Abstract
Computer-Aided Analysis of Blood Smear Images helps to identify several cell features which cannot be analyzed with the existing manual techniques. For this purpose, segmentation of required cell component is very important. The motive of this work is to segment the Cell Periphery, which holds the cytoplasm, from the Blood Smear Images. Primarily, these images are enhanced to increase the observability of various cell components. The enhancement is done using Dark Contrast Algorithm (DCA). This enhanced image is further segmented using K-Medoid Clustering, a technique based on spatial clustering. The number of clusters obtained as output are predefined. This technique clusters the data with the help of Similarity Index, to distribute it according to their similarities or dissimilarities by updating the medoids. The image hence obtained gives us the segmented Cell Periphery. The assessment of this proposed approach is done using parameters like Second Derivative like Measure of Enhancement (SDME) and Measure of Enhancement (EME) for enhancement and Dice Coefficient (DC) for segmentation.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Cell Periphery is important in identifying several contrasting features of leukocytes (commonly known as White Blood Cells). They help us in identifying the type of cell whether blast or not and in also identifying various types of WBCs. The conventional methods to identify type of cell are through manual observations which is both tedious and is subjected to inaccuracy [1]. Now, to rectify these problems, Computer-Aided Diagnosis is used for analysis of Blood Smear Images. These images are obtained by mounting digital camera on microscope, either optical or electron, used for observing blood slides. These Blood Smear Images are processed to extract cells’ characteristics to categorize them into their types [2]. Harun et al. [3] proposed a technique of nucleus segmentation from Blood Smear Images with a median filter and Dark Contrast Algorithm (DCA). Segmentation is done through thresholding and clustering. Classification of WBCs done by Dasariraju et al. [4] used Multi-Otsu Thresholding for segmentation of nucleus and cell boundary. The preprocessing step in this technique was the conversion from RGB to lab scale. The accuracy of this whole proposed work was moderate to high. A technique of segmentation through clustering was proposed by Su et al. [5] for bone marrow smear images. The segmentation here was done in two stages: firstly, K-Means clustering algorithm was used to divide the image into two clusters further and Hidden Markov Random Field (HMRF) was used for final segmentation. This method of segmentation was able to label six types of WBCs. A similar clustering approach was proposed by Acharya et al. [6] for segmentation of Blood Smear Images with the help of K-Medoid Clustering. This method is observed to be much effective than K-Means clustering. Another method of segmentation by morphological filters [7] to extract nucleus from Blood Smear Images is quite effective, but was not able to segment cell boundary. Eckardt et al. [8] proposed a deep learning approach to detect NPM1 mutation with the help of bone marrow smears. It was done using Faster Region-based Convolutional Neural Net (FRCNN) to improvise cell borders and an image annotator tool. These approaches conclude that enhancement preceded by segmentation gives best results in order to identify cell components. Experimentally, the accuracy of K-Medoid when compared to K-Means for segmentation is quite high. K-Means is technique which works on iterative clustering, while K-Medoid works on spatial clustering. The approach opted for this paper uses DCA for contrast enhancement as it works on darker regions of the image and K-Medoid Clustering for segmentation of the Cell Periphery. The organization of the paper in the following parts is as follows: Sect. 2 contains the proposed approach of enhancement and segmentation to extract Cell Periphery, Sect. 3 contains experimental results and analysis and Sect. 4 consists of conclusion.
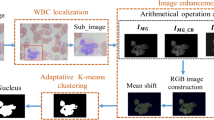
2 Proposed Approach of Enhancement and Segmentation to Extract Cell Periphery
The proposed approach to segment Cell Periphery from Blood Smear Images follows enhancement preceded by segmentation. To perform enhancement, firstly, the RGB Blood Smear Image is converted to grayscale image so that there is no error to define Cell Periphery. The enhancement done using Dark Contrast Algorithm (DCA) highlights the Cell Periphery further making it easy to segment. The segmentation done by K-Medoid Clustering converts this enhanced image into clusters of similar data.
2.1 Enhancement Using Dark Contrast Algorithm (DCA)
The enhancement approach used in this experimental work is through DCA which is a contrast stretching technique and stretches the contrast of darker regions. The working of DCA is dependent on its parameter which is Threshold Value (TH) and Stretching Factor (NTH). For DCA, the value of TH is less than that of NTH. The control function on which DCA works is given in Eq. (1) [3].
Here, p (x, y) and O (x, y) represent the input and output pixels, respectively. The maximum and minimum intensities of input image are represented by max_p and min_p, respectively. Equation (1) shows both the cases of DCA, that is, stretching and compression. The assessment of the enhanced image is done using Image Quality Assessment (IQA) parameters like Second Derivative like Measure of Enhancement (SDME) and Measure of Enhancement (EME) [9, 10].
2.2 Segmentation of Cell Periphery Using K-Medoid Clustering
Further, to segment the Cell Periphery from the enhanced image, K-Medoid Clustering is used. This is a type of spatial clustering where the clusters are divided according to the similarity in the data. The difference between K-Means [5] and K-Medoid, both being clustering algorithms, is K-Medoid being more sensitive to dissimilarities. K-Medoid Clustering takes medoids into consideration to cluster the data [6]. Figure 1 shows the graphical representation of data clustered by K-Medoid Clustering into two clusters.
In K-Medoid Clustering, the clusters are defined as inputs and medoid is first chosen as any random value. Medoid refers to a point in the cluster whose dissimilarities are minimum. These medoids are further updated according to the dissimilarity or similarity of data. The similarity measure on which this technique works is given by Eq. (2) [6].
Here, k is the number of clusters, Ci is the medoid, xj is the object and Si is the cluster. With the help of this similarity, the medoid is updated and is further compared with the medoid which is predefined. No change in medoid indicates similar data, while change in medoid indicates the data of other category [11]. In the proposed approach, the number of clusters are set to be two since the contrast-enhanced image is a gray scale. This clustering helps us to distinguish Cell Periphery from nucleus since both have different levels of intensity. Segmentation of Cell Periphery using K-Medoid gives us information regarding the disorientations of the same which helps us to identify blast, malignant cells. The segmented image of Cell Membrane hence is evaluated using Dice Coefficient (DC) [12]. The complete algorithm for the proposed approach of enhancement and segmentation to extract the Cell Periphery of Blood Smear Images is given in Algorithm I.

3 Experimental Results
3.1 Dataset
The database of Blood Smear aspirates for this experiment is obtained from the online repository of Cancer Imaging Archive [4], for single-cell images, and ASH [6], for multi-cell images. These images are acquired by a digital camera with dimensions 400 × 400 and 720 × 960, respectively. Two images one from each dataset were taken to analyze the results of the proposed approach to segment Cell Periphery. As initial steps of preprocessing, these images were converted from their original color model to gray scale for better identification in segmentation.
3.2 Simulation Results and Analysis
Two images chosen from the dataset were used to observe the proposed approach of enhancement using DCA and segmentation of Cell Periphery using K-Medoid Clustering Algorithm [3, 6]. The effectiveness of the DCA-enhanced image was compared with the original image on the basis of parameters like SDME and EME [9, 10], while the assessment of segmented image was done using DC by comparing it with the ground truth. The results from both the images are shown in Fig. 2.
The visuality of images (c) and (g) is better than that of (b) and (f) as shown in above Fig. 2. The cell components and periphery are much visible after enhancement. Further, the segmented image (d) and (h), when observed visually, shows us the successfully segmented Cell Periphery. These enhanced and segmented images are also evaluated qualitatively. The IQA metrics of enhanced images are given in Table 1.
On observing these values, it shows a clear increase in both SDME and EME by enhancement using DCA, thus increasing its metrics qualitatively. This indicates that the enhanced image has better contrast and brightness. Similarly, the assessment of segmented image of Cell Periphery is done using Dice Coefficient (DC) [12] as shown in Table 2.
The value of DC nears to 1 in the both types of images, which clearly indicates that the Cell Periphery is segmented successfully when compared with the ground truth results. So, visually and qualitatively, the performance of segmentation is evaluated and is successful. Therefore, this approach of enhancing the Blood Smear Images for segmenting Cell Periphery is successful in giving the desired results.
4 Conclusion
This paper presents an approach to segment the cell membrane/Cell Periphery from Blood Smear Images. Primarily, DCA was used for enhancement of these images to increase visibility of Cell Membrane. This was successful since the Cell Periphery can be seen in the enhanced image. This image was evaluated using SDME and EME, which shows that the enhancement was successful. Now, for segmenting our required ROI, K-Medoid Clustering was used. Here, the number of clusters were initialized to 2. The clusters hence obtained from segmentation contain the Cell Periphery. The results of segmentation were appraised using DC, which clearly show successful segmentation of Cell Periphery. This segmented image can be further used to calculate features like Nucleus-to-Cytoplasm Ratio.
References
Jagadev P, Virani HG (2018) Detection of Leukemia and its types using image processing and machine learning. In: Proceeding of international conference on trends in electronics and informatics (ICTEI 2017). IEEE, Tirunelveli, India, pp 522–526
Rezatofighi SH, Zadeh HS (2011) Automatic recognition of five types of white blood cells in peripheral blood. Comput Med Imaging Graph 35(4):333–343
Harun NH, Bakar JA, Hambali HA, Khair NM, Mashor MY, Hassan R (2018) Fusion noise—removal technique with modified algorithm for robust segmentation of acute leukemia cell images. Int J Adv Intell Inf 4(3):202–211
Dasariraju S, Huo M, McCalla S (2020) Detection and classification of immature leukocytes for diagnosis of acute myeloid leukemia using random forest algorithm. Bioengineering 7(4):120–131
Su J, Liu S, Song J (2017) A segmentation method based on HMRF for the aided diagnosis of acute myeloid leukemia. Comput Methods Prog Biomed 152(7):115–123
Acharya V, Ravi V, Pham TD, Chakraborty C (2021) Peripheral blood smear analysis using automated computer-aided diagnosis system to identify acute myeloid Leukemia. IEEE Trans Eng Manage 1–14
Hegde RB, Prasad K, Hebbar H, Singh BMK (2018) Development of a robust algorithm for detection of nuclei and classification of white blood cells in peripheral blood smear images. J Med Syst 42:110
Eckardt JN, Middeke JM et al (2022) Deep learning detects acute myeloid leukemia and predicts NPM1 mutation status from bone marrow smears. Leukemia 36:111–118
Trivedi M, Jaiswal A, Bhateja V (2013) A No-reference image quality index for contrast and sharpness measurement. In: 3rd IEEE international advance computing conference (IACC). IEEE, India, pp 1234–1239
Prajapati P, Narmawala Z, Darji NP, Moorthi SM, Ramakrishnan R (2015) Evaluation of perceptual contrast and sharpness measures for meteorological satellite images. In: Soni AK, Lobiyal DK (eds) 3rd International conference on recent trends in computing (ICRTC), procedia computer science, vol 57. Springer, India, pp 17–24
Halder A, Dasgupta A, Ghosh S (2022) Image segmentation using rough—Fuzzy K—medoid algorithm. In: Proceeding of international conference on communications, devices and intelligent systems. IEEE, India, pp 105–108
Kumar SN, Lenin Fred A, Ajay Kumar H, Sebastin Varghese P (2018) Performance metric evaluation of segmentation algorithms for gold standard medical images. In: Sa P, Bakshi S, Hatzilygeroudis I, Sahoo M (eds) Recent findings in intelligent computing techniques. Advances in intelligent systems and computing, vol 709. Springer. Singapore (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Verma, S., Bhateja, V., Singh, S., Gupta, S., Dogra, A., Nhu, N.G. (2023). Segmentation of Cell Periphery from Blood Smear Images Using Dark Contrast Algorithm and K-Medoid Clustering. In: Chakravarthy, V., Bhateja, V., Flores Fuentes, W., Anguera, J., Vasavi, K.P. (eds) Advances in Signal Processing, Embedded Systems and IoT . Lecture Notes in Electrical Engineering, vol 992. Springer, Singapore. https://doi.org/10.1007/978-981-19-8865-3_24
Download citation
DOI: https://doi.org/10.1007/978-981-19-8865-3_24
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-8864-6
Online ISBN: 978-981-19-8865-3
eBook Packages: EngineeringEngineering (R0)