Abstract
Cyanobacteria are the oldest group of prokaryotes with oxygen-evolving photosynthesis. They are supposed to have evolved in an atmosphere with little or no oxygen and therefore no protecting stratospheric ozone layer. Since cyanobacteria have to utilize sunlight for photosynthesis, they are simultaneously exposed to deleterious solar UV radiation. In order to survive, they had to develop countermeasures. One strategy is fast reproduction in order to make up for losses due to radiation damage. Another mechanism is mat and crust formation, which protects the organisms in lower levels while sacrificing the ones in the top layer. Vertical migration in the water column using changing buoyancy helps to bring the organisms out of the danger zone. Likewise, gliding cyanobacteria have been found to move to a position deeper in the water to avoid excessive UV exposure. Efficient repair mechanisms have been developed to replace damaged proteins in the photosynthetic apparatus and to repair damage in the cellular DNA. Many cyanobacteria synthesize UV-absorbing pigments such as mycosporine-like amino acids and scytonemin, deposited in the outer cell layers or extracellularly, which absorb UV photons before they can damage vital biomolecules within the cell.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
6.1 Introduction
Bacteria were the first organisms on earth using solar light to harvest energy. Most modern photosynthetic bacteria use a single photosystem (of two possible ones), which operates under anoxygenic conditions since oxygen is toxic to many prokaryotes. The active pigment is one of several bacteriochlorophylls. In contrast, cyanobacteria were the first organisms to develop oxygenic photosynthesis based on two photosystems, which operate in tandem. Oxygen is produced by splitting water. This process is thought to have started about 2.3 billion years ago and since then oxygen accumulates in the atmosphere (Soo et al. 2017). Cyanobacterial photosynthesis uses chlorophyll a in both reaction centers which eukaryotic plants also utilize; therefore, the latter are thought to have evolved from the prokaryotic ancestors, which have been taken up in the form of endosymbiosis (Cihlář et al. 2019).
Cyanobacteria can be unicellular, floating in fresh or marine water, or growing on terrestrial or underwater surfaces. While most have diameters of a few micrometers, some are so minute that they have long been overlooked in marine plankton communities because of the too large pore size of the common plankton nets. However, during the past few decades it was found that some of them, especially the picoplanktonic genera Prochlorococcus and Synechococcus, which are in the 0.1–1 μm diameter size class, form major components of the marine ecosystems (Casey et al. 2019). Assessments of the contribution of picoplankton to the total biomass in the top 150 m of the water column indicated that they may account for up to 50% or more with Prochlorococcus being the most abundant and responsible for 70% of the picoplankton population (Linacre et al. 2019). Figure 6.1 represents the morphological structure of some unicellular and filamentous cyanobacteria.
Unicellular cyanobacteria may form colonies, which are held together by the extracellular polysaccharide slime which the cells produce and excrete (Sato et al. 2017). Microcystis cells collected from Lake Mead, Nevada, USA, were found to produce an outer sheath up to 30 μm thick (He and Wert 2016). Other genera form unbranched, pseudo-branched, or truly branched uniseriate filaments, which are covered by a cylindrical slime tube (Kabirnataj et al. 2018; Singh 2017).
Cyanobacteria are found in almost all habitats on earth. Aquatic forms live both in fresh and marine waters. Terrestrial cyanobacteria are found from the tropics to polar regions; they can cover rocks, salt marshes and even the barks of trees. They are adapted to low temperatures and desiccation (Jimel 2020), while others can survive in hot thermal springs (Cheng et al. 2020). Several species of cyanobacteria have been reported from a hypersaline desert (Patel et al. 2019). A wide variety of cyanobacteria lives in symbioses with all kinds of plants and animals. Lichens are a symbiosis of algae and/or cyanobacteria with fungi (Pankratov et al. 2017), and the aquatic fern Azolla harbors cyanobacteria in thallus cavities (Sánchez-Baracaldo and Cardona 2020). Cyanobacteria have been found to form symbioses with diatoms, bryophytes, gymnosperms and angiosperms, and they are even found in symbiosis with animals such as marine sponges and worms (Rai 2018).
In contrast to some earlier reports, cyanobacteria cannot swim in water (Menon et al. 2020). They do not have cilia, flagella or other moving organelles such as bacteria, flagellates and other eukaryotes (Miyata et al. 2020). But many are motile using a slow gliding movement. Some uniseriate filaments such as Anabaena or Phormidium glide within their sheath, which they may shed at the rear. They may also reverse their direction of movement triggered by external light or chemical stimuli. Motility has been studied in the model cyanobacterium Synechocystis sp., PCC 6803. In this organism, motility has been identified to be based on the presence of thick TFP pili, which can be extended, retracted and adhered to the substratum (Chen et al. 2020). Even though not capable of active swimming, planktonic cyanobacteria can undergo vertical migrations in the water column by changing their buoyancy (Kai and Lan 2020). This can be achieved by the production and collapse of gas vesicles (Dyer and Needoba 2020).
6.2 Exposure to Solar UV Radiation
Solar radiation can be subdivided into ultraviolet (UV, <400 nm), visible (400–700 nm), and infrared (IR, >700 nm). Infrared radiation can hardly be used for photosynthesis; however, there is one example of a cyanobacterium (Synechococcus PCC7335) which has a second core-membrane linker (ApcE2) of the phycobilisome which is noncovalently bound which allows the organism to utilize near IR (Miao et al. 2016).
The UV wavelength range can be subdivided into UV-C (<280 nm), UV-B (280–315 nm), and UV-A (315–400 nm) (Aphalo 2017). Photosynthesis is mainly supported by visible radiation, but under certain conditions (low radiation under cloud cover) UV-A can be utilized by some macroalgae (Xu and Gao 2016). Generally speaking, UV radiation is detrimental for organisms, especially at excessive intensities. Today, UV-C is quantitatively absorbed by oxygen and ozone in the atmosphere. UV-B is also significantly filtered out mainly by stratospheric ozone. But before the atmospheric oxygenation, organisms were exposed to and had to cope with much higher surface UV-B and in addition UV-C than today. This was the situation for cyanobacteria during their Achaean evolution even though the presence of Fe(III)-Si precipitates absorbed up to 70% of the incoming UV-C radiation. However, it is assumed that the remaining UV-C caused high mortality rates and limited cyanobacterial expansion in marine habitats (Mloszewska et al. 2018).
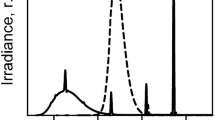
Solar irradiances strongly depend on a number of physical conditions on our planet. The solar zenith angle (SZA) determines the irradiance in all wavelength bands, which are highest in the tropics and gradually decrease toward the poles as monitored by 17 stations of the Eldonet network (Table 6.1). Much higher irradiances have been measured during a recent 1-year campaign in the high Andes near Laguna Lejia (Chile, latitude 23° 26′ 23.30″ S, longitude 67° 38′ 14.29″ W) at an elevation of 4715 m (Häder and Cabrol 2020). Figure 6.2 shows the mean monthly irradiances of PAR, UV-A, UV-B, and short-wavelength UV-B (295–310 nm).
Mean monthly irradiances of PAR, UV-A, UV-B, and short-wavelength UV-B (295–310 nm) monitored over a year in the high Andes (Laguna Lejia, Chile, latitude 23° 26′ 23.30″ S, longitude 67° 38′ 14.29″ W at an elevation of 4715 m) (Häder and Cabrol 2020)
Since today solar UV-C radiation does not hit the surface of the earth, UV-B is the most detrimental wavelength band for organisms exposed to solar radiation. In addition to latitude, UV-B radiation at ground level is controlled by the atmospheric water content (especially clouds), albedo and aerosols, and total column ozone (Häder and Cabrol 2020). At the same latitude, irradiances in the Southern Hemisphere are higher than in the Northern Hemisphere, because of the different earth–sun distances (Cordero et al. 2014). The stratospheric ozone concentration is lower in the tropics than at mid- and higher latitudes, resulting in higher solar UV-B irradiances. UV radiation increases with elevation (Blumthaler et al. 1997). In Northern Chile, IR increases by 27%, PAR by 6%, and UV by 20% from sea level to 5100 m altitude (Cordero et al. 2016). Clouds can reduce or enhance solar irradiation by absorption or scattering, quantified by the cloud modification factor (CMF) (Feister et al. 2015).
Stratospheric ozone depletion by chlorofluorocarbons (CFCs) and other anthropogenic trace gases such as organobromides and chlorocarbons has increased terrestrial UV-B radiation, but due to the Montreal Protocol and its amendments this effect is stopped and slowly reverses (Bais et al. 2018). But a recovery to pre-1980 levels is predicted only for or after mid-century due to the long lifetimes of CFCs in the stratosphere, which can be decades (Hoffmann et al. 2014). Global climate change alters total column ozone and therefore UV irradiances (Williamson et al. 2014; Schnell et al. 2016; Meul et al. 2016).
6.3 UV Effects on Cyanobacteria
There are a several nonphotosynthetic cyanobacteria whose diversity, distribution, and ecology are currently hardly known (Monchamp et al. 2019). Some are found in dark, deep terrestrial habitats such as rocks using a hydrogen-based lithoautotrophic metabolism (Puente-Sánchez et al. 2018). In contrast, all photosynthetic cyanobacteria require solar radiation for their energy harvesting. Therefore, they are inevitably exposed to solar UV radiation. Solar UV radiation affects several key cellular biomolecules and machinery (e.g., DNA and proteins), cellular morphology, photosynthesis, growth, survival, pigmentation, and nitrogen metabolism enzymes in cyanobacteria (Sinha et al. 1995a, 1998; Kumar et al. 1996; Rastogi et al. 2014a, b).
6.3.1 Damage and Repair of DNA
The most deleterious UV-B radiation is absorbed by important biomolecules including proteins, nucleic acids, and lipids, resulting in considerable damage of exposed organisms and affecting physiological, biochemical, and ecological functions, such as morphology, differentiation, growth, development, pigmentation, and motility and orientation (Häder 1993a, b; Pathak et al. 2018). Absorption of UV-B photons by the cellular DNA results in the formation of cyclobutane pyrimidine dimers (CPDs), which are the most notable lesions (about 75–80%) induced by solar UV radiations (Pathak et al. 2019b; Rastogi et al. 2010a) (Fig. 6.3). Besides CPDs, 6-4 photoproducts (6-4PPs), are the second most frequently occurring DNA lesions (about 20–25%), which are formed mainly under UV-C and readily converted into their Dewar valence isomers upon exposure to UV radiation (Rastogi et al. 2010a) (Fig. 6.4).
Formation of cyclobutane–pyrimidine dimers (CPDs) induced mainly by UV-B on DNA having adjacent thymine/cytosine bases. (a) Thymine–thymine cyclobutane–pyrimidine dimer (T<>T CPD) and (b) thymine–cytosine cyclobutane–pyrimidine (T<>C CPD) dimer. Both T<>T and T<>C CPDs split to form two canonical thymine/cytosine bases by means of photoreactivation in the presence of the photolyase enzyme (Rastogi et al. 2010a)
Formation of DNA lesion 6-4 photoproducts (6-4PPs) and their Dewar valence isomers (Rastogi et al. 2010a)
These dimers are induced between two adjacent pyrimidine bases (thymine, cytosine, and uracil). This defect seems like a minor change in the structure of the DNA, but may have far-reaching consequences for the biochemical processes in the cell since the DNA reproduction and transcription into RNA are stopped there.
CPDs are repaired by the cells in a process called photoreactivation, which involves the enzyme DNA photolyase (Rastogi et al. 2011). This enzyme possesses two noncovalently linked cofactors such as FADH2 and absorbs blue or UV-A photons and uses their energy to split the dimer (Pathak et al. 2019b; Rastogi et al. 2020). If the lesion is not repaired, it results in s signature mutation (Brash and Seidman 2020). Photolyases are very old enzymes found in bacteria all the way to vertebrates (Sinha and Häder 2002; Zhang et al. 2013). In cyanobacteria, this process has been studied, e.g., in Anacystis nidulans, and the enzyme has been purified (Eker et al. 1990). However, it is interesting to note that placental mammals including humans lack this repair mechanism and must rely on other DNA repair mechanisms such as nucleotide excision repair (see below) (Jans et al. 2005). UV-C mainly induces the formation of thymine–pyrimidone (6-4) dimers. These lesions are also repaired by a photolyase (Kavakli et al. 2019).
Other UV-induced DNA damages include DNA–protein crosslinks (Rastogi 2010; Richa et al. 2015; Rajneesh et al. 2018) and 8-oxo-7,8-dihydroguanyl, 8-oxo-Ade, 2,6-diamino-4-hydroxy-5-formamidoguanine and oxazolone, which result from oxidations products of purine bases of the DNA (Doetsch et al. 1995; Hall et al. 1996).
DNA lesions, which are not repaired by a photolyase during photoreactivation, can be mended by excision repair (see review by Pathak et al. 2019b). This mechanism is independent of light and uses several enzymes (Bergi and Trivedi 2020). It is based on the removal of a small number of bases, e.g., after a single-strand break, which are subsequently resynthesized and inserted using the complementary strand. One form of excision repair is the base excision repair pathway in which one or two bases are removed and substituted after, e.g., desiccation or radiation stress (Singh 2018). The alternative is nucleotide excision repair, which removes DNA lesions including CPDs or 6,4 photoproducts (6,4 PPs), DNA intrastrand crosslinks, chemical adducts, or by oxidative damage by reactive oxygen species (Sinha 2017).
Recombinational repair is a powerful mechanism to restore the correct DNA sequence after single- or double-strand DNA breaks. This pathway is fairly complex involving more than 20 gene products in E. coli. Initially, an exonuclease enlarges the DNA break and the gap is identified by RecFOR proteins. Subsequently, RecBCD and RecFOR perform the repair by homologous recombination (Rastogi et al. 2015). If all fails, cells retreat to the last resort, called SOS repair. This is initiated by different and substantial DNA damages or when the DNA replication is inhibited as studied in the cyanobacterium Anabaena sp. (Kumar et al. 2018). This pathway relies on the interaction of several repressor proteins including RecA and LexA, which block the 40 or so SOS response genes. Once the blockage is released, the SOS genes, each consisting of a 20-nucleotide-long SOS box, start their work. One of them codes for the SulA protein, which delays the cell division until all damages are repaired. However, many differences in the components of the SOS repair mechanism exist between bacteria and cyanobacteria and between species (Kumar et al. 2018).
6.3.2 Reactive Oxygen Species
Solar UV-B does not have to exert direct effects on cellular targets. It may be absorbed by proteins or other biomolecules in the cell upon which the excitation energy of the UV photon is transferred to, e.g., oxygen, which results in the formation of reactive oxygen species (ROS). The reduction of molecular oxygen results in superoxide, which may lead to the production of most other ROS (Turrens 2003).
Dismutation of superoxide results in hydrogen peroxide
which may be partially reduced to a hydroxide ion and a hydroxyl radical or may be fully reduced to water
Another pathway transfers the excited energy of an absorbing molecule, such as chlorophyll, to a nearby oxygen (the ground state of which is a triplet state, 3O2) which is converted to singlet oxygen (1O2) which is highly reactive and destructs nearby biomolecules and structures, even though its lifetime is rather short (on the order of 10–40 ns) (Moan and Berg 1991). In this case, the chlorophyll acts as a photosensitizer (Ph). This response is a major damaging mechanism in photosynthesis (Krieger-Liszkay 2005) but also occurs in mitochondria (Thomas et al. 1992). As an aside: This photodynamic reaction induced by introduced photosensitizers such as hematoporphyrin is used in medical treatment of superficial cancers in humans (Lv et al. 2016).
Oxygen is toxic at higher concentrations. After the development of an oxygenic atmosphere on our planet, many early life forms such as bacteria had to find protection from the increasing oxygen concentration. Today, many of these bacteria are confined to anoxic environments such as sediments (Valentine 2002). All other organisms were forced by the environmental pressure to develop mechanisms to protect themselves from ROS. This is mainly achieved by two different mechanisms. One is the employment of passive antioxidants such as ascorbic acid, α-tocopherol, glutathione, lycopene, lutein, and isoflavones (Sindhi et al. 2013). Such ROS scavengers are also found in many cyanobacteria (Radyukina et al. 2019; He and Häder 2002b). The alternative strategy to counter the stress of ROS is the involvement of antioxidant enzymes such as superoxide dismutase (SOD), catalase, glutathione peroxidase, ascorbate peroxidase, monodehydroascorbate reductase, dehydroascorbate reductase, or glutathione reductase found, e.g., in Anabaena sp. (He and Häder 2002a) and other cyanobacteria (Aráoz and Häder 1999a). The production of ROS induced by UV-B radiation can be shown and monitored by employing the ROS-sensitive, oxygen-sensing probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) (He and Häder 2002a; Rastogi et al. 2010b).
6.4 UV Damage of the Photosynthetic Apparatus and Repair
The photosynthetic apparatus is a key target of damaging solar UV radiation. In addition to unspecific effects on proteins, lipids, membranes, and other biologically important molecules and structures, UV-B affects the photosynthetic electron transport, quantum yield, and oxygen production (Xue et al. 2005). Exposure to UV radiation bleaches the photosynthetic pigments in the cyanobacterium Phormidium uncinatum (Donkor and Häder 1997; Sinha et al. 2005). Phycobilisomes containing the phycobiliprotein accessory pigments are broken down into smaller components by increasing exposure (Sinha et al. 1995c, b). In the initial phase of exposure, the phycobiliprotein fluorescence increases indicating that they can no longer transfer the excitation energy to the photosynthetic reaction centers (Donkor and Häder 1996). After prolonged exposure to UV, the fluorescence of the accessory phycobiliproteins decreases (Rastogi et al. 2015). Photodegradation of phycobilisomes by UV radiation was also confirmed in Nostoc sp. and Aulosira fertilissima (Aráoz García 1998; Banerjee et al. 1998). Exposure to UV radiation also impairs the translation activity in the cyanobacterium Nostoc sp. (Araoz et al. 1998). In contrast, low-level UV-B irradiances induce phycoerythrin synthesis in Nostoc (Aráoz and Häder 1999b).
As indicated above, ROS generated by photodynamic reactions are a potential mechanism to damage biomolecules and structures within the photosynthetic apparatus. Solar energy is absorbed by accessory pigments such as phycobilins in the phycobilisomes of cyanobacteria (as well as by chlorophyll b, chlorophyll c, or chlorophyll d in algae and higher plants) and transferred to the photosynthetic reaction centers of photosystems I and II (Jaiswal et al. 2018). An excited electron from the special chlorophyll a dimer P680 in PS II is transferred to a primary acceptor (pheophytin) (Khaing et al. 2019) from where it is handed along a chain of redox components via P700, the reaction center PS I (where the electron is again excited to a higher energetic level) until it is finally utilized to reduce NADP (Tikhonov and Subczynski 2019). The missing electron in P680 is subsequently replaced by an electron generated by the photolytic splitting of water on the inside of the thylakoids by an enzymatic Mn complex (Böhmer et al. 2017).
The oxygen is released as a waste product and the protons are used to drive an ATPase, which generates ATP, which is used together with the reduction equivalents (reduced NADP) to reduce CO2 to sugar in the Calvin cycle (Michelet et al. 2013; Janasch et al. 2019). Under high light conditions, the electron transport chain is fully reduced and cannot accept any more electrons from P680 (Lea-Smith et al. 2016). Therefore, the excitation energy can be transferred to a nearby oxygen resulting in singlet oxygen (s. above) (Lee and Min 2010). In order to avoid this, potential damaging situation-specific carotenoids are arranged in close vicinity to the chlorophyll so that the excitation energy can be transferred to the carotenoids, which relax the energy in the form of heat (Pospíšil and Prasad 2014; Schäfer et al. 2005).
Another target of solar UV is the D1 protein located in PS II encoded by the psbAI gene (Rexroth et al. 2017). This protein is responsible to transfer the excited electrons from P680 to pheophytin (Khaing et al. 2019). This protein is easily kinked by excessive visible or UV radiation, which stops the electron transport. This lesion is rapidly repaired by proteolytic removal of the damaged protein and subsequent replacement by a newly synthesized protein (Campbell et al. 1998; Ehling-Schulz and Scherer 1999).
6.5 Motility and Orientation
As indicated above, many unicellular and multicellular cyanobacteria are motile. Some show a gliding motility, others use changes in buoyancy to realize vertical migration. These mechanisms are used to avoid areas of deleterious solar radiation. Picoplanktons, such as Synechococcus and Prochlorococcus, which are major biomass producers in the oceans, use fast reproduction to overcome the population losses due to excessive UV (Häder and Gao 2018). In addition, there is a pronounced seasonal variability and changes in vertical distribution (Al-Otaibi et al. 2020). In the Red Sea, Synechococcus was found close to the surface, while Prochlorococcus was responsible for a chlorophyll maximum between 40 and 76 m. Prochlorococcus populations had a maximum in summer and a minimum in winter, while Synechococcus showed the opposite temporal distribution. In addition, there are low light- and high light adapted genetically different populations dwelling at different depths (Linacre et al. 2019).
Cyanobacteria with gliding motility respond to visible and UV radiation to select a suitable habitat. The coccoid Synechocystis sp. secretes a mixture of complex polysaccharides to drive their motility (Varuni et al. 2017) and shows a pronounced phototaxis, which can be positive (toward the light source) at low irradiances or negative (away from the light source) at high irradiances, while the rod-shaped Synechococcus elongatus PCC 7942 has no phototactic motility (Yang et al. 2018). In Synechocystis sp., PCC6803 a blue light-dependent signal cascade controls positive and negative phototaxis (Sugimoto et al. 2017). The cyanobacterial phytochrome 2 regulates the expression of motility-related genes via the second messenger cyclic-GMP (Wallner et al. 2020).
Also, some filamentous cyanobacteria show a gliding motility when in contact with a surface (Qiu et al. 2019). While the exact mechanism has not yet been revealed, several distinct structural features such as specifically arranged protein fibrils and organelle-like structures have been identified, which are thought to be involved in the secretion of mucilage (Hoiczyk 2000). The unbranched heterotrichous Anabaena variabilis has been found to move either as straight filaments or in a U-shaped form (Nultsch and Wenderoth 1983; Nultsch et al. 1979). When irradiated, the trichomes bend toward the light at low fluence rates (1.35 W m−2) and away from the light source at higher (27 W m−2). These authors assumed that the switch from positive to negative phototaxis is controlled by the intracellular level of singlet oxygen since gassing the moving filaments with N2 or Ar shifts the transition from positive to negative phototaxis to higher irradiances (Nultsch and Schuchart 1985). The photoreceptor for photoorientation is assumed to consist of a superfamily of tetrapyrrole-binding molecules, cyanobacteriochromes (Ikeuchi and Ishizuka 2008). Exposure to UV-B radiation delays differentiation of vegetative cells into heterocysts and akinetes (Blakefield and Harris 1994), induces bleaching of the phycobilins (Agel et al. 1987), and affects productivity and nitrogen fixation in Anabaena (Lesser 2008). It also causes a significant decrease in the quantum yield of PSII. The effects on photosynthesis are thought to be due to the production of ROS, since exposure to UVR results in an increase in the level of superoxide dismutase.
The nonheterocystous filamentous Phormidium uncinatum does not show phototaxis, but orients with respect to light using photophobic responses. When a trichome enters a bright light field from a dark area, e.g., when it leaves the shade under a leaf, it reverses the direction of movement and glides back; this response is called a step-up photophobic reaction. In contrast, when leaving a low irradiance area moving into a dark area, it may also reverse the direction of movement (step-down photophobic response) (Nultsch and Häder 1970). The organisms respond even to small differences in the irradiances of two adjacent light fields as low as 4%. This can be demonstrated by projecting a photographic negative onto a population of Phormidium trichomes, which accumulate in areas of appropriate irradiance, forming a photographic positive (Häder 1984) (Fig. 6.5). The photosynthetic pigments are responsible for the photophobic responses in this cyanobacterium (Nultsch and Häder 1974; Häder 1974). The direction of movement is controlled by an electric potential gradient along the length of the trichome. During a photophobic reversal of movement, this gradient inverts (Häder 1978; Häder and Burkart 1978).
Phormidium trichomes accumulate in low irradiance light fields projected onto a Petri dish (A). A photographic negative of the Münster in Freiburg has been projected onto a suspension of Phormidium trichomes, which accumulate in areas of appropriate irradiances forming a photographic positive (B) (after (Häder 1984))
Motility of Phormidium is strongly impaired by solar and monochromatic UV irradiation (Häder et al. 1986). The action spectrum shows a strong response in the UV-B. In contrast, the photophobic response was not impaired by solar or artificial UV radiation (Häder and Häder 1990). Bleaching kinetics indicate that the accessory phycobilins, D-phycoerythrin, is easily bleached by UV radiation, followed by the carotenoids, while chlorophyll a was found to be the most resistant pigment to bleaching.
Several filamentous cyanobacteria (Phormidium uncinatum, two strains, Anabaena variabilis and Oscillatoria tenuis) protect themselves by vertical migration. The filaments were suspended in an agar layer inside a slanting groove made from dark PVC, which was placed in a pond reaching from 10 to 100 cm. After 4-h exposure to solar radiation, the organisms had moved to a position at about 50–60 cm below the water surface (Donkor and Häder 1995). Mat-forming Oscillatoria on Antarctica’s McMurdo Ice Shelf have also been found to show vertical migration controlled by solar visible and UV radiation: At low irradiances (<8 W m−2, no UV), the filaments migrated completely to the surface, while higher irradiances (>60 W m−2, including UV-A and UV-B) induced downward migration (Nadeau et al. 1999). Similar vertical migrations were also found in Microcoleus and Halomicronema in microbial mats in the French Camargue (Fourçans et al. 2006) and in coastal microbial mats (Lichtenberg et al. 2020).
6.6 UV-Screening Pigments
In response to the pressure of solar UV radiation, many cyanobacteria (but also eukaryotic phytoplankton and macroalgae) have developed UV-absorbing pigments such as mycosporine-like amino acids (MAAs) and scytonemin (Scy) to screen out deleterious radiation before it can hit essential biomolecules and cellular structures (Sinha et al. 2007). Picoplanktons, such as the marine Synechococcus and Prochlorococcus, are too small (<1 μm) to use UV-screening pigments since the concentration would have to be too high to be effective over very small transmission distances (Garcia-Pichel 1994); therefore, these organisms rely on vertical migration, repair mechanisms, and fast replication to counter the challenge of solar UV radiation.
Mycosporine-like amino acids (MAAs) are small, hydrophilic, and colorless molecules with a cyclohexenimine or cyclohexenone chromophore attached to the nitrogen substituent of an amino acid or its imino alcohol (Pathak et al. 2019a). More than 20 different MAAs are known today, which are characterized by their high molar extinction coefficients (28,100–50,000 M−1 cm−1) and a strong absorption in the UV between 310 and 362 nm (Pathak et al. 2017a; Rastogi et al. 2020) (Fig. 6.6). The absorbed UV photon energy is dissipated as heat and does not result in ROS generation (Conde et al. 2007). Some MAAs have even be found to possess free radical scavenging capacity (Rastogi et al. 2016).
Some cyanobacteria—and only this group of organisms—have developed another group of UV-absorbing pigments, i.e., scytonemins (Rastogi et al. 2012, 2014c). These molecules are heterocyclic phenolic dimers, which are excreted into the extracellular polysaccharide sheath (Pathak et al. 2017a, b) (Fig. 6.7). In addition to a major peak at 386 nm, the oxidized form shows peaks at 252 and 300 nm. The UV-C peak may be a reminder of the life history of cyanobacteria in an anoxygenic atmosphere with no stratospheric ozone layer. In Nostoc punctiforme, the molecule is coded by a gene cluster of 18 ORFs (Soule et al. 2007), and a possible biosynthesis pathway has been suggested by Balskus and Walsh (2009).
6.7 Conclusions
Photosynthetic organisms require solar energy for their metabolism. Simultaneously, they are exposed to deleterious UV-A and UV-B radiation, since cyanobacteria have started their development on our planet when the atmosphere contained only traces of oxygen, and consequently, no stratospheric ozone layer existed to protect them from the even more energetic UV-C radiation. At moderate irradiances, UV-A can be utilized to drive photosynthesis in some phytoplankton, but shorter wavelengths are always detrimental for living organisms. UV photons are absorbed by lipids, proteins, and other biologically important molecules and are consequently prone to modify these components and destroy cellular structures. In addition, absorption of solar UV radiation can induce reactive oxygen species (ROS).
In order to protect the cells from deleterious solar UV radiation, organisms have developed a plethora of mechanisms and strategies against induced damage. The DNA is a key target of solar UV-B radiation, which is of vital importance since its integrity warrants the correct transmission of information to the next generation. Therefore, a large number of concepts have been developed to repair any UV-induced damage and modification including the involvement of photolyases, which remove dimers in the nucleotide strand. Other mechanisms include excision, recombination, and SOS repair pathways. A likewise important target of solar UV radiation is the photosynthetic apparatus. Short-wavelength photons bleach accessory pigments and chlorophyll a and induce ROS such as singlet oxygen, which in turn destroys biologically important structures. Cells have developed enzymatic and nonenzymatic strategies to quench ROS production. Damage of the redox elements of the photosynthetic electron transport chain is repaired by removal of mutilated proteins and replacement by newly synthesized molecules.
Other strategies to avoid excessive exposure to solar radiation include mat formation and vertical migration to bring organisms out of the danger zone. This can be achieved by using phototaxis or photophobic responses. One important mechanism is the production and incorporation of UV-absorbing pigments such as mycosporine-like amino acids and scytonemins, which prevent the transmission of damaging photons to vital biomolecules and structures in the center of the cell. In addition, some of these substances have antioxidant properties. These molecules have a potential to serve humans as UV protectants in suntan lotions as a replacement of artificial organic molecules (Guillerme et al. 2017; Richa and Sinha 2013).
References
Agel G, Nultsch W, Rhiel E (1987) Photoinhibition and its wavelength dependence in the cyanobacterium Anabaena variabilis. Arch Microbiol 147:370–374
Al-Otaibi N, Huete-Stauffer TM, Calleja ML, Irigoien X, Morán XAG (2020) Seasonal variability and vertical distribution of autotrophic and heterotrophic picoplankton in the Central Red Sea. PeerJ 8:e8612
Aphalo PJ (2017) Quantification of UV radiation. In: Jordan BR (ed) UV-B radiation and plant life: molecular biology to ecology. CAB International, Boston, MA
Aráoz García LR (1998) Tolerance mechanisms against ultraviolet-B radiation in phytoplankton organisms. Dissertation, Friedrich-Alexander University Erlangen-Nürnberg, Germany
Aráoz R, Häder D-P (1999a) Enzymatic antioxidant activity in two cyanobacteria species exposed to solar radiation. Recent Res Dev Photochem Photobiol 3:123–132
Aráoz R, Häder D-P (1999b) Phycoerythrin synthesis is induced by solar UV-B in the cyanobacterium Nostoc. Plant Physiol Biochem 37:223–229
Araoz R, Lebert M, Häder D-P (1998) Translation activity under ultraviolet radiation and temperature stress in the cyanobacterium Nostoc sp. J Photochem Photobiol B Biol 47:115–120
Bais AF, Lucas RM, Bornman JF, Williamson CE, Sulzberger B, Austin AT, Wilson SR, Andrady AL, Bernhard G, McKenzie RL et al (2018) Environmental effects of ozone depletion, UV radiation and interactions with climate change: UNEP Environmental Effects Assessment Panel, update 2017. Photochem Photobiol Sci 17:127–179
Balskus EP, Walsh CT (2009) An enzymatic cyclopentyl [b] indole formation involved in scytonemin biosynthesis. J Am Chem Soc 131:14648–14649
Banerjee M, Sinha RP, Häder D-P (1998) Biochemical and spectroscopic changes in phycobiliproteins of the cyanobacterium, Aulosira fertilissima, induced by UV-B radiation. Acta Protozool 37:145–148
Bergi J, Trivedi R (2020) Bioremediation of saline soil by cyanobacteria. Microbial bioremediation & biodegradation. Springer, New York
Blakefield MK, Harris DO (1994) Delay of cell differentiation in Anabaena aequalis caused by UV-B radiation and the role of photoreactivation and excision repair. Photochem Photobiol 59:204–208
Blumthaler M, Ambach W, Ellinger R (1997) Increase of solar UV radiation with altitude. J Photochem Photobiol B Biol 39:130–134
Böhmer S, Köninger K, Gómez-Baraibar Á, Bojarra S, Mügge C, Schmidt S, Nowaczyk MM, Kourist R (2017) Enzymatic oxyfunctionalization driven by photosynthetic water-splitting in the cyanobacterium Synechocystis sp. PCC 6803. Catalysts 7:240
Brash DE, Seidman MM (2020) Defective post-replication repair of UV photoproducts in melanoma: a mutator phenotype. Mol Oncol 14:5–7
Campbell D, Eriksson MJ, Öquist G, Gustafsson P, Clarke AK (1998) The cyanobacterium Synechococcus resists UV-B by exchanging photosystem II reaction-center D1 proteins. Proc Natl Acad Sci U S A 95:364–369
Casey JR, Björkman KM, Ferrón S, Karl DM (2019) Size dependence of metabolism within marine picoplankton populations. Limnol Oceanogr 64:1819–1827
Chen Z, Li X, Tan X, Zhang Y, Wang B (2020) Recent advances in biological functions of thick pili in the cyanobacterium Synechocystis sp. PCC 6803. Front Plant Sci 11:241
Cheng Y-I, Chou L, Chiu Y-F, Hsueh H-T, Kuo C-H, Chu H-A (2020) Comparative genomic analysis of a novel strain of Taiwan hot-spring cyanobacterium Thermosynechococcus sp. CL-1. Front Microbiol 11:82
Cihlář J, Füssy Z, Oborník M (2019) Evolution of tetrapyrrole pathway in eukaryotic phototrophs, Advances in botanical research. Elsevier, Amsterdam
Conde FR, Churio MS, Previtali CM (2007) Experimental study of the excited-state properties and photostability of the mycosporine-like amino acid palythine in aqueous solution. Photochem Photobiol Sci 6:669–674
Cordero RR, Seckmeyer G, Damiani A, Riechelmann S, Rayas J, Labbe F, Laroze D (2014) The world’s highest levels of surface UV. Photochem Photobiol Sci 13:70–81
Cordero R, Damiani A, Seckmeyer G, Jorquera J, Caballero M, Rowe P, Ferrer J, Mubarak R, Carrasco J, Rondanelli R (2016) The solar spectrum in the Atacama Desert. Sci Rep 6:22457
Doetsch PW, Zastawny TH, Martin AM, Dizdaroglu M (1995) Monomeric base damage products from adenine, guanine, and thymine induced by exposure of DNA to ultraviolet radiation. Biochemistry (USA) 34:737–742
Donkor VA, Häder D-P (1995) Protective strategies of several cyanobacteria against solar radiation. J Plant Physiol 145:750–755
Donkor VA, Häder D-P (1996) Effects of ultraviolet irradiation on photosynthetic pigments in some filamentous cyanobacteria. Aquat Microbial Ecol 11:143–149
Donkor VA, Häder D-P (1997) Ultraviolet radiation effects on pigmentation in the cyanobacterium Phormidium uncinatum. Acta Protozool 36:49–55
Dyer SW, Needoba JA (2020) Use of high-resolution pressure nephelometry to measure gas vesicle collapse as a means of determining growth and turgor changes in planktonic cyanobacteria. Appl Environ Microbiol 86(2):e01790–e01719
Ehling-Schulz M, Scherer S (1999) UV protection in cyanobacteria. Eur J Phycol 34:329–338
Eker A, Kooiman P, Hessels J, Yasui A (1990) DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J Biol Chem 265:8009–8015
Feister U, Cabrol N, Häder D-P (2015) UV irradiance enhancements by scattering of solar radiation from clouds. Atmosphere 5:1211–1228
Fourçans A, Solé A, Diestra E, Ranchou-Peyruse A, Esteve I, Caumette P, Duran R (2006) Vertical migration of phototrophic bacterial populations in a hypersaline microbial mat from Salins-de-Giraud (Camargue, France). FEMS Microbiol Ecol 57:367–377
Garcia-Pichel F (1994) A model for internal self-shading in planktonic organisms and its implications for the usefulness of ultraviolet sunscreens. Limnol Oceanogr 39:1704–1717
Guillerme J-B, Couteau C, Coiffard L (2017) Applications for marine resources in cosmetics. Cosmetics 4:35
Häder D-P (1974) Participation of two photosystems in the photo-phobotaxis of Phormidium uncinatum. Arch Microbiol 96:255–266
Häder D-P (1978) Evidence of electrical potential changes in photophobically reacting blue-green algae. Arch Microbiol 118:115–119
Häder D-P (1984) Wie orienteren sich Cyanobakterien im Licht. Biologie in unserer Zeit 14:78–83
Häder D-P (1993a) Effects of enhanced solar ultraviolet radiation on aquatic ecosystems. In: Tevini M (ed) UV-B radiation and ozone depletion. Effects on humans, animals, plants, microorganisms, and materials. Lewis Publ., Boca Raton, FL
Häder D-P (1993b) Risks of enhanced solar ultraviolet radiation for aquatic ecosystems. In: Round FE, Chapman DJ (eds) Progress in phycological research. Biopress Ltd., Bristol
Häder D-P, Burkart U (1978) Mathematical model for photophobic accumulations of blue-green algae in light traps. J Math Biol 5:293–304
Häder DP, Cabrol NA (2020) Monitoring of solar irradiance in the high Andes. Photochem Photobiol 96(5):1133–1139
Häder D-P, Gao K (2018) Phytoplankton responses to ocean climate change drivers. In: Aquatic ecosystems in a changing climate, vol 62. CRC Press, Boca Raton, FL
Häder D-P, Häder M (1990) Effects of solar radiation on motility, photomovements and pigmentation in two strains of the cyanobacterium, Phormidium uncinatum. Acta Protozool 29:291–303
Häder D-P, Watanabe M, Furuya M (1986) Inhibition of motility in the cyanobacterium, Phormidium uncinatum, by solar and monochromatic UV irradiation. Plant Cell Physiol 27:887–894
Häder D-P, Lebert M, Schuster M, del Ciampo L, Helbling EW, McKenzie R (2007) ELDONET - a decade of monitoring solar radiation on five continents. Photochem Photobiol 83:1348–1357
Hall DB, Holmlin RE, Barton JK (1996) Oxidative DNA damage through long-range electron transfer. Nature 382:731–735
He YY, Häder D-P (2002a) Involvement of reactive oxygen species in the UV-B damage to the cyanobacterium Anabaena sp. J Photochem Photobiol B Biol 66:73–80
He YY, Häder D-P (2002b) Reactive oxygen species and UV-B: effect on cyanobacteria. Photochem Photobiol Sci 1:729–736
He X, Wert EC (2016) Colonial cell disaggregation and intracellular microcystin release following chlorination of naturally occurring Microcystis. Water Res 101:10–16
Hoffmann L, Hoppe C, Müller R, Dutton G, Gille J, Griessbach S, Jones A, Meyer C, Spang R, Volk C (2014) Stratospheric lifetime ratio of CFC-11 and CFC-12 from satellite and model climatologies. Atmos Chem Phys 14:12479–12497
Hoiczyk E (2000) Gliding motility in cyanobacteria: observations and possible explanations. Arch Microbiol 174:11–17
Ikeuchi M, Ishizuka T (2008) Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol Sci 7:1159–1167
Jaiswal A, Koli DK, Kumar A, Kumar S, Sagar S (2018) Pigments analysis of cyanobacterial strains. Int J Chem Stud 6:1248–1251
Janasch M, Asplund-Samuelsson J, Steuer R, Hudson EP (2019) Kinetic modeling of the Calvin cycle identifies flux control and stable metabolomes in Synechocystis carbon fixation. J Exp Bot 70:973–983
Jans J, Schul W, Sert Y-G, Rijksen Y, Rebel H, Eker AP, Nakajima S, van Steeg H, de Gruijl FR, Yasui A (2005) Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol 15:105–115
Jimel M (2020) Algal and cyanobacterial adaptations to low temperature and desiccation. Bachelor, Karlova
Kabirnataj S, Nematzadeh GA, Talebi AF, Tabatabaei M, Singh P (2018) Neowestiellopsis gen. nov, a new genus of true branched cyanobacteria with the description of Neowestiellopsis persica sp. nov. and Neowestiellopsis bilateralis sp. nov., isolated from Iran. Plant Syst Evol 304:501–510
Kai W, Lan SS (2020) Vertical migration by bulk phytoplankton sustains biodiversity and nutrient input to the surface ocean. Sci Rep 10:1142
Kavakli IH, Ozturk N, Gul S (2019) DNA repair by photolyases, Advances in protein chemistry and structural biology. Elsevier, Amsterdam
Khaing EP, Zhong V, Eaton-Rye JJ (2019) Impairment of photosystem II assembly and acceptor side electron transfer following mutation of Thr243 and Lys264 of D2 in cyanobacteria. Photosynth Hydrogen Energy Res Sustain 46
Krieger-Liszkay A (2005) Singlet oxygen production in photosynthesis. J Exp Bot 56:337–346
Kumar A, Sinha RP, Häder D-P (1996) Effect of UV-B on enzymes of nitrogen metabolism in the cyanobacterium Nostoc calcicola. J Plant Physiol 148:86–91
Kumar A, Kirti A, Rajaram H (2018) Regulation of multiple abiotic stress tolerance by LexA in the cyanobacterium Anabaena sp. strain PCC7120. Biochim Biophys Acta Gene Regul Mech 1861:864–877
Lea-Smith DJ, Bombelli P, Vasudevan R, Howe CJ (2016) Photosynthetic, respiratory and extracellular electron transport pathways in cyanobacteria. Biochim Biophys Acta Bioenergetics 1857:247–255
Lee J, Min DB (2010) Analysis of volatile compounds from chlorophyll photosensitized linoleic acid by headspace solid-phase microextraction (HS-SPME). Food Sci Biotechnol 19:611–616
Lesser MP (2008) Effects of ultraviolet radiation on productivity and nitrogen fixation in the cyanobacterium, Anabaena sp. (Newton’s strain). Hydrobiologia 598:1–9
Lichtenberg M, Cartaxana P, Kühl M (2020) Vertical migration optimizes photosynthetic efficiency of motile cyanobacteria in a coastal microbial mat. Front Mar Sci 7:359
Linacre L, Durazo R, Camacho-Ibar V, Selph K, Lara-Lara J, Mirabal-Gómez U, Bazán-Guzmán C, Lago-Lestón A, Fernández-Martín E, Sidón-Ceseña K (2019) Picoplankton carbon biomass assessments and distribution of Prochlorococcus ecotypes linked to Loop Current Eddies during summer in the southern Gulf of Mexico. J Geophys Res Oceans 124(11):8342–8359
Lv W, Zhang Z, Zhang KY, Yang H, Liu S, Xu A, Guo S, Zhao Q, Huang W (2016) A mitochondria-targeted photosensitizer showing improved photodynamic therapy effects under hypoxia. Angew Chem Int Ed 55:9947–9951
Menon SN, Varuni P, Menon GI (2020) Information integration and collective motility in phototactic cyanobacteria. PLoS Comput Biol 16:e1007807
Meul S, Dameris M, Langematz U, Abalichin J, Kerschbaumer A, Kubin A, Oberländer-Hayn S (2016) Impact of rising greenhouse gas concentrations on future tropical ozone and UV exposure. Geophys Res Lett 43:2919–2927
Miao D, Ding W-L, Zhao B-Q, Lu L, Xu Q-Z, Scheer H, Zhao K-H (2016) Adapting photosynthesis to the near-infrared: non-covalent binding of phycocyanobilin provides an extreme spectral red-shift to phycobilisome core-membrane linker from Synechococcus sp. PCC7335. Biochim Biophys Acta Bioenergetics 1857:688–694
Michelet L, Zaffagnini M, Morisse S, Sparla F, Pérez-Pérez ME, Francia F, Danon A, Marchand C, Fermani S, Trost P (2013) Redox regulation of the Calvin–Benson cycle: something old, something new. Front Plant Sci 4:470
Miyata M, Robinson RC, Uyeda TQ, Fukumori Y, Si F, Haruta S, Homma M, Inaba K, Ito M, Kaito C (2020) Tree of motility–a proposed history of motility systems in the tree of life. Genes Cells 25:6–21
Mloszewska AM, Cole DB, Planavsky NJ, Kappler A, Whitford DS, Owttrim GW, Konhauser KO (2018) UV radiation limited the expansion of cyanobacteria in early marine photic environments. Nat Commun 9:1–8
Moan J, Berg K (1991) The photodegradation of porphyrins in cells can be used to estimate the lifetime of singlet oxygen. Photochem Photobiol 53:549–553
Monchamp M-E, Spaak P, Pomati F (2019) Long term diversity and distribution of non-photosynthetic cyanobacteria in peri-alpine lakes. Front Microbiol 9:3344
Nadeau T-L, Howard-Williams C, Castenholz RW (1999) Effects of solar UV and visible irradiance on photosynthesis and vertical migration of Oscillatoria sp.(Cyanobacteria) in an Antarctic microbial mat. Aquat Microb Ecol 20:231–243
Nultsch W, Häder D-P (1970) Bestimmungen der photo-phobotaktischen Unterschiedsschwelle bei Phormidium uncinatum. Berichte der Deutschen Botanischen Gesellschaft 83:185–192
Nultsch W, Häder D-P (1974) Über die Rolle der beiden Photosysteme in der Photo-phobotaxis von Phormidium uncinatum. Berichte der Deutschen Botanischen Gesellschaft 87:83–92
Nultsch W, Schuchart H (1985) A model of the phototactic reaction chain of the cyanobacterium Anabaena variabilis. Arch Microbiol 142:180–184
Nultsch W, Wenderoth K (1983) Partial irradiation experiments with Anabaena variabilis (Kütz). Z Pflanzenphysiol 111:1–7
Nultsch W, Schuchart H, Höhl M (1979) Investigations on the phototactic orientation of Anabaena variabilis. Arch Microbiol 122:85–91
Pankratov T, Kachalkin A, Korchikov E, Dobrovol’skaya T (2017) Microbial communities of lichens. Microbiology 86:293–309
Patel HM, Rastogi RP, Trivedi U, Madamwar D (2019) Cyanobacterial diversity in mat sample obtained from hypersaline desert, Rann of Kachchh. 3 Biotech 9:304
Pathak J, Ahmed H, Sinha RP (2017a) Metabolomic profiling of cyanobacterial UV-protective compounds. Curr Metabolomics 5:138–163
Pathak J, Sonker AS, Richa, Rajneesh R, Kannaujiya VK, Singh V, Ahmed H, Sinha RP (2017b) Screening and partial purification of photoprotective pigment scytonemin from cyanobacterial crusts dwelling on the historical monuments in and around Varanasi, India. Microbiol Res 8(1):4–12
Pathak J, Häder D-P, Sinha RP (2018) Impacts of ultraviolet radiation on certain physiological and biochemical processes in cyanobacteria inhabiting diverse habitats. Environ Exp Bot 161:375–387
Pathak J, Ahmed H, Singh PR, Singh SP, Häder D-P, Sinha RP (2019a) Mechanisms of photoprotection in cyanobacteria, Cyanobacteria. Elsevier, Amsterdam
Pathak J, Singh PR, Häder DP, Sinha RP (2019b) UV-induced DNA damage and repair: a cyanobacterial perspective. Plant Gene 19:100194
Pospíšil P, Prasad A (2014) Formation of singlet oxygen and protection against its oxidative damage in photosystem II under abiotic stress. J Photochem Photobiol B Biol 137:39–48
Puente-Sánchez F, Arce-Rodríguez A, Oggerin M, García-Villadangos M, Moreno-Paz M, Blanco Y, Rodríguez N, Bird L, Lincoln SA, Tornos F (2018) Viable cyanobacteria in the deep continental subsurface. Proc Natl Acad Sci U S A 115:10702–10707
Qiu Y, Tian S, Gu L, Hildreth M, Zhou R (2019) Identification of surface polysaccharides in akinetes, heterocysts and vegetative cells of Anabaena cylindrica using fluorescein-labeled lectins. Arch Microbiol 201:17–25
Radyukina N, Mikheeva L, Karbysheva E (2019) Low molecular weight antioxidants in cyanobacteria and plant cells. Biol Bull Rev 9:520–531
Rai AN (2018) CRC handbook of symbiotic cyanobacteria. CRC Press, Boca Raton, FL
Rajneesh, Chatterjee A, Pathak J, Ahmed H, Singh V, Singh DK, Pandey A, Singh SP, Richa, Häder D-P, Sinha RP (2018) Ultraviolet radiation-induced DNA damage and mechanisms of repair in cyanobacteria: an overview. In: Sinha RP, Shrivastava UP (eds) Biotechnology in agriculture, industry and medicine. Trends in life science research. Nova Biomedical, New York
Rastogi RP (2010) UV-B-induced DNA damage and repair in cyanobacteria. PhD, Banaras Hindu University
Rastogi RP, Kumar A, Tyagi MB, Sinha RP (2010a) Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucleic Acids 2010:592980
Rastogi RP, Singh SP, Häder D-P, Sinha RP (2010b) Detection of reactive oxygen species (ROS) by the oxidant-sensing probe 2′,7′-dichlorodihydrofluorescein diacetate in the cyanobacterium Anabaena variabilis PCC 7937. Biochem Biophys Res Commun 397:603–607
Rastogi RP, Singh SP, Häder D-P, Sinha RP (2011) Ultraviolet-B-induced DNA damage and photorepair in the cyanobacterium Anabaena variabilis PCC 7937. Environ Exp Bot 74:280–288
Rastogi RP, Kumari S, Richa, Han T, Sinha RP (2012) Molecular characterization of hot spring cyanobacteria and evaluation of their photoprotective compounds. Can J Microbiol 58:719–727
Rastogi RP, Incharoensakdi A, Madamwar D (2014a) Responses of a rice-field cyanobacterium Anabaena siamensis TISTR-8012 upon exposure to PAR and UV radiation. J Plant Physiol 171:1545–1553
Rastogi RP, Sinha RP, Moh SH, Lee TK, Kottuparambil S, Kim Y-J, Rhee J-S, Choi E-M, Brown MT, Häder D-P (2014b) Ultraviolet radiation and cyanobacteria. J Photochem Photobiol B Biol 141:154–169
Rastogi RP, Sonani RR, Madamwar D (2014c) The high-energy radiation protectant extracellular sheath pigment scytonemin and its reduced counterpart in the cyanobacterium Scytonema sp. R77DM. Bioresour Technol 171:396–400
Rastogi RP, Sonani RR, Madamwar D (2015) Effects of PAR and UV radiation on the structural and functional integrity of phycocyanin, phycoerythrin and allophycocyanin isolated from the marine cyanobacterium Lyngbya sp. A09DM. Photochem Photobiol 91(4):837–844
Rastogi RP, Sonani RR, Madamwar D, Incharoensakdi A (2016) Characterization and antioxidant functions of mycosporine-like amino acids in the cyanobacterium Nostoc sp. R76DM. Algal Res 16:110–118
Rastogi RP, Madamwar D, Nakamoto H, Incharoensakdi A (2020) Resilience and self-regulation processes of microalgae under UV radiation stress. J Photochem Photobiol C Photochem Rev 43:100322
Rexroth S, Nowaczyk MM, Rögner M (2017) Cyanobacterial photosynthesis: the light reactions, Modern topics in the phototrophic prokaryotes. Springer, New York
Richa, Sinha RP (2013) Biomedical applications of mycosporine-like amino acids. In: Kim SK (ed) Marine microbiology, bioactive compounds and biotechnological applications. Wiley-VCH Publishers, Germany
Richa, Sinha RP, Häder D-P (2015) Physiological aspects of UV-excitation of DNA. In: Barbatto M, Borin AC, Ullrich S (eds) Topics in current chemistry: photoinduced phenomena in nucleic acids II: DNA fragments and phenomenological aspects. Springer, Berlin
Sánchez-Baracaldo P, Cardona T (2020) On the origin of oxygenic photosynthesis and cyanobacteria. New Phytol 225:1440–1446
Sato M, Omori K, Datta T, Amano Y, Machida M (2017) Influence of extracellular polysaccharides and calcium ion on colony formation of unicellular Microcystis aeruginosa. Environ Eng Sci 34:149–157
Schäfer L, Vioque A, Sandmann G (2005) Functional in situ evaluation of photosynthesis-protecting carotenoids in mutants of the cyanobacterium Synechocystis PCC6803. J Photochem Photobiol B Biol 78:195–201
Schnell JL, Prather MJ, Josse B, Naik V, Horowitz LW, Zeng G, Shindell DT, Faluvegi G (2016) Effect of climate change on surface ozone over North America, Europe, and East Asia. Geophys Res Lett 43:3509–3518
Sindhi V, Gupta V, Sharma K, Bhatnagar S, Kumari R, Dhaka N (2013) Potential applications of antioxidants–a review. J Pharm Res 7:828–835
Singh K (2017) Taxonomy and morphology of cyanobacteria the genus Hapalosiphon (Stigonematales). J Nat Resour Dev 12:5–10
Singh H (2018) Desiccation and radiation stress tolerance in cyanobacteria. J Basic Microbiol 58:813–826
Sinha RP (2017) Role of photosynthetically active radiation and dark conditions on the repair of ultraviolet-B radiation-induced damages in the cyanobacterium Nostoc sp. strain HKAR-2. Focus Med Sci J 3
Sinha RP, Häder D-P (2002) UV-induced DNA damage and repair: a review. Photochem Photobiol Sci 1:225–236
Sinha RP, Kumar HD, Kumar A, Häder D-P (1995a) Effects of UV-B irradiation on growth, survival, pigmentation and nitrogen metabolism enzymes in cyanobacteria. Acta Protozool 34:187–192
Sinha RP, Lebert M, Kumar A, Kumar HD, Häder D-P (1995b) Disintegration of phycobilisomes in a rice field cyanobacterium Nostoc sp. following UV irradiation. Biochem Mol Biol Int 37:697–706
Sinha RP, Lebert M, Kumar A, Kumar HD, Häder D-P (1995c) Spectroscopic and biochemical analyses of UV effects on phycobiliproteins of Anabaena sp. and Nostoc carmium. Bot Acta 108:87–92
Sinha RP, Krywult M, Häder D-P (1998) Effects of ultraviolet, monochromatic and PAR waveband on nitrate reductase activity and pigmentation in a rice field cyanobacterium, Anabaena sp. Acta Hydrobiol 40:105–112
Sinha RP, Kumar A, Tyagi MB, Häder D-P (2005) Ultraviolet-B-induced destruction of phycobiliproteins in cyanobacteria. Physiol Mol Biol Plants 11:313–319
Sinha RP, Singh SP, Häder D-P (2007) Database on mycosporines and mycosporine-like amino acids (MAAs) in fungi, cyanobacteria, macroalgae, phytoplankton and animals. J Photochem Photobiol B Biol 89:29–35
Soo RM, Hemp J, Parks DH, Fischer WW, Hugenholtz P (2017) On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science 355:1436–1440
Soule T, Stout V, Swingley WD, Meeks JC, Garcia-Pichel F (2007) Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. J Bacteriol 189:4465–4472
Sugimoto Y, Nakamura H, Ren S, Hori K, Masuda S (2017) Genetics of the blue light-dependent signal cascade that controls phototaxis in the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol 58:458–465
Thomas C, MacGill RS, Miller GC, Pardini RS (1992) Photoactivation of hypericin generates singlet oxygen in mitochondria and inhibits succinoxidase. Photochem Photobiol 55:47–53
Tikhonov AN, Subczynski WK (2019) Oxygenic photosynthesis: EPR study of photosynthetic electron transport and oxygen-exchange, an overview. Cell Biochem Biophys 77:47–59
Turrens JF (2003) Mitochondrial formation of reactive oxygen species. J Physiol 552:335–344
Valentine DL (2002) Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: a review. Antonie Van Leeuwenhoek 81:271–282
Varuni P, Menon SN, Menon GI (2017) Phototaxis as a collective phenomenon in cyanobacterial colonies. Sci Rep 7:1–10
Wallner T, Pedroza L, Voigt K, Kaever V, Wilde A (2020) The cyanobacterial phytochrome 2 regulates the expression of motility-related genes through the second messenger cyclic di-GMP. Photochem Photobiol Sci 19(5):631–643
Williamson CE, Zepp RG, Lucas RM, Madronich S, Austin AT, Ballaré CL, Norval M, Sulzberger B, Bais A, McKenzie RL, Robinson SA, Häder D-P, Paul ND, Bornman JF (2014) Solar ultraviolet radiation in a changing climate. Nat Clim Chang 4:434–441
Xu J, Gao K (2016) Photosynthetic contribution of UV-A to carbon fixation by macroalgae. Phycologia 55:318–322
Xue L, Zhang Y, Zhang T, An L, Wang X (2005) Effects of enhanced ultraviolet-B radiation on algae and cyanobacteria. Crit Rev Microbiol 31:79–89
Yang Y, Lam V, Adomako M, Simkovsky R, Jakob A, Rockwell NC, Cohen SE, Taton A, Wang J, Lagarias JC (2018) Phototaxis in a wild isolate of the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci U S A 115:E12378–E12387
Zhang F, Scheerer P, Oberpichler I, Lamparter T, Krauß N (2013) Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore. Proc Natl Acad Sci U S A 110:7217–7222
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Häder, D.P., Rastogi, R.P. (2021). UV Stress Responses in Cyanobacteria. In: Rastogi, R.P. (eds) Ecophysiology and Biochemistry of Cyanobacteria. Springer, Singapore. https://doi.org/10.1007/978-981-16-4873-1_6
Download citation
DOI: https://doi.org/10.1007/978-981-16-4873-1_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-4872-4
Online ISBN: 978-981-16-4873-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)