Abstract
Finger millet, grown on about 5 Mha globally under semi-arid environments of East Africa and South Asia, serves as an important dual-purpose crop to address food, forage, and nutritional needs in these marginal regions. Despite the tremendous yield potential, the area cultivated for small millets, including finger millet, decreased by 25.7% globally between 1961 and 2018. Finger millet improvement program began in 1913 in India; however, concentrated efforts to realize genetic gains in this climate-resilient crop are yet to be deployed compared to the efforts invested in improving other major cereals. This has resulted in lower productivity of finger millet in farmer’s fields than its potential yield even after more than 100 years of breeding. However, significant genetic variability is available for traits of importance. The breeding programs in Asia and Africa have refined the hybridization techniques and breeding objectives as per local needs. ICRISAT, an international center with finger millet as one of its mandate crops, is engaged with partners to generate new germplasm to enhance the productivity of this crop in marginal regions. This program, based in India and Kenya, has developed and distributed germplasm and breeding lines globally in the last few decades. Many promising and widely adapted cultivars have been released and adopted in many countries. Hybridization between the Indian and African gene pools of finger millet in the 1990s brought a paradigm shift in finger millet production in India. Now, breeding pipelines have been strengthened with the identification of newly identified germplasm for traits of importance, especially for blast resistance. Recently, finger millet genome sequencing was accomplished, and with the availability of advanced phenotyping protocols for various traits of importance, it has opened new opportunities to enhance genetic gains in this crop. This chapter informs about historical breeding efforts and discusses the prospects and challenges of finger millet breeding to enhance breeding efficiency and genetic gains in finger millet. International collaborative efforts toward improving agronomic traits, value addition, and the trade value of finger millet would help marginal farmers of southeast Asia and Africa but will also help enhance the commercial value of this underutilized millet.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
7.1 Introduction
Finger millet (Eleusine coracana) is an important component of low input agriculture prevalent in semi-arid tropics of South Asia (India, Nepal, and Sri Lanka) and the drylands of Africa (Uganda, Kenya, Zimbabwe, Zambia, Malawi, Tanzania, Rwanda, Zaire, Democratic Republic of the Congo, and South Africa). In terms of area and production, finger millet is the third-most important millet worldwide, after sorghum and pearl millet (Meena et al. 2021). Currently, the crop is cultivated across 25 countries, semi-arid regions, and tropical regions, up to an altitude of 2300 m. The major producing countries of finger millet are Uganda, India, Nepal, and China (Onyango 2016). In Africa, finger millet is mainly cultivated in minimal-scale cereal farming systems, mostly in the upland areas of Eastern Africa (Uganda, Ethiopia, Tanzania, and Kenya). It is cultivated on around 3–4 million ha in several Eastern and Southern African (ESA) countries, while on about 1.2 million ha with a production of 1.82 mt in India, Karnataka, Andhra Pradesh, Tamil Nadu, Maharashtra, Odisha, Jharkhand, etc. being the main cultivating states (ICAR-AICRP on small millets report, 2018; Reddy et al. 2008). Archaeological evidence suggests that it originated approximately 5000 years ago in the highlands of Ethiopia and western Uganda, whereas in India, records of its cultivation trace back to 3000–4000 BC in the western Ghats (Hilu and Dewet 1976; Hilu et al. 1979). Among all the known species of genus Eleusine, E. coracana spp. africana and E. coracana spp. coracana are the only two cultivated subspecies (Rawat et al. 2022).
Recognizing its immense health benefits, the consumption of finger millet in various functional food formulations, such as pasta, cookies, bread, cake, and noodles can be observed in Africa, Asia, Europe, and the USA (Deshpande et al. 2021). Traditionally, tribal people consume finger millet in the form of porridge, malt, and beverages, while straw is fed to the farming animals. The nutritional superiority over major cereals (i.e., wheat and rice) in terms of the gluten-free nature of the protein, exceptionally high calcium content, low glycemic index, and bioactive secondary metabolites of diverse therapeutic uses makes it a highly valuable crop. Furthermore, its wide adaptation to drought-prone environments, low input dryland agriculture, and marginal and fragile hilly agroecosystems make it a crop of the future. However, despite its high agricultural value, the global area of finger millet production and cultivation has declined. The largest reduction was observed in Asia, whereas the smallest was observed in Africa (Meena et al. 2021). The productivity of finger millet doubled from 0.7 t ha−1 (1950–1951) to 1.6 t ha−1 (1976–1980) in India owing to the cultivation of high-yielding blast-tolerant varieties (http://www.aicrpsm.res.in/). However, after that, the crop productivity stagnated at 1.6 t ha−1 in India with minor improvement despite the crop’s high nutritive properties and excellent sustainability in semi-arid systems (http://data.icrisat.org/dld/). Padulosi et al. (2015) reported the potential productivity of finger millet as 10 t ha−1. However, the actual productivity of finger millet is very low in Uganda (0.4–0.8 t ha−1; Tenywa et al. 1999), India (1.6 t ha−1; ICAR-AICRP on small millets report, 2018), Nepal (1.1 t ha−1; Khadka et al. 2016), and Asia (1.3 t ha−1; Onyango 2016). Kurosaki and Wada (2015) presented spatial patterns of long-term changes in finger millet cultivation in India from 1965 to 2007. The study also reported the declined area of finger millet in Tamil Nadu, Karnataka, Andhra Pradesh, and Odisha as opposed to the fine cereals (rice, wheat, and maize). This downfall may be due to the lack of focused research and policy support compared to major cereals.
Large germplasm collections have been maintained in international and national genebanks, and adequate genetic variation has been reported for various agronomic traits (Upadhyaya et al. 2006). However, appreciable genetic gains through harnessing the available genetic variation for agronomic and nutraceutical traits have not been achieved due to inherent problems such as cumbersome floral biology, small seed size, seed shattering, and unsynchronized maturity (Sood et al. 2019). The latest trends in cutting-edge biotechnological and omics tools, particularly the availability of reference genome sequences (Hittalmani et al. 2017; Hatakeyama et al. 2018) and their integration with conventional breeding, hold immense potential to overcome these limitations. In fact, acquiring high-density genomic data coupled with high-dimensional phenotypic records will certainly improve our understanding of genetic control of complex traits of agronomic and nutraceutical importance.
In this chapter, we summarize the importance of finger millet in diversifying the future cropping systems as well as its origin, phylogeny, genetic resources, production constraints, breeding achievements, and genomic advancements. We also provide perspectives and a roadmap on utilizing emerging genomic tools like gene editing and next-generation genotyping to make finger millet a viable and competitive crop in contemporary agroecosystems.
7.2 Taxonomy, Biology, and Genetic Resources
7.2.1 Taxonomy
The cultivated finger millet (Eleusine coracana (L.) Gaertn.) is an allotetraploid belonging to the family Poaceae, subfamily Chloridoideae, and tribe Chloride (Vetriventhan et al. 2020). E. coracana subsp. africana is considered an assumed progenitor to the cultivated finger millet, and it is completely cross-compatible with the cultivated finger millet and produces fertile hybrids (Mehra 1963; Hiremath and Salimath 1992). The genus Eleusine comprises about ten species, including annuals and perennials, with three basic chromosome numbers 8, 9, and 10. The cultivated species E. coracana can be classified into races and subraces (Prasada Rao et al. 1993). The species E. coracana contains of two subspecies, africana (wild type) and coracana (cultivated type). The subsp. africana is again divided into two wild races, africana and spontanea.
7.2.2 Biology
Finger millet is a robust, tufted annual-growing crop from about 30–150 cm tall and takes 3–6 months to complete the seed cycle. The stems are erect, slender, compressed, glabrous, and capable of producing many tillers and nodal branches. At maturity, the stems are somewhat laterally flattened. The inflorescence is an arrangement of many spikelets, which are known as fingers. The inflorescence consists of a variable number of spikes ranging from 3 to 20 arranged in a bird’s foot style. It resembles fingers on a hand, hence its common name “finger millet.” Spikes are straight or slightly incurved and up to 11 cm in length. Each spike contains serially arranged four to ten florets on the finger. Two large barren leaves cover the florets, each enclosed between a pair of scales known as palea. The flowerets are in the axil of the lower flowering glumes, known as a lemma; near the base of the ovary, two little scaly lodicules are present (Gupta et al. 2011; Dodake and Dhonukshe 1998). The three stamens are 0.5–0.8 mm long, not penicillate (Nanda and Agarwal 2008). The gynoecium is bicarpellary and unilocular, with a larger ovary having two styles with plumose stigma (Seetharam et al. 2003). The androecium mostly surrounds the stigma. Anthers are bigger than filaments (Gupta et al. 2010); spikelets are usually 5–8 mm long and 3–4 mm wide. Spikelets are arranged alternately on the rachis, and each spikelet contains about four to seven seeds. The seeds vary in diameter from 1 to 2 mm (Reddy et al. 2008). Except for the terminal ones, which can occasionally be sterile, all florets are excellent flowers. The caryopsis is globose and smooth; the color can be white, light brown, reddish-brown, ragi brown, and dark brown. The seed pericarp is easily removed from the seed coat because it is independent of the kernel. The shape of the grain varies from oval and round to oblong. In cultivate, seed shattering at maturity is not that common, while in wild species, it is common (Sood et al. 2019; De Wet et al. 1984) (Fig. 7.1).
7.2.3 Genetic Resources
A large number of finger millet germplasm accessions are available for the scientific society. Globally, >37,000 germplasm accessions of finger millet have been conserved in various genebanks (Vetriventhan et al. 2016; Dwivedi et al. 2012). The major collections of finger millet accessions are conserved in India, Kenya, Ethiopia, Uganda, and Zambia. The National Bureau of Plant Genetic Resources, New Delhi, India, has the largest germplasm collection, which maintains >10,500 accessions under long-term conservation. Most of them are indigenous in nature. The ICRISAT genebank in Patancheru, India, comprises a total of 7519 germplasm accessions from 26 countries, of which 205 are wild species, 7121 traditional cultivars/landraces, 143 advanced/improved cultivars, and 50 breeding/research material. The concept of core and mini-core collections has been proposed for better utilization of diversity in crop improvement programs. Following this approach, the ICRISAT has developed core and mini-core collections in finger millet. The finger millet core collection contains 622 accessions (~10% of the total collection), and the mini-core collection contains 80 accessions (10% of core collection or 1% of the total collection). In addition, a composite collection of germplasm consisting of 1000 accessions has been developed under the Generation Challenge Program (Upadhyaya et al. 2006). The core and/or mini-core collections established at the ICRISAT genebank have been evaluated for agronomic, grain nutrients (Upadhyaya et al. 2011), salinity (Krishnamurthy et al. 2014), drought (Krishnamurthy et al. 2016), and fodder quality traits (Backiyalakshmi et al. 2021a, b) and identified promising trait-specific sources for use in crop improvement.
7.3 Target Traits and Their Relationships
Despite finger millet’s enormous potential attempts to enhance its genetics lag well behind those of other main crops. Breeding targets for finger millet improvement may be classified as must-have and long-term traits. It is possible to increase yield by improving its components like plant height, days to flowering, synchronous maturity, inflorescence length and number of productive tillers, grain size, and threshability while taking the must-have traits into account (Sood et al. 2019). Besides these basic traits, breeding for blast resistance is the most important objective of finger millet genetic improvement programs across the globe (Kumar et al. 2021a). The blast caused by the fungus Pyricularia grisea is the most important biotic constraint which severely affects the production of finger millet worldwide. It affects the finger millet plant at all the growth stages and expresses symptoms in the form of leaf blast (LB), neck blast (NB), and finger blast (FB), with neck blasts causing the greatest yield losses. In endemic and hotspot areas, 70–80% yield loss has been reported (Mbinda and Masaki 2021). Like blast, the infestation by a parasitic weed Striga is a serious biotic constraint that severely affects finger millet production in sub-Saharan Africa (Teka 2014).
Drought is the main abiotic stress of finger millet, especially in the low rainfall, low altitude areas of sub-Saharan Africa. Finger millet is mainly affected by terminal drought after flowering at the grain filling stage. Producing short-duration varieties that escape terminal drought is the main measure for drought. Therefore, breeding for stable blast and striga resistance and drought escape/tolerance is one of the primary breeding objectives and must-have traits of all the finger millet breeding programs in Asia and Africa. Breeding for snapping varieties for the ease of harvesting, medium height (≤90 cm), good plant aspect and strong stem to prevent lodging, compact heads as an indicator for high yield, and three to four productive heads are the traits to be considered for popularization and commercialization of finger millet. With the improvement of yield, usually leading to increased head size, plant lodging is becoming an inherent problem, with two main negative effects: (1) finger millet grain usually germinates the moment it gets in contact with soil leading to great yield losses and (2) lodging complicates machine harvesting. Efforts to breed for stronger or stiff stalks are addressing this problem. Enhancing fodder yield by selecting and including genotypes with high basal tiller numbers in hybridization programs is another important target of finger millet breeding programs. Emphasis on breeding for high fodder nutrient digestibility and high threshability is required for sustainable food and food security in semi-arid areas of Asia and Africa.
Besides these basic traits, enhancing the seed size coupled with synchronized and early maturity of the tillers is a major long-term trait of the finger millet genetic improvement program. Very small seed size and unsynchronized maturity of most of the available finger millet cultivars are causing difficulties in mechanical planting and harvesting of the crop (Meena et al. 2021). Some inherent problems like high seed shattering also need to be addressed in the long-term breeding goals of finger millet. The poor initial vigor of finger millet leads to a heavy infestation of weeds bringing in more competition for light and nutrients, leading to a poor crop stand and significant yield losses. Seedling vigor is highly correlated to drought tolerance and has been used as an early selection criterion. It is also highly correlated to high yield and higher 1000 grain weight. Further, manual weeding increases quality seed production costs without an effective pre- and post-emerging herbicide. Therefore, breeding for herbicide-tolerant finger millet through modern approaches like transgenic development and genome editing is an important long-term target for finger millet genetic improvement (Joshi et al. 2018).
Quality and market-driven traits of finger millet include grain color (red and white), high puffing percentage, and taste. Due to the high quantities of tannins and phenolics, finger millet grains are typically dark brown, making the product’s appearance unappealing. Sometimes the plentiful tannins and phenolics give a bitter taste to the improved products, thereby reducing their consumer acceptability. Therefore, breeding for white-seeded finger millet is an effective approach for adding its value and enhancing market demand for the products (Joshi et al. 2021a). Traits for consideration for yield are normally correlated and can be assessed together with yield, or a number of them can be assessed as yield indicators, especially in early generations where yield per se is not assessed or in the early vegetative period of the crop. Correlation analysis on phenotypic characterization data of trials conducted at Kiboko, Kenya, showed yield to be highly correlated (P < 0.01) to all agronomic and yield-related traits, viz., grain yield, agronomic aspects, days to 50% flowering, days to maturity, plant height, productive tillers, ear weight, ears harvested, 1000-grain weight, blast resistance, lodging estimate, and seedling vigor evaluated (Table 7.1), implying that the traits can be used to indirectly select for yield. Ear weight, days to flowering, and plant height were highly correlated to all or most of the traits implying they are good traits for selection per se.
There’s immense potential for enrichment in finger millet. The exploitation of yield parameters might lead to weakening impacts for numerous nutritionally valuable components within the seeds, which are available in different cereals. Therefore, such dilution effects need to be considered while breeding for higher yield. However, improving the major nutrient contents in the finger millet grain has been shown not to affect yield significantly. Correlation of yield with grain nutrient content from 480 accessions evaluated in Kiboko showed nonsignificant associations with yield, implying that breeding for high nutrient content will not have any significant effect on yield and vice versa. Similarly, Gupta et al., (2009) also reported no penalty on grain yield and seed size while breeding for grains rich in these micronutrient. Calcium (Ca), the main nutrient in finger millet, was highly correlated to the other important nutrients in finger millet such as iron (Fe), zinc (Zn), and nitrogen (protein), while Fe, Zn, and nitrogen (protein) were highly correlated (P < 0.001) to all nutrients in the grain, implying that it is possible to improve the different mineral contents in the grain simultaneously. Studies suggest that grain yield and Ca have a low correlation or negative correlation—the same in grain yield and Fe, Zn, and protein (Ojulong et al. 2021). Previous studies on finger millet have also suggested a low or negative correlation between grain yield and grain nutrient traits (Upadhyaya et al. 2010; Kumar et al. 2010; Ng’uni et al. 2011), and iron and zinc have a low negative correlation with yield in sorghum. Ojulong et al. (2021) suggest a highly significant (P < 0.001) correlation among the yield and calcium, copper, iron, potassium, magnesium, manganese, potassium, sulfur, zinc, and protein.
Although finger millet has a lot of room for development, higher yield parameters might dilute some of the nutritionally important components of seeds, as shown in several cereals. Therefore, such dilution effects need to be considered while breeding for higher yield. Knowing the genetic architecture of crucial breeding targets like flowering, early duration, yield, resistance to disease/pest, and nutritional quality is a must to execute suitable breeding strategies for enhancing genetic gain. However, very limited studies have been conducted on finger millet to understand the genetics of these important breeding targets. Therefore, genetic mapping studies need to be implemented to learn more about the underlying genes for the traits of economic importance.
7.4 Target Product Profile and Market Segments for Africa and Asia
For the success of breeding programs, it is very important to work closely toward the trait-specific requirements of its stakeholders. The breeding programs in Africa and Asia are well-aligned with the farmer and consumer needs in the finger millet-growing countries. For instance, ICRISAT’s East African breeding program has identified five different market segments, while the Indian program has identified two segments. Product profiles have been developed considering the trait-specific requirements for each segment. The type of cultivar requirement, area, target regions, and regions of different product profiles (segments) are shown in Table 7.2 as an example of these two breeding programs. Must-have traits are the ones that can be addressed with the available trait variability and tools. They are immediately needed in the current-day cultivars, while long-term traits are the ones which are visioned for the future, and efforts are required to strengthen them in the breeding pipeline (Table 7.2).
7.5 Genetic Variability for Traits of Importance
The global germplasm of finger millet conserved at the ICRISAT genebank shows a large variability for morpho-agronomic, grain and fodder quality and stress tolerance traits (Vetriventhan et al. 2016). For example, a huge variability trait is for important agronomic traits such as days to 50% flowering that varied from 40 to 120 days, plant height from 30 to 240 cm, number of basal tillers from 1 to 70 (wild species accessions produce a large number of tillers), and inflorescence length from 40 to 320 mm (http://genebank.icrisat.org/), and germplasm diversity representative subset called core collection (Upadhyaya et al. 2006) and mini-core collection (Upadhyaya et al. 2010) were established to enhance the use of diverse germplasm in crop improvement. Evaluation for grain nutrient content of the finger millet core collection revealed a substantial variability for Fe (21.71 mg/kg), Zn (16.58–25.33 mg/kg), Ca (1.84–4.89 g/kg), and protein (6.0–11.09%) and also reported a weaker and nonsignificant correlations of grain yield with Fe, Zn, Ca, and protein indicating better prospects for combining higher grain nutrients with higher yield background (Upadhyaya et al. 2011). Finger millet is primarily grown as a food crop in Asia and Africa, but its stover serves as an important source of fodder, producing excellent hay and green forage for cattle, sheep, and goats (Sampath 1986; Gupta et al. 2017). The finger millet diversity panel conserved at the ICRISAT genebank was assessed for fodder quality traits, and the study showed a substantial variability for fodder quality traits (2.8) to 10.7 t/ha of dry fodder yield, 6.47–8.15% of crude protein, >90% of dry matter content, and 45.21–49.09% of in vitro organic matter digestibility (IVOMD) and identified promising accessions for developing dual-purpose cultivars (Backiyalakshmi et al. 2021a, b). Similarity, a large variability for salinity (Krishnamurthy et al. 2014) and drought (Krishnamurthy et al. 2016) were reported in the international collection (core/mini-core) of finger millet, and promising sources were identified for use in crop improvement.
7.5.1 Genetic Variability
Significant genetic variability for different traits has been reported in finger millet crop. For instance, a very high variation was observed among the agronomic and yield-related traits in a study conducted on 480 accessions constituted from collections and farmers and improved varieties from Eastern and Southern African countries and the finger millet mini core (Table 7.3). Days to 50% flowering ranged from 46 to 92, indicating that sources for short-, medium-, and long-duration varieties were available. Very high variation in productive tillers (2–21) highlighted the great chances of improving this important trait for yield, and so was the number of heads harvested (2–21). Numbers of fingers and other important traits contributing to yield had high variability (4–12). Grain spikes (3–8) were variable too. Grain yield, the main trait for improvement, was highly variable (0.7–4.6 t/ha), and so was thresh percentage (47.2–94.8%). All these show high prospects of improving from the available germplasm using conventional means.
Nutrient profiling showed high diversity in the materials evaluated. Calcium values ranged from 115.5 to 540 mg/100 g, Fe from 1.4 to 24.5 mg/100 g, and Zn from 0.1 to 10.1 mg/100 g, again showing the promising aspects of improving the nutrient content from the germplasm. An ICRISAT genebank trial in 2018 and 2019 quantified for Ca, Fe, Zn, and Aluminium (Al) showed large genetic variation micronutrients, which could be further exploitable in nutrition-inclusive breeding programs (Fig. 7.2). Ojulong et al. (2021) also established high variability among the different nutrient traits in the region. Finger millet cluster analysis studies suggested two main clusters. The first cluster contains varieties from countries of finger millet origin, Uganda and Ethiopia (Hilu and DeWet 1976; Dida et al. 2008), and the major finger millet-growing countries in the East and Southern Africa region, Tanzania, Kenya, Malawi, and Zimbabwe. The second cluster contains germplasm from countries in the diverse region that are among the largest finger millet producers: India and Nepal (Hilu and DeWet 1976; Dida et al. 2008). Cluster analysis studies suggested that the highest diversity for the different nutrient traits for the enrichment of finger millet exists in centers of origin. Earlier studies also observed that the domesticated varieties were low in maximum nutrient content, most likely a result of farmer selection by the farmers, who preferred brown grain, which is relatively lower in nutrient content compared to dark brown. Studies on finger millet found it to be rich in protein (8–10%) which is associated with seed color (Vadivoo et al. 1998), and lower in fat (2.5–4%) which makes it a healthy option for the modern diet.
Manyasa et al. (2014) conducted genetic diversity studies on 340 finger millet germplasm from Kenya, Tanzania, and Uganda, and 15 mini-core accession using single-sequence repeat markers and qualitative traits found explained the diversity by variability within the countries and subregions than that among the countries and subregions. The low variability among the countries explained the shared gene pool, as the crop originated from the East African region. Studies suggest that farmer’s selection for adaptation and end-use could have contributed to the high diversity within the countries. The genetic diversity studies explained that finger millet was domesticated in Africa and later introduced to India (Dida et al. 2008). It is observed that Asian accessions are earlier in maturity with short plant height and small flag leaf length when compared to African germplasm, which has high plant height and longer and wider flag leaves with higher intraspecific diversity (Dida et al. 2008; Bharathi 2011; Babu et al. 2014d). Further, as compared to the African gene pool, it is reported that the Asian gene pool was created from limited founder populations lacking unique genes. Heritability of the different traits is high in finger millet.
7.5.2 Breeding Methods
Hybridization in finger millet started around the early 2000s in many African countries, with the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) in Nairobi, Kenya; the Agricultural and Livestock Research Organization (KALRO) in Kenya; and the National Semi-Arid Resources Research Institute of the National Agricultural Research Organization (NaSARRI-NARO) in Uganda taking the lead. Pedigree breeding was the most used method in both African and Asian countries. The progenies advanced based on the combination of several highly heritable traits as per the needs of different segments, like selecting for seedling vigor, plant aspect, plant height, number of productive tillers, tillering type, days to flowering, days to maturity, head shape and size, finger number and size, thresh percent and ease of threshing, grain yield, 1000 grain weight, and grain color and shape. In India, finger millet improvement work started in 1913, but hybridization started in 1951.
7.5.3 Historical Breeding Efforts in India
Crop improvement efforts for finger millet were initiated in India at Zonal Agricultural Research Station, V.C. Farm, Mandya, Karnataka state, by Dr. Leslie C. Coleman in 1913. Productivity was very low at that time due to a lack of high-yielding varieties, improved crop management practices, soil health issues, and new technological interventions. According to the available literature, efforts for finger millet improvement in India can be divided into five stages.
7.5.3.1 Stage I (1913–1938): Pure Line Selections—Indigenous Varieties
During this period, the varietal improvement work was conducted at different research centers in Karnataka states of India. Pure line selections were made from indigenous varieties. During this period, Dr. Leslie noted that the complete emergence of inflorescence required about 10 days, and flowering takes 7–8 days while working on floral biology, anthesis, and pollination in finger millet. He also observed that cross-pollination is very rare because the period of anthesis is very short.
7.5.3.2 Stage II (1938–1963): Initiation of Recombination Breeding
During this period, pure line selection work was continued from indigenous germplasm lines and landraces. To enhance the genetic base of the crop, hybridization was initiated through the contact method in 1951. In the contact method, panicles from the plant of the recipient parent and from the plant of the desired/donor parent were chosen as they were both about to start the anthesis process. To prevent unintended cross-pollination, both panicles were joined together and covered with butter paper bags. The varieties released using this method exhibited a yield advantage of approximately 50% over the existing pure line varieties with wide agroecological adaptation capable of fulfilling the need of different growing seasons of finger millet.
7.5.3.3 Stage III (1964–1988): Widening the Genetic Base by Combining Divergent Gene Pools
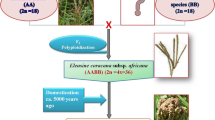
This period is regarded as the most significant period in finger millet improvement resulting in a quantum jump in area and crop production. Hybridization was initiated between the two divergent gene pools of finger millet by Dr. C.H. Lakshmanaiah in 1964. The locally adapted, early maturing but low-yielding Indian genotypes were crossed with late maturing, high-yielding, and stress-tolerant African genotypes (Sood et al. 2022). This effort resulted in the developing of 16 Indo-African varieties designated as “INDAF” varieties.
7.5.3.4 Stage IV (1988–2013)
During this period, emphasis was given to the development of dual-purpose varieties (high straw and grain yield) along with resistance to blast adaptability to rainfed and irrigated conditions.
7.5.3.5 Stage V (2013 to Date) Genomic Interventions
During this period, the finger millet’s draft genome was sequenced by Hittalmani et al. in 2017 and Hatakeyama et al. in 2018, creating opportunities to use genome-level information to accelerate the improvement of finger millet. Forward breeding or marker-assisted selections are being used to fast track the varietal development. A set of SNPs (49) was developed to refine the crossing technique and identify true hybrids. A set of SNPs has also been developed for forward breeding of blast resistance (Table 7.4).
7.5.4 Breeding for Traits of Importance
7.5.4.1 Climate Adaptation
Finger millet production is limited majorly by two critical constraints: high-temperature stress and terminal drought. This is linked to the low and erratic rainfall (150–800 mm) of production environments where finger millet is traditionally grown in eastern South Africa and India.
Heat Stress
High-temperature stress has been reported as another most important cause of change in physiology by arresting the cell expansion, which causes a reduction in plant growth and development, leading to loss of productivity (Sato et al. 2002; Abdelmageed et al. 2003). The optimum temperature for the growth of finger millet is 28–32 °C and can be well sustained up to 36 °C (Yogeesh et al. 2016). It has been evident from the literature (Sato et al. 2002; Abdelmageed et al. 2003) that finger millet deviates from its normal morpho-physiology when temperatures cross the cardinal thresholds (day = 36 °C; night = 26 °C), which affects the stable physiological functions resulting in yield reduction. When seedlings are exposed for 5 h to temperatures between 38 and 54 °C, shoot and root growth is affected (Venkatesh Babu et al. 2013). It has been reported that yield and yield-contributing traits like flowering, maturity, ear head length, finger length, number of branches, and grain size are severely affected when the crop is exposed to temperature stress (42–44 °C) (Yogeesh et al. 2016). A suitable crop management strategy for finger millet would be to avoid heat stress during the most vulnerable reproductive stages by selecting the right genotypes (phenology and duration) and planting dates.
7.5.4.2 Drought Stress
Drought is known to affect finger millet in many ways and depends on the crop’s stage. The soil moisture stress during flowering and grain filling stages is a very frequent form of drought in finger millet, contributing to a significant yield loss (Maqsood and Ali 2007). This is also referred to as terminal drought stress. It is mainly caused by cessation of rain toward the end of the rainy season in semi-arid tropics where the cropping period is limited. Breeders define a drought-adapted variety as having the ability to give a high or reasonable yield under drought conditions. The variety can achieve this through drought escape-short duration varieties or being tolerant to drought. Screening the germplasm and farmer-preferred varieties resulted in the identification of such varieties. In the African region, short duration varieties U15, Ekama, and Gulu E from Uganda and KNE 741 from Kenya have been used to reduce the days to flowering of a number of lines, and currently ICRISAT-Nairobi has a pipeline of short duration lines, a number at advanced stages for release in Africa. A number of lines have also stayed green characteristics and remain green under drought conditions giving reasonable yields. We now have kits of materials that flower before 60 days and give a high yield.
7.5.4.3 Biotic Stress Resistance
Finger millet is affected by numerous diseases caused by fungal, bacterial, and viral pathogens, including blast, seedling blight, wilt or foot rot, Cercospora leaf spot, downy mildew, smut, bacterial blight, ragi mottle streak, and ragi severe mosaic. Most of these diseases are region-specific and of minor importance. However, blast, caused by an ascomycete fungus Magnaporthe oryzae (anamorph: Pyricularia oryzae), is the most destructive and widespread disease that affects the yield, utilization, and trade of finger millet within East Africa and South Asia (Mgonja et al. 2007). The disease affects the crop at all growth stages leading to leaf, neck, and finger blasts; neck and finger blasts are the most destructive forms of the disease. The average yield losses due to finger millet blast have been reported to be around 30%; however, yield losses could be as high as 80–90% in the susceptible cultivars under favorable conditions of disease development (Vishwanath et al. 1986; Rao 1990; Nagaraja et al. 2007).
The pathogen is known to infect more than 50 graminaceous hosts, including food security crops such as rice, wheat, finger millet, pearl millet, and foxtail millet. Blast is largely managed through host plant resistance, the most economical, efficient, and ecologically sustainable method of disease management. For the development of durable disease-resistant varieties, information on diversity in the pathogen populations is essentially required. The pathogen causing a blast of finger millet is highly variable, which necessitates generating information on virulence diversity in the pathogen populations adapted to finger millet. This can help in developing finger millet varieties with durable resistance to blast disease. Though efforts have been made to study the genetic diversity and aggressiveness of the pathogen populations (Kiran Babu et al. 2013a; Takan et al. 2012), limited information is available on the virulence diversity in finger millet-infecting populations of the pathogen. Kiran Babu et al. (2015) developed the host differential set and reported the pathogenic variation in the isolates collected from finger millet grown in different states in India. This differential set is being used at ICRISAT to monitor variation in the pathogen population and select diverse pathotypes for greenhouse screening of a finger millet lines for blast resistance. Kiran Babu et al. (2013b) screened finger millet mini-core collection for blast resistance. Nine accessions (IE 1055, −2821, −2872, −4121, −4491, −4570, −5066, −5091, and −5537) with desirable agronomic traits, such as early flowering (<65 days), medium plant height (105–125 cm), and semi-compact to compact inflorescence were identified for use in a breeding program. The africana type mini-core accession IE 4709, with a high level of resistance to blast, agronomically desirable characters, and high content of grain nutrients such as Fe, Ca, Zn, and protein, was identified as a promising source for use in finger millet breeding. An African cultivar IE 1012 has been extensively used in India as a source of blast resistance (Gowda et al. 1986).
Resistance sources have been identified through multilocation screening and are used in breeding programs. Multilocation evaluation of 29 genotypes at hot spots led to the identification of five promising genotypes (IE Nos. 2883, 2871, 6240, 2710, and GE3767) with stable resistance to both finger and neck blasts for further use in breeding programs (Das et al. 2021). Of the 81 finger millet germplasm accessions from East Africa evaluated for blast resistance, three accessions (G18, G43, and G67) were identified as resistant to all three stages: leaf, neck, and panicle blasts (Manyasa et al. 2019). Similarly, Dida et al. (2021) identified one improved variety (KACIMMI22), and four landraces (TZ1637, BKFM0031, ACC214988, ACC203544) in Kenya with high resistance to the blast isolate for use in breeding programs. Resistance to multiple pathogen isolates was observed in IE 2911, IE 2957, and GPU 28 in the greenhouse screening at ICRISAT, India (Kiran Babu et al. 2015). GPU 28, the cultivar, occupies a maximum area of about 80% of the total area cultivated in India and has shown resistance to blast over time in different states of India and exhibited lineage-wide resistance to M. oryzae populations as well (Nagaraja et al. 2008; Kiran Babu et al. 2015).
Of late, this cultivar has started showing susceptibility to blast. GPU 28 was released for cultivation in Karnataka in 1996, and after that GPU 26, GPU 45, and GPU 48 were released for cultivation on farmers’ fields in India. Information on blast-resistant varieties identified and released for cultivation in different finger millet-growing areas in India has been compiled by Palanna et al. (2021). As virulence change in the pathogen populations has been cited as the main cause of the breakdown of resistance in the released cultivars to blast disease, monitoring virulence shift in the pathogen population, identifying new virulent pathotypes, screening breeding lines for resistance against the new virulent pathotypes under controlled conditions, further screening of promising lines at hotspots to identify stable sources of resistance, and strategically using these resistance sources in the breeding programs form the strategy of management of finger millet blast.
7.5.4.4 Nutrition-Inclusive Breeding
Grain quality
A large proportion of the population in developing countries is deficient in essential nutrients like iron (Fe), zinc (Zn), and calcium (Ca) (Maharajan et al. 2021). Finger millet is especially rich in Ca (~350 mg per 100 g), which could be the potential crop to combat Ca deficiency. Apart from Ca, finger millet grains have a protein content of 6–13%, which is better balanced with sulfur-containing amino acids, such as methionine and cystine, as well as lysine, threonine, and valine, than other millets (Shobana et al. 2013; Saleh et al. 2013; Sharma et al. 2017; Rodríguez et al. 2020). Large variability exists for grain nutrient content in the core collection (Upadhyaya et al. 2010) and identified 15 promising accessions each for grain Fe, Zn, Ca, and protein and 24 accessions were identified which are superior for two or more nutrients and provide an opportunity for breeding nutrient-dense cultivars. The ICRISAT product profiles included grain nutrient improvement, particularly Ca improvement as a target trait in the breeding program.
Fodder quality
Finger millet is used as an important forage to some extent but not extensively used, like sorghum and pearl millet, due to a lack of scientific research on the quantity and quality of finger millet crop residues. The recent study on the fodder quality of finger millet germplasm conserved at the ICRISAT genebank indicated considerable variability. It provided evidence that finger millet crop residues have higher forage quality than rice and wheat, comparable with sorghum and pearl millet (Backiyalakshmi et al. 2021a, b). Thus, the promising lines identified could be used in the breeding program for breeding dual-purpose finger millet cultivars. With food security and nutrition-sensitive agriculture gaining momentum, this nutria-cereal is finding demand in urban food markets.
7.6 Novel Breeding Methods
7.6.1 Prebreeding: Widening the Gene Pool
Wild and weedy relatives of the genus Eleusine are the treasure troves for various economic traits, which are lacking in the primary gene pool of the finger millet. Introgression of novel traits like drought tolerance, blast and striga resistance, plant vigor, and superior nutritional quality from unadapted wild species to locally adapted popular cultivars of finger millet through a prebreeding approach will be an effective strategy for its genetic enhancement. Finger millet has two subspecies: africana and coracana. Subspecies africana is a diploid (2n = 18), while subspecies coracana is a tetraploid that evolved from the diploid subspecies (Paschapur et al. 2021). The diploid species E. indica, E. floccifolia, and E. tristachya form the secondary gene pool and E. intermedia, E. gaegeri, E. kigeziensis, E. multiflora, and E. semisterlis (E. compressa) from tertiary gene pool holds a great potential to address major production constraints of finger millet (Joshi et al. 2021b). However, incompatibility barriers must be investigated for developing interspecific hybrids between cultivated finger millet and its distant gene pool. Advancements in molecular breeding applications like advanced backcross and QTL analysis (Tanksley et al. 1996) enhance the possibility of utilizing a wild gene pool in the genetic improvement of finger millet.
Hybridization between Indian (E. coracana subspecies coracana) and African gene pool (E. coracana subspecies africana) of finger millet in the 1990s brought a paradigm shift in finger millet production in India, and the Indaf (Indian × African accessions) varieties replaced almost all the earlier released varieties. Apart from high grain yield, these varieties are known to possess unique traits like drought tolerance, lodging and enhanced protein quality acquired from the African gene pool. The ICRISAT breeding program is focused on widening the genetic base of the crop by combining the better stress tolerance traits of E. africana in Indian genotypes to enhance the genetic gain and identify the best heterotic combinations through multilocation testing in collaboration with NARS partners across Asia.
7.6.2 Improving Crossing Efficiency
Variability plays a vital role in crop improvement, but inducing new variability is a daunting challenge in highly self-fertilized crops like finger millet with cleistogamous flowers. In general, there are about 280–1330 spikelets per panicle (4–7 seeds per spikelet), and a spike is reported to be 8–15 cm long and 1.3 cm broad, and it takes 5–7 days to complete anthesis in finger millet. Therefore, ensuring male sterility through hand emasculation in such small florets is a cumbersome and time-consuming task. Further, growing seeds for identifying a few hybrid plants in a traditional contact method (Sood et al. 2019) requires more resources, time, space, and labor. Therefore, ICRISAT, Patancheru, Hyderabad (17.3° N, 78.5° E), has done a good amount of work and recommended temperature and duration on a particular anthesis stage for effective emasculation in finger millet. Few seeds are set in the female panicle using this technique, and most are true hybrid plants. In addition, the hot water emasculation method was also studied by the ICRISAT breeding team (data unpublished). Compared to chemical treatment, hot water treatment is more efficient in emasculating female lines and enhancing the breeding process. After developing F1s by hot water treatment method, we can quickly identify hybrid plants in F1 generation using knowledge of identifiable morphological markers (e.g., pigmentation and panicle shape) in the case of male/donor line should have a dominant character. In the absence of a dominant pigmented marker on the nodes and panicles of the donor parent, the F2 generation is raised and critically observed for the segregation of panicle or other plant traits. Recently, ICRISAT has performed whole-genome resequencing, and a set of 48 SNPs were identified for quality control and identification of putative F1s.
Some programs are using the plastic bag technique. This method, adapted from the sorghum technique, involves covering the florescent with a plastic bag of the right gauge and leaving it overnight or until the stigmas open. Covering with a bag leads to the condensing of the water due to respiration, which will soak the anthers, making them not to disperse the pollen. The plastic bag is then removed, and the plant stalk is tapped gently to let the anthers fall. Pollen from the desirable donor is brought and dusted over the flower, the inflorescent which have not opened are removed, and the flower is covered. This technique has been very successful and is now universally used in many African breeding programs. As a result, thousands of lines have been developed by ICRISAT-Nairobi and shared with NARS partners in the region, west Africa, and with ICRISAT-Hyderabad.
7.6.3 Advanced Phenotyping Methods
The interaction of genotypes with the environment restrains genetic gain and insights into adaptation to different environmental constraints (abiotic stress). Therefore, it is important to characterize the environment in which the crop is grown (G×E) and design the phenotyping strategy relevant to the environment to empower the breeding programs for better selection.
ICRISAT has developed innovative methods and a high-throughput phenotyping platform (HTP) to facilitate precise characterization and screening for abiotic stress adaptation. It has helped NARS researchers from national programs to screen for several cereal and legume genetic materials (elite lines, national checks, advanced breeding lines, breeding populations) for crop improvement programs for changing climate adaptation (drought, heat, and salinity adaptation) using high-throughput phenotyping platform (LeasyScan, http://gems.icrisat.org/leasyscan/; Lysimeter facility, http://gems.icrisat.org/lysimetric-facility/). LeasyScan is “camera to plant”-based technology to characterize component traits of adaptation in just 4–6 weeks. A Lysimetric system with a rainout shelter facility is designed to impose various kinds of stress and evaluate the plant’s performance. Efforts are underway to use AI technology for UAV-based field phenotyping to digitalize the field phenotyping of breeding trials and multilocation trials. There is also robust development in sensor-based technology for quick assessment of nutritional traits like macronutrient (Benchtop NIRS and mobile NIRS), micronutrient (XRF), and post-harvest traits (HarvestMaster, computer tomography) to support the nutrition inclusive breeding programs (Fig. 7.3).
7.6.4 Speed Breeding
Over the last ten decades, plant breeders developed and released crop varieties through conventional approaches in many crops, but the conventional process is time-consuming because it involves crossing in between parental lines and generating progenies, followed by four to six generations of selfing or maintaining homogeneity to advance/fix the lines to evaluate productivity traits and agronomic performance. This is a time-consuming breeding approach for crop improvement that is often limited to only one to two generations per year, depending on the crops (Hickey et al. 2019). Speed breeding is a swift technique to enhance genetic gain and accelerate the breeding program/crop improvement in a shorter time with limited resources, manpower, and space compared to conventional breeding.
The generation period for finger millet cultivars in the field is around 4–5 months (Kumar et al. 2021b). However, under completely controlled conditions, the rapid generation advancement (RGA) technique may produce up to three to four generations of finger millet each year. The RGA protocol will accelerate the plant life cycle, and, on the other hand, it shortens the generation/breeding cycle time in light-, temperature-, and humidity-controlled conditions. In the case of short-day plants like finger millet, the protocol has already been developed based on light-emitting diode for some other short-day crops (soybean, rice, and amaranth) (Jähne et al. 2020), and efforts are underway to standardize speed breeding for finger millet. For rapid generation turnover, the rapid single-seed descent (rSSD) method applies to get near-homozygous lines in a year or two, depending on the crop species and duration. Five generations per year can be achieved in the case of soybean by using the protocol of Jähne et al. (2020). This is an economically and scientifically important and useful method compared to the conventional generation advancement method and shuttle breeding. Speed breeding allows and has significance in the development of populations, biparental populations (RILs and NILs), and mapping populations via robust phenotyping for trait specificity using X-ray fluorescence (XRF), near-infrared reflectance spectroscopy (NIRS), and computed tomography (CT) imaging, the marker-assisted selection (MAS), genomic selection (GS) models, and genome editing (Fig. 7.4).
7.7 Finger Millet Improvement Using Genomic Tools for Prospects of Accelerating Genetic Gain
7.7.1 Genomic Resources
Compared to major crops such as rice, wheat, maize, etc., few reports are available on genomic resources in small millets, including finger millet. Genomes of five small millets, namely, foxtail millet, finger millet, proso millet, teff, and Japanese barnyard millet, have been made available (Antony-Ceasar et al. 2018). Of these small millets, the genome of foxtail millet is the smallest (423–510 Mb), while finger millet has the largest one (1.5 Gb). Recently, the DArTseq approach was employed to assess finger millet genetic diversity and population structure. Analysis of about 33,884 high-quality single-nucleotide polymorphism (SNP) markers on 318 accessions revealed considerable genetic diversity (Backiyalakshmi et al. 2021a, b). As limited genomic resources are available until recently in finger millet, comparative genomics has played an important role with high genomic co-linearity reported between finger millet and rice (Srinivasachary et al. 2007). The SSR markers were correlated to the genetic relatedness among the species with the cross-transferability of these markers to finger millet. For example, it has been reported that 71% of SSRs in rice (Babu et al. 2018) and 73–95% in foxtail millet SSRs (Pandey et al. 2013) were cross-transferable. The finger millet EST sequences showed homology with rice blast-resistant genes which suggested that genes responsible for rice blast resistance play an important role in finger millet blast resistance (Babu et al. 2014b, 2018). Further, as mentioned earlier in this chapter, under the subheading biotic stress, finger millet accessions from African countries are highly resistant to blast disease, whereas most of the Indian subcontinent accessions are susceptible, as revealed by SSR markers (Babu et al. 2014a, b, c).
7.7.1.1 Reference Genome
The whole-genome sequence of finger millet genotype ML-365 (drought-tolerant and blast-resistant genotype) was sequenced on the platform Illumina and sequencing by oligonucleotide ligation and detection (SOLiD) technologies (Hittalmani et al. 2017). In the sequencing, about 45 Gb paired-end and 21 Gb mate-pair data were generated with a genome assembly consisting of 525,759 scaffolds (>200 bp) and N50 length of 23.73 Kb. In another study by Hatakeyama et al. (2018), genome assembly of the genotype PR 202 (IC: 479099) was reported using a novel polyploidy genome assembly workflow. Their analysis identified the genome size of finger millet as 1.5 Gb, and the genome assembled was 1189 Mb covering 78.2% of the genome. The whole genome consisted of 2387 scaffolds with the N50 value of 905.318 Kb with a maximum sequence length of 5 Mb and an overall gene number of 62,348, of which nearly 91% genes were functionally annotated and 96.5% were single-copy genes (Hatakeyama et al. 2018).
7.7.1.2 Trait Discovery and Mapping
Although next-generation sequencing technologies for genomic studies are now available, progress in identifying and tapping genes for important traits has been slow in finger millet until recently. The use of genetic markers to characterize functional traits diversity in finger millet has accelerated in recent years. The first genetic map using genomic SSRs, RFLP, AFLP, and EST markers was reported by Dida et al. (2007). Based on the genotype-phenotype association data, significant quantitative traits loci (QTLs) responsible for agronomic traits, as well as resistance for blast diseases, were identified, which showed strong associations with SSR primers designed from the blast genes (Babu et al. 2014b, 2018). Blast resistance gene homologs from rice and genes responsible for nutritional traits from other cereal crops have been developed and used invariably. Recently, the -omics approaches have efficiently been used in several studies to identify candidate genes responsible for nutritional variation as well as biotic/abiotic stress tolerance in finger millet (Rahman et al. 2014; Gupta et al. 2013). The identified markers are to be validated and fine mapped for use in marker-assisted breeding (MAB) programs of finger millet. In summary, the development of markers and comparative genomics paved the way for marker-assisted breeding. However, limited studies reported characterization of abiotic stress tolerance in finger millet using molecular markers.
7.7.2 Genomics-Assisted Breeding in Finger Millet
Biparental QTL mapping approach has been rarely initiated in finger millet due to the difficulty in crossing, variable synchronization in flowering, unavailability of stable contrasting parental lines, etc. for important quantitative traits. Further, fine mapping of the QTL region is unlikely due to high linkage disequilibrium (LD) in populations (Sood et al. 2019). Likely, the first biparental mapping population developed is an interspecific mapping population of E. coracana subsp. coracana cv. Okhle 1 (a landrace from Nepal) and its wild progenitor E. coracana subsp. africana accession MD 20 to develop the first linkage map in finger millet (Dida et al. 2007). In finger millet, the availability of diverse germplasm resources has allowed the use of LD-based association mapping to detect marker-phenotype associations and identify linked markers associated with agronomic traits and disease resistance (Babu et al. 2014a,b; Bharathi 2011).
Recently, the application of NGS in finger millet has resulted in genome sequencing and identification of thousands of SNP markers for use in trait mapping and molecular breeding (Gimode et al. 2016). Significant and promising marker-trait associations for five important agronomic traits were identified using a genome-wide association study (GWAS) (Sharma et al. 2018). Identified SNPs through the whole-genome resequencing (WGRS) approach of global finger millet collections would provide useful genomic resources for identifying QTLs and linked molecular markers for important biotic/abiotic stresses and quality traits that can be used in early-generation selection. In this direction, the finger millet research team at ICRISAT endeavored WGRS in approx. 170 important germplasm lines (unpublished). On the other hand, genomic resources are being attempted to optimize genomic selection (GS) and genomics-enabled prediction in finger millet. The GS approach combines genotypic as well as phenotypic data of training populations to estimate the genomic-estimated breeding values (GEBV) of each individual of test populations (Crossa et al. 2017).
Further, molecular markers distributed throughout the genome would be used to predict individuals’ GEBV, reducing the cost and time requirement of developing new crop varieties (Varshney et al. 2005). However, robust training populations and well-defined marker maps are the prerequisites for applying approaches such as GS in finger millet. Findings from these studies would facilitate rapid selection of superior genotypes overcoming the limitations of MAS (Fig. 7.5).
7.8 Summary and Outlook
Finger millet productivity in African and Asian countries is much below the real potential of this crop, even after 100 years of breeding. However, significant genetic variability is available for traits of importance. Germplasm exchange between Africa and Asia can be a game-changer. The challenge of crossing finger millet due to small-sized flowers can now be handled using recently devised new methods to increase crossing percentage, and a set of identified markers can be used to detect true crosses. With the availability of sequence data of the finger millet genome, important traits linked to productivity and biotic and abiotic stress tolerances have been mapped. With the improved understanding of the genetics of traits of importance, identification of donor lines for different traits, availability of improved methods of phenotyping, and the possibility of three to four crops in a year using speed breeding protocols, finger millet breeding programs across the world will have a major push to enhance genetic gains in this crop in the coming years.
References
Abdelmageed AH, Gruda N, Geyer B (2003) Effect of high temperature and heat shock on tomato (Lycopersicon esculentum M.) genotypes under controlled conditions. In: Conf Int Agr Res Develop. Deutscher Tropentag, Gottingen, 8–10 Oct
Antony-Ceasar S, Maharajan T, Ajeesh Krishna TP et al (2018) Finger millet [Eleusine coracana (L.) Gaertn.] improvement: current status and future interventions of whole genome sequence. Front Plant Sci 9:1054
Babu BK, Agrawal PK, Pandey D et al (2014a) Association mapping of agro-morphological characters among the global collection of finger millet genotypes using genomic SSR markers. Mol Biol Rep 41:5287–5297. https://doi.org/10.1007/s11033-014-3400-6
Babu BK, Pandey D, Agrawal PK et al (2014b) Comparative genomics and association mapping approaches for blast resistant genes in finger millet using SSRs. PLoS One 9(6):e99182. https://doi.org/10.1371/journal.pone.0099182
Babu BK, Pandey D, Agrawal PK et al (2014c) Molecular analysis of world collection of finger millet accessions for blast disease resistance using functional SSR markers. SABRAO J Breed Genet 46:202–216. http://eprints.icrisat.ac.in/14225/
Babu BK, Agrawal PK, Pandey D et al (2014d) Comparative genomics and association mapping approaches for opaque2 modifier genes in finger millet accessions using genic, genomic and candidate gene-based simple sequence repeat markers. Mol Breed 34:1261–1279. https://doi.org/10.1007/s11032-014-0115-2
Babu BK, Sood S, Chandrashekara C et al (2018) Mapping quantitative trait loci for important agronomic traits in finger millet (Eleusine coracana) mini core collection with genomic and genic SSR markers. J Plant Biochem Biotechnol 27:401. https://doi.org/10.1007/s13562-018-0449-7
Backiyalakshmi C, Babu C, Reddy DN et al (2021a) Assessing forage potential of the global collection of finger millet (Eleusine coracana (L.) Gaertn.) conserved at the ICRISAT genebank. Agronomy 11(9):1706
Backiyalakshmi C, Vetriventhan M, Deshpande S et al (2021b) Genome-wide assessment of population structure and genetic diversity of the global finger millet germplasm panel conserved at the ICRISAT genebank. Front Plant Sci 12:692463. https://doi.org/10.3389/fpls.2021.692463
Bharathi A (2011) Phenotypic and genotypic diversity of global finger millet (Eleusine coracana (L.) Gaertn) composite collection. PhD Dissertation, Tamil Nadu Agricultural University, Coimbatore, India. http://oar.icrisat.org/113/1/A.Bharathi_Thesis.pdf
Crossa J, Rodriguez PP, Cuevas J et al (2017) Genomic selection in plant breeding: methods, models and perspectives. Trends Plant Sci 22:961. https://doi.org/10.1016/j.tplants.2017.08.011
Das IK, Palanna KB, Patro TSSK et al (2021) A multilocational evaluation of blast resistance in a diverse panel of finger millet in India. Crop Prot 139:105401. https://doi.org/10.1016/j.cropro.2020.105401
De Wet JMJ, Prasada Rao KE, Brink DE, Mengesha MH (1984) Systematic and evolution of Eleusine coracana (Gramineae). Am J Bot 71:550–557
Deshpande S, Tripathi MK, Mohapatra D, Jadam RS (2021) Product development from millets. In: Kumar A, Tripathi MK, Joshi D, Kumar V (eds) Millets and millet technology. Springer, Singapore. https://doi.org/10.1007/978-981-16-0676-2_
Dida MM, Srinivasachary, Ramakrishnan S et al (2007) The genetic map of finger millet, Eleusine coracana. Theor Appl Genet 114:321–332. https://doi.org/10.1007/s00122-006-0435-7
Dida MM, Wanyera N, Dunn MLH et al (2008) Population structure and diversity in finger millet (Eleusine coracana) germplasm. Trop Plant Biol 1(2):131–141. https://doi.org/10.1007/s12042-008-9012-3
Dida MM, Oduori CA, Manthi SJ et al (2021) Novel sources of resistance to blast disease in finger millet. Crop Sci 61(1):250–262
Dodake SS, Dhonukshe BL (1998) Variability in floral structure and floral biology of finger millet (Eleusine coracana (L.) Gaertn.). Indian J Genet 58:107–112
Dwivedi S, Upadhyaya HD, Senthilvel S et al (2012) Millets: genetic and genomic resources. Plant Breed Rev 35:247–375
Ekwamu A (1991) Influence of head blast infection on seed germination and yield components of finger millet (Eleusine coracana L. Gaertn). Trop Pest Manag 37(2):122–123. https://doi.org/10.1080/09670879109371556
Gimode D, Odeny DA, de Villiers EP et al (2016) Identification of SNP and SSR markers in finger millet using next generation sequencing technologies. PLoS One 11(7):e0159437. https://doi.org/10.1371/journal.pone.0159437
Gowda BTS, Seetharam A, Vishwanath S et al (1986) Incorporation of blast resistance to Indian elite finger millet cultivars from African cultivar IE 1012. SABRO J 18:119–120
Gupta SK, Velu G, Rai KN et al (2009) Association of grain iron and zinc content with grain yield and other traits in pearl millet (Pennisetum glaucum (L.) R.Br.). Crop Improv 36(2):4–7. https://doi.org/10.1186/2193-1801-3-763
Gupta A, Mahajan V, Gupta HS (2010) Genetic resources and varietal improvement of small millets for Indian Himalaya. In: Tewari LM, Pangtey YPS, Tewari G (eds) Biodiversity potentials of the Himalaya. Gyanodaya Prakashan, India, pp 305–316
Gupta A, Sood S, Agrawal PK et al (2011) Floral biology and pollination system in small millets. Eur J Plant Sci Biotechnol 6:81–86
Gupta AK, Gaur VS, Gupta S et al (2013) Nitrate signals determine the sensing of nitrogen through differential expression of genes involved in nitrogen uptake and assimilation in finger millet. Funct Integr Genomics 13:179–190. https://doi.org/10.1007/s10142-013-0311-x
Gupta SM, Arora S, Mirza N et al (2017) Finger millet: a “certain” crop for an “uncertain” future and a solution to food insecurity and hidden hunger under stressful environments. Front Plant Sci 8:643. https://doi.org/10.3389/fpls.2017.00643
Hatakeyama M, Aluri S, Balachadran MT et al (2018) Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res 25:39–47. https://doi.org/10.1093/dnares/dsx036
Hickey LT, Hafeez AN, Robinson H et al (2019) Breeding crops to feed 10 billon. Nat Biotechnol 37:744–754. https://doi.org/10.1038/s41587-019-0152-9
Hilu KW, DeWet JMJ (1976) Racial Evolution in Eleusine coracana ssp. coracana (Finger millet). Am J Bot 63:1311–1318. https://doi.org/10.1002/j.1537-2197.1976.tb13216.x
Hilu KW, de Wet JMJ, Harlan JR (1979) Archaeobotanical Studies of Eleusine coracana ssp. coracana (Finger Millet). Am J Bot 66:330. https://doi.org/10.1002/j.1537-2197.1979.tb06231.x
Hiremath SC, Salimath SS (1992) The “A” genome donor of Eleusine coracana (L.) Gaertn. (Gramineae). Theor Appl Genet 84:747–754
Hittalmani S, Mahesh HB, Shirke MD et al (2017) Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn) provides insights into drought tolerance and nutraceutical properties. BMC Genomics 18:1–16. https://doi.org/10.1186/s12864-017-3850-z
Jähne F, Hahn V, Würschum T et al (2020) Speed breeding short-day crops by LED-controlled light schemes. Theor Appl Genet 133(8):2335–2342. https://doi.org/10.1007/s00122-020-03601-4
Joshi DC, Sood S, Hosahatti R et al (2018) From zero to hero: the past, present and future of grain amaranth breeding. Theor Appl Genet 131:1807–1823
Joshi DC, Sood S, Gupta A et al (2021a) VL Mandua 382: the first early maturing, white seeded finger millet cultivar suitable for rainfed organic agro-ecology of the Himalayan region. Electron J Plant Breed 12(4):1308–1313
Joshi DC, Meena RP, Chandora R (2021b) Genetic resources: collection, characterization, conservation and documentation. In: Singh M, Sood S (eds) Millets and pseudocereals. Woodhead Publishing, pp 25–31
Kiran Babu T, Sharma R, Upadhyaya HD et al (2013a) Evaluation of genetic diversity in Magnaporthe grisea populations adapted to finger millet using Simple Sequence Repeats (SSRs). Physiol Mol Plant Pathol 84:10–18
Kamal K, Rachana D, Asis S (2016) Constraints and opportunities for promotion of finger millet in Nepal. https://doi.org/10.13140/RG.2.2.13997.69606
Kiran Babu T, Thakur RP, Upadhyaya HD (2013b) Identification of sources of blast resistance in mini-core collection of finger millet. Eur J Plant Pathol 135:299–311
Kiran Babu T, Sharma R, Thakur RP et al (2015) Pathogenic variation in Magnaporthe grisea populations adapted to finger millet [Eleusine coracana (L.) Gaertn.]. Plant Dis 99:1784–1789
Krishnamurthy L, Upadhyaya HD, Purushothaman R et al (2014) The extent of variation in salinity tolerance of the minicore collection of finger millet (Eleusine coracana L. Gaertn.) germplasm. Plant Sci 227:51–59. https://doi.org/10.1016/j.plantsci.2014.07.001
Krishnamurthy L, Upadhyaya HD, Kashiwagi J et al (2016) Variation in drought-tolerance components and their interrelationships in the minicore collection of finger millet germplasm. Crop Sci 56:1914–1926. https://doi.org/10.2135/cropsci2016.03.0191
Kumar AA, Reddy BVS, Sahrawat KL et al (2010) Combating micronutrient malnutrition: identification of commercial sorghum cultivars with high grain iron and zinc. J SAT Agric Res 8(1). ejournal.icrisat.org
Kumar A, Tripathi MK, Joshi D et al (2021a) Millets and millet technology. Springer, Singapore, p 438
Kumar A, Sharma D, Pathak RK et al (2021b) Science-led innovation for searching and creating values in natural gene pool of millets for agri-food nutrition and health. In: Kumar A, Tripathi MK, Joshi D, Kumar V (eds) Millets and millet technology. Springer, Singapore. https://doi.org/10.1007/978-981-16-0676-2_10
Maharajan T, Antony Ceasar S, Ajeesh Krishna TP et al (2021) Finger millet [Eleusine coracana (L.) Gaertn]: an orphan crop with a potential to alleviate the calcium deficiency in the semi-arid tropics of Asia and Africa. Front Sustain Food Syst 5:684447. https://doi.org/10.3389/fsufs.2021.684447
Manyasa EO, Tongoona P, Shanahan P et al (2014) Genetic diversity in East African finger millet (Eleusine coracana (L.) Gaertn) landraces based on SSR markers and some qualitative traits. Characterization and utilization. Plant Genet Resour 1–11. https://doi.org/10.1017/S1479262114000628
Manyasa EO, Tongoona P, Shanahan P et al (2019) Exploiting genetic diversity for blast disease resistance sources in finger millet (Eleusine coracana). Plant Health Progress 20(3):180–186
Maqsood M, Ali SNA (2007) Effects of environmental stress on growth, radiation use efficiency, and yield of finger millet (Eleusinecoracona). Pak J Bot 39(2):463–474
Mbinda W, Masaki H (2021) Breeding strategies and challenges in the improvement of blast disease resistance in finger millet. A current review. Front Plant Sci 11:602882
Meena RP, Joshi DC, Bisht JK et al (2021) Global scenario of millets cultivation. In: Kumar A, Tripathi MK, Joshi D, Kumar V (eds) Millets and millet technology. Springer, Singapore
Mehra KL (1963) Differentiation of the cultivated and wild Eleusine species. Phyton 20:189–198
Mgonja MA, Lenne JM, Manyasa E, Sreenivasaprasad S (eds) (2007) Finger millet blast management in East Africa: creating opportunities for improving production and utilization of finger millet: proceedings of the first International finger millet stakeholder workshop, Nairobi. International Crops Research Institute for the Semi-Arid Tropics, Patancheru 502 324, AP, India, pp 1–192. ISBN: 978-92-9066-505-2
Mohanty B, Gupta SD, Ghosh P (1985) Callus initiation and plant regeneration in ragi (Eleusine coracana Gaertn.). Plant Cell Tissue Organ Cult 5:147–150. https://doi.org/10.1007/BF00040311
Nagaraja A, Jagadish PS, Ashok EG et al (2007) Avoidance of finger millet blast by ideal sowing time and assessment of varietal performance under rainfed production situations in Karnataka. J Mycopathol Res 45(2):237–240
Nagaraja A, Gowda J, Krishnappa M et al (2008) GPU 28: a finger millet variety with durable blast resistance. J Mycopathol Res 46:109–111
Nanda JS, Agarwal PK (2008) Botany of Field crops, vol 1. Kalyani Publisher, India, p 381
Ng’uni D, Geleta M, Johansson E et al (2011) Characterization of the Southern African sorghum varieties for mineral contents: prospects for breeding for grain mineral dense lines. Afr J Food Sci 5:436–445
Ojulong HF, Sheunda P, Kibuka J et al (2021) Characterization of finger millet germplasm for mineral contents: prospects for breeding. J Cereals Oilseeds 12(1):33–44. https://doi.org/10.5897/JCO2020.0222
Onyango AO (2016) Finger millet: food security crop in the Arid and Semi-Arid Lands (ASALs) of Kenya. World Environ 6:62–70. https://doi.org/10.5923/j.env.20160602.03
Padulosi S, Mal B, King OI et al (2015) Minor millets as a central element for sustainably enhanced incomes, empowerment, and nutrition in rural India. Sustainability 7(7):8904–8933. https://doi.org/10.3390/su7078904
Palanna KB, Hosahatti R, Ramesh GV et al (2021) Current scenario and integrated approaches for management of finger millet blast (Magnaporthe grisea). In: Chandra Nayaka S, Hosahatti R, Prakash G, Tara Satyavathi C, Sharma R (eds) Blast disease of cereal crops: evolution and adaptation in context of climate change. Springer International Publishing, Cham, pp 27–49
Pandey G, Misra G, Kumari K et al (2013) Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet (Setaria italica L.). DNA Res 20(2):197–207. https://doi.org/10.1093/dnares/dst002
Paschapur AU, Joshi D, Mishra KK et al (2021) Millets for life: a brief introduction. In: Kumar A, Tripathi MK, Joshi D, Kumar V (eds) Millets and millet technology. Springer, Singapore. https://doi.org/10.1007/978-981-16-0676-2_1
Prasada Rao KE, de Wet JMJ, Reddy VG, Mengesha MH (1993) Diversity in the small millets collection at ICRISAT. In: Riley KW, Gupta SC, Seetharam A, Mushonga JN (eds) Advances in small millets. Oxford & IBH Publishing Co., New Delhi, pp 331–346
Rahman H, Jagadeesh SN, Valarmathi R et al (2014) Transcriptome analysis of salinity responsiveness in contrasting genotypes of finger millet (Eleusine coracana L.) through RNA-sequencing. Plant Mol Biol 85:485–503. https://doi.org/10.1007/s11103-014-0199-4
Rao ANS (1990) Estimates of losses in finger millet (Eleusine coracana) due to blast disease (Pyricularia grisea). J Agric Sci 24:57–60
Rawat L, Bisht TS, Kukreti A (2022) Potential of seed biopriming with Trichoderma in ameliorating salinity stress and providing resistance against leaf blast disease in finger millet (Eleusine coracana L.). Indian Phytopathol 75:147–164. https://doi.org/10.1007/s42360-021-00441-0
Reddy VD, Rao KV, Reddy TP et al (2008) Finger millet. In: Koleand C, Hall TC (eds) Compendium of transgenic crop plants: transgenic cereals and forage grasses. Blackwell, London, pp 191–198
Rodríguez JP, Rahman H, Thushar S et al (2020) Healthy and resilient cereals and pseudo-cerels for marginal agriculture: molecular advances for improving nutrient bioavailability. https://doi.org/10.3389/fgene.2020.00049
Saleh ASM, Zhang Q, Chen J et al (2013) Millet grains: nutritional quality, processing, and potential health benefits. Compr Rev Food Sci Food Saf 12:281–295. https://doi.org/10.1111/1541-4337.12012
Sampath SR (1986) Scope for using small millets as forage in India. In: Seetharam A, Riley KW, Harinarayana G (eds) Small millets in global agriculture. Proceedings of the 1st International Small Millets Workshop Bangalore, India, October 29–November 2, 1986
Sato S, Peet MM, Thomas JF (2002) Determining critical pre and post-anthesis periods and physiological process in Lycopersicon esculentum Mill. Exposed to moderately elevated temperatures. J Exp Bot 53:1187–1195
Seetharam A, Gowda J, Halaswamy JH (2003) Small millets. In: Chowdhury SK, Lal SK (eds) Nucleus and breeder seed production manual. Indian Agricultural Research Institute, New Delhi, pp 54–67
Sharma D, Jamra G, Singh UM et al (2017) Calcium biofortification: three pronged molecular approaches for dissecting complex trait of calcium nutrition in finger millet (Eleusine coracana) for devising strategies of enrichment of food crops. Front Plant Sci 7:2028. https://doi.org/10.3389/fpls.2016.02028
Sharma D, Tiwari A, Sood S et al (2018) Genome wide association mapping of agromorphological traits among a diverse collection of finger millet (Eleusine coracana L.) genotypes using SNP markers. PLoS One 13:e0199444. https://doi.org/10.1371/journal.pone.0199444
Shobana S, Krishnaswamy K, Sudha V et al (2013) Finger millet (Ragi, Eleusine coracana L.). A review of its nutritional properties, processing, and plausible health benefits, 1st edn. Copyright & Copy; 2013 Elsevier Inc. All rights reserved
Sood S, Joshi DC, Chandra AK, Kumar A (2019) Phenomics and genomics of finger millet: current status and future prospects. Planta 250:731–751. https://doi.org/10.1007/s00425-019-03159-6
Sood S, Babu BK, Joshi D (2022) History, botanical and taxonomic description, domestication, and spread. In: Kumar A, Sood S, Babu BK, Gupta SM, Rao BD (eds) The finger millet genome. Compendium of plant genomes. Springer, Cham. https://doi.org/10.1007/978-3-031-00868-9_1
Srinivasachary, Dida MM, Gale MD et al (2007) Comparative analyses reveal high levels of conserved co-linearity between the finger millet and rice genomes. Theor Appl Genet 115:489–499. https://doi.org/10.1007/s00122-007-0582-5
Swati P, Sahu PP, Beynon S et al (2020) Genome-wide association mapping and comparative genomics identifies genomic regions governing grain nutritional traits in finger millet (Eleusine coracana L. Gaertn.). Plants People Planet 2:649–662. https://doi.org/10.1002/ppp3.10120
Takan JP, Chipili J, Muthumeenakshi S et al (2012) Magnaporthe oryzae populations adapted to finger millet and rice exhibit distinctive patterns of genetic diversity, sexuality and host interaction. Mol Biotechnol 50(2):145–158
Takashi K, Kazuya W (2015) Spatial characteristics of long-term changes in Indian agricultural production: district-level analysis, 1965-2007. Rev Agrar Stud 5(1) Foundation for Agrarian Studies
Tanksley S, Grandillo S, Fulton TM et al (1996) Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor Appl Genet 92:213–224
Teka HB (2014) Advance research on striga control: a review. Afr J Plant Sci 8(11):492–506
Tenywa JS, Nyende P, Kidoido M et al (1999) Prospects and constraints for finger millet production in eastern Uganda. Afr Crop Sci J 7:31–35. https://doi.org/10.4314/acsj.v7i4.27751
Tiwari A, Sharma D, Sood S et al (2020) Genome-wide association mapping for seed protein content in finger millet (Eleusine coracana) global collection through genotyping by sequencing. J Cereal Sci 91:102888. https://doi.org/10.1016/j.jcs.2019.102888
Tyagi DVS, Rawat RS (1989) Two new ragi var. for rainfed areas. Indian Farm 38:3
Upadhyaya HD, Gowda CLL, Pundir RPS et al (2006) Development of core subset of finger millet germplasm using geographical origin and data on 14 quantitative traits. Genet Resour Crop Evol 53:679–685. https://doi.org/10.1007/s10722-004-3228-3
Upadhyaya H, Gowda C, Reddy VG (2007) Morphological diversity in finger millet germplasm introduced from Southern and Eastern Africa. J SAT Agric Res 3:1–3
Upadhyaya HD, Sarma NDRK, Ravishankar CR et al (2010) Developing a mini-core collection in finger millet using multilocation data. Crop Sci 50(5):1924–1931. https://doi.org/10.2135/cropsci2009.11.0689
Upadhyaya HD, Ramesh S, Sharma S et al (2011) Genetic diversity for grain nutrients contents in a core collection of finger millet (Eleusine coracana (L.) Gaertn.) germplasm. Field Crops Res 121(1):42–52. https://doi.org/10.1016/j.fcr.2010.11.017
Vadivoo AS, Joseph R, Ganesan NM (1998) Genetic variability and diversity for protein and calcium contents in finger millet (Eleusine coracana (L.) Gaertn) in relation to grain color. Plant Foods Hum Nutr 52:353–364
Varshney RK, Graner A, Sorrells NE (2005) Genomics assisted breeding for crop improvement. Trends Plant Sci 10(12):621–630. https://doi.org/10.1016/j.tplants.2005.10.004
Venkatesh Babu D, Sudhakar P, Kumar RS (2013) Screening of thermo tolerant ragi genotypes at seedling stage using TIR technique. Int Qua J Sci 8(4):1493–1495
Vetriventhan M, Upadhyaya HD, Dwivedi SL et al (2016) Finger and foxtail millets. In: Genetic and genomic resources for grain cereals improvement. Academic, pp 291–319
Vetriventhan M, Azevedo VCR, Upadhyaya HD et al (2020) Genetic and genomic resources, and breeding for accelerating improvement of small millets: current status and future interventions. Nucleus 63:217–239. https://doi.org/10.1007/s13237-020-00322-3
Vishwanath S, Sanne Gowda S, Seetharam A et al (1986) Reaction to blast disease of released and pre-released varieties of finger millet from different states. Millet Newslett 5:31
Yadav S, Kumar A, Sood S (2020) Unraveling the genetics of calcium content in finger millet grains through association mapping. Indian J Genet 80:432–440. https://doi.org/10.31742/IJGPB.80.4.9
Yogeesh LN, Naryanareddy AB, Nanjareddy YA et al (2016) High temperature tolerant genotypes of finger millet (Eleusine coracana L.). Nat Environ Pollut Technol 15(4):1293–1296
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Joshi, P. et al. (2023). Finger Millet Improvement in Post-genomic Era: Hundred Years of Breeding and Moving Forward. In: Sharma, D., Singh, S., Sharma, S.K., Singh, R. (eds) Smart Plant Breeding for Field Crops in Post-genomics Era . Springer, Singapore. https://doi.org/10.1007/978-981-19-8218-7_7
Download citation
DOI: https://doi.org/10.1007/978-981-19-8218-7_7
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-8217-0
Online ISBN: 978-981-19-8218-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)