Abstract
Plants contain associated microbiomes, which facilitate the plant’s productivity by supporting their healthy growth in native niche. Globally, sustainable food production is quintessential, wherein the growth-promoting abilities of plant-associated microbes become vital. Recent literatures reveal significant structure and dynamics on plant microbiome, which recognizes the regulation of plant fitness by native microbiome. Henceforth, it is essential to practice modern microbial developments for strengthening crop production. Many of the environmental factors modulate the microbiome organization. This chapter emphasizes the comprehensive characterization of different niche’s microbiome and impact of differential influential factors such as genotype, microbial interactions, agricultural habits, bioinoculants, and abiotic factors to microbial diversity and sustainable plant health. Moreover, conventional and advanced approaches (omics/multi-omics) for studying plant microbiome diversity and plant microbiome interactions are also discussed, which would be helpful in developing novel microbial formulations for sustainable agriculture.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
8.1 Introduction
Term “plant microbiome” describes “the microorganism’s community with the potential of harboring a space in plant body as symbionts, commensal or pathogen.” Plants harbor microbial hotspots along with soil as major natural microbial resource. This microbial plethora is vital in promoting plant’s growth, health, and productivity. However, food productivity enhancement strategies (use of fertilizers, pesticides, and agrochemicals), industrialization, and anthropogenic interventions led to an enforced soil and microenvironment deterioration and attenuation in agriculture lands (Compant et al. 2019). These factors cumulatively exert pressure on agro-ecosystems and challenge the high-yielding economical and sustainable agriproducts. In this background, plant microbiomes are potential tool for enhancing agricultural productivity while maintaining sustainable ecosystem. The plant microbiomes not only promote the growth of plants but also enhance the soil fertility for facilitating sustainable agriculture. Moreover, microbiome aids in seed germination, growth, productivity, yield, adaptation, and stress resistance/tolerance of plants, which impacts the fitness of the plant leading to the sustainable agriculture (Aamir et al. 2021).
8.2 Plant Microbiome: Diversity, Composition, and Distribution
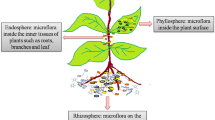
Microbiomes possess high degree of diversity and are dispersed on soil, water, and air, which can associate with the plant organs (Gupta et al. 2021). The microbiome distribution on plant is majorly categorized into phyllospheric (present on aerial parts such as stem, leaves, and flowers), endospheric (present on inner plant tissues), and rhizospheric (present on below ground components such as roots and surrounding soil) (Fig. 8.1) (Aamir et al. 2021; Santos and Olivares 2021). In addition, microbes also reside as epiphyte on plant surface (Compant et al. 2019). As endophyte microbes penetrate the epidermis and colonize inter- and intracellularly. In general, ectomycorrhizal fungi penetrate root spaces intracellularly after developing a mantle, which surrounds root tip. However, endomycorrhizal fungi develop arbuscules for intracellular colonization (Santos and Olivares 2021). Naturally, healthy plant harbors multifaceted diversity of microorganisms such as bacteria, and fungi are foremost constituent associated with plants (Trivedi et al. 2020). Some examples of plant-associated bacterial and fungal strains and their functional role in normal/stressed environment are listed in Tables 8.1 and 8.2, respectively.
Generally, microbial dynamics is associated with the types of plant species, its developmental stages, or genetic makeup. And, changes in host plant environment during the different life stages play a crucial role and arbitrate the microbial diversity by innate defense system stimulation of plant (Dastogeer et al. 2020; Gupta et al. 2021). Additionally, abiotic and biotic stresses-driven and -compromised defense system can also alter microbiome diversity. Henceforth, to study the biotic/abiotic factors influencing microbial matrix at phylogenetic and functional level, it is essential to estimate the plant microbiome’s heterogeneity individually and in understanding different niches (Santos and Olivares 2021).
Recent studies and advances in the knowledge of plant microbiome have demonstrated that plant-associated heterogenetic microbial consortia influence their growth, development, and productivity (Bhatt et al. 2020; Singh et al. 2019; Trivedi et al. 2020). The microbiome consists of some microorganisms, which can interact with other species/host and influence theorganization microbial community. Such microorganisms are known as “hub-microorganisms” of the microbiome (Agler et al. 2016). Microbiome diversity and composition are majorly affected by soil profile and physiochemical characteristics (texture, moisture proportion, pH, temperature, salinity, organic matter, etc.). Moreover, microbe–microbe/plant–microbe interaction, microbial/host secretome, anthropogenic factors, cultivation exercises, and environmental changes also influence at a certain extent (Bhatt et al. 2020; Li et al. 2020; Oyserman et al. 2021).
8.3 Approaches for Studying Plant Microbiome Diversity
Plant microbiome exhibits extensively wide-ranging diversity depending upon habitat and plant organs such as phyllo/endo/rhizospheric regions. Plant microbiome diversity investigation aims to unravel the structural/functional diverseness of a specific host plant-associated microbial community in a definite habitat (Bhatt et al. 2020; Dastogeer et al. 2020; Liu et al. 2019). The colonization of microbiome is facilitated by the host’s secondary metabolite secretome and specialized structures such as hairs and trichomes (Gupta et al. 2021).
Conventionally, identification and characterization of plant microbiomes (bacteria and fungi) consists of sequencing of universally conserved sequences and bioinformatic analysis. Briefly, this method consists of different sequential steps as: (1) microbial isolation from phyllo/endo/rhizosphere, (2) morphological characterization (for phenotypic and microscopic characteristics), (3) biochemical characterization (for biomolecules and enzymes), (4) Sanger’s sequencing (for 16S rNRA/18S rRNA), and (5) Basic Local Alignment Search Tool (BLAST) analysis using National Centre for Biological Information (NCBI) database.
These conventional methods give the outlook information only, henceforth holistic approaches using modern biotechnological techniques to unwrap complete multi-dimension information to understand the mechanistic component are essential. In this context, during last few years various studies implemented advance techniques to unwrap the microbiome exclusively and understand their interaction mechanism with host (Fitzpatrick et al. 2020). It has been established that along with the host and habitat, microbiomes are also affected by their environment and developmental stages. In recent years, advanced modified Sanger’s sequencing techniques including 454-pyrosequencing, Oxford nanopore, Illumina, Ion- torrent, PacBio, and hybrid-platform were used to reveal rare microbial taxa in definite niche (Bhatt et al. 2020; Gupta et al. 2021). These techniques extract complete genomic information leading to comprehensive microbial diversity study in a given population through phylogenetic and comparative genomics studies. Nevertheless, these techniques fail to differentiate dead/live cells, which raises the possibility of errors in estimation or hiked microbiome diversity, which are being addressed by managing PCR error using specifically tagged DNA (Gupta et al. 2021).
Moreover, microbiome also contains enormous diversity in uncultivable microbes, which makes it difficult to select a suitable method for their identification. However, in recent years advanced culture-independent direct-environmental DNA cloning, next generation sequencing, single strand conformation polymorphism, denaturing/temperature gradient gel-electrophoresis, terminal restriction/restriction fragment length polymorphism, and fatty acid methyl esters were reported for plant microbiome studies (Bodor et al. 2020; Gupta et al. 2021; del Orozco-Mosqueda et al. 2018).
Omics technologies (genomics/metagenomics, transcriptomics/meta-transcriptomics, proteomics/meta-proteomics, and metabolomics) give a comprehensive genetic, structural, and functional information of the plant microbiomes (Sharma et al. 2020). This information, coupled with the metabolic engineering at cellular and/or molecular level, enhances the knowledge on dynamics of microbiomes.
8.4 Factors Affecting Plant Microbiome Diversity
Microbiome consists of several microbes; however, various factors including genome organization, microbial interactions, biotic/abiotic components, local agricultural practices, environmental factors, and physiochemical factors greatly affect plant microbiome organization in local habitat. Some important factors are described below:
8.4.1 Impact of Genomic Organization
Plants benefit from capacities of their microbiome, which is the reason they devote some fraction of their carbon sources toward the development and sustenance of the microbiota. Plants with different genotypes perform distinctively toward metabolism of roots, composition of root secretome, recognition system, and native defense mechanism (Santos and Olivares 2021). Plant genotype alters the root metabolism by modulating the metabolic intermediates (pyruvic acid, citric acid, malic acid, lactic acid, etc.), nitrogenase activity, and reactive oxygen species (ROS) generation (Shcherbakova et al. 2017). Genotype also modulates composition of root secretome in terms of organic acids (citric, malic, and succinic), sugars (ribose, glucose, galactose, and fructose), amino acids (glycine, serine, lysine, and histidine), isoflavones, enzymes (chitinases, proteases, phytases), and phytoalexins (Shcherbakova et al. 2017). Genotype mediates the localized/systemic immune responses of plants by transcriptional changes leading to the elaboration of physical barriers, synthesis of antimicrobial compounds, defense phytohormones, jasmonic acid, salicylic acid, and gaseous ethylene (Lebeis et al. 2015).
Studies demonstrate that varieties of this nature can modify the design and action of the plant microbiome, which thus follows up on development, sustenance, and protection from biotic and abiotic stresses (Santos and Olivares 2021). Curlango-Rivera et al. demonstrated the impact of genotype toward composition of root microbiome, wherein cotton cultivars showed structural differences in their border cells (Curlango-Rivera et al. 2013). These cells are shaped from meristematic cells and were at first thought to be “dead” and answerable for the mechanical security of the root tip. Today, it is realized that line cells are connected with colonization of the root by microorganisms. Essentially, plant roots store high and low sub-atomic weight compounds at the root–soil interface, the rhizodeposition, which might shift as per their genotype. Along these lines, plants with various genotypes can deliver compounds with various creations, advancing explicit microbial flagging and colonization. While examining the seed microbiome, various creators distinguished the effect of the plant genotype on the microbial synthesis of the seed. In the tomato phyllosphere, a review uncovered that four of the nine genotypes tried had an alternate microbial arrangement, which demonstrates that the hereditary elements of the host plant might shape the related microbiota (Morella et al. 2020). In the rhizosphere, microorganisms from the families Solibacteraceae, Pseudomonadaceae, Cytophagaceae, and Bacillaceae were more bountiful in Fusarium-safe bean cultivars (Mendes et al. 2019). Fusarium opposition formed the microbial gathering of the rhizosphere and chose bacterial taxa with biocontrol action (Santos and Olivares 2021). In recent study, transplantation of rhizospheric soil from a Ralstonia solanacearum-resistant tomato plant to a prone plant genotype stifled the susceptibility (Kwak et al. 2018).
8.4.2 Impact of Agricultural Activities
Agricultural activities alter soil’s nutrient and physiochemical properties. Nutrient properties act as selection pressure for selection of plant microbiome and altered nutrition influences plant’s microbiome profile in negative way and reduces their productivity. Several studies reported the change in microbiome in different agricultural practices using green manure (Bergottini et al. 2017), organic manure (Lupatini et al. 2017), mulch (Qin et al. 2017), nitrogen fertilizers (Cai et al. 2017; Zhu et al. 2016), and crop rotation (Hong et al. 2020). Therefore, for achieving high agricultural productivity, it is essential to comprehend the impact of feasible practices on the design of the plant’s microbiome referring native plants and vegetable/animal residues (Santos and Olivares 2021).
8.4.3 Impact of Bioinoculants
Bioinoculant is a practical strategy to build crop efficiency while lessening the utilization of synthetic chemicals, though information on its effect on the plant microbiome is extremely scant. Subsequently, understanding the connection among inoculated and native microbial networks in plants will add to the improvement of natural products.
Microbial inoculants proficiency relies upon the inoculum characteristics and its capacity to bridge local microbial contest or its layout in the rhizosphere. Rhizospheric fitness (in terms of efficient microbial contents) is an essential factor for promising plant responses (plant-microbe interactions), however, has been explored below par. The cooperation between local miniature life forms of the plant and miniature organic entities (called “transients”) can be competitive, parasitic, mutualistic, or predatory (Santos and Olivares 2021), wherein secretomes containing probiotic/antimicrobial compounds selectively stimulate/inhibit the growth of microorganisms (Pieterse et al. 2016). Taken together, the native microbial organization of the host plant is urgent for the achievement of its colonization by transient microbes. Different signalling routes between plants and microorganisms as well as among microorganisms make up the base of the microorganisms in the root area. During plant-microbe interactions the population density is managed by releasing low atomic weight quorum sensing compounds such as acylated-homoserine-lactones (Hartmann et al. 2014).
The impact of the bioinoculant is regarded as indirect when it causes changes in the native microbiome structure and function by producing probiotic or antimicrobial compounds (Santos and Olivares 2021). Several reports stated the alteration in endophytic population’s structure while using inoculants (Andreote et al. 2014; Conn and Franco 2004). In another study, changes in the phenotype of the endophytic potato were reported after Methylobacterium sp. inoculation to it, wherein relative proportion of biocontrol regulators (Acinetobacter sp., Massilia sp., Phoma sp., and Entyloma sp.) were also increased (Ardanov et al. 2016).
The bioinoculants effects are considered as direct if inoculum directly alters/improves the metabolism of plant or root exudates profile, which in turn modulates microbiome’s structure and function (Santos and Olivares 2021). Molecular advanced technologies permit portraying the microbiome of various plants or contrasting microbiome of plants treated with bio-inoculants. This information will be helpful in developing novel bioinoculants, or shaping it as per the agricultural need.
8.4.4 Impact of Pathogens
The endophytic plant community can harbor useful and destructive microbes in an inactive state. These microbes can be “reactivated” by extrinsic factors including pathogen’s attack (Santos and Olivares 2021). The endophytic community encourages plant resistance either with the presence of resistance inducing microbes or reactivation of endophytic population by entry of a microbial biocontrol strain/abiotic stimulus (Podolich et al. 2015). Nevertheless, reactivation of inactive endophytic microbes after pathogenic attack is more proficient (Santos and Olivares 2021). Agler et al. reported phyllosphere’s reformed colonization of endophytic and endophytic bacteria by pathogenic fungi Albugo sp. and Dioszegia sp. (Agler et al. 2016). Likewise, a decrease in the number of Bacillus species in roots, and Streptomyces & Flavisolibacter in the rhizosphere of Chinese cabbage was observed with the presence of pathogenic Plasmodiophora brassicae (Lebreton et al. 2019). These findings show that the microbiome can protect plants against pathogenic attacks, hence it is conceivable that it can change the microbiota of the host plant.
8.4.5 Impacts of Abiotic Factors
Plants and their microbiome are naturally exposed to different environmental variations including humidity, pH, temperature, salinity, and ultraviolet rays, which straightforwardly or by implication alter the microbial composition. The microbiome profiles of soil from various climate zones shows enormous taxonomic and functional variety, although arid conditions revealed the least functional variation (Tripathi et al. 2017). Lower functional diversity in arid regions is associated with the up-regulation of sporulation/dormancy-associated genes and down-regulation of nutrient (N, P, and S) cycling and stress-associated genes (Tripathi et al. 2017). This study has significant ramifications for understanding the effects of environmental change on various microbiomes. In comparison of soil rhizospheric microbiome of eight different countries, highly significant variation was displayed and credited to difference in pH of different country’s soil (Simonin et al. 2020).
Water-stress/drought has maximum impact on plant microbiome (Santos and Olivares 2021). Santos-Medellín et al. studied rice plants exposed to water pressure had their root microbiome rebuilt, which can add to the endurance of plants in this condition (Santos-Medellín et al. 2017). Soil heat (50–80 °C) exposure influences microbiome and disease resistance due to the reduction in antagonistic bacterial loads of Mycobacteriaceae, Micrococcaceae, and Streptomycetaceae (van der Voort et al. 2016). Likely, with routine exposure of plants to UV radiation due to DNA damage phyllospheric microbiome stratification occurs; however, some bacteria acquire UV protection by endospore formation, exo-polysaccharides, and pigments such as melanin, xanthomonadine (Kumar et al. 2019).
Taken together, abiotic factor greatly influences the structure and diversity of plant microbiomes.
8.5 Role of Plant Microbiome in Sustainable Agriculture
Plant microbiome influences agriculture via enhancing soil fertility and promotion of plant growth. Microbes are diverse in nature and constitute of beneficial, harmful, and pathogenic microorganisms.
Plant growth-promoting bacteria (PGPB) synthesize phytohormones (cytokinin, auxin: indole-3-acetic acid, gibberellin, jasmonic acid, and salicylic acid), which act as phytostimulant and enhances plant growth (Kaur 2020; Rastegari 2020; Santos and Olivares 2021). Moreover, ethylene (a stress hormone) is regulated by the enzyme 1-aminocyclopropane-1-carboxylate deaminase (ACC deaminase), which breaks down the molecule ACC and stimulates plant development by lowering ethylene levels (Glick 2014; Santos and Olivares 2021; Shahid et al. 2021). Some common ACC deaminase producers are Pseudomonas spp., Bacillus spp., Paraburkholderia spp., Arthrobacter spp., and Pantoea spp. (Rastegari 2020; Santos and Olivares 2021). These genera exhibit various properties including phytohormones, phosphate solubilization, and nitrogen fixation which enhances nutrient uptake and stress tolerance. In contrast, some bacteria produce phytoactive compounds, which cause disease symptoms in plants. Some common examples of pathogenic bacteria are Pseudomonas syringae (infects tomato, tobacco, olive, and green bean), Xylella fastidiosa (infects potato and banana), Erwinia amylovora (infects ornamental plants), Xanthomonas spp. (infects banana), Ralstonia solanacearum (infects banana and potato) (Gupta et al. 2021; Rastegari 2020). Several bacteria directly facilitate essential nutrients (iron, nitrogen, phosphorus, etc.) acquisition and promote plant development (del Orozco-Mosqueda et al. 2018).
Plant resistance is influenced by microorganisms which are present above and below the soil due to alteration in plant defense system or commensal interactions (Igiehon and Babalola 2018). Various biocontrol mechanism are involved in plant resistance induction such as production of siderophores (chelating agents with potential of insoluble ferric-ions sequestration), antibiotics (2,4-diacetylphloroglucinol), enzymes (proteases, chitinases, phytases), lipopeptides (bacillomycin-D, mycosubtilin, and iturin-A), volatile organic compounds, and bacteriocins (del Orozco-Mosqueda et al. 2018; Santos and Olivares 2021).
Of note, microbiome not only induces plant growth by these biocontrol mechanisms, but also these perform bio-fertilization (by controlled availability and attainment of nutrients for plants) for regulating plant growth. Some typical examples of biofertilization are symbiotic associations between Rhizobium (nitrogen-fixing bacteria), arbuscular-mycorrhizal fungi, and phosphate solubilizing bacteria to deliver nitrogen and phosphorus, to plant, respectively (Santos and Olivares 2021). Some examples of phosphate solubilizing bacteria are Rhizobium, Bacillus, Microbacterium, Azotobacter, Erwinia, Serratia, Burkholderia, Enterobacter, Beijerinckia, Flavobacterium, and Pseudomonas (Chhabra 2019; Chhabra et al. 2013; Chhabra and Dowling 2017; Kumar et al. 2019).
Rhizobium evolves with leguminous plants and has potential of directly fixing atmospheric nitrogen, hence facilitates self-establishment of plants in low nitrogen soils (Santos and Olivares 2021). Notably, along with Rhizobium, leguminous plants also contain some nitrogen-fixing endophytic bacteria such as Azospirillum, Azotobacter, Gluconacetobacter, and cyanobacteria (Calothrix, Nostoc, Anabena) (Kumar et al. 2019). Moreover, there are some nitrogen-fixing microorganisms (Bacillus, Beijerinckia and Klebsiella), which can fix nitrogen in non-leguminous plant in their free form (Santos and Olivares 2021).
Taken together, above-mentioned characteristics of microbiome justify their importance toward sustainable agriculture in terms of inducing crop’s nutrient intake, disease resistance, harsh environment tolerance, growth, yield, and productivity.
8.6 Current Trends and Future Perspectives
Continuously increasing global demand of food is challenging for the farmers, wherein they have to tackle with various stresses such as changing hostile climate, nutrient, water scarcity, and pollution. In this background, plant growth-promoting potential natural resources such as plant microbiome become a viable alternative tool for supporting plant health and sustainable agriculture. In plant microbiome establishment microbial diversity, their inter/intra generic interactions and environmental factor are critical. Despite extensive microbiome knowledge to scientists, still advances are needed for comprehensive information. In recent years, advanced biotechnological tools, bioinformatics, and meta-omics approaches together have established pipelines for structural, genomic, and functional organization of plant microbiomes and their interaction with plants. With such all-inclusive information, it would be easy to improve native microorganisms at cellular or molecular level leading to the more efficient bioinoculants development for improving crop’s yield and productivity.
References
Aamir M, Samal S, Rai A, Kashyap SP, Singh SK, Ahmed M, Upadhyay RS (2021) Plant microbiome: diversity, distribution, and functional relevance in crop improvement and sustainable agriculture. In: Microbiome stimulants for crops. Elsevier, Amsterdam, pp 417–436. https://doi.org/10.1016/b978-0-12-822122-8.00001-7
Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM (2016) Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol 14:1–31. https://doi.org/10.1371/journal.pbio.1002352
Alok D, Annapragada H, Singh S, Murugesan S, Singh NP (2020) Symbiotic nitrogen fixation and endophytic bacterial community structure in Bt-transgenic chickpea (Cicer arietinum L). Sci Rep 10:1–12. https://doi.org/10.1038/s41598-020-62199-1
Andreote FD, Gumiere T, Durrer A (2014) Exploring interactions of plant microbiomes. Sci Agric 71:528–539. https://doi.org/10.1590/0103-9016-2014-0195
Ardanov P, Lyastchenko S, Karppinen K, Häggman H, Kozyrovska N, Pirttilä AM (2016) Effects of Methylobacterium sp. on emergence, yield, and disease prevalence in three cultivars of potato (Solanum tuberosum L.) were associated with the shift in endophytic microbial community. Plant Soil 405:299–310. https://doi.org/10.1007/s11104-015-2500-y
Babu AG, Kim SW, Yadav DR, Hyum U, Adhikari M, Lee YS (2015) Penicillium menonorum: a novel fungus to promote growth and nutrient management in cucumber plants. Mycobiology 43:49. https://doi.org/10.5941/MYCO.2015.43.1.49
Bergottini VM, Hervé V, Sosa DA, Otegui MB, Zapata PD, Junier P (2017) Exploring the diversity of the root-associated microbiome of Ilex paraguariensis St. Hil. (Yerba mate). Appl Soil Ecol 109:23–31. https://doi.org/10.1016/J.APSOIL.2016.09.013
Bhatt P, Verma A, Verma S, Anwar MS, Prasher P, Mudila H, Chen S (2020) Understanding Phytomicrobiome: a potential reservoir for better crop management. Sustainability 12:5446. https://doi.org/10.3390/SU12135446
Bodor A, Bounedjoum N, Vincze GE, Erdeiné Kis Á, Laczi K, Bende G, Szilágyi Á, Kovács T, Perei K, Rákhely G (2020) Challenges of unculturable bacteria: environmental perspectives. Rev Environ Sci Bio/Technol 191(19):1–22. https://doi.org/10.1007/S11157-020-09522-4
Cai F, Pang G, Miao Y, Li R, Li R, Shen Q, Chen W (2017) The nutrient preference of plants influences their rhizosphere microbiome. Appl Soil Ecol 110:146–150. https://doi.org/10.1016/J.APSOIL.2016.11.006
Chhabra S (2019) Phosphorus management in agroecosytems and role and relevance of microbes in environmental sustainability. In: Sustainable green technologies for environmental managment. Springer, Singapore, pp 53–66. https://doi.org/10.1007/978-981-13-2772-8_3
Chhabra S, Dowling DN (2017) Endophyte-promoted nutrient acquisition: phosphorus and iron. In: Functional importance of the plant microbiome. Implications for agriculture, forestry and bioenergy. Springer, Cham, pp 21–42. https://doi.org/10.1007/978-3-319-65897-1_3
Chhabra S, Brazil D, Morrissey J, Burke JI, O’Gara F, Dowling ND (2013) Characterization of mineral phosphate solubilization traits from a barley rhizosphere soil functional metagenome. Microbiology 2:717–724. https://doi.org/10.1002/MBO3.110
Chitarra W, Pagliarani C, Maserti B, Lumini E, Siciliano I, Cascone P, Schubert A, Gambino G, Balestrini R, Guerrieri E (2016) Insights on the impact of arbuscular mycorrhizal symbiosis on tomato tolerance to water stress. Plant Physiol 171:1009–1023. https://doi.org/10.1104/pp.16.00307
Compant S, Samad A, Faist H, Sessitsch A (2019) A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J Adv Res 19:29–37. https://doi.org/10.1016/j.jare.2019.03.004
Conn VM, Franco CMM (2004) Effect of microbial inoculants on the indigenous actinobacterial endophyte population in the roots of wheat as determined by terminal restriction fragment length polymorphism. Appl Environ Microbiol 70:6407–6413. https://doi.org/10.1128/AEM.70.11.6407-6413.2004
Curlango-Rivera G, Huskey DA, Mostafa A, Kessler JO, Xiong Z, Hawes MC (2013) Intraspecies variation in cotton border cell production: rhizosphere microbiome implications. Am J Bot 100:1706–1712. https://doi.org/10.3732/ajb.1200607
Dastogeer KMG, Tumpa FH, Sultana A, Akter MA, Chakraborty A (2020) Plant microbiome—an account of the factors that shape community composition and diversity. Curr Plant Biol 23:100161. https://doi.org/10.1016/j.cpb.2020.100161
De La Torre-Ruiz N, Ruiz-Valdiviezo VM, Rincón-Molina CI, Rodríguez-Mendiola M, Arias-Castro C, Gutiérrez-Miceli FA, Palomeque-Dominguez H, Rincón-Rosales R (2016) Effect of plant growth-promoting bacteria on the growth and fructan production of Agave Americana L. Brazilian J Microbiol 47:587–596. https://doi.org/10.1016/j.bjm.2016.04.010
del Orozco-Mosqueda MC, del Rocha-Granados MC, Glick BR, Santoyo G (2018) Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol Res 208:25–31. https://doi.org/10.1016/J.MICRES.2018.01.005
Durán P, Tortella G, Viscardi S, Barra PJ, Carrión VJ, De La Luz Mora M, Pozo MJ (2018) Microbial community composition in take—all suppressive soils. Front Microbiol 9:1–15. https://doi.org/10.3389/fmicb.2018.02198
Fitzpatrick CR, Salas-González I, Conway JM, Finkel OM, Gilbert S, Russ D, Teixeira PJPL, Dangl JL (2020) The plant microbiome: from ecology to reductionism and beyond. Annu Rev Microbiol 74:81–100. https://doi.org/10.1146/annurev-micro-022620-014327
Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169:30–39. https://doi.org/10.1016/J.MICRES.2013.09.009
Gupta R, Anand G, Gaur R, Yadav D (2021) Plant–microbiome interactions for sustainable agriculture: a review. Physiol Mol Biol Plants 27:165–179. https://doi.org/10.1007/s12298-021-00927-1
Hara S, Matsuda M, Minamisawa K (2019) Growth stage-dependent bacterial communities in soybean plant tissues: Methylorubrum transiently dominated in the flowering stage of the soybean shoot. Microbes Environ 34:446–450. https://doi.org/10.1264/jsme2.ME19067
Hartmann A, Rothballer M, Hense BA, Schröder P (2014) Bacterial quorum sensing compounds are important modulators of microbe-plant interactions. Front Plant Sci 5:131. https://doi.org/10.3389/FPLS.2014.00131/BIBTEX
Hashem A, Abd-Allah EF, Alqarawi AA, Al-Huqail AA, Wirth S, Egamberdieva D (2016) The interaction between arbuscular mycorrhizal fungi and endophytic bacteria enhances plant growth of Acacia gerrardii under salt stress. Front Microbiol 7:1–15. https://doi.org/10.3389/fmicb.2016.01089
Hong S, Jv H, Lu M, Wang B, Zhao Y, Ruan Y (2020) Significant decline in banana Fusarium wilt disease is associated with soil microbiome reconstruction under chilli pepper-banana rotation. Eur J Soil Biol 97:103154. https://doi.org/10.1016/J.EJSOBI.2020.103154
Igiehon NO, Babalola OO (2018) Below-ground-above-ground plant-microbial interactions: focusing on soybean, rhizobacteria and mycorrhizal fungi. Open Microbiol J 12:261. https://doi.org/10.2174/1874285801812010261
Jodeh S, Alkowni R, Hamed R, Samhan S (2015) The study of electrolyte leakage from barley (Hordeum vulgare L) and pearlmillet using plant growth promotion (PGPR) and reverse osmosis. J Food Nutr Res 3:422–429. https://doi.org/10.12691/jfnr-3-7-3
Kaur T (2020) Fungal endophyte-host plant interactions: role in sustainable agriculture. In: Sustainable crop production. IntechOpen, London. https://doi.org/10.5772/INTECHOPEN.92367
Kinnunen-Grubb M, Sapkota R, Vignola M, Nunes IM, Nicolaisen M (2020) Wheat domestication has dramatically affected the root-associated microbiome 1–23. https://doi.org/10.21203/rs.3.rs-20588/v1
Konieczny A, Kowalska I (2016) The role of arbuscular mycorrhiza in zinc uptake by lettuce grown at two phosphorus levels in the substrate. Agric Food Sci 25:124–137. https://doi.org/10.23986/afsci.55534
Kumar I, Mondal M, Gurusamy R, Balakrishnan S, Natarajan S (2019) Plant-microbiome interaction and the effects of biotic and abiotic components in agroecosystem. In: Microbial interventions in agriculture and environment: volume 2: rhizosphere, microbiome and agro-ecology. Springer, Cham, pp 517–546. https://doi.org/10.1007/978-981-13-8383-0_18
Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ, Jung EJ, Park H, Roy N, Kim H, Lee MM, Rubin EM, Lee SW, Kim JF (2018) Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol 3611(36):1100–1109. https://doi.org/10.1038/nbt.4232
Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, Del Rio TG, Jones CD, Tringe SG, Dangl JL (2015) Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349:860–864. https://doi.org/10.1126/SCIENCE.AAA8764/SUPPL_FILE/PAPV2.PDF
Lebreton L, Guillerm-Erckelboudt AY, Gazengel K, Linglin J, Ourry M, Glory P, Sarniguet A, Daval S, Manzanares-Dauleux MJ, Mougel C (2019) Temporal dynamics of bacterial and fungal communities during the infection of Brassica rapa roots by the protist Plasmodiophora brassicae. PLoS One 14:e0204195. https://doi.org/10.1371/JOURNAL.PONE.0204195
Lee EH, Eom AH (2015) Growth characteristics of Rhizophagus Clarus strains and their effects on the growth of host plants. Mycobiology 43:444–449. https://doi.org/10.5941/MYCO.2015.43.4.444
Li R, Feng Y, Chen H, Zhang C, Huang Y, Chen L, Hao Q, Cao D, Yuan S, Zhou X (2020) Whole-genome sequencing of Bradyrhizobium diazoefficiens 113-2 and comparative genomic analysis provide molecular insights into species specificity and host specificity. Front Microbiol 11:1–17. https://doi.org/10.3389/fmicb.2020.576800
Liu F, Hewezi T, Lebeis SL, Pantalone V, Grewal PS, Staton ME (2019) Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol 19:1–19. https://doi.org/10.1186/s12866-019-1572-x
Lupatini M, Korthals GW, de Hollander M, Janssens TKS, Kuramae EE (2017) Soil microbiome is more heterogeneous in organic than in conventional farming system. Front Microbiol 7:2064. https://doi.org/10.3389/FMICB.2016.02064/BIBTEX
Mendes LW, de Chaves MG, de Fonseca MC, Mendes R, Raaijmakers JM, Tsai SM (2019) Resistance breeding of common bean shapes the physiology of the rhizosphere microbiome. Front Microbiol 10:1–10. https://doi.org/10.3389/fmicb.2019.02252
Mitter EK, de Freitas JR, Germida JJ (2017) Bacterial root microbiome of plants growing in oil sands reclamation covers. Front Microbiol 8. https://doi.org/10.3389/fmicb.2017.00849
Mohumad Tahat M (2012) The potential of endomycorrhizal fungi in controlling tomato bacterial wilt Ralstonia solanacearum under glasshouse condition. African J Biotechnol 11:13085–13094. https://doi.org/10.5897/ajb11.3629
Morella NM, Weng FCH, Joubert PM, Jessica C, Lindow S, Koskella B (2020) Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc Natl Acad Sci U S A 117:1148–1159. https://doi.org/10.1073/pnas.1908600116
Oyserman BO, Cordovez V, Flores SS, Leite MFA, Nijveen H, Medema MH, Raaijmakers JM (2021) Extracting the GEMs: genotype, environment, and microbiome interactions shaping host phenotypes. Front Microbiol 11:1–8. https://doi.org/10.3389/fmicb.2020.574053
Padmavathi T, Dikshit R, Seshagiri S (2015) Effect of Rhizophagus spp. and plant growth-promoting Acinetobacter junii on Solanum lycopersicum and Capsicum annuum. Rev Bras Bot 38:273–280. https://doi.org/10.1007/s40415-015-0144-z
Pieterse CMJ, de Jonge R, Berendsen RL (2016) The soil-borne supremacy. Trends Plant Sci 21:171–173. https://doi.org/10.1016/J.TPLANTS.2016.01.018
Podolich O, Ardanov P, Zaets I, Pirttilä AM, Kozyrovska N (2015) Reviving of the endophytic bacterial community as a putative mechanism of plant resistance. Plant Soil 388:367–377. https://doi.org/10.1007/S11104-014-2235-1/FIGURES/2
Qin S, Yeboah S, Xu X, Liu Y, Yu B (2017) Analysis on fungal diversity in rhizosphere soil of continuous cropping potato subjected to different furrow-ridge mulching managements. Front Microbiol 8:845. https://doi.org/10.3389/fmicb.2017.00845
Rastegari AA (2020) Microbiomes for sustainable. Agriculture. https://doi.org/10.1007/978-3-030-38453-1
Rodrigues AA, Forzani MV, de Soares RS, Sibov ST, Vieira JDG (2016) Isolation and selection of plant growth-promoting bacteria associated with sugarcane. Pesqui Agropecuária Trop 46:149–158. https://doi.org/10.1590/1983-40632016v4639526
Santos LF, Olivares FL (2021) Plant microbiome structure and benefits for sustainable agriculture. Curr Plant Biol 26:100198. https://doi.org/10.1016/j.cpb.2021.100198
Santos-Medellín C, Edwards J, Liechty Z, Nguyen B, Sundaresan V (2017) Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. MBio 8:e00764-17. https://doi.org/10.1128/MBIO.00764-17
Sapre S, Gontia-Mishra I, Tiwari S (2018) Klebsiella sp. confers enhanced tolerance to salinity and plant growth promotion in oat seedlings (Avena sativa). Microbiol Res 206:25–32. https://doi.org/10.1016/j.micres.2017.09.009
Shahid M, Khan MS, Syed A, Marraiki N, Elgorban AM (2021) Mesorhizobium ciceri as biological tool for improving physiological, biochemical and antioxidant state of Cicer arietinum (L.) under fungicide stress. Sci Rep 11(1):1–18. https://doi.org/10.1038/s41598-021-89103-9
Shahsavar AR, Refahi A, Zarei M, Aslmoshtaghi E (2016) Analysis of the effects of Glomus etunicatum fungi and Pseudomonas fluorescence bacteria symbiosis on some morphological and physiological characteristics of Mexican lime (Citrus aurantifolia L.) under drought stress conditions. Adv Hortic Sci 30:39–45. https://doi.org/10.13128/ahs-18700
Sharma M, Sudheer S, Usmani Z, Rani R, Gupta P (2020) Deciphering the omics of plant-microbe interaction: perspectives and new insights. Curr Genomics 21:343–362. https://doi.org/10.2174/1389202921999200515140420
Shcherbakova EN, Shcherbakov AV, Andronov EE, Gonchar LN, Kalenskaya SM, Chebotar VK (2017) Combined pre-seed treatment with microbial inoculants and Mo nanoparticles changes composition of root exudates and rhizosphere microbiome structure of chickpea (Cicer arietinum L.) plants. Symbiosis 73:57–69. https://doi.org/10.1007/S13199-016-0472-1/TABLES/4
Shi SM, Chen K, Gao Y, Liu B, Yang XH, Huang XZ, Liu GX, Zhu LQ, He XH (2016) Arbuscular mycorrhizal fungus species dependency governs better plant physiological characteristics and leaf quality of mulberry (Morus alba L.) seedlings. Front Microbiol 7:1–11. https://doi.org/10.3389/fmicb.2016.01030
Shuab R, Lone R, Naidu J, Sharma V, Imtiyaz S, Koul KK (2014) Benefits of inoculation of arbuscular mycorrhizal fungi on growth and development of onion (Allium cepa) plant. Environ Sci 14:527–535. https://doi.org/10.5829/idosi.aejaes.2014.14.06.12347
Simonin M, Dasilva C, Terzi V, Ngonkeu ELM, Diouf D, Kane A, Béna G, Moulin L (2020) Influence of plant genotype and soil on the wheat rhizosphere microbiome: evidences for a core microbiome across eight African and European soils. FEMS Microbiol Ecol 96:1–18. https://doi.org/10.1093/femsec/fiaa067
Singh D, Raina TK, Kumar A, Singh J, Prasad R (2019) Plant microbiome: a reservoir of novel genes and metabolites. Plant Gene. https://doi.org/10.1016/j.plgene.2019.100177
Tripathi BM, Moroenyane I, Sherman C, Lee YK, Adams JM, Steinberger Y (2017) Trends in taxonomic and functional composition of soil microbiome along a precipitation gradient in Israel. Microb Ecol 74:168–176. https://doi.org/10.1007/S00248-017-0931-0/FIGURES/4
Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK (2020) Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol:607–621. https://doi.org/10.1038/s41579-020-0412-1
van der Voort M, Kempenaar M, van Driel M, Raaijmakers JM, Mendes R (2016) Impact of soil heat on reassembly of bacterial communities in the rhizosphere microbiome and plant disease suppression. Ecol Lett 19:375–382. https://doi.org/10.1111/ELE.12567
Viscardi S, Ventorino V, Duran P, Maggio A, De Pascale S, Mora ML, Pepe O (2016) Assessment of plant growth promoting activities and abiotic stress tolerance of Azotobacter chroococcum strains for a potential use in sustainable agriculture. J Soil Sci Plant Nutr 16:848–863. https://doi.org/10.4067/s0718-95162016005000060
Zahir ZA, Munir A, Asghar HN, Shaharoona B, Arshad M (2008) Effectiveness of rhizobacteria containing ACC deaminase for growth promotion of peas (Pisum sativum) under drought conditions. J Microbiol Biotechnol 18:958–963
Zhang QM, Gong M, Liu K, Chen Y, Yuan J, Chang Q (2020) Rhizoglomus intraradices improves plant growth, root morphology and Phytohormone balance of Robinia pseudoacacia in arsenic-contaminated soils. Front Microbiol 11:1428. https://doi.org/10.3389/FMICB.2020.01428/BIBTEX
Zhu S, Vivanco JM, Manter DK (2016) Nitrogen fertilizer rate affects root exudation, the rhizosphere microbiome and nitrogen-use-efficiency of maize. Appl Soil Ecol 107:324–333. https://doi.org/10.1016/J.APSOIL.2016.07.009
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Pandey, A.K., Mehta, S., Bhati, P., Chhabra, S. (2023). Microbiome of Plants: The Diversity, Distribution, and Their Potential for Sustainable Agriculture. In: Chhabra, S., Prasad, R., Maddela, N.R., Tuteja, N. (eds) Plant Microbiome for Plant Productivity and Sustainable Agriculture . Microorganisms for Sustainability, vol 37. Springer, Singapore. https://doi.org/10.1007/978-981-19-5029-2_8
Download citation
DOI: https://doi.org/10.1007/978-981-19-5029-2_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-5028-5
Online ISBN: 978-981-19-5029-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)