Abstract
Medical image plays a key role in the analysis and treatment of the patient’s condition in today’s medicine, but the clinical processing of medical image is still largely dependent on the subjective experience of doctors, and the process of manual extraction of information is time-consuming and labor-consuming. With the development of deep learning, it has become a trend to apply deep learning to medical image processing. However, due to the characteristics of medical images, many advanced segmentation algorithms fail to achieve good results in medical images. In addition, medical image data generally have the problem of insufficient or too small data sets, and there are few ways to obtain samples, mainly from some hospitals and medical institutions. Besides, the labeling work is not competent for ordinary people. So how to realize the automatic analysis and processing of medical images has always been a hot topic in the field of computer science. In this paper, we designed an experiment of prostate segmentation algorithm, built a model using a U-Net network that performs well in the field of medical image segmentation, and tried to improve its performance by combining a variety of structures, such as ASPP, ResNext, and attention module. The results show that our proposed model achieves better segmentation performance.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 The Introduction
Due to the highly developed modern computer technology and the continuous development of medical imaging technology, medical imaging technology can play an important auxiliary role in a variety of research in the medical field, as well as in clinical treatment and diagnosis. Medical imaging can realize the non-invasive examination of patients in human anatomy and visibility of the development status of lesions, thus providing important reference and guidance for surgery, carrying out a comprehensive and in-depth analysis of lesions for tissues and organs in the human body, and carrying out a rigorous monitoring of the whole process and the treatment related to surgery. For medical analysis, it is of critical importance whether the organ tissue has a clear outline and whether there is adhesion to other tissues.
Medical image segmentation includes manual, semi-automatic and automatic segmentation [1]. Among them, the artificial segmentation method consumes a lot of time, and has high requirements on the clinician's own experience judgment, and the repeatability is very low, unable to meet a variety of clinical requirements. Semi-automatic segmentation needs human–computer interaction as the core. Generally speaking, the core methods of edge segmentation include region extraction method and other methods [2,3,4,5,6], which can optimize the segmentation speed to some extent, but it still needs the observers as the core for analysis and judgment. And due to the lack of self learning ability, anti-interference ability remains a low level. Therefore, it can not be applied well in clinic. Automatic segmentation method relies on the automatic extraction of the edge of the region of interest by computer, which can ensure that it is not subject to various influences of the observer on the subjective level, so as to realize more rapid data processing and have relatively ideal repeatability. Therefore, medical image segmentation with deep learning as the core is one of the core research directions for image processing at this stage [7].
The introduction of artificial intelligence in the treatment of medical influence can not only optimize the analysis and judgment of the patient's condition, promote the scientificity of treatment, but also provide scientific reference for doctors to grasp and handle the condition. At the present stage, the distribution of medical resources in China is generally uneven. If the artificial way can be introduced to improve the scientificity and rationality of medical diagnosis, the dependence on doctors’ experience can be reduced, so as to avoid further aggravation of the phenomenon of medical difficulties. Therefore, the introduction of deep learning in the medical field can also make good research progress in this discipline.
With the joint efforts of a large number of researchers at home and abroad, many image segmentation methods have been widely used in the processing of medical images. Some algorithms are the optimization of some existing algorithms, or organic combination through several structures, etc. The theory of image segmentation algorithm has significant diversity. For example, the edge detection as the core [8] mainly includes parallel differential operator, the deformation model as the core and the surface fitting as the core method [9], the statistical theory as the core and the random field as the core method, etc. Generally speaking, medical image segmentation algorithms must be organically combined with a variety of existing segmentation methods at the present stage to achieve more ideal results. Whether the results are accurate or not is of critical significance for the analysis and judgment of doctors. If the results are wrong, the diagnosis of the condition by doctors will also produce errors, and even cause medical accidents. Therefore, accuracy is a crucial factor. In addition, real-time performance and stability of the algorithm need to be continuously optimized.
In recent years, medical image segmentation with deep learning as the core can be divided into two different types, namely deep convolutional neural networks (DCNNs) as the core method and full convolutional neural networks (FCNs) as the core method. FCN [10] has important performance for image segmentation, especially U-NET [11], an optimized full convolutional neural network with FCN as the core, is further transformed into the core network used for medical image segmentation. Based on IEEE retrieval, the number of papers related to it can be found to be about 200. At the same time, some of them play an important role in medical imaging and can be recognized by a large number of relevant scholars. The segmentation objects of this kind of article have high diversity, such as lung, liver and so on. The components of U-NET include an up-and-down sampling encoder and a skip connection between them. Encoding and decoding can combine local and global information organically. Up to now, U-NET has gradually produced many types of variants, all of which can play a relatively ideal role in medical image segmentation. At the same time, U-NET has been further transformed from the original 2D to 3D, thus forming new 3D U-NET [12], V-NET and other models.
2 Introduction to the Foundation and Additional Structure of Improved U-NET Neural Network
2.1 Improve U-Net Neural Network
The core of convolutional neural network (CNN) is to learn and utilize the feature mapping of images to develop a more detailed feature mapping. It plays an obvious role in the classification problem, because the image is transformed into a vector after processing, and then the subsequent classification is expanded. However, at the same time of image segmentation, the feature image should be processed to make it into a vector, and the vector should be taken as the core to complete image reconstruction. This task is difficult because the process of turning a vector into an image is more complex than vice versa. The core concepts of U-Net are all generated in response to this problem.
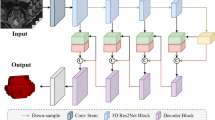
Contract the path, and there will be a window of pooling equal to 2 × 2 after two convolution layers, and the step length is equal to the largest pool of 2 layers. The dimension of a convolution is equal to 3 × 3, and the step length is equal to 1. All the backsides of the convolution layer needs to play the role of activation through Relu activation functions. When each one is completed under a sampling process, the number of channels increases. As for the upper sampling in the decoding path, there is a convolution layer with a size equal to 2 × 2 in each round of sampling, and the activation process needs to be further completed by ReLU function (Fig. 1).
At the same time, there are also two convolutional layers, the size of the convolutional kernel is equal to 3 × 3, and the step size is equal to 1. In addition, each upsampling is organically combined with the feature graph obtained from the symmetric contraction path, that is, jump link. Finally, the final classification process is completed by means of a convolutional layer of size equal to 1 × 1.
This time, through the organic combination of the subsampling part contained in the U-Net network and the residual network ResNet-Layer, the corresponding transformation is carried out for U-Net.
2.2 ASPP
When it comes to ASPP, Dilated Convolution needs to be emphasized. Because the existing convolutional neural network has two core problems in the process of completing the segmentation task, the first is the loss of information caused by subsampling, and the second is that the convolutional neural network has the characteristic of spatial invariance. Therefore, a method is derived which can not only avoid the process of subsampling, but also effectively amplify the receptive field, namely, void convolution. In the process of convolutional pooling of the pyramid with void space, ASPP is the process of parallel sampling with the correct sampling rate and void convolution for specific inputs. In essence, it has the same property as the global information contained in the image captured by several proportions (Fig. 2).
2.3 SE
SE block belongs to the category of the cell, and to all the specific transform structure:\({ }F_{tr} = X \to U,X \in R^{{H^{\prime} \times W^{\prime} \times C^{\prime}}} ,{\text{U}} \in R^{H \times W \times C}\). For the purpose of simplifying the flow, in the following discussion, \({\text{F}}_{{{\text{tr}}}}\) is proposed as a convolution operator. Set \({\text{V}} = \left( {v_{1} ,v_{2} , \ldots ,v_{c} } \right)\) refers to the learning set of the filter kernel, and \({\text{v}}_{{\text{c}}}\) refers to the parameters of the c filter. The output of \(F_{tr}\) is expressed as \({\text{U}} = \left( {u_{1} ,u_{2} , \ldots ,u_{c} } \right)\), and \(u_{c} = v_{c} *X = \mathop \sum \nolimits_{s = 1}^{{C^{\prime}}} v_{c}^{s} *x^{s}\).
Extrusion: global information embedding. In order to overcome the problem of channel dependence, a comprehensive analysis of the signal characteristics of all channels is needed. All the filters learned have the corresponding local acceptance domain. So none of the elements of the transformation output U can make use of the global information outside this region. This problem becomes more and more obvious in the lower layers of neural networks with small acceptance domains. Therefore, this study proposes to compress the global spatial information and further transfer it to the channel descriptor. Channel statistics are obtained using global average pooling. In terms of form, The statistic \({\text{z}} \in R^{C}\) is generated by the contraction of U through the spatial dimension H × W, and the Cth element in Z is expressed as follows:
Stimulation: adaptive adjustment. In order to make use of the aggregation information in the extrusion process, a second scrubbing was carried out to thoroughly collect the dependencies related to the channel. Therefore, the function is required to meet two criteria. First, it has high flexibility. Second, you need to learn a non-exclusive relationship. Therefore, the Sigmoid activation mechanism with low complexity was chosen in this study: \({\text{s}} = F_{ex} \left( {z,W} \right) = \sigma \left( {g\left( {z,W} \right)} \right) = \sigma \left( {W_{2} \delta \left( {W_{1} z} \right)} \right)\), where δ refers to the Relu function. \(W_{1} \in R^{{\frac{C}{r} \times C}} ,W_{2} \in R^{{C \times \frac{C}{r}}}\). Final output of the block needs to refer to scaling transformation further will be output, U finally get activation \(\widetilde{{X_{c} }} = F_{scale} \left( {u_{c} ,s_{c} } \right) = s_{c} \cdot u_{c}\), \(\tilde{X} = \left[ {\widetilde{{x_{1} }},\widetilde{{x_{2} }}, \ldots ,\widetilde{{x_{C} }}} \right]\) (Fig. 3).
3 Experimental and Structural Analysis
3.1 Loss Function
Use BCE With Logits Loss as a Loss function, which is expressed as follows:
3.2 Analysis and Comparison of Experimental Results
The data set. For the prostate data set used in this study, the number of training images was equal to 1549 and the number of test images was equal to 683. The processor used in the experiment was I5, the operating system was Linux, NVIDIA GeForce GTX 1650 GPU was selected, and all experimental processes were carried out based on PyTorch framework.
Training algorithm. The commonly used methods include stochastic gradient descent (SGD) and some adaptive training approaches, such as adaptive gradient descent (ADAGRAD), adaptive momentum estimation (ADAM), etc. The ADAM algorithm, which converges rapidly, is used in this experiment (Fig. 4 and Table 1).
References
Qian, Z.: Research and Application of Medical Image Segmentation Method. Southern Medical University, Guangdong (2014)
Xueming, W.: Research on Algorithms of Image Segmentation. Chengdu University of Technology (2006)
Hongying, H., Tian, G., Tao, L.: Overview of image segmentation methods. Comput. Knowl. Technol. 15(5), 176–177 (2019)
Songjin, Y., Helei, W., Yongfen, H.: Research status and prospects of image segmentation methods. J. Nanchang Water Conservancy Hydropower Coll. 2004(02), 15–20 (2004)
Lili, Z., Feng, J.: Overview of image segmentation methods. Appl. Res. Comput. 34(07), 1921–1928 (2017)
Weibo, W., Zhenkuan, P.: Overview of Image segmentation methods. World Sci. Technol. Res. Dev. 2009(6), 1074–1078 (2009)
Xiaowei, X., Qing, L., Lin, Y., et al.: Quantization of fully convolutional networks for accurate biomedical image segmentation. In: Proceedings of the 36th International Conference on Computer Vision and Pattern Recognition, pp. 8300–8308. IEEE Press, NJ (2018)
Djemel, Z., Salvatore, T.: Edge detection techniques—an overview. Int. J. Pattern Recogn. Image Anal. 8(4), 537–559 (1998)
Alvarez, L., Morel, L.: Image selective smoothing and edge detection by nonlinear diffusion. II. Siam J. Numer. Anal. 29(3), 845–866 (1992)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(4), 640–651 (2015)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer International Publishing, Berlin (2015)
Iek, Z., Abdulkadir, A., Lienkamp, S.S., et al.: 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. Springer, Cham (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Sun, M. (2022). Study on Prostate Image Segmentation Using Improved U-NET. In: Yao, J., Xiao, Y., You, P., Sun, G. (eds) The International Conference on Image, Vision and Intelligent Systems (ICIVIS 2021). Lecture Notes in Electrical Engineering, vol 813. Springer, Singapore. https://doi.org/10.1007/978-981-16-6963-7_22
Download citation
DOI: https://doi.org/10.1007/978-981-16-6963-7_22
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-6962-0
Online ISBN: 978-981-16-6963-7
eBook Packages: EngineeringEngineering (R0)