environ Microbiol 78(8):2660–2668
Dunahay TG (1993) Transformation of Chlamydomonas reinhardtii with silicon carbide whiskers. BioTechniques 15(3):452–455
Elhai J, Wolk CP (1988) [83] Conjugal transfer of DNA to cyanobacteria. In: Methods enzymol, vol 167. Academic, New York, pp 747–754
Energy Information Administration (US) (2012) Annual energy review 2011. Government Printing Office, Washington, DC
Golden SS, Brusslan J, Haselkorn R (1987) [12] Genetic engineering of the cyanobacterial chromosome. In: Methods enzymol, vol 153. Academic, New York, pp 215–231
Goncalves EC, Wilkie AC, Kirst M, Rathinasabapathi B (2016) Metabolic regulation of triacylglycerol accumulation in the green algae: identification of potential targets for engineering to improve oil yield. Plant Biotechnol J 14(8):1649–1660
Gong Y, Hu H, Gao Y, Xu X, Gao H (2011) Microalgae as platforms for production of recombinant proteins and valuable compounds: progress and prospects. J Ind Microbiol Biotechnol 38(12):1879–1890
Gouveia L, Oliveira AC (2009) Microalgae as a raw material for biofuels production. J Ind Microbiol Biotechnol 36(2):269–274
Grobbelaar JU (2003) Algal nutrition–mineral nutrition. In: Handbook of microalgal culture: biotechnology and applied phycology. Blackwell Science, Oxford, pp 95–115
Gustafsson C, Govindarajan S, Minshull J (2004) Codon bias and heterologous protein expression. Trends Biotechnol 22(7):346–353
Heiser W (1992) Optimization of biolistic transformation using the helium-driven PDS-1000/He system. Bio-Rad Bull 1688
Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327(5962):167–170
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157(6):1262–1278
Huang HH, Lindblad P (2013) Wide-dynamic-range promoters engineered for cyanobacteria. J Biol Eng 7(1):10
Huang CH, Shen CR, Li H, Sung LY, Wu MY, Hu YC (2016) CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942. Microb Cell Factories 15(1):196
Ihara M, Kawano Y, Urano M, Okabe A (2013) Light driven CO2 fixation by using cyanobacterial photosystem I and NADPH-dependent formate dehydrogenase. PLoS One 8(8):e71581
Ikeuchi M, Tabata S (2001) Synechocystis sp. PCC 6803—a useful tool in the study of the genetics of cyanobacteria. Photosynth Res 70(1):73–83
Illman AM, Scragg AH, Shales SW (2000) Increase in chlorella strains calorific values when grown in low nitrogen medium. Enzym Microb Technol 27(8):631–635
Jacobsen JH, Frigaard NU (2014) Engineering of photosynthetic mannitol biosynthesis from CO2 in a cyanobacterium. Metab Eng 21:60–70
Jagadevan S, Banerjee A, Banerjee C, Guria C, Tiwari R, Baweja M, Shukla P (2018) Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production. Biotechnol Biofuels 11(1):185
Jarvis EE, Brown LM (1991) Transient expression of firefly luciferase in protoplasts of the green alga Chlorella ellipsoidea. Curr Genet 19(4):317–321
Kaczmarzyk D, Anfelt J, Särnegrim A, Hudson EP (2014) Overexpression of sigma factor SigB improves temperature and butanol tolerance of Synechocystis sp. PCC6803. J Biotechnol 182:54–60
Kerr RA (2007) Global warming is changing the world. Science 316(5822):188–190
Kilian O, Benemann CS, Niyogi KK, Vick B (2011) High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc Natl Acad Sci U S A 108(52):21265–21269
Kindle KL (1998) [3] High-frequency nuclear transformation of Chlamydomonas reinhardtii. In: Methods enzymol, vol 297. Academic, New York, pp 27–38
Knoop H, Zilliges Y, Lockau W, Steuer R (2010) The metabolic network of Synechocystis sp. PCC 6803: systemic properties of autotrophic growth. Plant Physiol 154(1):410–422
Koksharova OA, Wolk CP (2002) Novel DNA-binding proteins in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 184(14):3931–3940
Kumar SV, Rajam MV (2007) Induction of agrobacterium tumefaciens vir genes by the green alga, Chlamydomonas reinhardtii. Curr Sci 92:1727–1729
La Russa MF, Qi LS (2015) The new state of the art: Cas9 for gene activation and repression. Mol Cell Biol 35(22):3800–3809
Lan EI, Liao JC (2011) Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab Eng 13(4):353–363
Lan EI, Ro SY, Liao JC (2013) Oxygen-tolerant coenzyme A-acylating aldehyde dehydrogenase facilitates efficient photosynthetic n-butanol biosynthesis in cyanobacteria. Energy Environ Sci 6(9):2672–2681
Li Y, Han D, Hu G, Dauvillee D, Sommerfeld M, Ball S, Hu Q (2010) Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab Eng 12(4):387–391
Li H, Shen CR, Huang CH, Sung LY, Wu MY, Hu YC (2016) CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab Eng 38:293–302
Lin CH, Tsai ZTY, Wang D (2013) Role of antisense RNAs in evolution of yeast regulatory complexity. Genomics 102(5–6):484–490
Machado IM, Atsumi S (2012) Cyanobacterial biofuel production. J Biotechnol 162(1):50–56
Mata TM, Martins AA, Caetano NS (2010) Microalgae for biodiesel production and other applications: a review. Renew Sust Energ Rev 14(1):217–232
Matos ÂP (2017) The impact of microalgae in food science and technology. J Am Oil Chem Soc 94(11):1333–1350
Melis A (2009) Solar energy conversion efficiencies in photosynthesis: minimizing the chlorophyll antennae to maximize efficiency. Plant Sci 177(4):272–280
Miyak M, Nagai H, Shirai M, Kurane R, Asada Y (1999) A high-copy-number plasmid capable of replication in thermophilic cyanobacteria. In: Twentieth symposium on biotechnology for fuels and chemicals. Humana Press, Totowa, NJ, pp 267–275
Mukhopadhyay A, Redding AM, Rutherford BJ, Keasling JD (2008) Importance of systems biology in engineering microbes for biofuel production. Curr Opin Biotechnol 19(3):228–234
Nasir BM, Nor-Anis N, Islam AKMA, Anuar N, Yaakob Z (2019) Genetic improvement and challenges for cultivation of microalgae for biodiesel: a review. Mini Rev Org Chem 16(3):277–289
Niederholtmeyer H, Wolfstädter BT, Savage DF, Silver PA, Way JC (2010) Engineering cyanobacteria to synthesize and export hydrophilic products. Appl Environ Microbiol 76(11):3462–3466
Oliver JW, Machado IM, Yoneda H, Atsumi S (2013) Cyanobacterial conversion of carbon dioxide to 2, 3-butanediol. Proc Natl Acad Sci U S A 110(4):1249–1254
Panda B, Jain P, Sharma L, Mallick N (2006) Optimization of cultural and nutritional conditions for accumulation of poly-β-hydroxybutyrate in Synechocystis sp. PCC 6803. Bioresour Technol 97(11):1296–1301
Patel A, Matsakas L, Rova U, Christakopoulos P (2019) A perspective on biotechnological applications of thermophilic microalgae and cyanobacteria. Bioresour Technol 278:424–434
Peters JM, Silvis MR, Zhao D, Hawkins JS, Gross CA, Qi LS (2015) Bacterial CRISPR: accomplishments and prospects. Curr Opin Microbiol 27:121–126
Porter RD (1986) Transformation in cyanobacteria. Crit Rev Microbiol 13(2):111–132
Purton S, Szaub JB, Wannathong T, Young R, Economou CK (2013) Genetic engineering of algal chloroplasts: progress and prospects. Russ J Plant Physiol 60(4):491–499
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152(5):1173–1183
Ramazanov A, Ramazanov Z (2006) Isolation and characterization of a starchless mutant of Chlorella pyrenoidosa STL-PI with a high growth rate, and high protein and polyunsaturated fatty acid content. Phycol Res 54(4):255–259
Robert LS, Donaldson PA, Ladaique C, Altosaar I, Arnison P, Fabijanski SF (1990) Antisense RNA inhibition of β-glucuronidase gene expression in transgenic tobacco can be transiently overcome using a heat-inducible β-glucuronidase gene construct. Bio/Technology 8(5):459
Rude MA, Schirmer A (2009) New microbial fuels: a biotech perspective. Curr Opin Microbiol 12(3):274–281
Saha R, Verseput AT, Berla BM, Mueller TJ, Pakrasi HB, Maranas CD (2012) Reconstruction and comparison of the metabolic potential of cyanobacteria Cyanothece sp. ATCC 51142 and Synechocystis sp. PCC 6803. PLoS One 7(10):e48285
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32(4):347
Sanz-Luque E, Ocaña-Calahorro F, Galván A, Fernández E, de Montaigu A (2016) Characterization of a mutant deficient for ammonium and nitric oxide signalling in the model system Chlamydomonas reinhardtii. PLoS One 11(5):e0155128
Savakis P, Hellingwerf KJ (2015) Engineering cyanobacteria for direct biofuel production from CO2. Curr Opin Biotechnol 33:8–14
Shimizu Y (1996) Microalgal metabolites: a new perspective. Annu Rev Microbiol 50(1):431–465
Sizova I, Greiner A, Awasthi M, Kateriya S, Hegemann P (2013) Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J 73(5):873–882
Srivastava A, Brilisauer K, Rai AK, Ballal A, Forchhammer K, Tripathi AK (2017) Down-regulation of the alternative sigma factor SigJ confers a photoprotective phenotype to Anabaena PCC 7120. Plant Cell Physiol 58(2):287–297
Stucken K, Ilhan RM, Dagan T, Martin WF (2012) Transformation and conjugal transfer of foreign genes into the filamentous multicellular cyanobacteria (subsection V) Fischerella and Chlorogloeopsis. Curr Microbiol 65(5):552–560
Surzycki R, Greenham K, Kitayama K, Dibal F, Wagner R, Rochaix JD, Ajam T, Surzycki S (2009) Factors effecting expression of vaccines in microalgae. Biologicals 37(3):133–138
Sydney EB, Sturm W, de Carvalho JC, Thomaz-Soccol V, Larroche C, Pandey A, Soccol CR (2010) Potential carbon dioxide fixation by industrially important microalgae. Bioresour Technol 101(15):5892–5896
Tsong TY (1989) Electroporation of cell membranes. In: Electroporation and electrofusion in cell biology. Springer, Boston, pp 149–163
Ungerer J, Pakrasi HB (2016) Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci Rep 6:39681
Van Der Meer IM, Stam ME, van Tunen AJ, Mol JN, Stuitje AR (1992) Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4(3):253–262
Vermaas WF (1998) [20] Gene modifications and mutation mapping to study the function of photosystem II. In: Methods enzymol, vol 297. Academic Press, New York, pp 293–310
Wang C, Yoon SH, Jang HJ, Chung YR, Kim JY, Choi ES, Kim SW (2011) Metabolic engineering of Escherichia coli for α-farnesene production. Metab Eng 13(6):648–655
Wang B, Wang J, Zhang W, Meldrum DR (2012) Application of synthetic biology in cyanobacteria and algae. Front Microbiol 3:344
Wang B, Pugh S, Nielsen DR, Zhang W, Meldrum DR (2013) Engineering cyanobacteria for photosynthetic production of 3-hydroxybutyrate directly from CO2. Metab Eng 16:68–77
Wang Q, Lu Y, Xin Y, Wei L, Huang S, Xu J (2016) Genome editing of model oleaginous microalgae Nannochloropsis spp. by CRISPR/Cas9. Plant J 88(6):1071–1081
Wannathong T, Waterhouse JC, Young RE, Economou CK, Purton S (2016) New tools for chloroplast genetic engineering allow the synthesis of human growth hormone in the green alga Chlamydomonas reinhardtii. Appl Microbiol Biotechnol 100(12):5467–5477
Wendt KE, Ungerer J, Cobb RE, Zhao H, Pakrasi HB (2016) CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb Cell Factories 15(1):115
Weyman PD, Beeri K, Lefebvre SC, Rivera J, McCarthy JK, Heuberger AL, Peers G, Allen AE, Dupont CL (2015) Inactivation of Phaeodactylum tricornutum urease gene using transcription activator-like effector nuclease-based targeted mutagenesis. Plant Biotechnol J 13(4):460–470
Wu Q, Vermaas WF (1995) Light-dependent chlorophyll a biosynthesis upon chlL deletion in wild-type and photosystem I-less strains of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 29(5):933–945
Xu W, Chen H, He CL, Wang Q (2014) Deep sequencing-based identification of small regulatory RNAs in Synechocystis sp. PCC 6803. PLoS One 9(3):e92711
Yang J, Pan Y, Bowler C, Zhang L, Hu H (2016) Knockdown of phosphoenolpyruvate carboxykinase increases carbon flux to lipid synthesis in Phaeodactylum tricornutum. Algal Res 15:50–58
Zajac G (2008) Influence of FAME addition to diesel fuel on exhaust fumes opacity of diesel engine. Int Agrophys 22:179–183
Zeman FS, Keith DW (2008) Carbon neutral hydrocarbons. Philos Trans A Math Phys Eng Sci 366(1882):3901–3918
Zhang X, Wang J, Cheng Q, Zheng X, Zhao G, Wang J (2017) Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov 3:17018
Zhou J, Li Y (2010) Engineering cyanobacteria for fuels and chemicals production. Protein Cell 1(3):207–210
Acknowledgement
AS is supported by Ministry of Education, Youth and Sports of the Czech Republic (Project: LO1416).
Conflict of Interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Srivastava, A., Villalobos, M.B., Singh, R.K. (2020). Engineering Photosynthetic Microbes for Sustainable Bioenergy Production. In: Singh, P., Singh, R., Srivastava, V. (eds) Contemporary Environmental Issues and Challenges in Era of Climate Change. Springer, Singapore. https://doi.org/10.1007/978-981-32-9595-7_10
Download citation
DOI: https://doi.org/10.1007/978-981-32-9595-7_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-32-9594-0
Online ISBN: 978-981-32-9595-7
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)