Abstract
Congenital heart disease (CHD) has a strong genetic etiology, making it a likely candidate for therapeutic intervention using genetic editing. Complex genetics involving an orchestrated series of genetic events and over 400 genes are responsible for myocardial development. Cooperation is required from a vast series of genetic networks, and mutations in such can lead to CHD and cardiovascular abnormalities, affecting up to 1% of all live births. Genome editing technologies are becoming better studied and with time and improved logistics, CHD could be a prime therapeutic target. Syndromic, nonsyndromic, and cases of familial inheritance all involve identifiable causative mutations and thus have the potential for genome editing therapy. Mouse models are well-suited to study and predict clinical outcome. This review summarizes the anatomical and genetic timeline of myocardial development in both mice and humans, the potential of gene editing in typical CHD categories, as well as the use of mice thus far in reproducing models of human CHD and correcting the mutations that create them.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Overview
The heart is the first visceral organ to be formed during organogenesis [1]. Myocardial development involves orchestrated series of genetic networks turning on and off cooperatively to execute molecular, cellular, and morphogenetic events to form a normal heart [2, 3]. Mutations in these genes result in congenital heart disease (CHD) at birth or cardiovascular abnormalities later in life [1, 2]. CHD is the most common congenital abnormality, which occurs in about 1 in 100 live births [4] and in 10% of aborted fetuses [5]. More than 400 genes have been associated with CHD [6]. Oyen and associates [7, 8] investigated the overall CHD risks among family members of the proband and found that the risks for concordant [7, 8] and discordant [8, 9] defects among the first-degree relatives are 3–80 [7, 8] and 2 [8, 9], respectively. This is indicative of the genetic and genomic underpinning of CHD [2, 10] (Excellent recent reviews please see Williams et. al. [6, 11], Zaidi and Brueckner [6, 11]).
Should the enormous scientific, logistical, clinical, ethical, and regulatory issues regarding the use of therapeutic genome editing technologies be resolved, CHD would be prime targets. The syndromic CHDs are typically due to large chromosomal defects, such as microdeletions and microduplications, and associated with significant abnormalities apart from the cardiovascular system. Nonsyndromic CHD cases outnumber syndromic cases, and are mainly due to monogenic or digenic mutations, which are small (point mutations mostly), and vis-à-vis state-of-the-art genome editing, correctable.
Familial inheritance of most CHD cases favors the use of genome editing in treatment. The causative mutated gene is often known or can be easily identified, particularly as the list of genes associated with CHD increases (for comprehensive gene lists, see [6]). As well, phenotypic abnormalities of nonsyndromic cases are usually confined to the heart, their developmental onset and progression may be known, and the severity of the anomaly and thus the need for some form of intervention predictable. Lastly, the position of the heart in the vascular system may mean the delivery of editing tools and their effectiveness would be outstanding.
Mouse models of CHD would be essential to formulating a CHD gene editing plan of known efficacy and a strong prediction of a beneficial clinical outcome. Mice are a favorable model system as many of their genes, gene modifiers, and molecular pathways are conserved with those of humans. Biological processes, anatomy, and physiology are likewise conserved with humans, examples being four-chambered hearts, vascular systems, and visceral organs, which share similar structure and function [12,13,14,15].
Mouse models of specific human molecular variants can be easily constructed to determine the best developmental time and approach to intervene and correct the gene defect.
This review begins with a discussion of the anatomical development of the human and mouse hearts (Table 1 and Figs. 1 and 2), including a list of the genes (Table 2) and their roles at anatomical positions and gestational times. We follow this with a discussion of the potential roles of gene editing in different traditional categories of CHD. We complete our review with examples of how mice have been used so far, both in generating models of human CHD and in correcting mutations in orthologs of human CHD genes.
Schematic of human cardiac morphogenesis. Illustrations depict cardiac development with color coding of morphologically related regions, seen from a ventral view. Cardiogenic precursors form a crescent (left-most panel) that is specified to form specific segments of the linear heart tube, which is patterned along the anterior–posterior axis to form the various regions and chambers of the looped and mature heart. Each cardiac chamber balloons out from the outer curvature of the looped heart tube in a segmental fashion. Neural crest cells populate the bilaterally symmetrical aortic arch arteries (III, IV, and VI) and aortic sac (AS) that together contribute to specific segments of the mature aortic arch, also color coded. Mesenchymal cells form the cardiac valves from the conotruncal (CT) and atrioventricular valve (AVV) segments. Corresponding days of human embryonic development are indicated. A atrium, Ao aorta, DA ductus arteriosus, LA left atrium, LCC left common carotid, LSCA left subclavian artery, LV left ventricle, PA pulmonary artery, RA right atrium, RCC right common carotid, RSCA right subclavian artery, RV right ventricle, V ventricle. (Re-use with copyright permission granted from [2])
Mouse cardiac development. The heart originates from mesodermal cells in the primitive streak. During gastrulation, cardiac progenitors migrate to the splanchnic mesoderm to form the cardiac crescent. At E7.5 in the mouse, the cardiac crescent can be divided into two heart field lineages based on differential gene expression and their respective contribution to heart, a first heart field (red) and a second heart field (yellow), which is located posteriorly and medially to the first heart field. At E8.0, the linear heart tube is present. At E8.5, the looping is associated with uneven growth of cardiac chambers. The outflow tract is at the arterial pole and the inflow tract and primitive atria are at the venous pole. By E9.5, the common atrium has moved superior to the ventricles and is separated by a distinct atrio-ventricular canal. By E10.5, cardiac neural crest cells from the dorsal neural tube migrate via the pharyngeal arches to the cardiac outflow tract. Further cardiac development involves a series of septation events and myocardial trabeculation that result in a mature four-chambered heart integrated with the circulatory system depicted at E15.5. (Re-use with copyright permission granted from [16])
2 Cardiac Development
2.1 Early Development and Cardiac Crescent
Heart development begins [20] when cells in the anterior lateral mesoderm move from the primitive streak and give rise to cardiomyocytes, beginning the process that allows the heart to form and contract. These mesodermal cells gather in shape known as the cardiac crescent, made of the first and second heart fields. It is thought that this movement is facilitated by an influx of transcription factors and secreted molecules [10]. Cells in the first heart field go on to form the linear tube; meanwhile, cells in the second heart field, which is medial and dorsal to the first heart field, become the right ventricle, outflow tract, and parts of the atria [16].
2.2 Cardiac Looping
One of the first cells to be specified are cardiac muscle cells, which quickly find their way to the ventral midline of the embryo and form a beating heart tube. This tube has an endocardial and myocardial layer with a layer of extracellular matrix (ECM) in between. [2]. The following process to occur is known as cardiac looping and it takes place around embryonic day E9.5 and E10.5 in mice and weeks 6 4/7 and 7 5/7 in humans [17]. In the lateral mesoderm, uneven gene expression will cause the linear heart tube to loop to the right. This event is essential for heart chamber formation, and the proper alignment between heart chambers and vasculature [2]. Structurally on the left side, blood will go from, what at this point is made up of the atrial cavity, atrial ventricular junction, and what will be the left ventricle to the interventricular foramen. On the right side, blood will flow from the early right ventricle to the truncus arteriosus. In addition, the arterial ventricular junction is enveloped in endocardial cushion tissue [18]. During this time of development, the ventricular and atrial chambers grow in size and start to become distinct on the left and right. Along with this distinction, the interventricular foramen narrows as the ventricles grow, allowing the ventricles to continue communicating. Also, the atrial ventricular junction becomes denser [18].
2.3 Atrium, Sino Atrial Node, and Atrial Ventricular Node Development
Beginning in the primary heart tube, the atrial septum divides into the septum primum and the septum secundum and looks similar in both mice and humans. The process spans E10.5 to E13.5 in mice and days 48–56 in humans [17]. Starting at the posterior wall of the atrium, the septum primum extends out and eventually meets the endocardial cushions surrounding the atrial ventricular junction. This spine that extends from the wall of the atrium is made of mesenchymal cells that later become muscle cells. This process helps to close the interventricular foramen [18]. Later, the septum secundum formation begins in the dorsal wall of the atrium, when it in folds and grows to the right of where the pulmonary vein will develop [17].
In addition to septation, between E10.5 and E11.5 in mice and around week 5 in humans, some atrial mesenchymal cells will become the sinoatrial node. The comma-shaped node’s characteristic head and tail regions develop independently, in order of functions. As it develops, the left-sided sinoatrial node joins the surrounding myocardium and begins to look like characteristic working myocardium. But unlike the left-sided atrial node, the right side does not look like its working myocardium surroundings. Instead, it keeps a more primitive function, enabling it to be tracked even in late development [19].
2.4 Ventricular Development
Like atrial development, ventricular development looks very similar between mice and humans. It begins on day E11.5 in mice and week 8 in humans, near the end of the looped heart stage [17]. During ventricular development, the ventricular septum protrudes from the ventral part of the ventricular chamber floor and grows toward the atria. Closing the ventricular septum is thought to be mediated by cells moving from the secondary heart field in the dorsal direction.
2.5 Atrioventricular Valve Development
Atrioventricular valve development spans over E10.5-E17.5 in mice and days 48–66 in humans. The valves’ structure is complete by E12.5 and day 56, in mice and humans, respectively, but continues to develop into their polished versions until E17.5 and day 66 [17]. Endocardial cushions, or extracellular matrix areas, that line the atrial ventricular canals divide this area into left and right, and create the atrial ventricular valves. There forms the mitral valve on the left and the tricuspid valve on the right, though at this stage they are thick. In the next 10 days of human gestation, these valves thin and are developed. In mice though, the tricuspid valve takes until E17.5 to develop, and even further development occurs postnatally [17].
2.6 Outflow Track Development
The truncus arteriosus is the site of many events in cardiac development. Cells from the second heart field interacting with neural crest cells will create a septum and become arteries [18]. In humans and mice, at days 50 and 11.5, respectively, neural crest cells facilitate the formation of two ridges. As the truncus arteriosus cushions begin twisting, they separate into the aorta and pulmonary arteries. At least this is true distally. At this point, proximally the truncus arteriosus is still connected as one [18]. By day E12.5 in mice, the outflow tracts are separate proximally as well. In general, the development of the outflow tracts looks similar between humans and mice [17]. This process is delicate in that 30% of congenital heart defects are due to this neural crest cell process. ET-1, dHAND, and neurophilin-1 are known to regulate neural crest cell development [2]. Neural crest cells are proven to be needed to properly close the ductus, separate the outflow tract, form the aortic arch, and form the ventricular septation [10]. Neural crest cells are also thought to induce the development of the cardiac conduction system, though it is not known precisely how [19].
The semilunar valve is made in a process similar to that of the atrial ventricular valves, in that it comes from cushion tissue, this time truncal, and is thick to start. Semilunar valves then thin out over time. This process begins in mice at day E12.5 and week 8 in humans [17].
2.7 Conductance System
Development of accessory pathways, or accessory bundles of cardiomyocytes, are essential as the atria and ventricles develop, because they conduct action potentials in both the atrial ventricular direction and the ventricular-atrial direction. When the atrial ventricular junction forms, conduction between the atria and ventricles is one of the only connections the two areas have. They are located both endocardially and epicardially to start, and thus have different cellular morphologies. However, it is also imperative that these decrease in number and size as the heart, specifically the atrioventricular junction, develop, or else it can lead to cardiomyopathies later on in development [19].
When it comes to the cardiac conduction system, the posterior end of the heart field will give rise to the sinoatrial node. Some suspect it will also contribute to the formation of the atrioventricular node. However, it is still debated [19]. Furthermore, at E9.5 in mice, epicardial cells from the venous pole of the heart migrate over the developing heart to create the outer layer of the epicardium. Epicardium-derived cells go on to help form smooth muscle cells, coronary vasculature, the atrial ventricular valves, and the compact myocardium. Still, they are also thought to play a role in developing the peripheral conduction system through Purkinje fiber cells [19]. When it comes to the atrioventricular node, it starts to develop at week 5 in humans and day E11.5 in mice. The atrioventricular node develops from the myocardium and begins as an anterior and posterior node, the posterior node eventually playing the more significant role. After all, it is the node that connects to the His bundle. Ultimately, the anterior and posterior atrioventricular nodes fuse [19]. There are many theories as to which cells exactly give rise to the atrioventricular node, but for now, it is only agreed upon that it has multiple cell sources [19]. During early development, the atrioventricular canal conducts slowly, which is known as the atrioventricular delay. As the heart continues to develop, the annulus fibrosus forms and interrupts the myocardial continuity, which would interrupt conduction to the ventricles. As a result, the common bundle begins conducting the electrical impulse to the now working ventricular myocardium and, these electrical impulses speed up [19].
3 Genetic Archetypes in Cardiac Development
As a beginning step, the heart tube formation initiates with the help of the progenitor cells within the anterior lateral plate mesoderm, which becomes committed to a cardiogenic fate around embryonic day (E) 15 in humans. Specific signaling molecules such as bone morphogenetic proteins, fibroblast growth factors (Fgfs), and Wnts are responsible for this step [21,22,23]. Cardiac precursors bilaterally come together and fuse at the cephalic portion of the primitive streak and forms the cardiac tube. This straight heart tube contains an outer myocardium and an inner endocardium separated by an extracellular matrix (ECM) known as the cardiac jelly. This process is shown to be under GATA transcription factors control [24]. The tubular heart initiates rhythmic contractions at approximately E23.0 in humans. The linear heart tube is segmentally shaped along the cranial (arterial pole) to caudal (venous pole) end into precursors of the aortic sac, conotruncus (outflow tracts), and primitive ventricle, primitive atria, and sinus venosus. The original upside-down heart tube lies in the cranial part of the embryo and needs to be curved. This critical development is called cardiac looping. In all vertebrates, the linear heart tube at first undergoes rightward, C shape looping and next S shape looping, which is essential for proper orientation of the pulmonary (right) and systemic (left) ventricles, and remodeling of the heart chambers with the vasculature [25]. With this looping process, the heart tube changes its orientation. From cranial to caudal direction, the structures lie as the aortic sac, primitive atrium to the primitive ventricle, and bulbus cordis.
The molecular mechanisms controlling the cardiac looping remain unknown but transforming growth factor-ß (TGF-ß) seems to be playing the role. The creation of a looped heart tube then enables the structure of four chambers and the arterial venous poles. After proper looping, the heart tube is ready to be divided into four chambers. This is followed by symmetrical atrial septation into the left and right atria, which governs NKX2.5 and TBX5 genes. The formation of heart valves and sequential ventricular septation into the left and right ventricles with a formation of the primitive interventricular septum between them, is mainly controlled by the TBX5 gene [2]. As we improve our understanding of cardiac development and the role of genetics in this process, the more underlying pathological processes depend on genetic abnormalities. We have enough evidence to assume that a significant portion of the CHD’s originates from errors or disruption in heart development’s genetic control.
The etiology of congenital heart defects is recently becoming a more interesting topic in the literature. When we look at the genetic determinants of CHD, we can identify almost 30% of the genetic abnormalities behind CHD’s. The majority of the genetic determinants, nearly 70%, are still unrecognized [11]. However, as we improve our understanding of the genetic contribution to heart development and improve genetic technology, the undetermined portion will be less in the near future. Gene therapy is becoming a compelling treatment option when genetic etiology is apparent in many diseases. In this review, we will divide the genetic archetypes behind the CHD, and we will review the gene therapy options based on the genetic model of the CHD.
4 Genetic Archetypes for Syndromic Congenital Heart Defects
This subset of CHD cases has an exact genetic etiology, including various chromosome abnormalities, microdeletion/microduplication syndromes or, single-gene disorders, some of which are syndromic and some of which are nonsyndromic. Recurrence risk estimation is much easier for these cases with a clear genetic etiology; the magnitude of the risk depends on the specific cause. Except for the nonsyndromic single-gene causes of CHD, genetic causes of CHD often involve clinical or developmental features in addition to CHD. People with Down syndrome often have a higher than average number of abnormalities, such as intellectual disabilities, hypotonia, dysmorphic features, and other extracardiac symptoms [26]. Deletion 22q syndrome is commonly associated with oral clefting, velopharyngeal insufficiency, learning disabilities, calcium regulation issues, and thymus hypoplasia [27]. People with Holt–Oram syndrome due to mutations in TBX5 are often characterized by abnormalities in the limb and heart, such as atrial and ventricular septal defects [28]. Costello syndrome occurs due to HRAS mutations and can cause pulmonary stenosis and hypertrophic cardiomyopathy in the heart. People with Alagille syndrome typically have bile duct paucity, typical facies, and vertebral and heart anomalies. In this syndrome, the mutations are generally in JAG1and NOTCH2, and most cardiac defects are pulmonary stenosis, hypoplasia, and Tetrology of Fallot [29]. Currently, prevention and treatment for this group have not been the focus of the current era. We are doing an excellent job counseling the recurrence risk and diagnosing them prenatally to inform the patients.
5 Genetic Archetypes of Nonsyndromic Isolated Congenital Heart Defects
In recent years, we have learned about several single genes that, when mutated, are associated with nonsyndromic familial CHD. Unlike syndromic CHD, where individuals often have various other medical and developmental concerns in addition to their cardiac problems, nonsyndromic familial CHD is associated with isolated heart defects. Many point mutations of NKX2.5 have been found in families with atrial septal defects and arrhythmias [30]. Sporadic mutations of the NKX2.5 can cause tetralogy of Fallot, an outflow tract alignment defect, and tricuspid valve defects. The exact mechanism of these mutations resulting in cardiac defect is not precise yet. The linkage of particular loss of function with mutations in some distinct abnormalities suggests that different aspects of NKX2.5 functions can be altered in other developmental portions of the heart. Typically, these conditions exhibit both reduced penetrance and variable expressivity. For example, some people in a family who inherit an NKX2.5 mutation will have completely normal hearts, some will have Tetralogy of Fallot, some may have atrial septal defects, etc., but everyone who inherits the mutation, regardless of their phenotype, can transmit that mutation to the next generation [31]. Mutations in GATA4 can cause atrial septal defects, atrioventricular septal defects, and great artery abnormalities, specifically pulmonary artery abnormalities. NOTCH1 mutations go with the bicuspid aortic valve, aortic stenosis, aortic coarctation, and hypoplastic left heart syndromes [32]. The currently known genes to cause nonsyndromic familial CHD’s are listed in Table 2. These new developments demonstrate that single-gene defects can lead to isolated congenital heart disease and reveal more about molecular pathways important in cardiac morphogenesis.
6 Genetic Archetypes for Left–Right Patterning
The left-right asymmetry of the heart is required for proper oxygenation of the body, with the left side of the heart holding the responsibility of systemic circulation in order to provide oxygenated blood throughout the body. In contrast, the right side of the heart is responsible for pulmonary circulation to the lung for gas exchange. The abnormal left-right patterning, or laterality defect, is highly associated with CHD [33, 34] indicating the importance of left-right patterning in cardiac development.
This left-right asymmetry established with the rightward cardiac looping (the 4th week of human gestation and E8.5-E10.5 in mice) reflects the left–right body axis. The human body is highly asymmetric, with the body plan following the three axes (anteroposterior [A/P], dorsoventral [D/V], left-right [L/R]) that are established very early in embryonic development (human embryonic day E23 and mouse embryonic day E8.5) [35]. The major visceral organs are packed into the human body with a striking left-right asymmetry. The vast majority of the human population has developed this asymmetric thoracoabdominal organ arrangement, known as the normal situs, called situs solitus (SS). Comparative studies revealed that this directionality of the situs asymmetry is vertebrate-conserved, from fish, frog, mouse, to higher mammals, including humans [36]. However, when the asymmetry fails to develop correctly, it results in a pathogenic condition, heterotaxy, also known as situs ambiguous, which is generally associated with a spectrum of intra-cardiac defects, found in 1 of 10,000 births, and is associated with at least 3% of CHD cases [37]. Over the past few decades, it has been recognized that cilia, the highly conserved microtubule-based structures found in almost all cell types, play central roles in left-right asymmetry in development. Ciliary abnormality, primary ciliary dyskinesia (PCD) [38], accounts for a host of human diseases such as cystic kidney disease, retinal degeneration, and Bardet-Biedl and Meckel-Gruber syndromes [39,40,41,42,43,44,45,46]. The left–right patterning [47] is established by nodal motile cilia rotating clockwise to generate a leftward flow of morphogens resulting in an asymmetrical gradient around the node, thus breaking the initial embryonic symmetry and establishing left and right asymmetry [41, 45]. Intra-ciliary calcium oscillation dynamics [41,42,43, 48] are identified as a key signaling pathway that initiates cascades of subsequent events in left–right development. Growing numbers of genetic analyses in both humans [37, 49,50,51,52] and mice [53,54,55,56,57,58] have uncovered arrays of PCD genes. Among all the PCD patients examined [49], about 48% developed SIT, 6% heterotaxy, and 46% had normal SS. How the bodily left–right axis established during nodal development affects the left–right patterning in the heart is not completely clear. A recent discovery of intrinsic cellular chirality [59] showed that the developing chick cardiomyocytes are intrinsically chiral and exhibit dominant clockwise rotation in vitro. Furthermore, the developing myocardium is chiral as evident by a rightward bias of cell alignment and a rightward polarization of the Golgi complex, correlating with the direction of cardiac looping. It is possible that the intrinsic cellular chirality regulates the left–right patterning from a cellular level, cardiac looping, to the overall body plan.
7 Genetic Archetypes of Inherited Arrhythmias
The most common inherited primary arrhythmia syndromes are Long QT syndromes, catecholaminergic polymorphic ventricular tachycardia, Brugada syndromes, and short QT syndromes. The inherited primary arrhythmia syndromes are mainly caused by cardiac channelopathies. Genetic mutations that cause inherited primary arrhythmia syndromes are mostly in genes encoding ion channels and associated regulatory proteins in the heart [60]. The inheritance pattern for primary arrhythmia syndromes is usually Mendelian. The onset of the disorders appears early in life. Epidemiological studies on the spectrum of etiologies of sudden cardiac death (SCD) indicated that the primary arrhythmia disorders are one major culprit of SCD among young and healthy individuals [61]. However, clinical features and phenotypical expression of the inherited primary arrhythmia syndromes resulting from cardiac channelopathies can be variable [62]. The complex interplay of the mutation characteristics, epigenetic, and environmental factors can influence the vulnerability to arrhythmias as well as the disease progression [63]. Many of these diseases exhibit overlapping symptoms. Therefore, precise genetic testing can be of merit in diagnosis, prognosis, guiding clinical management and more specific therapy [62].
7.1 Long QT Syndromes
The congenital long QT syndrome (LQTS) is one of the most common inherited arrhythmias in structurally normal hearts. It is usually diagnosed with prolongation of the QT interval on the electrocardiogram (ECG). Clinically it can cause syncope, seizures, polymorphic ventricular tachycardia (VT) (torsades de pointes), cardiac arrest, and sudden death.
To date, 16 genes have been associated with LQTS. The most common genes seen in 90% of all genotype-positive cases are KCNQ1 (LQTS1), KCNH2 (LQTS2), and SCN5A (LQTS3). The diagnostic yield of these mutations is high. It allows us to understand the penetrance type and determine the risk for the upcoming generations in the same family before being discovered by their clinical symptoms. As a treatment option, the type of mutation will help us start prophylactic treatments, which have already been proven to reduce cumulative mortality [64].
7.2 Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT)
Catecholaminergic polymorphic ventricular tachycardia (CPVT) is an inherited arrhythmic disorder. It is characterized by adrenergic mediated polymorphic or bidirectional ventricular tachycardia (VT) that may degenerate into ventricular fibrillation (VF) which can cause cardiac arrest or sudden cardiac death in patients with structurally normal hearts. As indicated by Drs. Asatryan and Mederios-Domingo [65], for patients with clinical CPVT is suspected, the presence of pathogenic mutations of RyR2 or CASQ2 can be diagnosed by genetic testing, which can have almost 60% diagnostic yield. Besides confirming the diagnosis, a positive result is very useful to identify other affected family members at risk for sudden cardiac death [65, 66].
7.3 Brugada Syndromes (BrS)
Brugada syndrome (BrS) is characterized by a typical ECG pattern of coved-type ST-segment elevation with successive negative T waves in the right precordial leads with or without cardiac conduction delays. Ventricular tachyarrhythmias and sudden death in sleep are the most common clinical findings and manifest between 30 and 40 years of life. The prevalence is higher in males. Although it is rare compared to Long QT syndrome, it is a silent killer due to silent course and intermittent ECG patterns. Although the genetic test’s diagnostic yield is around 30%, mutation-specific genetic testing is recommended for family members after identifying a causative mutation. It allows for presymptomatic diagnosis in relatives at risk who need further clinical follow-up, prophylactic treatment, etc. At least 20 genetic mutations have been found to account for 30–35% of BrS cases [65, 67], with loss-of-function mutations in SCN5A contributing to about 30% of said cases [65, 68, 69]. Therefore, global as well as specific SCN5A genetic testing is an expected course of action for any patient suspected of having BrS. It is generally recommended that family members of BrS patients have genetic testing as well in order to allow for early diagnosis and presymptomatic clinical and treatment plans.
7.4 Short QT Syndrome (SQTS)
While exceedingly rare, short QT syndrome (SQTS) is a heritable, grave, deadly cardiac channelopathy. ECG reveals short QT intervals in these patients, making them increasingly vulnerable to atrial and ventricular arrhythmias and sudden death [65, 70]. While some patients present first with these arrhythmias in the form of heart palpitations or syncope, 40% of cases present cardiac arrest as their first symptom, with any survivors showing a high rate of recurrence. KCNH2 (SQTS1) was the first gene discovered in relation to SQTS, with about an 80% penetrance, though data is limited. Therefore those with any clinical suspicion or family history of SQTS should undergo genetic screening for three major genes associated with SQTS, KCNH2, KCNQ1, and KCNJ2, the yield of which is around 40% [65, 71, 72].
8 Genetic Archetypes of Inherited Cardiomyopathy
Cardiomyopathy is a form of heart disease affecting the cardiac muscle and can cause major cardiac-related morbidity in almost all ages. A significant portion of them has a genetic origin. Advances in molecular genetics allowed us to identify multiple genes responsible for cardiomyopathies. Surprisingly, different mutations in the same gene can cause different types of cardiomyopathies. Cardiomyopathies can be classified as dilated, hypertrophic, arrhythmogenic right ventricular, restrictive, or left ventricular non-compaction cardiomyopathies. As a neuromuscular disorder, especially Duchenne and Becker muscular dystrophies, cardiomyopathy is also characterized by skeletal myopathy [73].
8.1 Dilated Cardiomyopathy (DCM)
Dilated cardiomyopathy (DCM) is characterized by left ventricular dilatation and abnormality in systolic function. DCM is the most common indication for cardiac transplant. Inheritance patterns are generally autosomal dominant in 30–50% of cases. Small percentages can be autosomal recessive, X-linked, and mitochondrial inheritance. More than 40 genes have been described in DCM. Defects with LMNA-encoded lamin mutations, myosin heavy chain beta mutations, ribonucleic acid-binding protein mutations, and many other complex molecular deficits have been implicated in the pathogenesis of DCM [74]. Most genetic mutations associated with DCM have extremely low prevalence and high heterogeneity. Therefore, it is often necessary to sequence large numbers of genes in order for effective genetic testing. If the DCM is together with conduction disease and/or arrhythmia and strong family history, then focused testing for LMNA, desmosomal, and SCN5A mutations may have a substantial clinical impact. Identification of a genetic mutation in the setting of family history allows early screening, appropriate monitoring, and prophylactic treatments [75].
8.2 Hypertrophic Cardiomyopathy (HCM)
The inheritance pattern of hypertrophic cardiomyopathy (HCM) is autosomal dominant, characterized by concentric hypertrophy of the left ventricle and the septum [76]. The genes that encode sarcomeric proteins are involved in the pathogenesis of HCM. The most common of them that accounts for 20–30% of the HCM is mutations in MYH7 (encoding the β-myosin heavy chain), MYBPC3 (encoding the cardiac myosin binding protein C), and cardiac troponin T (TNNT1and 2) [77]. These mutations, in general, cause decreased myocyte relaxation and increased myocyte growth with prominent involvement of the interventricular septum. Approximately 10% of the patients may carry multiple sarcomeric mutations, presenting with more severe diseases at younger ages. This indicates the need for detailed genetic evaluation of the family for early diagnosis and treatment. The genetic diagnosis can go up to 70% if there is a family history of HCM. The yield is lower when sporadic diseases are considered. For effective screening, it is important to know the pathogenicity of the mutation. Many mutations that cause HCM can be unique to the individual family; therefore, careful genetic counseling and family assessment are needed in this type of inherited cardiomyopathy [78].
8.3 Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC)
The arrhythmogenic right ventricular cardiomyopathy (ARVC) is a form of heart disease characterized by fibrosis and fatty infiltration of the myocardium, mainly in the right ventricle. The inheritance pattern for ARVC is autosomal dominant with incomplete penetrance. Mutations in the genes cause encoding desmosomal proteins are the main etiology. These mutated genes disorganized desmosomal integrity, making muscle fibers more fragile, sensitive to tearing, fragmentation, and eventually cell death in the course of the cardiac cycle. As a result, the desmosomal function, the gap junction remodeling, sodium channel function, and electrocardiographic parameters in cardiomyocytes are also compromised. Besides, disturbance of desmosomal proteins promotes adipogenesis in mesodermal precursors by suppressing the Wnt/β-catenin signaling pathway. This particular pathway is known for its role in cardiac myogenesis [79].
As a consequence of this abnormal process, the fibro-fatty replacement of the ventricular myocardium becomes more prominent in the RV. Multiple variants in the mutation cause early presentation and more severe presentation of the disease. The presence of more than one variant was associated with a nearly fivefold increase in odds of penetrant disease. This information is essential during the genetic evaluation, and recommendation is usually needed to sequence all five desmosomal genes [80].
8.4 Restrictive Cardiomyopathy (RCM)
Restrictive cardiomyopathy (RCM) is a form of heart disease in which the heart chambers gradually become stiff over time. The initial findings are increased ventricular stiffness that impairs ventricular filling without ventricular hypertrophy or systolic dysfunction [81]. The most common inheritance is autosomal dominant. Alterations in genes encoding for sarcomeric proteins (e.g., TNNT2), Z-disc proteins (e.g., MYPN), or transthyretin (TTR) have been identified in patients with RCM [82]. Familial RCM is increasingly recognized as a specific phenotype within the HCM spectrum and can be seen in those who share mutations expressed as classic hypertrophic cardiomyopathy in other family members.
8.5 Left Ventricular Non-compaction Cardiomyopathy
This is a heterogeneous disorder characterized by prominent trabeculae, a thin compacted layer, and deep intertrabecular recesses most evident in the left ventricle apex. Non-compaction may involve the right ventricle, presenting as either a biventricular or isolated right ventricular non-compaction phenotype. The genetic form is commonly inherited as an X-linked recessive or autosomal dominant condition [83]. Mutations that affect the compaction of the endomyocardial layer progress from the base to the apex of the heart during embryogenesis. The genes encoding for sarcomeric (e.g., MYH7), Z-disc (e.g., LDB3), nuclear envelope (e.g., LMNA), mitochondrial (e.g., TAZ), and ion channel proteins (e.g., SCN5A) are found to be responsible for this type of cardiomyopathy.
8.6 Cardiomyopathy in Other Disorders
Duchene muscular dystrophy (DMD), Beker’s muscular dystrophy, Marfan syndrome, and Barth syndrome are the other disorders where different types of CMP can be observed. In DMD, three stages are present, usually starting with hypertrophic CMP and some diastolic dysfunction with no heart failure symptoms. Later the heart dilates and accumulates fibrosis and, as the last stage represents, the end-stage heart failure findings such as diastolic dysfunction and arrhythmias. The female carriers of DMD mutations may also manifest dilated cardiomyopathy. This has the potential to progress to heart failure in some cases; therefore, the appropriate genetic counseling and close monitoring of the carriers are also needed. Marfan syndrome is caused by mutations in the FBN1 gene that codes for fibrillin-1. The inheritance is autosomal dominant [84]. Fibrillin-1 is an extracellular protein that plays a role in microfibril formation and provides elastic properties to tissues [85]. Dilated cardiomyopathy is typically associated with Marfan syndrome. The Barth syndrome is an X-linked autosomal recessive disorder is caused by mutations in the tafazzin (TAZ) gene [86]. The loss of tafazzin and increased cardiolipin results in changes in energy stores decreased contractility, and increased heart damage. Barth syndrome cardiomyopathy is usually dilated cardiomyopathy, but cases of hypertrophic and left ventricle non-compaction cardiomyopathies have been described.
9 Genome Editing in Modeling Inheritable Heart Diseases in Model Organisms
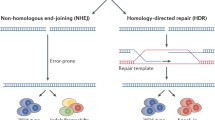
Model organisms, such as mice, are indispensable tools for understanding the etiology of inheritable heart diseases and gene functions [87]. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated Cas9-based gene editing technology [88,89,90] allowing efficient generation of mutant mice in one step [91], mutant mice can be systematically generated for interrogating and modeling human inheritable heart diseases, as well as for dissecting mechanisms for gene functions.
Pre-B cell leukemia factor 1 (PBX1) is a transcription factor essential for development and associated with CAKUTHED syndrome, characterized by multiple congenital defects including CHD. Alankarage and associates identified a de novo missense variant, PBX1:c.551G>C p.R184P, in a syndromic CHD patient with tetralogy of Fallot with absent pulmonary valve [92]. Using CRISPR-Cas9 gene editing to generate a mouse model with this mutation, Alankarage et al. [92] conducted functional and phenotypical analysis of Pbx1 in mice to show that p.R184P is disease-causal. Wnt/β-catenin signaling cascade [93,94,95] is important transcriptional regulation for morphogenesis. Combining zebrafish and mouse genetics, Cantù et al. [96] used CRISPR-Cas9 gene editing to demonstrate that tissue-selective perturbation of Bcl9 and Pygo as selective β-catenin cofactors in a subset of canonical Wnt responses caused severe CHD.
Sufficiently sizeable cohorts of probands when searching for causative genes in CHD can be challenging to assemble [8] because of large numbers of causative variants, low frequency of causative variants for individual genes, and diverse genetic backgrounds of the human population. Forward genetic screening with N-ethyl-N-nitrosourea (ENU) mutagenesis [97] in model organisms, such as inbred mouse strains, with very highly induced rates of mutation throughout the genome in the homogeneous genetic background, is therefore invaluable to uncover causative genes in CHD [53, 55, 98, 99]. However, ENU mutagenesis can cause point mutations in ~100 genetic loci per genome; consequently, it is imperative to rule out non-causative mutations for the CHD phenotypes. Gene editing to knock in or knock out specific gene mutations in the inbred wild-type (WT) background can authenticate the causality of the gene in question. Leveraging forward genetic screening with ENU mutagenesis, Liu et al. [98] have identified causative digenic mutations in Sap130 and Pcdha9 that can synergistically cause hypoplastic left heart syndrome (HLHS) in double homozygous mutants with incomplete penetrance and non-mendelian complex genetic inheritance [98, 99]. Gene editing using the CRISPR/Cas9 system to generate the same point mutations in these two genes in the WT background produced identical phenotypes with similar penetrance and substantiated that Sap130 and Pcdha9 are causative genes for HLHS.
Furthermore, the CRISPR/Cas9 system has shown tremendous potential to correct genetic defects in zygotes or postnatal mice [100]. Using adeno-associated virus (AAV9 or AAV8) to deliver CRISPR/Cas9-mediated gene editing components, in vivo somatic genome editing has been shown to correct the disease-causing gene mutation of Duchenne muscular dystrophy (DMD) in mice and improve phenotypical outcomes in postnatal mice [101, 102]. Sarcoplasmic reticulum Ca2+-ATPase 2a (SERCA2a) and its inhibitory protein called phospholamban (PLN) are pivotal for Ca2+ handling in cardiomyocytes. Their expression levels and activities were changed in heart failure patients. Using the CRISPR-cas9 system, Kaneko et al. [103] showed that PLN inhibition could significantly improve cardiac function and survival in calsequestrin overexpressing mice, a severe heart failure mouse model, suggesting PLN deletion could be a promising approach to improve both mortality and cardiac function in the heart failure.
One challenge for using the gene editing approach to dissect functional mechanisms for cardiac development and CHD is the presence of functionally redundant genes in the genetic network for cardiac development. It is necessary to knock out multiple genes in the same functional network to exhibit phenotypes. Conventionally, mutant mice carrying multiple genetic mutations were generated by time-consuming intercrossing of mice with different single genetic mutations. Wang et al. [104] have shown the feasibility of multiplex gene editing with the CRISPR/cas-9 system. Coinjection of Cas9 mRNA and single-guide RNAs (sgRNAs) targeting both Tet1 and Tet2 genes into zygotes generated mice with biallelic mutations in both genes with an efficiency of 80%. The CRISPR/Cas system allows the one-step generation of animals carrying mutations in multiple genes, an approach that will accelerate the in vivo study of functionally redundant genes and epistatic gene interactions [104].
With the availability of the Mouse Genome Database (MGD) [105, 106], large-scale efforts such as the Knockout Mouse Project (KOMP) [107] and the European Conditional Mouse Mutagenesis (EUCOMM) Program [108] are systematically generating knockout mice for dissecting mechanisms for gene functions. International collaborations combining European Mouse Mutant Archive (EMMA), Infrastructure for Phenotyping, Arching and Distribution of Mouse Diseases Models (IPAD-MD), and International Mouse Phenotyping Consortium (IMPC) [109, 110] are pursuing efforts for detailed phenotypic characterization to gain mechanistic insights into gene function. The challenges remain that over 30% of the genes in mice are essential for development and cause embryonic lethality or neonatal survivability when deleted [109], consequently, it is not feasible to analyze postnatal gene functions. Conditional knockouts [111] with Cre/loxP system can overcome the embryonic lethality by knocking out the gene later in life. Conditional knockouts with CRISPR/cas systems [112, 113] can facilitate efficient conditional knockouts for dissecting gene functions in viable adult animals.
10 Other Considerations
Although strong genetic underpinning regulates cardiac development and CHD, extreme locus heterogeneity, incomplete penetrance, and lack of a genotype-phenotype correlation [114, 115] indicate other non-genetic factors [116] can impede cardiogenesis and contribute to the development of CHD. The penetrance of CHD is incomplete and highly variable. Probands [8] and their relatives carrying the same genetic variants can exhibit different cardiac outcomes, ranging from nearly normal to complex CHD with different CHD lesions. Mechanical perturbation [115, 117,118,119,120,121,122,123] of ventricular preload pressure and shear stress, as well as exposure to alcohol [124] and environmental toxins [125] can cause CHD. Epidemiology studies have identified maternal risk factors associated with CHD, such as cardiometabolic disorders, stress, preeclampsia, obesity, and diabetes mellitus [126,127,128,129]. The risk of congenital anomalies in infants of diabetic mothers is estimated to be between 2.5 and 12%, with an over-representation of CHD [130]. Better understanding of the gene–environment interactions in myocardial development and the pathogenesis of CHD is needed to facilitate effective genome editing as a therapeutic intervention. Additionally, ethical concerns of germline genome editing to correct developmental diseases need to be addressed before the genome editing technologies can treat curable CHD and other cardiovascular diseases [131,132,133].
11 Conclusions
Reverse and forward genetics will continue to enhance and refine our models of heart development, associate mutated genes with abnormal phenotypes, be used to screen embryos, fetuses, parents, and family for mutations in these genes, and provide highly informed genetic counseling. If and when gene editing is available for treatment of inheritable disorders, the accumulated knowledge of heart development and disease will guide details on when, where, and how to apply gene editing.
References
van den Brink L, Grandela C, Mummery CL, Davis RP (2020) Inherited cardiac diseases, pluripotent stem cells, and genome editing combined-the past, present, and future. Stem Cells 38(2):174–186
Srivastava D, Olson EN (2000) A genetic blueprint for cardiac development. Nature 407(6801):221–226
Srivastava D (2006) Genetic regulation of cardiogenesis and congenital heart disease. Annu Rev Pathol 1:199–213
van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, Roos-Hesselink JW (2011) Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol 58(21):2241–2247
Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB (2012) Heart disease and stroke statistics—2012 update: a report from the American Heart Association. Circulation 125(1):e2–e220
Williams K, Carson J, Lo C (2019) Genetics of congenital heart disease. Biomolecules 9(12):879
Oyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M (2009) Recurrence of congenital heart defects in families. Circulation 120(4):295–301
Triedman JK, Newburger JW (2016) Trends in congenital heart disease: the next decade. Circulation 133(25):2716–2733
Oyen N, Poulsen G, Wohlfahrt J, Boyd HA, Jensen PK, Melbye M (2010) Recurrence of discordant congenital heart defects in families. Circ Cardiovasc Genet 3(2):122–128
Bruneau BG (2008) The developmental genetics of congenital heart disease. Nature 451(7181):943–948
Zaidi S, Brueckner M (2017) Genetics and genomics of congenital heart disease. Circ Res 120(6):923–940
Wu YL, Lo CW (2017) Diverse application of MRI for mouse phenotyping. Birth Defects Res 109(10):758–770
Georgi B, Voight BF, Bucan M (2013) From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet 9(5):e1003484
Nadeau JH (2001) Modifier genes in mice and humans. Nat Rev Genet 2(3):165–174
Nguyen D, Xu T (2008) The expanding role of mouse genetics for understanding human biology and disease. Dis Model Mech 1(1):56–66
Epstein JA, Aghajanian H, Singh MK (2015) Semaphorin signaling in cardiovascular development. Cell Metab 21(2):163–173
Krishnan A, Samtani R, Dhanantwari P, Lee E, Yamada S, Shiota K, Donofrio MT, Leatherbury L, Lo CW (2014) A detailed comparison of mouse and human cardiac development. Pediatr Res 76(6):500–507
Dhanantwari P, Lee E, Krishnan A, Samtani R, Yamada S, Anderson S, Lockett E, Donofrio M, Shiota K, Leatherbury L, Lo CW (2009) Human cardiac development in the first trimester: a high-resolution magnetic resonance imaging and episcopic fluorescence image capture atlas. Circulation 120(4):343–351
Jongbloed MR, Vicente Steijn R, Hahurij ND, Kelder TP, Schalij MJ, Gittenberger-de Groot AC, Blom NA (2012) Normal and abnormal development of the cardiac conduction system; implications for conduction and rhythm disorders in the child and adult. Differentiation 84(1):131–148
Brade T, Pane LS, Moretti A, Chien KR, Laugwitz KL (2013) Embryonic heart progenitors and cardiogenesis. Cold Spring Harb Perspect Med 3(10):a013847
Schultheiss TM, Burch JB, Lassar AB (1997) A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev 11(4):451–462
Schneider VA, Mercola M (2001) Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev 15(3):304–315
Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB (2001) Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev 15(3):316–327
Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M (1997) The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J 16(18):5687–5696
Männer J (2004) On rotation, torsion, lateralization, and handedness of the embryonic heart loop: new insights from a simulation model for the heart loop of chick embryos. Anat Rec A Discov Mol Cell Evol Biol 278(1):481–492
Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S (2004) Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet 5(10):725–738
Goldmuntz E (2005) DiGeorge syndrome: new insights. Clin Perinatol 32 (4):963-978:ix–x
Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE (1997) Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 15(1):30–35
McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB (2006) NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet 79(1):169–173
McElhinney DB, Geiger E, Blinder J, Benson DW, Goldmuntz E (2003) NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol 42(9):1650–1655
Sarkozy A, Conti E, Neri C, D’Agostino R, Digilio MC, Esposito G, Toscano A, Marino B, Pizzuti A, Dallapiccola B (2005) Spectrum of atrial septal defects associated with mutations of NKX2.5 and GATA4 transcription factors. J Med Genet 42(2):e16
Koenig SN, Bosse KM, Nadorlik HA, Lilly B, Garg V (2015) Evidence of aortopathy in mice with haploinsufficiency of Notch1 in Nos3-Null background. J Cardiovasc Dev Dis 2(1):17–30
Lin AE, Krikov S, Riehle-Colarusso T, Frías JL, Belmont J, Anderka M, Geva T, Getz KD, Botto LD (2014) Laterality defects in the national birth defects prevention study (1998-2007): birth prevalence and descriptive epidemiology. Am J Med Genet A 164a(10):2581–2591. https://doi.org/10.1002/ajmg.a.36695
Gabriel GC, Lo CW (2020) Left-right patterning in congenital heart disease beyond heterotaxy. Am J Med Genet C Semin Med Genet 184(1):90–96
Delhaas T, Decaluwe W, Rubbens M, Kerckhoffs R, Arts T (2004) Cardiac fiber orientation and the left-right asymmetry determining mechanism. Ann N Y Acad Sci 1015:190–201
Cooke J (2004) Developmental mechanism and evolutionary origin of vertebrate left/right asymmetries. Biol Rev Camb Philos Soc 79(2):377–407
Brueckner M (2007) Heterotaxia, congenital heart disease, and primary ciliary dyskinesia. Circulation 115(22):2793–2795
Zariwala MA, Knowles MR, Leigh MW (1993) Primary ciliary dyskinesia. GeneReviews® [Internet], University of Washington, Seattle
Pennekamp P, Menchen T, Dworniczak B, Hamada H (2015) Situs inversus and ciliary abnormalities: 20 years later, what is the connection? Cilia 4(1):1
Berdon WE, Willi U (2004) Situs inversus, bronchiectasis, and sinusitis and its relation to immotile cilia: history of the diseases and their discoverers-Manes Kartagener and Bjorn Afzelius. Pediatr Radiol 34(1):38–42
Patel A, Honore E (2010) Polycystins and renovascular mechanosensory transduction. Nat Rev Nephrol 6(9):530–538
Fliegauf M, Benzing T, Omran H (2007) When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8(11):880–893
Yuan S, Zhao L, Brueckner M, Sun Z (2015) Intraciliary calcium oscillations initiate vertebrate left-right asymmetry. Curr Biol 25(5):556–567
Babu D, Roy S (2013) Left-right asymmetry: cilia stir up new surprises in the node. Open Biol 3(5):130052
Kennedy MP, Plant BJ (2014) Primary ciliary dyskinesia and the heart: cilia breaking symmetry. Chest 146(5):1136–1138
Sharma N, Berbari NF, Yoder BK (2008) Ciliary dysfunction in developmental abnormalities and diseases. Curr Top Dev Biol 85:371–427
Dykes IM (2014) Left right patterning, evolution and cardiac development. J Cardiovasc Dev Dis 1(1):52–72
Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE (2013) Primary cilia are specialized calcium signalling organelles. Nature 504(7479):311–314
Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, Robinson BV, Minnix SL, Olbrich H, Severin T, Ahrens P, Lange L, Morillas HN, Noone PG, Zariwala MA, Knowles MR (2007) Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation 115(22):2814–2821
Leigh MW, Pittman JE, Carson JL, Ferkol TW, Dell SD, Davis SD, Knowles MR, Zariwala MA (2009) Clinical and genetic aspects of primary ciliary dyskinesia/Kartagener syndrome. Genet Med 11(7):473–487
Brueckner M (2012) Impact of genetic diagnosis on clinical management of patients with congenital heart disease: cilia point the way. Circulation 125(18):2178–2180
Nakhleh N, Francis R, Giese RA, Tian X, Li Y, Zariwala MA, Yagi H, Khalifa O, Kureshi S, Chatterjee B, Sabol SL, Swisher M, Connelly PS, Daniels MP, Srinivasan A, Kuehl K, Kravitz N, Burns K, Sami I, Omran H, Barmada M, Olivier K, Chawla KK, Leigh M, Jonas R, Knowles M, Leatherbury L, Lo CW (2012) High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. Circulation 125(18):2232–2242
Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, Chang C, Yagi H, San Agustin JT, Thahir M, Anderton S, Lawhead C, Vescovi A, Pratt H, Morgan J, Haynes L, Smith CL, Eppig JT, Reinholdt L, Francis R, Leatherbury L, Ganapathiraju MK, Tobita K, Pazour GJ, Lo CW (2015) Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 521(7553):520–524
Rao Damerla R, Gabriel GC, Li Y, Klena NT, Liu X, Chen Y, Cui C, Pazour GJ, Lo CW (2014) Role of cilia in structural birth defects: insights from ciliopathy mutant mouse models. Birth Defects Res C Embryo Today 102(2):115–125
Liu X, Francis R, Kim AJ, Ramirez R, Chen G, Subramanian R, Anderton S, Kim Y, Wong L, Morgan J, Pratt HC, Reinholdt L, Devine W, Leatherbury L, Tobita K, Lo CW (2014) Interrogating congenital heart defects with noninvasive fetal echocardiography in a mouse forward genetic screen. Circ Cardiovasc Imaging 7(1):31–42
Miller KA, Ah-Cann CJ, Welfare MF, Tan TY, Pope K, Caruana G, Freckmann ML, Savarirayan R, Bertram JF, Dobbie MS, Bateman JF, Farlie PG (2013) Cauli: a mouse strain with an Ift140 mutation that results in a skeletal ciliopathy modelling Jeune syndrome. PLoS Genet 9(8):e1003746
Francis RJ, Christopher A, Devine WA, Ostrowski L, Lo C (2012) Congenital heart disease and the specification of left-right asymmetry. Am J Physiol Heart Circ Physiol 302(10):H2102–H2111
Tan SY, Rosenthal J, Zhao XQ, Francis RJ, Chatterjee B, Sabol SL, Linask KL, Bracero L, Connelly PS, Daniels MP, Yu Q, Omran H, Leatherbury L, Lo CW (2007) Heterotaxy and complex structural heart defects in a mutant mouse model of primary ciliary dyskinesia. J Clin Investig 117(12):3742–3752
Ray P, Chin AS, Worley KE, Fan J, Kaur G, Wu M, Wan LQ (2018) Intrinsic cellular chirality regulates left-right symmetry breaking during cardiac looping. Proc Natl Acad Sci U S A 115(50):E11568–e11577
Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, Hershberger RE, Judge DP, Le Marec H, McKenna WJ, Schulze-Bahr E, Semsarian C, Towbin JA, Watkins H, Wilde A, Wolpert C, Zipes DP (2011) HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm 8(8):1308–1339
Chugh SS, Reinier K, Teodorescu C, Evanado A, Kehr E, Al Samara M, Mariani R, Gunson K, Jui J (2008) Epidemiology of sudden cardiac death: clinical and research implications. Prog Cardiovasc Dis 51(3):213–228
Shen WK, Sheldon RS, Benditt DG, Cohen MI, Forman DE, Goldberger ZD, Grubb BP, Hamdan MH, Krahn AD, Link MS, Olshansky B, Raj SR, Sandhu RK, Sorajja D, Sun BC, Yancy CW (2017) 2017 ACC/AHA/HRS guideline for the evaluation and management of patients with syncope: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Circulation 136(5):e25–e59
Abi Khalil C (2014) The emerging role of epigenetics in cardiovascular disease. Ther Adv Chronic Dis 5(4):178–187
Schwartz PJ, Crotti L, Insolia R (2012) Long-QT syndrome: from genetics to management. Circ Arrhythm Electrophysiol 5(4):868–877
Asatryan B, Medeiros-Domingo A (2019) Emerging implications of genetic testing in inherited primary arrhythmia syndromes. Cardiol Rev 27(1):23–33
Priori SG, Wilde AA, Horie M, Cho Y, Behr ER, Berul C, Blom N, Brugada J, Chiang CE, Huikuri H, Kannankeril P, Krahn A, Leenhardt A, Moss A, Schwartz PJ, Shimizu W, Tomaselli G, Tracy C (2013) HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm 10(12):1932–1963
Schulze-Bahr E, Eckardt L, Breithardt G, Seidl K, Wichter T, Wolpert C, Borggrefe M, Haverkamp W (2003) Sodium channel gene (SCN5A) mutations in 44 index patients with Brugada syndrome: different incidences in familial and sporadic disease. Hum Mutat 21(6):651–652
Kapplinger JD, Tester DJ, Alders M, Benito B, Berthet M, Brugada J, Brugada P, Fressart V, Guerchicoff A, Harris-Kerr C, Kamakura S, Kyndt F, Koopmann TT, Miyamoto Y, Pfeiffer R, Pollevick GD, Probst V, Zumhagen S, Vatta M, Towbin JA, Shimizu W, Schulze-Bahr E, Antzelevitch C, Salisbury BA, Guicheney P, Wilde AA, Brugada R, Schott JJ, Ackerman MJ (2010) An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm 7(1):33–46
Wang QI, Ohno S, Ding WG, Fukuyama M, Miyamoto A, Itoh H, Makiyama T, Wu J, Bai J, Hasegawa K, Shinohara T, Takahashi N, Shimizu A, Matsuura H, Horie M (2014) Gain-of-function KCNH2 mutations in patients with Brugada syndrome. J Cardiovasc Electrophysiol 25(5):522–530
Gussak I, Brugada P, Brugada J, Wright RS, Kopecky SL, Chaitman BR, Bjerregaard P (2000) Idiopathic short QT interval: a new clinical syndrome? Cardiology 94(2):99–102
Brugada R, Hong K, Dumaine R, Cordeiro J, Gaita F, Borggrefe M, Menendez TM, Brugada J, Pollevick GD, Wolpert C, Burashnikov E, Matsuo K, Wu YS, Guerchicoff A, Bianchi F, Giustetto C, Schimpf R, Brugada P, Antzelevitch C (2004) Sudden death associated with short-QT syndrome linked to mutations in HERG. Circulation 109(1):30–35
Giustetto C, Schimpf R, Mazzanti A, Scrocco C, Maury P, Anttonen O, Probst V, Blanc JJ, Sbragia P, Dalmasso P, Borggrefe M, Gaita F (2011) Long-term follow-up of patients with short QT syndrome. J Am Coll Cardiol 58(6):587–595
Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB (2006) Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 113(14):1807–1816
Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, McDonough B, Smoot L, Mullen MP, Woolf PK, Wigle ED, Seidman JG, Seidman CE (2000) Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med 343(23):1688–1696
Watkins H, Ashrafian H, Redwood C (2011) Inherited cardiomyopathies. N Engl J Med 364(17):1643–1656
Towbin JA (2014) Inherited cardiomyopathies. Circ J 78(10):2347–2356
Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet JP, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M (2003) Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 107(17):2227–2232
Millat G, Bouvagnet P, Chevalier P, Dauphin C, Jouk PS, Da Costa A, Prieur F, Bresson JL, Faivre L, Eicher JC, Chassaing N, Crehalet H, Porcher R, Rodriguez-Lafrasse C, Rousson R (2010) Prevalence and spectrum of mutations in a cohort of 192 unrelated patients with hypertrophic cardiomyopathy. Eur J Med Genet 53(5):261–267
Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS, Marian AJ (2006) Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Investig 116(7):2012–2021
Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA, Calkins H, Corrado D, Cox MG, Daubert JP, Fontaine G, Gear K, Hauer R, Nava A, Picard MH, Protonotarios N, Saffitz JE, Sanborn DM, Steinberg JS, Tandri H, Thiene G, Towbin JA, Tsatsopoulou A, Wichter T, Zareba W (2010) Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur Heart J 31(7):806–814
Sen-Chowdhry S, Syrris P, McKenna WJ (2010) Genetics of restrictive cardiomyopathy. Heart Fail Clin 6(2):179–186
Caleshu C, Sakhuja R, Nussbaum RL, Schiller NB, Ursell PC, Eng C, De Marco T, McGlothlin D, Burchard EG, Rame JE (2011) Furthering the link between the sarcomere and primary cardiomyopathies: restrictive cardiomyopathy associated with multiple mutations in genes previously associated with hypertrophic or dilated cardiomyopathy. Am J Med Genet A 155A(9):2229–2235
Towbin JA (2010) Left ventricular noncompaction: a new form of heart failure. Heart Fail Clin 6(4):453–469, viii
Ramirez F, Caescu C, Wondimu E, Galatioto J (2018) Marfan syndrome; A connective tissue disease at the crossroads of mechanotransduction, TGFβ signaling and cell stemness. Matrix Biol 71-72:82–89
Godwin ARF, Singh M, Lockhart-Cairns MP, Alanazi YF, Cain SA, Baldock C (2019) The role of fibrillin and microfibril binding proteins in elastin and elastic fibre assembly. Matrix Biol 84:17–30
Dudek J, Maack C (2017) Barth syndrome cardiomyopathy. Cardiovasc Res 113(4):399–410
Lee D, Threadgill DW (2004) Investigating gene function using mouse models. Curr Opin Genet Dev 14(3):246–252
Motta BM, Pramstaller PP, Hicks AA, Rossini A (2017) The impact of CRISPR/Cas9 technology on cardiac research: from disease modelling to therapeutic approaches. Stem Cells Int 2017:8960236
Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213):1258096
Moore JD (2015) The impact of CRISPR-Cas9 on target identification and validation. Drug Discov Today 20(4):450–457
Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154(6):1370–1379
Alankarage D, Szot JO, Pachter N, Slavotinek A, Selleri L, Shieh JT, Winlaw D, Giannoulatou E, Chapman G, Dunwoodie SL (2020) Functional characterization of a novel PBX1 de novo missense variant identified in a patient with syndromic congenital heart disease. Hum Mol Genet 29(7):1068–1082
Mosimann C, Hausmann G, Basler K (2009) Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10(4):276–286
Nakamura Y, de Paiva AE, Veenstra GJ, Hoppler S (2016) Tissue- and stage-specific Wnt target gene expression is controlled subsequent to β-catenin recruitment to cis-regulatory modules. Development 143(11):1914–1925
Ding Y, Ploper D, Sosa EA, Colozza G, Moriyama Y, Benitez MD, Zhang K, Merkurjev D, De Robertis EM (2017) Spemann organizer transcriptome induction by early beta-catenin, Wnt, Nodal, and Siamois signals in Xenopus laevis. Proc Natl Acad Sci U S A 114(15):E3081–e3090
Cantù C, Felker A, Zimmerli D, Prummel KD, Cabello EM, Chiavacci E, Méndez-Acevedo KM, Kirchgeorg L, Burger S, Ripoll J, Valenta T, Hausmann G, Vilain N, Aguet M, Burger A, Panáková D, Basler K, Mosimann C (2018) Mutations in Bcl9 and Pygo genes cause congenital heart defects by tissue-specific perturbation of Wnt/β-catenin signaling. Genes Dev 32(21-22):1443–1458
Stottmann R, Beier D (2014) ENU Mutagenesis in the Mouse. Curr Protoc Hum Genet 82:15.14.11-10
Liu X, Yagi H, Saeed S, Bais AS, Gabriel GC, Chen Z, Peterson KA, Li Y, Schwartz MC, Reynolds WT, Saydmohammed M, Gibbs B, Wu Y, Devine W, Chatterjee B, Klena NT, Kostka D, de Mesy Bentley KL, Ganapathiraju MK, Dexheimer P, Leatherbury L, Khalifa O, Bhagat A, Zahid M, Pu W, Watkins S, Grossfeld P, Murray SA, Porter GA Jr, Tsang M, Martin LJ, Benson DW, Aronow BJ, Lo CW (2017) The complex genetics of hypoplastic left heart syndrome. Nat Genet 49(7):1152–1159
Yagi H, Liu X, Gabriel GC, Wu Y, Peterson K, Murray SA, Aronow BJ, Martin LJ, Benson DW, Lo CW (2018) The genetic landscape of hypoplastic left heart syndrome. Pediatr Cardiol 39(6):1069–1081
Koch L (2016) In vivo genome editing—growing in strength. Nat Rev Genet 17(3):124–124
Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN (2016) Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351(6271):400–403
Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351(6271):403–407
Kaneko M, Hashikami K, Yamamoto S, Matsumoto H, Nishimoto T (2016) Phospholamban ablation using CRISPR/Cas9 system improves mortality in a murine heart failure model. PLoS One 11(12):e0168486
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153(4):910–918
Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE (2014) The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res 42(Database issue):D810–D817
Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA (2008) The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res 36(Database issue):D724–D728
Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, Grieder FB, Heintz N, Hicks G, Insel TR, Joyner A, Koller BH, Lloyd KC, Magnuson T, Moore MW, Nagy A, Pollock JD, Roses AD, Sands AT, Seed B, Skarnes WC, Snoddy J, Soriano P, Stewart DJ, Stewart F, Stillman B, Varmus H, Varticovski L, Verma IM, Vogt TF, von Melchner H, Witkowski J, Woychik RP, Wurst W, Yancopoulos GD, Young SG, Zambrowicz B (2004) The knockout mouse project. Nat Genet 36(9):921–924
Friedel RH, Seisenberger C, Kaloff C, Wurst W (2007) EUCOMM—the European conditional mouse mutagenesis program. Brief Funct Genomic Proteomic 6(3):180–185
Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie SM, Fuchs H, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu CW, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Newbigging S, Nutter LM, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, Tamura M, Trainor AG, Tudose I, Wakana S, Warren J, Wendling O, West DB, Wong L, Yoshiki A, MacArthur DG, Tocchini-Valentini GP, Gao X, Flicek P, Bradley A, Skarnes WC, Justice MJ, Parkinson HE, Moore M, Wells S, Braun RE, Svenson KL, de Angelis MH, Herault Y, Mohun T, Mallon AM, Henkelman RM, Brown SD, Adams DJ, Lloyd KC, McKerlie C, Beaudet AL, Bućan M, Murray SA (2016) High-throughput discovery of novel developmental phenotypes. Nature 537(7621):508–514
Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie Helmut Fuchs SM, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu CW, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Nutter S, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, Tamura M, Trainor AG, Tudose I, Wakana S, Warren J, Wendling O, West DB, Wong L, Yoshiki A, Wurst W, MacArthur DG, Tocchini-Valentini GP, Gao X, Flicek P, Bradley A, Skarnes WC, Justice MJ, Parkinson HE, Moore M, Wells S, Braun RE, Svenson KL, de Angelis MH, Herault Y, Mohun T, Mallon AM, Henkelman RM, Brown SDM, Adams DJ, Lloyd KCK, McKerlie C, Beaudet AL, Murray M (2017) Corrigendum: High-throughput discovery of novel developmental phenotypes. Nature 551(7680):398
Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474(7351):337–342
Gurumurthy CB, O’Brien AR, Quadros RM, Adams J Jr, Alcaide P, Ayabe S, Ballard J, Batra SK, Beauchamp MC, Becker KA, Bernas G, Brough D, Carrillo-Salinas F, Chan W, Chen H, Dawson R, DeMambro V, D’Hont J, Dibb KM, Eudy JD, Gan L, Gao J, Gonzales A, Guntur AR, Guo H, Harms DW, Harrington A, Hentges KE, Humphreys N, Imai S, Ishii H, Iwama M, Jonasch E, Karolak M, Keavney B, Khin NC, Konno M, Kotani Y, Kunihiro Y, Lakshmanan I, Larochelle C, Lawrence CB, Li L, Lindner V, Liu XD, Lopez-Castejon G, Loudon A, Lowe J, Jerome-Majewska LA, Matsusaka T, Miura H, Miyasaka Y, Morpurgo B, Motyl K, Nabeshima YI, Nakade K, Nakashiba T, Nakashima K, Obata Y, Ogiwara S, Ouellet M, Oxburgh L, Piltz S, Pinz I, Ponnusamy MP, Ray D, Redder RJ, Rosen CJ, Ross N, Ruhe MT, Ryzhova L, Salvador AM, Alam SS, Sedlacek R, Sharma K, Smith C, Staes K, Starrs L, Sugiyama F, Takahashi S, Tanaka T, Trafford AW, Uno Y, Vanhoutte L, Vanrockeghem F, Willis BJ, Wright CS, Yamauchi Y, Yi X, Yoshimi K, Zhang X, Zhang Y, Ohtsuka M, Das S, Garry DJ, Hochepied T, Thomas P, Parker-Thornburg J, Adamson AD, Yoshiki A, Schmouth JF, Golovko A, Thompson WR, Lloyd KCK, Wood JA, Cowan M, Mashimo T, Mizuno S, Zhu H, Kasparek P, Liaw L, Miano JM, Burgio G (2019) Reproducibility of CRISPR-Cas9 methods for generation of conditional mouse alleles: a multi-center evaluation. Genome Biol 20(1):171
Yang H, Wang H, Jaenisch R (2021) Response to “Reproducibility of CRISPR-Cas9 methods for generation of conditional mouse alleles: a multi-center evaluation”. Genome Biol 22(1):98
Yuan S, Zaidi S, Brueckner M (2013) Congenital heart disease: emerging themes linking genetics and development. Curr Opin Genet Dev 23(3):352–359
Lindsey SE, Butcher JT, Yalcin HC (2014) Mechanical regulation of cardiac development. Front Physiol 5:318
Russell MW, Chung WK, Kaltman JR, Miller TA (2018) Advances in the understanding of the genetic determinants of congenital heart disease and their impact on clinical outcomes. J Am Heart Assoc 7(6):e006906
Hogers B, DeRuiter MC, Gittenberger-de Groot AC, Poelmann RE (1999) Extraembryonic venous obstructions lead to cardiovascular malformations and can be embryolethal. Cardiovasc Res 41(1):87–99
Sedmera D, Pexieder T, Rychterova V, Hu N, Clark EB (1999) Remodeling of chick embryonic ventricular myoarchitecture under experimentally changed loading conditions. Anat Rec 254(2):238–252
Sedmera D, Hu N, Weiss KM, Keller BB, Denslow S, Thompson RP (2002) Cellular changes in experimental left heart hypoplasia. Anat Rec 267(2):137–145
Miller CE, Wong CL, Sedmera D (2003) Pressure overload alters stress-strain properties of the developing chick heart. Am J Physiol Heart Circ Physiol 285(5):H1849–H1856
Reckova M, Rosengarten C, deAlmeida A, Stanley CP, Wessels A, Gourdie RG, Thompson RP, Sedmera D (2003) Hemodynamics is a key epigenetic factor in development of the cardiac conduction system. Circ Res 93(1):77–85
Groenendijk BC, Hierck BP, Vrolijk J, Baiker M, Pourquie MJ, Gittenberger-de Groot AC, Poelmann RE (2005) Changes in shear stress-related gene expression after experimentally altered venous return in the chicken embryo. Circ Res 96(12):1291–1298
deAlmeida A, McQuinn T, Sedmera D (2007) Increased ventricular preload is compensated by myocyte proliferation in normal and hypoplastic fetal chick left ventricle. Circ Res 100(9):1363–1370
Denny L, Coles S, Blitz R (2017) Fetal alcohol syndrome and fetal alcohol spectrum disorders. Am Fam Physician 96(8):515–522
Sun R, Liu M, Lu L, Zheng Y, Zhang P (2015) Congenital heart disease: causes, diagnosis, symptoms, and treatments. Cell Biochem Biophys 72(3):857–860
Helle E, Priest JR (2020) Maternal obesity and diabetes mellitus as risk factors for congenital heart disease in the offspring. J Am Heart Assoc 9(8):e011541
Wren C, Birrell G, Hawthorne G (2003) Cardiovascular malformations in infants of diabetic mothers. Heart 89(10):1217–1220
Lisowski LA, Verheijen PM, Copel JA, Kleinman CS, Wassink S, Visser GH, Meijboom EJ (2010) Congenital heart disease in pregnancies complicated by maternal diabetes mellitus. An international clinical collaboration, literature review, and meta-analysis. Herz 35(1):19–26
Loffredo CA, Wilson PD, Ferencz C (2001) Maternal diabetes: an independent risk factor for major cardiovascular malformations with increased mortality of affected infants. Teratology 64(2):98–106
Narchi H, Kulaylat N (2000) Heart disease in infants of diabetic mothers. Images Paediatr Cardiol 2(2):17–23
German DM, Mitalipov S, Mishra A, Kaul S (2019) Therapeutic genome editing in cardiovascular diseases. JACC Basic Transl Sci 4(1):122–131
Seeger T, Porteus M, Wu JC (2017) Genome editing in cardiovascular biology. Circ Res 120(5):778–780
Hernandez-Benitez R, Martinez-Martinez ML, Lajara J, Magistretti P, Montserrat N, Izpisua Belmonte JC (2018) At the heart of genome editing and cardiovascular diseases. Circ Res 123(2):221–223
Acknowledgments
The authors like to thank Drs. Jonathan A. Epstein, Deepak Srivastava, and Eric N. Olson for providing figures. This work was supported in part by the grants to YLW from the National Institute of Health (1R21EB023507, R21 NS121706), American Heart Association (18CDA34140024), Department of Defense (W81XWH1810070, W81XWH-22-1-0221), and the Children’s Hospital of Pittsburgh of the UPMC Health System (RAC-CHP00-CY19-16212).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
The authors declare no competing financial interests.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Turan, S., Chaillet, J.R., Stapleton, M.C., Wu, Y.L. (2023). Genome Editing and Myocardial Development. In: Xiao, J. (eds) Genome Editing in Cardiovascular and Metabolic Diseases. Advances in Experimental Medicine and Biology, vol 1396. Springer, Singapore. https://doi.org/10.1007/978-981-19-5642-3_4
Download citation
DOI: https://doi.org/10.1007/978-981-19-5642-3_4
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-5641-6
Online ISBN: 978-981-19-5642-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)