Abstract
To feed the ever-increasing human population globally, continuous attempts have been made to enhance the agricultural productivity significantly through the application of modern breeding techniques. Agronomic approaches such as increased abiotic/biotic stress tolerance, reduced toxicity, superior nutritional quality, delayed ripening, better post-harvest quality, etc. were attempted. However, loss of agricultural lands due to urbanization in both developing and under-developed countries, drastic climate changes, plant diseases, and farmer’s non-compliance to modern agricultural methods are some of the major hindrances to these strategies. Plant-epigenetic modifications play vital roles in acclimatization, stress tolerance, adaptation, and evolution processes. The agronomically important traits of crop life such as flowering time, fruit development, risk avoidance from environmental factors, and crop immunity are attained by the epigenetic modifications such as DNA methylation, histone post-translational modifications, and small non-coding RNAs modification. This chapter emphasizes on the epigenetic changes and its effect on crop growth, yield, and herbicide resistance. As well as develop plants that are resistant to harsh weather conditions such as cold, heat, and rain and also to increase the crop yield in poorly-arable places like deserts, marshy lands, backwaters, high altitudes and areas with high/low soil pH. Hence, the potential of epigenetics in crop improvement and sustained agriculture is highly recognized by researchers both in academia and industry and by policy makers and government.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Plant Epigenetics
- Sustained agriculture
- Crop improvement
- Epigenetic regulation
- Plant immunity
- Plant breeding
14.1 Introduction
14.1.1 Epigenetics
The term epigenetics was coined by Conrad Hal Waddington in 1942, it is defined as phenotypic changes due to heritable pattern of alteration in chromatin without any changes in the underlying DNA sequences. The mechanism by which the gene expression turned on or off was poorly understood at that time. However in the last few decades numerous scientific studies revealed the mechanism of epigenetic modifications and its influence on gene expression profile in normal development and disease conditions in an organism. Unlike the genetic mutations the epigenetic modifications can be reversed, as well can be manipulated to obtain the desired phenotype, this phenomenon allows epigenesist to understand the differentiation and development process of a seed to plant and has high potential to generate phenotypic variations that could well adapt to abiotic and biotic stress and favorable agronomical traits such as disease resistant and high yielding quality crops. Plants being sessile developed several sophisticated mechanisms of gene regulation including epigenetic regulation to respond, adapt, and thrive in a constantly changing ecosystem. The three major epigenetic marks are methylation of fifth carbon of cytosine in Guanine–Cytosine (GC) rich sequence in a gene, acetylation /deacetylation of histone proteins and small RNAs. To alter the gene expression all these can act alone or in combination with each other. DNA-methylation in plants is done by three different enzymes, namely (1) MET1 DNA methyltransferase at GC rich sequence; (2) CHROMOMETHYLASE3—CMT3 at CHG (H = A, C, or T); (3) DOMAINS REARRANGED METHYLTRANSFERASE—DRM1/DRM2 or CMT2 methyltransferase are responsible for methylating CHH (Weinhold 2006; Simmons 2008; Gibney and Nolan 2010). DNA methylation frequently happens in repetitive regions such as transposable elements (TEs), centromeric repeats and in rDNA—genes that encode ribosomal RNA. The TE transposition can disrupt genomic integrity and could alter the gene expression if it integrates within or near the gene; hence plants epigenetically silence them by methylation. TE silencing is commonly found in epigenome of almost all the investigated plants that ranges from moss -Physcomitrella to gymnosperms and angiosperms.
Manipulation of DNA methylation in plants can alter its phenotype, for example, rice treated with DNA demethylating agents results in dwarf progeny. Reduction in global DNA methylation was observed in vernalization treatment and it might be one of the factors for initiating flowering in plants. Similarly, DNA methylation pattern changes were observed in cold stressed-maize and Arabidopsis plants. Constant response to the environmental stress induces methylation changes in plants; such epigenetic modifications can be either reversed and the gene expression returns to its near normal levels or it is carried as epigenetic stress memory to its progeny. Thus the DNA methylation plays significant role in various stages of plant development which will be discussed in several following sub-headings.
The regulation of gene activity has been achieved by reversible histone acetylation and deacetylation at the N-terminus of histone tails catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. Regulation of gene expression in plant development and its responses to environmental stresses can be also achieved by HATs and HDACs due to its interaction with various chromatin-remodeling factors and transcription factors. Around 80% of proteins in plants undergo acetylation.
14.1.2 RNAs—miRNA, ShRNA, si RNA, Non-coding RNA
Precursors of miRNAs or siRNAs are generated in several ways. DNA-dependent RNA polymerase II (RNA Pol II) transcripts with extensive self-complementarity fold back on themselves to form stem-loop structures in the case of miRNAs. The double-stranded precursors can be generated via convergent, bidirectional transcription by a DNA-dependent RNA polymerase such as RNA Pol II, thereby generating transcripts that overlap and base-pairing the case of siRNAs. RNA transcripts can be used as templates for an RNA-dependent RNA polymerase (RdRP), to generate a complementary strand. The criticality for plant development requires miRNAs which supports the functions of complementary mRNAs to a specific subset of cells by bringing about their posttranscriptional degradation or translational repression in adjacent cells. In the context of a multi-protein RISC complex miRNAs silence gene expression by base-pairing to target mRNAs, which facilitates mRNA cleavage, “slicing,” or the inhibition of mRNA translation. Transcriptional gene silencing by inducing DNA methylation and histone H3K9 demethylation are regulated by heterochromatic siRNAs.
14.1.3 Small RNAs Can Trigger DNA Methylation and Chromatin Modification
Large number of long non-coding RNAs (lncRNAs) are produced by many eukaryotes, including plants which holds regulatory roles in various developmental processes. lncRNAs are versatile regulatory molecules.
14.1.4 Chromatin Remodeling/Condensation
The basal state of chromatin remodeling can be altered by promoting either an “open” (activation of transcription) or a “closed” (repression of transcription) chromatin configuration. According to the size of transcripts NcRNAs can be classified into two categories they are Nc RNAs with more than 200 nucleotides are considered as long ncRNAs (lncRNAs), whereas short ncRNAs are less than 200 nucleotides. The micro RNA (miRNAs), small interfering RNAs (siRNAs), and Piwi-interacting RNAs (piRNAs) are included in the short ncRNAs. The stable inheritance of chromatin structures can be invoked by transcriptional states to daughter cells through mitosis or even meiosis. The existence of post-translational modifications (mainly acetylation, methylation, and phosphorylation) of the amino-terminal tails of the histones is demonstrated in various plants based studies. In plants, histone H4 can be acetylated at Lys 5, 8, 12, 16, and 20, whereas histone H3 is acetylated at positions 9, 14, 18, and 23. To a lower extent histone H2A and H2B are also acetylated.
14.1.5 Polycomb Proteins
The formation of conserved regulatory structures that can suppress genes through a variety of physiological roles and types of epigenetic patterning is the major role of polycomb proteins (PcG). PcG proteins bind nucleosomes thereby alters the intrinsic structure of chromatin to initiate epigenetic modifications and maintain these modifications during development. These multi-protein complexes modify chromatin structure to form flexible, repressive chromatin configurations that include numerous targeted genes and maintain silencing. The paradigmatic model for epigenetic regulation of gene silencing has been controlled by the PcG of proteins. PcG proteins are a collection of transcriptional regulatory factors that can control gene expression, whose transcriptional imposed silencing can be transmitted from embryos to adulthood.
14.1.6 Fungal Prions and Epigenetics
Due to self-perpetuating abilities which enable to record and reproduce the memory of acquired alteration that initially caused prion formation prions are built as perfect machines of molecular memory. Prion-like elements trigger environmental signals to cellular and organismal processes. Two-level regulatory mechanism, with a prion-like domain in the regulatory protein for initial sensing and recording a signal can be achieved by epigenetic pathways with regulatory change in the chromatin organization or in the mode of gene transcription.
14.1.7 Global Hunger and Crop Production
Chronic hunger and malnutrition are undue burden on humanity; global statistics are alarming with 821 million people who are in hunger. WHO’s Global Hunger Index (GHI) shows that the many of the African countries are in the serious (GHI; 20–34.9) to extremely alarming (GHI ≥ 50) category. Developing economies like India is also in serious condition with the GHI of 30.3. Hence urgent measures are needed to address this global crisis. One of the major factors for the hunger is crop failure due to natural disasters, unpredictable rainfall, drought, drastic climate changes. It also hinders livestock management and adds up to food shortage. As well the on-farm losses due to pests, rodents, birds, diseases, weather, harvesting and storage methods (Chen 1990; Wu et al. 2014). Additionally, human activities such as urbanization, deforestation lead to reduction in crop land. The environmental factors such as soil erosion, fall of water tables, and aquifer pose threat to sustained agriculture. Hence, governments all over the world make policies and programs to protect the agricultural lands and prevent on-farm loss to maximize crop yield and utilization of agricultural inputs to combat famine (Acevedo 2011). Genetic and epigenetic approaches resulted in crops with high nutritional values, drug and drought varieties and increased the crop yield; which in-turn could reduce the global hunger.
14.2 Plant Epigenetics
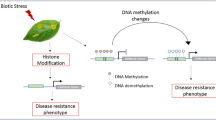
The study of genes, genetic variation, and heredity specifically in plants is the plant genetics. Plant epigenetics based technologies dramatically increase our capacity to understand the molecular basis of traits and utilize the available resources for accelerated development of stable high-yielding, nutritious, input-use efficient and climate-smart crop varieties. The highly diversified group of sessile organisms, plants gets evolved with extreme ultraviolet light, salinity, and hypoxia interactors. Some biotic interactors are beneficial for the plant (pollinators, rhizobia, mycorrhiza), whereas some interactors (herbivores, pathogens, or strong competitors) are detrimental to plants. Molecular level changes of plants constantly are required to adapt themselves to the changing environments via epigenetic regulations which may improve the survivability of plants in terms of tolerance toward external stress. Gene expression changes in plants may respond to environmental stimuli/chromatin-based gene regulation. The chromatin modification in plants through DNA methylation is the major part of plant epigenetics. Direct modification of DNA genes in plants occurs through mitosis and amitosis (Chen et al. 2010). DNA methylation and histone modification cause the direct phenotypic characterization of epigenetic mutants which possess the ability to characterize whole-plant traits and organ traits (Alonso et al. 2019). Specific genetic and epigenetic variants dynamic interplay in case of plant stress responses (Eriksson et al. 2020). The epigenetic modified plant is shown in Fig. 14.1.
Epigenetic regulation in plants reflects their mode of development, lifestyle, and evolutionary history. Plants grow by continuously producing new organs from self-sustaining stem cell populations known as meristems. Unlike growth in mammals, in which organ and tissue formation is largely specified during embryonic development. The high degree of phenotypic plasticity can be achieved by the postembryonic development in plants in a continuous process shaped by environmental influences. Because plants are unable to escape their surroundings, they are forced to cope with changeable and often unfavorable growth conditions. Epigenetic regulatory mechanisms can facilitate metastable changes in gene activity and fine-tune gene expression patterns, thus enabling plants to survive and reproduce successfully in unpredictable environments. Genome size, genome complexity, and the ratio of heterochromatin to euchromatin in seed plants are generally comparable to mammals. In case of gene regulation plants and mammals make similar use of DNA methylation and histone post-translational modifications (PTMs).
DNA methylation consists mostly in adding a methyl group at the fifth carbon position of a cytosine ring, and, different to what happens in animals, plants have three sites that frequently can suffer methylation: CG, CHG (where H is A, C, or T), and CHH. Modification of DNA methylation profiles in plant can cause phenotypic variation. For instance, demethylation of rice genomic DNA causes an altered pattern of gene expression, inducing dwarf plants. Thus, the knowledge of epigenetic contributions in phenotypic plasticity and hereditable variation is important to understand how natural population can adapt in different environmental condition, especially in a world context of climate change.
14.3 Epigenetics to Increase the Crop Yield and Sustained Agriculture
Improved crop yield with a goal for sustained agriculture may be achieved through epigenetic regulation (Fig. 14.1) through epigenetic mechanisms leading to changes in chromatin architecture, such as DNA methylation, post-translational histone modifications, and the action of non-coding RNA molecules which are either small RNAs (small interfering RNAs, siRNAs and microRNAs, miRNAs) or long non-coding RNAs (long snRNAs) (Kapazoglou et al. 2018). The understanding of epigenetics supports towards the crop improvement and ultimately this will get replicated in the nutritional management and plant breeding. The molecular and mechanistic basis of genotype along with the environmental interactions facilitates the epigenetic mechanisms. Epigenetics thus supports plant cellular/physiological processes during its development with increased fitness/adaptability to the plant in the changing environmental conditions (Kumar 2019). Recently the newer technologies of gene editing accomplish things in shorter periods of time than a natural breeding process. The genetically modified crops may open path for convenient and more economical production practices for farmers.
14.4 Epigenetics for Crop Growth
Prevailing conditions for epigenetic changes for sustained agriculture was shown in Fig. 14.2. The two major types of small RNAs, small interfering RNAs (siRNAs) and microRNAs (miRNAs) in plants are likely to be associated with the silencing of gene expression. miRNAs regulate the plant growth, development, organogenesis, and responses to various biotic and abiotic stresses. The critical step for successful grain production in rice is the flowering time control with criticality in the day length as a key factor for rice flowering. Shorter day length will greatly induce rice flowering with criticality in the photoperiod. Liu et al. 2014 reported that the accurate control of flowering based on photoperiod is regulated by key mechanisms that involve the regulation of flowering genes such as Heading date1 (Hd1), Early hd1 (Ehd1), Hd3a, and RFT1. Rice enhancer of zeste [E(z)] genes SDG711 and SDG718, which encode the polycomb repressive complex2 (PRC2) key subunit is required for trimethylation of histone H3 lysine 27 (H3K27me3) which supports the long day (LD) and short day (SD) regulation of key flowering genes (Liu et al. 2014).
Kapazoglou et al. 2013 emphasized that epigenetic factors such as DNA methylation and histone modifications may regulate plant development. Gene silencing or activation occurs by cytosine methylation and demethylation. They reported that in seed development of Arabidopsis, cytosine demethylation is achieved by specific DNA glycosylases, including AtDME (DEMETER) and AtROS1 (REPRESSOR OF SILENCING1). They studied the DME homologue from barley (HvDME), during seed development and drought conditions. Their phylogenetic analysis revealed high degree of homology to other monocot DME glycosylases, and sequence divergence from the ROS1, DML2, and DML3 orthologs. They concluded that expression analysis during seed development and under dehydration conditions provides role for HvDME in endosperm development, seed maturation, and in response to drought.
The daily protein requirements have been increased due to wheat consumption and wheat becomes one of the most important cereal crops having global production of >700 million tones (Kumar et al. 2017). Limited success in growing wheat on salt-affected soils has been achieved because only a few salt-tolerant bread wheat genotypes have been identified. The accumulation of excessive salt contents in the soil may lead to the salt stress which eventually results in the crop growth inhibition and ultimately leads to crop death which creates very dangerous for agricultural productivity. The impairment of water potential cells, ion toxicity, membrane integrity and function, and delayed uptake of essential mineral nutrients occurs in crop growth due to salt stress. This ultimately affects the metabolic processes in plants. Understandings on biochemical, physiological, genetic, and epigenetic aspects of salt tolerance along with cloning of the genes involved in salt tolerance and development of transgenic may support toward the better breeding strategy for the crops growing under saline conditions. Accumulation of sodium ion (Na+) in plant tissues inhibits uptake of essential macronutrients like potassium (K+) and calcium (Ca2+) from soil which creates the one of the detrimental effects in crop growth (Very and Sentenac 2003; Shi et al. 2003).
Hamamoto et al. (2015) investigated the physio-biochemical responses of four wheat cultivars under salt stress, and with response to the genotypes for their salt tolerance level to identify the most contrasting salt-responsive genotypes. Further they investigated the mechanisms responsible for genotype and tissue specific differential expression of TaHKTs genes and found better antioxidant potential, membrane stability, increased accumulation of osmolytes/phytophenolics, and higher K+/Na+ ratio under 200 mMNaCl stress-induced condition of Kharchia-65 and found the same to be the most salt-tolerant cultivar. The sensitivity towards stress may be ensured by reduced soluble sugar, proline, total chlorophyll, total phenolics contents and lower antioxidant potential in HD-2329. Salt-sensitive (HD-2329) genotype may be revealed by genetic and bioinformatic analyses of HKT1;4 of contrasting genotypes (Kharchia-65 and HD-2329) revealed deletions, transitions, and transversions resulting into altered structure and loss of conserved motifs (Ser-Gly-Gly-Gly and Gly-Arg).
In order to gain insights into the genomic basis of apple (Malus domestica) evolution and domestication a newer high quality apple WGS, GDDH13 v1.1, was released which serves as the reference genome for apple. WGSs apple shown enormous impact on apple biological functioning, trait physiology, and inheritance, with the valued outcome of high quality crop. Here in WGS apple shown improvement associated with innovative approach to obtain durable, environmentally sound, productive, and consumer desirable apples (Peace et al. 2019).GDDH13 based apple reference genome supports in the understanding of epigenetic mechanisms involved in fruit size regulation which helps in the fruit size regulation. Kumar et al. (2017) investigated the physio-biochemical, molecular indices, and defense responses of wheat cultivars to identify the most contrasting salt-responsive genotypes and the mechanisms associated with their differential responses. For multivariate analysis in order to identify the most contrasting genotypes the physio-biochemical traits specifically membrane stability index, antioxidant potential, osmoprotectants, and chlorophyll contents are measured at vegetative stage. They observed that Kharchia-65 to be the most salt-tolerant cultivar based on the genetic and epigenetic analyses with mechanisms associated with differential response of the wheat genotypes under salt stress such as improved antioxidant potential, membrane stability, increased accumulation of osmolytes/phytophenolics, and higher KC/NaC ratio under 200 mM NaCl stress. Sensitivity to the stress associated with HD-2329 strain was elicited by increased MDA level, reduced soluble sugar, proline, total chlorophyll, total phenolics contents, and lower antioxidant potential. The contrasting genotypes can be revealed by deletions, transitions, and transversions resulting into altered structure, loss of conserved motifs (Ser-Gly-Gly-Gly and Gly-Arg) and function in salt-sensitive (HD-2329) (Kharchia-65 and HD-2329). Further the tissue- and genotype-specific changes were explained by their epigenetic variations in cytosine methylation. Overall they indicated that abiotic stress exerts significant impact on plant’s growth, development, and productivity, which can be overcomed by epigenetic and molecular mechanisms of salt tolerance in plants.
Recognization and respond of plants to pathogens have been evolved by its innate immune systems which includes pathogen-associated molecular pattern (PAMP)-triggered immunity and effector-triggered immunity. The key signaling molecules involved in defense and growth, including jasmonic acid, salicylic acid, and reactive oxygen species which includes the plant–pathogen interactions and neighboring plants perception via light-quality receptors influence affects the profile of plant secondary metabolites and emitted volatile organic compounds shown greater impact on plant herbivore and plant pollinator interactions which in turn affects the fitness of the plant (Alonso et al. 2019). Cai et al. (2018) reported that host Arabidopsis cells secrete exosome-like extracellular vesicles to deliver sRNAs into fungal pathogen Botrytis cinerea which accumulate at the infection sites and are taken up by the fungal cells. These sRNA-containing vesicles accumulate at the infection sites and are taken up by the fungal cells. The silencing of fungal genes critical for pathogenicity has been induced by transferred host sRNAs. They reported that Arabidopsis has adapted immune response based exosome-mediated cross-kingdom RNA interference during the evolutionary arms race with the pathogen. They emphasized that functional studies of host transferred sRNAs support the identification of important virulence genes in interacting pathogens and pests.
Crespo-Salvador et al. (2018) determined the histone marks’ profile in two differentially expressed genes in response to B. cinerea, as well as to oxidative stress, given its relevance in this infection. Here in both the induced CYP71A13 essential against this necrotroph and the repressed EXL7 (Exordium-like 1), which encodes a cytochrome P450 involved in camalexin synthesis. On both the promoter and the body of the highly induced PR1 in Arabidopsis plants infected with B. cinerea at 24 and 33 h after inoculation they performed chromatin immunoprecipitation analysis associated with activating marks H3K4me3, H3K9ac, and the repressor one H3K27me3. They observed that at 24 hpi, H3K4me3 level increased on the promoter associated with different locations of the body of the genes induced upon B. cinerea, including DES (divinyl ethyl synthase), LoxD (lipoxygenase D), DOX1 (α-dioxygenase 1), PR2 (pathogenesis-related protein2), WRKY53 and WRKY33. They further analyzed the genes as B. cinerea potential biomarkers for infection in crops. Pathogen infection induces the expression of histone deacetylase in Arabidopsis which indicates that histone acetylation/deacetylation has an important role in the plant pathogenic response. In this regimen De-La-Pena et al. (2012) observed that the loss of function of ASHH2 and ASHR1 resulted in faster hypersensitive responses to both mutant (hrpA) and pathogenic (DC3000) strains of P. syringae, whereas control (Col-0) and ashr3 mutants appeared to be more resistant to the infection after 2 days with highest expression level of PR1 gene on infection with DC3000 with increased resistance against this pathogen. The histone H3 lysine 4 dimethylation (H3K4me2) levels decreased at the promoter region of PR1 on infection with the DC3000 strain in both the ashr1 and ashh2 backgrounds, suggesting that an epigenetically regulated PR1 expression is involved in the plant defense. They concluded that histone methylation is essential in the signaling and defense processes of microbes against plants (De-La-Pena et al. 2012). They hypothesized that microorganism upon contact with a plant switch on different signaling pathways. If the microorganism is nonpathogenic, JA-related genes, such as JAR1, are induced.
Plants are continuously exposed to different biotic and abiotic pressures. Newer tools based plants to build resistance or tolerance to environmental stress to plants can be achieved by DNA mutations. The epigenetic mark DNA methylation is based on the covalent and reversible modifications to DNA/histone proteins, may alter the chromatin structure and, in specific cases, can be inherited to the next generation. By controlling the expression of several resistance genes plant response to biotic stressors can be activated by epigenetic modulations. Here in upon the first stress exposure, plants can be primed and subsequently activate defense genes more efficiently against the next stress encounter based on the DNA methylation and histone modifications of immune memory state of plants. Non-coding RNAs (ncRNAs) such aslnc RNAs, siRNA, and miRNAs can act as direct and indirect modulators in epigenetics research. The restoration of the stress-induced epigenetic modifications to initial levels when the stress is removed will occurs. But in certain conditions inheritance across mitotic or even meiotic cell divisions will occur and makes the stress to be stable. Epigenetic modifications may support the plants to relief the stable stress (Fig. 14.3). Even though plants have a peculiar characteristics of priming system which may offer enhanced resistance in which the previous localized attack can be memorized by the plants at distal sites (Ramirez-Prado et al. 2018).
In order to detect the effectors plants use transmembrane/intracellular receptors known as “resistance proteins” for the effector recognition which may support the effector-triggered immunity. The concept of transcriptional gene silencing also supports towards the more stable layer of defense against DNA viruses. Development of herbicide-resistant weed populations is the major problem affecting the agricultural productivity. Herbicide resistance occurs due to mutation which reduces the herbicide binding at the protein target of the herbicide. A strong abiotic stress associated with the herbicides will be achieved in which the weeds respond by activating stress-signaling networks that reprogram gene expression.
Kim et al. (2017) performed methyl C sequencing of Arabidopsis thaliana leaves that developed after either mock treatment or two different sub-lethal doses of the herbicide glyphosate treatments which causes herbicide injury of 9205 differentially methylated regions across the genome among which 5914 of these differentially methylated regions were induced in a dose-dependent manner with positive correlation of methylation levels with that of the herbicide injury. They observed that out of 3680 genes associated with glyphosate induced differentially methylated regions only 7% were implicated in methylation changes following biotic or salinity stress. They concluded that plants responding to herbicide stress through changes in methylation patterns are associated with dose-sensitivity and stress specificity. Globally horseweed (Conyza canadensis) is one of the most commonly encountered weed species that developed resistance to herbicide glyphosate. Wherein glyphosate application, to horseweed shown phenotypic plasticity in response to selection pressure. In this strategy Gunjune Kim et al. (2017) observed phenotypic alterations and differential expression of the EPSPS gene could be attributed to epigenetic changes. They observed that DNA methylation plays a pivotal role in many biological procedures (gene expression, differentiation, and cellular proliferation). Their results shown that differential methylation pattern between the two populations wherein the increased resistance of C. canadensis to glyphosate and future development strategies that restrict weed resistance to herbicides may be elucidated (Margaritopoulou et al. 2018).
Synthetic herbicides manage the weeds affecting the plants. The control of weeds has been achieved by approaches of laborious manual weeding and environmentally damaging tillage. Improved agricultural productivity can be achieved by adopting synthetic herbicides with increased efficacy of weeding. Synthetic weeds killers are affected by wide spread evolution of resistance to commercial herbicides. The mechanisms of target-site resistance (TSR) and non-target-site resistance (NTSR) have been associated with most herbicide classes. Mutations (non-synonymous single-nucleotide polymorphisms, polymorphisms in more than one codon or entire codon deletions in genes) encoding the protein targets of herbicides are encountered with TSR which affects the binding of the herbicide (near catalytic domains or in regions affecting access), whereas NTSR target-site mechanisms involve absorption or translocation and increased sequestration or metabolic degradation (Gaines et al. 2011). The black-grass (Alopecurus myosuroides) and annual rye-grass (Lolium perenne) hold a global problem of loss of chemical weed control due to multiple-herbicide resistance. In this strategy Cummins et al. 2013 found that in both annual rye-grass and black-grass, multiple-herbicide resistance was observed with the increased expression of an evolutionarily distinct plant phi (F) GSTF1 that had a restricted ability to detoxify herbicides. They emphasized that upon black-grass A. myosuroides (Am) AmGSTF1 expression in Arabidopsis thaliana, the transgenic plants acquired resistance to multiple herbicides and showed similar changes in their secondary, xenobiotic, and antioxidant metabolism to those determined in MHR weeds. Their results of transcriptome array experiments showed that these changes in biochemistry were not due to changes in gene expression. Here in the AmGSTF1 showed a direct regulatory control on metabolism that led to an accumulation of protective flavonoids. They conclude that specific GSTFs in multiple-herbicide resistance in weeds with similar roles for its potentiality as targets for chemical intervention in resistant weed management (Cummins et al. 2013).The response of plant to environmental stresses has been achieved by variation in the expression of numerous genes. The major threat to grass weed is the non-target-site-based resistance to herbicides (NTSR). Duhoux and Delye (2013) identified a set of reference genes with a stable expression to be used as an internal standard for the normalization of quantitative PCR data in studies investigating NTSR to herbicides inhibiting aceto-lactate synthase (ALS) in the major grass weed Lolium sp., in these references gene has been used to check the herbicide response. In resistant plants their results indicate that herbicide application enhanced CYP gene expression with up-regulation of all CYP genes (Duhoux and Delye 2013)
14.5 Epigenetic Modifications to Sustain Agriculture for Crops Growing in Deserts
Plants get activated when they are exposed to the stressful conditions allowing to react under epigenetic regulation for improving crop productivity associated with stress conditions. Plant adaptation ability can be checked by analysis of candidate genes and studying their regulation in response to abiotic stresses. With response to abiotic stresses the analysis of stress related genes and their regulation of expression are commonly employed for enhanced understanding of the plants ability to adapt under abiotic stress environments. The different abiotic stress associated with plants is shown in Fig. 14.4. The exposure to abiotic stresses may cause demethylation of certain functionally inactive genes. Transgenerational inheritance can be achieved by the expression of certain genes modified by epigenetic mechanism.
Mousavi et al. (2019) performed a multidisciplinary approach, including physiological, epigenetic, and genetic studies to clarify the salt tolerance mechanisms in olive varieties (Koroneiki, Royal de Cazorla, Arbequina, and Picual) and a related form (O. europaea subsp. cuspidata) by growing in a hydroponic system under different salt concentrations from zero to 200 mM. They checked the photosynthesis, gas exchange, and relative water content at different time points, chlorophyll and leaf concentration of Na+, K+, and Ca2+ ions, quantified at 43 and 60 days after treatment. Their results indicate that several fragments differentially get methylated among genotypes, treatments, and time points. Significant expression changes related to plant response to salinity were observed by the real time quantitative PCR (RT-qPCR) analysis. Four genes (OePIP1.1, OePetD, OePI4Kg4, and OeXyla) were identified, as well as multiple retrotransposon elements usually targeted by methylation under stress conditions. Overall their studies concluded that olive cultivation may successfully face the new scenarios driven by the climate change through the selection of tolerant varieties to salt and drought stresses.
Guarino et al. (2015) investigated the epigenetic diversity of the poplar populations of 83 white poplar trees at different sample locations on the island of Sardinia by determining their DNA methylation status; to assess if and how methylation status influence population clustering; to shed light on the changes that occur in the epigenome of ramets of the same poplar clone. They checked the methylation sensitive amplified polymorphism on the genomic DNA extracted from leaves at the same juvenile stage. Their results showed that the genetic biodiversity of poplars is quite limited but it is counterbalanced by epigenetic inter-population molecular variability. Their results of variable epigenetic status of Sardinian white poplars shown a decreased number of population clusters. Ramets of the same clone were differentially methylated in relation to their geographic position as observed during the landscape genetics analyses. They concluded that genetic biodiversity of the Sardinian white poplar is limited, by epigenetic inter-population diversity, which supports white poplars to grow in very large areas of the island of Sardinia.
Salinity is the major environmental factor which limits agricultural productivity. Epigenetic modulations under environmental stresses cause rice to be underexplored. DNA methylation may regulate the gene expression with responses to environmental stresses. In this regimen Ratna Karan et al. (2012) checked the effect of salt stress on DNA methylation in four genotypes of rice differing in the degree of salinity tolerance. Important role in regulating gene expression in organ and genotype-specific manner under salinity stress was reported by gene body methylation. They concluded that natural genetic variation for salt tolerance observed in rice germplasm may be independent of the extent and pattern of DNA methylation which may have been induced by abiotic stress followed by accumulation through the natural selection process (Karan et al. 2012). The abiotic stress associated with plants are shown in Fig. 14.5.
The important fruit tree and cash crop is the Apple (Malus domestica) globally. The crucial role of low temperature of winter season is the great risk for apple tree. In order to survive the chilling temperature of winters apple tree undergoes dormancy. Kumar et al. (2016) studied the cytosine methylation based epigenetic regulation of chilling mediated dormancy release in apple by employing methylation sensitive amplified polymorphism (MSAP) approach to analyze the changes in cytosine methylation pattern during dormancy break and subsequent fruit set. They found that under high chill conditions, total methylation gets decreased from 27% in dormant bud to 21% in fruiting stage, with no significant reduction under low chill conditions. Higher expression of DNA methyl transferases and histone methyl transferases during dormancy and fruit set, and lower expression of DNA glycosylases during active growth under low chill conditions were elicited during RNA-Seq analysis. Their results shown significant association between chilling and methylation changes which suggesting chilling acquisition during dormancy in apple is likely to affect the epigenetic regulation through DNA methylation.
The drought tolerant, low input, and high yielding sweet potato (Ipomoea batatas), produces more nutrients and has higher edible energy than most staples such as rice, cassava, wheat, and sorghum. The devastating loss of sweet potato productivity has been reported to get improved by micro-propagation techniques (meristem or nodal tip culture, coupled with thermotherapy or cryotherapy). Akomeah et al. (2019) checked the extent of in vitro culture induced soma-clonal variation, at a phenotypic, compositional, and genetic/epigenetic level, by comparing field-maintained and micropropagated lines of three elite Ghanaian sweet potato genotypes grown in a common garden. Their results indicates that micropropagated plants shown no observable morphological abnormalities compared to field-maintained plants which shown significantly lower levels of iron, total protein, zinc, and glucose. Their results of methylation sensitive amplification polymorphism analysis shown higher level of in vitro culture induced molecular variation in micropropagated plants. Overall they concluded that clonal fidelity of the micropropagated bio-fortified lines may reduce potential losses in the nutritional value. Benoit et al. (2019) developed a bioinformatics approach for the functional annotation of retrotransposons containing long terminal repeat and defined all full-length Rider elements in the tomato genome. They reported that accumulation of Rider transcripts and transposition intermediates in the form of extrachromosomal DNA is triggered by drought stress and relies on abscisic acid signaling. They concluded that Rider as an environment-responsive element and a potential source of genetic and epigenetic variation in plants. Herrera et al. (2013) investigated the transgenerational constancy of epigenetic structure in three populations of the perennial herb stinking hellebore (Helleborus foetidus). Their extensive epigenetic differentiation between sporophyte populations was revealed by single-locus and multilocus analyses. They observed 75% of epigenetic markers persisted unchanged through gametogenesis during locus-by-locus comparisons of methylation status in individual sporophytes and descendant gametophytes. Their findings indicate that individuals and populations of H. foetidus indicate that epigenetic marks acquired during the sporophyte life stage in response to biotic or abiotic stress would enhance parental fitness if passed unchanged to the germline.
Du et al. (2020) studied 91 bud mutations of “Fuji” apple using the genetic variation within “Fuji” as the control and examined the characteristics of epigenetic variation at different levels in both varieties and mutant groups. They observed a global genomic DNA methylation level of the 91 bud mutants of “Fuji” ranged from 29 to 45%, with an average of 36%. The main DNA methylation pattern is the internal cytosine methylation. They observed methylation level variation in the color mutant group; however, variation in methylation pattern was more obvious in both the early maturation and spur mutant groups. Their study concluded that abundant changes in methylation levels and patterns between bud mutants and their mother “Fuji” indicate the possibility of epigenetics mediated DNA methylation in “Fuji” bud mutation line.
14.6 Epigenome Engineering Novel Techniques for Crop Improvement
The novel techniques based on epigenome engineering for crop improvement include the genetic modification with Agrobacterium tumefaciens (Agrobacterium), which introduces a piece of its own DNA into the plant genome, in terms of tumor-inducing (Ti) plasmid into the plant cell causing genome integration. Hence the possibility of incorporation in to plant genome from distantly related/related organisms in terms of transgenesis/cisgenesis has been achieved (Sedeek et al. 2019). Epigenome editing using mobile RNA has the potential to allow breeding of artificial sport cultivars in vegetative crop propagation (Kasai et al. 2016). The clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 system offers Cas9 nuclease complex direction to specific sites in the genome as determined by complementary base-pairing between the DNA and a short single guide RNA (sgRNA) (Lee et al. 2019). Genome editing technologies improve crops and ensure global food security. Genome-edited crops of natural/artificial mutagenesis based may support biotechnology companies to adopt genome editing (Sedeek et al. 2019). By adopting several approaches the plant genome can be altered which includes targeted introduction of nucleotide changes, deleting DNA segments, introducing exogenous DNA fragments, and epigenetic modifications. Targeted changes are mediated by sequence specific nucleases (SSNs), such as zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR (clustered regularly interspersed short palindromic repeats)-Cas (CRISPR associated protein) systems. Improvement in the delivery of exogenous DNA into plant cells also increases successful gene editing events rate. Crop breeding based newer approaches for enhancing the resistance to abiotic stress, as knowledge of stability and heritability features of epigenetic marks and epigenetic regulatory mechanisms are crucial for breeding applications.
14.7 Epigenetics in Agricultural Sector Patents
Oliver et al. (1994) patented a technology of creating transgenic plant containing gene whose expression can be controlled by application of an external stimulus. Gene expression with positive control may be achieved by an external stimulus to maintain gene expression. Here in technology of gene expression in case of plant phenotype which is possible to grow favorable or unfavorable conditions may be selected, based on the selection plant may be grown accordingly. Bucher and Thieme (2016) patented a method for the mobilization of a transposable element by providing an inhibitor of DNA methylation, and/or an inhibitor of transcription, and by contacting the inhibitor(s) with a cell comprising inactivated transposable elements, yielding a cell with mobilized transposable elements using inhibitor of DNA methylation and/or an inhibitor of transcription. During 1990s, the first epigenetic patents has been granted. These patents claimed laboratory methods that lay the foundation for future epigenetic advances by describing how to detect and manipulate DNA methylation. Nari, a company announced that it has secured exclusive patent licenses for epigenetics from the University of California, with Los Angeles (UCLA) the revolutionizing plant breeding by tapping natural genetic diversity. The agreement, through UCLA’s Technology Development Group, gives Inari access to tools that will positively influence crop performance without altering a plant’s genetic code. U.S. Patent No. 5,871,917 claims methods of detecting hypomethylation (decreased DNA methylation) or hypermethylation in a CpG sequence. Prior to 2000 only nine epigenetic patents were granted. Epigenetic patenting expanded rapidly in recent years. During the last 5-year period from 2000 to 2004 a sharp increase in the number of successful epigenetic patent applications filed has been recorded.
14.8 Challenges and Opportunities in Phyto-Epigenetics
The genetic diversity is associated with wild-type plants collected from different geographical origins. The environmental conditions such as photoperiod/temperature changes provoke changes causing RNA- or chromatin-based transcriptional regulation. Altered chromatin and gene expression states also produce challenges even if the plant returns to the original environmental condition, as in the case of vernalization. Degree of epigenetic variation between ecotypes is also substantial, which supports to explore whether epigenetic adaptations contribute to plant form, survival, and performance under different conditions. Switches in epigenetic states such as mutations allow read-through transcripts to bring about the silencing of adjacent genes, including tumor-suppressor genes in humans. There is also the possibility that environmentally or pathogen-induced epigenetic states might be transmitted to progeny if the changes occur in meristems and can be maintained through meiosis. Numerous changes at the level of the chromatin that lead to activating or repressing specific gene expression are caused by environmental factors. The epigenetic changes may be inherited over the generation that often results in phenotypic variations. It is becoming evident that epigenetic changes play important roles in acclimatization, stress tolerance, adaptation, and evolution processes. Epigenetic variations suggest their effect on gene expression, with epigenetic machinery of gene regulation in plants, and its possible use in epigenome engineering/editing for crop improvement is found to be the crucial steps. Defining the molecular basis of transgenerational epigenetic inheritance could ultimately lead to development of epialleles designed for specific environmental conditions through targeted epigenetic modifications in genes of interest.
14.9 Conclusion
The innate capability to survive plants under drastic climatic conditions has been hold by plants. Improvement of plant capacity to produce more nutritious food and capacity to survive under drastic climatic conditions are helpful. Here in the gene expression profile of plants offers major impact towards the transformation of epigenotype to phenotypes. In the forthcoming years monitoring and manipulation of crop epigenome may offer wider scope which may support the development of superior crops with improved agricultural productivity. Even though research has paid way for better understanding for stress tolerance novel genetic engineering tools application may still provide wider scope in future.
References
Acevedo MF (2011) Interdisciplinary progress in food production, food security and environment research. Environ Conserv 38(2):151–171
Akomeah B, Quain MD, Ramesh SA, Anand L, Rodrıguez-Lopez CM (2019) Common garden experiment reveals altered nutritional values and DNA methylation profiles in micropropagated three elite Ghanaian sweet potato genotypes. PLoS One 14(4):e0208214
Alonso C, Cruz DR, Becker C (2019) The role of plant epigenetics in biotic interactions. New Phytol 221:731–737
Benoit M, Drost HG, Catoni M, Gouil Q, Lopez-Gomollon S, Baulcombe D et al (2019) Environmental and epigenetic regulation of Rider retrotransposons in tomato. PLoS Genet 15(9):e1008370
Etienne Bucher, Michael Thieme (2016) Mobilisation of transposable elements to enhance genetic and epigenetic variability in a population. WO2017093317A1
Cai Q, Qiao L, Wang M, He B, Lin FM, Palmquist J, Huang SD, Jin H (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360(6393):1126–1129
Chen RS (1990) Global agriculture, environment, and hunger: past, present, and future links. Environ Impact Assess Rev 10(4):335–358
Chen M, Lv S, Meng Y (2010) Epigenetic performers in plants. Develop Growth Differ 52:555–566
Crespo-Salvador Ó, Escamilla-Aguilar M, López-Cruz J, López-Rodas G, González-Bosch C (2018) Determination of histone epigenetic marks in Arabidopsis and tomato genes in the early response to Botrytis cinerea. Plant Cell Rep 37(1):153–166
Cummins I, Wortley DJ, Sabbadin F, He Z, Coxon CR, Straker HE, Sellars JD, Knight K, Edwards L, Hughes D et al (2013) Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc Natl Acad Sci USA 110:5812–5817
De-La-Pena C, Rangel-Cano A, Alvarez-Venegas R (2012) Regulation of disease responsive genes mediated by epigenetic factors: interaction of Arabidopsis–Pseudomonas. Mol Plant Pathol 13:388–398
Du X, Wang Y, Liu M, Liu X, Jiang Z, Zhao L et al (2020) The assessment of epigenetic diversity, differentiation, and structure in the ‘Fuji’ mutation line implicates roles of epigenetic modification in the occurrence of different mutant groups as well as spontaneous mutants. PLoS One 15(6):e0235073
Duhoux A, Delye C (2013) Reference genes to study herbicide stress response in Lolium sp.: up-regulation of P450 genes in plants resistant to acetolactate-synthase inhibitors. PLoS One 8(5):e63576
Eriksson MC, Szukala A, Tian B, Paun O (2020) Current research frontiers in plant epigenetics: an introduction to a virtual issue. New Phytol 226:285–288
Gaines TA, Duke SO, Morran S, Rigon CAG, Tranel AK, Dayan FE (2011) Mechanisms of evolved herbicide resistance. J Agric Food Chem 59(11):5886–5889
Gibney E, Nolan C (2010) Epigenetics and gene expression. Heredity 105:4–13
Guarino F, Cicatelli A, Brundu G, Heinze B, Castiglione S (2015) Epigenetic diversity of clonal white poplar (Populus alba L.) populations: could methylation support the success of vegetative reproduction strategy. PLoS One 10(7):e0131480
Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder J, Uozumi N (2015) HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr Opin Biotechnol 32:113–120
Herrera CM, Medrano M, Bazaga P (2013) Epigenetic differentiation persists after male gametogenesis in natural populations of the perennial herb Helleborus foetidus (Ranunculaceae). PLoS One 8(7):e70730
Kapazoglou A, Drosou V, Argiriou A, Tsaftaris AS (2013) The study of a barley epigenetic regulator, HvDME, in seed development and under drought. BMC Plant Biol 13:172. https://doi.org/10.1186/1471-2229-13-172
Kapazoglou A, Ganopoulos I, Tani E, Tsaftaris A (2018) Epigenetics, epigenomics and crop improvement. Adv Bot Res 86:287–324
Karan R, DeLeon T, Biradar H, Subudhi PK (2012) Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting Rice genotypes. PLoS One 7(6):e40203
Kasai A, Bai S, Hojo H, Harada T (2016) Epigenome editing of potato by grafting using transgenic tobacco as siRNA donor. PLoS One 11(8):e0161729
Kim G, Clarke CR, Larose H, Tran TH, Haak DC, Zhang L, Askew S, Barneyand J, Westwood JH (2017) Herbicide injury induces DNA methylome alterations in Arabidopsis. PeerJ 5:e3560
Kumar S (2019) Epigenomics for crop improvement: current status and future perspectives. J Genet Cell Biol 3(1):128–134
Kumar G, Rattan UK, Singh AK (2016) Chilling-mediated DNA methylation changes during dormancy and its release reveal the importance of epigenetic regulation during winter dormancy in apple (malus x domestica Borkh.). PLoS One 11(2):e0149934
Kumar S, Beena AS, Awana M, Singh A (2017) Physiological, biochemical, epigenetic and molecular analyses of wheat (Triticum aestivum) genotypes with contrasting salt tolerance. Front Plant Sci 8:1151
Lee JE, Neumann M, Duro DI, Schmid M (2019) CRISPR-based tools for targeted transcriptional and epigenetic regulation in plants. PLoS One 14(9):e0222778
Liu X, Zhou C, Zhao Y, Zhou S, Wang W, Zhou DX (2014) The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time. Front Plant Sci 5:1–9
Margaritopoulou T, Tani E, Chachalis D, Travlos I (2018) Involvement of epigenetic mechanisms in herbicide resistance: the case of Conyza Canadensis. Agriculture 8:17
Mousavi S, Regni L, Bocchini M, Mariotti R, Cultrera NM, Mancuso S, Googlani J, Chakerolhosseini MR, Guerrero C, Albertin E, Baldoni L, Proietti P (2019) Physiological, epigenetic and genetic regulation in some olive cultivars under salt stress. Sci Rep 9:1093, 1–17
Oliver MJ, Quisenberry JE, Trolinder NLG, Keim DL (1994) Control of plant gene expression. US5723765A
Peace CP, Bianco L, Troggio M, Weg EV, Howard NP, Cornille A et al (2019) Apple whole genome sequences: recent advances and new prospects. Hort Res 6(59):1–24
Ramirez-Prado JS, Abulfaraj AA, Rayapuram N, Benhamed M, Hirt H (2018) Plant immunity: from signaling to epigenetic control of defense. Trends Plant Sci 23(9):833–844
Sedeek KEM, Mahas A, Mahfouz M (2019) Plant genome engineering for targeted improvement of crop traits. Front Plant Sci 10:114
Shi H, Lee BH, Wu SJ, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21:81–85
Simmons D (2008) Epigenetic influence and disease. Nature. Education 1(1):6
Very AA, Sentenac H (2003) Molecular mechanisms and regulation of Na+ transport in higher plants. Annu Rev Plant Biol 54:575–603
Weinhold B (2006) Epigenetics: the science of change. Environ Health Perspect 114(3):A160–A167
Wu SH, Ho C, Nah SL, Chau CF, Global Hunger (2014) A challenge to agricultural, food, and nutritional sciences. Crit Rev Food Sci Nutr 54(2):151–162
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Krishnaswami, V., Kumar, M., Vijayaraghavalu, S. (2022). Advances in Epigenetics for Crop Improvement and Sustainable Agriculture. In: Singh, R.L., Mondal, S., Parihar, A., Singh, P.K. (eds) Plant Genomics for Sustainable Agriculture. Springer, Singapore. https://doi.org/10.1007/978-981-16-6974-3_14
Download citation
DOI: https://doi.org/10.1007/978-981-16-6974-3_14
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-6973-6
Online ISBN: 978-981-16-6974-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)