Abstract
Cyanobacteria are known to have unique capability of nitrogen fixation in their specialized cell known as heterocyst. However, differentiation of vegetative cell toward heterocyst reduces competitive ability of cyanobacteria because it led to a shift of energy allocation from carbon to nitrogen metabolism. Therefore, heterocyst formation is regulated to avoid the differentiation commitment due to short-term nitrogen fluctuation. Once nitrogen deficiency signal is sensed by the cyanobacteria, pattern of heterocyst formation is determined that ensures equidistance formation of heterocyst cells with about one heterocyst per ten vegetative cells. After differentiation, heterocyst provides anaerobic condition that is prerequisite for the nitrogenase complex to fix the atmospheric dinitrogen. Microoxic condition inside the heterocyst is attained by elimination of oxygen-producing photosystem II activity, increasing respiration rate, and by formation of thick heterocyst-specific exopolysaccharide and glycolipid layer. Nitrogen-fixing machinery is assembled and activated during heterocyst differentiation. The nitrogenase complex is encoded by nif gene family. Many of these genes are interrupted in the vegetative cells by interruption elements and these are excised during differentiation of heterocyst by a site-specific recombinase, leading to the activation of genes. In this chapter, we have outlined the molecular circuit of heterocyst differentiation and discussed the assembly of nitrogen-fixing machinery and role of key enzymes in the nitrogen metabolism in the cyanobacteria.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
12.1 Introduction
Nitrogen metabolism is the group of reactions, which includes the conversion of atmospheric dinitrogen (N2) into glutamate at the expense of energy. This conversion is necessary because of two reasons; one is the storage of ammonia that is quite toxic to a cell and another is the amino acid glutamate that acts as a precursor for many metabolic pathways like 5-aminolevulinate, phycobilin, and chlorophyll biosynthesis (Flores and Herrero 1994). Nitrogen fixation, which is characteristic feature of some prokaryotes, is an important phenomenon to sustain nutrient cycling. It occurs in symbiotic bacteria like Rhizobium or in cyanobacteria like Nostoc and Anabaena.
Cyanobacteria are prokaryotic autotrophs which play an imperative role in food chain and have a diverse ecological niche. Cyanobacteria are not confined to oceanic condition yet their assorted variety in biochemistry has enabled these groups of species to occupy almost any terrestrial and aquatic living space on earth (Schirrmeister et al. 2013). Cyanobacteria normally live in marine or fresh water, some cyanobacteria live in a place with earthly biological system and some even thrive in outrageous conditions like desert, the polar area or warm water (Muro-Pastor and Hess 2020). Cyanobacteria are not in every case free-living yet many are fundamental for organizing complex microbial networks in endolithic form for example, in stromatolites, microbial mats, coastal and desert biological soil, and as symbionts of certain higher plants and fungi (Muro-Pastor and Hess 2020). They are also called as diazotrophs because they have capability to fix the atmospheric nitrogen (Lee 2018). Diazotrophic cyanobacteria species contribute considerable quantity of combined nitrogen into the biosphere by changing dinitrogen into ammonia, a procedure known as biological nitrogen fixation (Muro-Pastor and Hess 2020).
These are the oldest multicellular organism on earth (Herrero et al. 2016). According to paleobotanical study, these organisms were thought to evolve about 2.7 billion years ago and have characteristic blue green color due to the principal pigment c-phycocyanin and c-phycoerythrin. The cells of cyanobacteria are covered with mucilaginous sheath called capsule. Cyanobacterial cell wall shows similarity with gram-negative bacteria, a peptidoglycan layer is present at the outer side of the cell membrane. This layer is composed of NAM (N-acetyl muramic acid), NAG (N-acetyl glucosamine), and tetrapeptides, which are further linked by amide bond. They have porin protein on outer membrane which is permeable for all macro- and micromolecules. But the plasma membrane acts as a permeability barrier for these biomolecules.
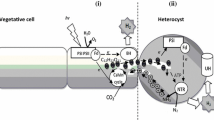
Cyanobacteria are for the most part described by their high protein content. In these, nitrogen metabolism is directed by a saved calibrating framework, which detects the cell balance among carbon and nitrogen level (Forchhammer and Lüddecke 2016). In Synechocystis PCC 6803, for example, photosynthetic rates are found to be positively correlated with amino acid and protein levels, but not with growth rates (Esteves-Ferreira et al. 2017). The growth of any cyanobacteria requires two interdependent cell types, viz., vegetative cells for oxygenic photosynthesis and heterocyst for dinitrogen fixation. The fix ratio of two macronutrients carbon and nitrogen (5:1) plays an important role in metabolic homeostasis. Vegetative cells supply reduced carbon to heterocyst, similarly heterocyst supply fixed nitrogen to vegetative cells and maintains the carbon-nitrogen pool. The balanced metabolism of C and N is essential for optimal growth. Heterocysts are connected with vegetative cells through microplasmodesmata or septosome for minerals and substrates, so it manifests the best example of cell-cell communication in cyanobacteria. Heterocyst itself is a modified vegetative cell, thick walled, pale yellow in color due the principal pigment carotenoid, lacks oxygen evolving PSII activity, and creates a microoxic environment for the key enzyme nitrogenase (Harish 2020).
Heterocysts develop from vegetative cells by decomposition of granular inclusions (carboxysomes and glycogen granules), disintegration of photosynthetic thylakoids, and formation of new membrane structures. They neither fix carbon dioxide nor produce oxygen, but have a high oxygen consumption rate via respiration, surrounded by thick layered laminated cell wall. A special system “Honey comb” is present close to heterocyst poles and has a role in respiration and photosynthesis. The differentiation of heterocyst is completed in two steps—first step is reversible in which the vegetative cell senses the nitrogen-deprived condition and converts it into proheterocyst and the next step is irreversible in which conversion of proheterocyst to heterocyst occurs and nif gene is activated. Proteins like NtcA, HetR, HetC, PatA, PatS, and PatB participate in heterocyst differentiation and pattern formation (Harish 2020). Nitrogen-deprived condition induces the vegetative filament for heterocyst differentiation, accumulation of 2-oxoglutarate (an intermediate of tricarboxylic acid (TCA) cycle), which acts as a signaling molecule for heterocyst differentiation and pattern formation (Esteves-Ferreira et al. 2018). The 2-OG provides a carbon framework for ammonia assimilation through GS-GOGAT cycle (Zhang et al. 2018; Forchhammer and Selim 2020). Heterocyst differentiation is the suitable example of remodeling and cell differentiation.
The key enzyme of biological nitrogen fixation is nitrogenase. There is a temporal and spatial separation in cyanobacteria to prevent denaturation of nitrogenase. Nitrogenase has two components, first is dinitrogenase reductase (a type of iron protein) and second is dinitrogenase (a type of molybdenum-iron protein) (Flores and Herrero 2005). Ammonia is the most preferable nitrogen source because it diffuses passively through the membrane. The ammonia is further converted into glutamate through GS-GOGAT pathway by succeeding reaction held by glutamine synthetase (GS) and glutamate synthase (GOGAT). The reaction catalyzed by GS is ATP-dependent and GOGAT is an amino-transferase which transfers amide group of glutamine to 2-OG resulting in formation of glutamate amino acid. Cyanobacteria can assimilate many organic and inorganic nitrogen-containing compounds other than atmospheric nitrogen, these may be nitrate, nitrite, ammonium, urea, cyanate, and amino acids such as arginine, glutamine, and glutamate, but ammonia is the favored nitrogen source (Esteves-Ferreira et al. 2017). The concentration of ammonia in a medium acts as a regulator (inducer or repressor) for the signal transduction pathway; this pathway is closely correlated with plants because the evolution of plastid is phylogenetically correlated with cyanobacteria by endosymbiosis theory. Nitrogen fixation is a metabolically expensive process because it involves 16 ATP for fixing each molecule of nitrogen. Like GS-GOGAT cycle, there are some amino acids like arginine and aspartate that combinedly form a nitrogen storage reservoir called cyanophycin. It is a nonribosomal synthesized protein like polymer which is arranged in a poly-aspartate form (Lee 2018).
12.2 Heterocyst Differentiation
Heterocyst differentiation is a quite complex mechanism and many proteins are involved in its regulation. Multiple layers of regulation ensure that cyanobacteria do not commit heterocyst formation due to short-term fluctuations in the soil nitrogen source content. When cyanobacterial filament receives a lasting signal for nitrogen depletion condition, it led to synthesis of 2-oxoglutarate. This is considered as first sensing signal for induction of heterocyst formation to overcome the nitrogen starvation condition. 2-oxoglutarate or α-ketoglutarate of the Krebs cycle and this metabolite connect C and N metabolism (Huergo and Dixon 2015). Increase in concentration of 2-oxoglutarate triggers the synthesis of NtcA protein also known as global nitrogen regulator, due to its role in overall regulation of nitrogen metabolism in the cyanobacterial filament. Further, in its downstream cascade signal perpetuation, synthesis of HetR protein occurs. This protein is known as specific master regulator, due to its specific role in heterocyst differentiation. HetR itself is regulated by Pkn22 Kinase, cyABrB1, some other genes like asl1930, alr3234, and alr2902. Multiple layers of regulation ensure fine tuning of development mechanism for heterocyst differentiation. Further, HetR itself has autocatalytic activity and phosphorylation of serine residue present at 130 positions in HetR protein is essential for its activity. HetR protein then interacts with HetP and HetZ proteins and developmental signal is passed to these proteins. HetP protein led to irreversible commitment toward heterocyst formation.
Number of heterocyst cells with respect to vegetative cells are regulated by pattern determination. Because too many heterocyst cells will incur huge energy cost in terms of entire filament and will reduce the competitive ability of the filament. Heterocyst differentiation is energy intensive phenomenon for the cyanobacteria. Therefore, equal distribution of heterocyst cells throughout the filament ensures the equal distribution of fixed nitrogen compound to neighboring vegetative cells, and also conserves the energy for carbon metabolism in the filament. Pattern distribution is therefore equally important aspect when considering the heterocyst differentiation. Proteins of Pat gene family regulate this aspect in the cyanobacteria. HetR protein is involved in synthesis of PatS, which is an inhibitor of the HetR protein itself, thereby controlling the number of heterocyst cells in the filament. PatS is processed to short peptide that acts as concentration-dependent manner to inhibit the heterocyst formation. HetC protein is known for its role in transport of short peptides of PatS. The concentration ratio of PatS and HetR determines the development of heterocyst and position of the heterocyst cell in the filament. PatX is another protein that inhibits heterocyst formation and there is functional overlap between PatS and PatX. PatC protein ultimately selects the cell for differentiation into heterocyst and thereby governs the spatial pattern determination of heterocyst in cyanobacterial filament. Other proteins are also identified that play role in regulation of heterocyst frequency like PatD, PatN, and PknH.
Cell wall of the heterocyst is thick to keep the oxygen concentration minimum in the interior of the cell. Therefore, entire remodeling of the cell wall is done during differentiation of vegetative cell to the heterocyst cell. Cyanobacteria are gram-negative as far as their cell wall organization is concerned, but the thickness of peptidoglycan layer is intermediate (15–35 nm) between gram-positive bacteria and gram-negative bacteria and cross-linking is also higher in cyanobacterial cell wall in comparison to gram-negative bacteria. During heterocyst differentiation, two additional layers are developed. The external polysaccharide layer is known as hep layer and internal glycolipid layer is known as hgl layer. HepA protein is involved in synthesis of hep layer and gene of this protein is regulated by HetR and HepK proteins. Some more genes involved in synthesis of hep layer are hepA, hepK, hepN, hepS, henR, murB, murC, hcwA, amiC1, amiC2, pbp6, sepJ (fraG), fraC, fraD, and sjcF1. Additionally, hgl layer prevents the entry of oxygen inside the cell and therefore ensures low oxygen concentration for functioning of nitrogenase complex. Some genes, which regulated the formation of hgl layer, have been identified like hgdB, hgdC, devBCA operon, devH, and hglE (Table 12.1).
The availability of ammonia in a medium, acts as a regulator (inducer or repressor) for the signal transduction pathway. Where global nitrogen regulator gene ntcA and signal transducer P-II (which is encoded by glnB) control the activity of many genes like henA, hetR, hetC, patA, patB, and patC which are responsible for the heterocyst differentiation and pattern formation. NtcA is a bacterial transcription factor which is a member of catabolic repressor protein. NtcA can inactivate GS-activity by coding inhibitory polypeptides (IF-7 and IF-17) by protein – protein interaction (Muro-Pastor and Florencio 2003). By this, cyanobacteria maintain the metabolic homeostasis. In nitrogen-starved condition, storage level of 2-OG is very high and NtcA self-regulates the expression of hetR for heterocyst differentiation (Muro-Pastor et al. 2001). HetR is a kind of serine type protease and also a DNA-binding protein. In in vivo condition, HetR performs as a homodimer and this homodimer is essential for DNA-binding activity and heterocyst differentiation (Huang et al. 2004). Another gene patS inhibits this DNA-binding affinity (Huang et al. 2004). The nitrogen regulatory protein PII (PII) interacts with 2-OG and brings conformational changes of PII leading to the release of the PII interacting protein X (PipX). PipX interacts with the nitrogen control factor (NtcA) of cyanobacteria.
12.3 Nitrogenase and Alternate Nitrogenase
In order to enter the biogeochemical cycle, atmospheric N2 must be first reduced to a form that can be readily assimilated by organisms in a process known as nitrogen fixation. In cyanobacteria and other N2-fixing prokaryotes, molecular dinitrogen (N2) is reduced in multiple electron transfer reactions requiring 16 ATPs per N2 fixed, resulting in the synthesis of ammonia and the release of hydrogen as a by-product. H2 generated during the N2 fixation process may be oxidized by a hydrogenase in a subsequent step (Esteves-Ferreira et al. 2018).
The reduction of molecular nitrogen to ammonium is catalyzed in all nitrogen-fixing organisms via the nitrogenase enzyme complex. Nitrogenase can also reduce many other substances, such as acetylene, hydrogen azide, hydrogen cyanide, or nitrous oxide. Of these, acetylene reduction to ethylene can be monitored because both acetylene and ethylene can be detected easily by gas chromatography (Fay 1992). Based on the type of metal center, there are three well-known types of nitrogenases: iron and molybdenum (Fe/Mo) nitrogenase, iron and vanadium (Fe/V) nitrogenase, and iron only (Fe) nitrogenase. The Fe/Mo-type is the most commonly found in cyanobacteria and rhizobia. The Fe-only and V-nitrogenases are referred as alternative nitrogenases and are considered as “backup” enzymes when Mo is limiting (McRose et al. 2017). The Fe/Mo-nitrogenase is encoded by nitrogen fixation genes (nifHDK), the V-nitrogenase by vanadium-dependent nitrogen fixation genes (vnfHDK), and the Fe-nitrogenase by alternative nitrogen fixation genes (anfHDK) (McRose et al. 2017).
In nonheterocystous cyanobacteria, nitrogenase enzyme is present in all vegetative cells, while in heterocystous form it is localized only in heterocysts. The enzyme nitrogenase that is expressed in heterocyst is Mo-dependent nitrogenase (Nif-1), which has two components—a Mo-Fe protein (molybdoferredoxin or dinitrogenase) and Fe protein (azoferredoxin or dinitrogenase reductase). The dinitrogenase (Mo-Fe protein) is an α2β2 tetramer and its subunits are encoded by nifD and nifK genes, respectively. The other component, dinitrogenase reductase (Fe protein) is a dimer of two identical subunits (γ) encoded by nifH gene. Fifteen nitrogen fixation-related genes are found clustered together in six transcriptional units: nifB-fdxN-nifS-nifU, nifHDK, nifEN, nifX-orf2, nifW-hesA-hesB, and fdxH. A gene-designated glbN is found positioned between nifU and nifH, which encodes monomeric hemoglobin called cyanoglobin. A second functional Mo-dependent nitrogenase Nif2 has been reported in Anabaena variabilis ATCC 29413 which is synthesized in the vegetative cells solely under anoxic conditions after the cells have been starved of nitrogen and long before heterocysts form (Schrautemeier et al. 1995; Thiel et al. 1997). Nif2 has also been observed in vegetative cells of nonheterocystous species (Berman-Frank et al. 2003).
Vanadium-containing nitrogenase was first reported in Anabaena variabilis, which significantly reduced acetylene (C2H2) to ethane (C2H6) under Mo deficiency and in the presence of vanadium (V). It was further identified that the V-nitrogenase is encoded by vnf genes cluster (vnfDGKEN) in A. variabilis (Thiel 1996). The V-dinitrogenase is actually encoded by vnfDGK gene cluster, while, vnfEN gene cluster located downstream of vnfDGK is found to be essential for V-nitrogenase activity. In addition to vnfDGKEN gene cluster, four other vnfH genes are located 23 kb downstream of vnfN and are responsible for encoding dinitrogenase reductase of V-nitrogenase.
The V-nitrogenase is a heterooctomer consisting of two α-subunit (VnfD), two β-subunit (VnfK), four δ-subunits (VnfG), and two Fe-V cofactors (Thiel and Pratte 2014). In comparison to Mo-nitrogenase, the V-nitrogenases have lower substrate-binding efficiency; therefore, it reduces less dinitrogen and produces three times more hydrogen than the Mo-nitrogenase (Thiel and Pratte 2014).
Nitrogenase is extremely oxygen sensitive. The oxygen is kept far away from nitrogenase by biochemical pathways like the Mehler-reaction or by special oxygen scavenging molecules such as cyanoglobin that binds oxygen reversibly, with high affinity and noncooperatively (Thorsteinsson et al. 1996). In addition, cyanobacteria are diverse group of gram-negative bacteria which coordinate two mutually exclusive process; O2-evolving photosynthesis and O2-sensitive nitrogenase-dependent nitrogen fixation. Cyanobacteria have an efficient way to protect O2-sensitive nitrogenase from O2−evolved during photosynthesis. In cyanobacteria, these processes are either separated temporally (as in nonheterocystous form/unicellular cyanobacteria, where alternate cycles of nitrogen fixation and photosynthesis take place) or spatially (as in heterocystous forms). Interestingly, heterocyst lacks photosystem II activity; therefore, they do not evolve oxygen that inhibits nitrogen fixation.
Numerous nonheterocystous cyanobacterial strains can fix and reduce atmospheric N2 to ammonium when confronting nitrogen hardship, for example, Synechocystis and Arthrospira (Spirulina) maxima (Esteves-Ferreira et al. 2017). N-fixation is a costly metabolic reaction catalyzed by nitrogenase, which is restrained by O2 (Esteves-Ferreira et al. 2018). To shield nitrogenase from O2, photosynthesis and N-fixation are transiently isolated. High nitrogenase activity peaks 12 h after the peak of photosynthesis, at the same time with higher respiratory rates. An alternate N-fixation methodology is solely seen in strains of the genera Trichodesmium (Bergman et al. 2013). In these genera, nitrogenase is situated in roughly 20% cells of the filament, and inquisitively these cells display high N−fixation rates at midday (Rodriguez and Ho 2014).
12.4 Uptake of Nitrogen Sources
The most commonly used nitrogen sources by cyanobacteria are nitrate, ammonium, urea, and dinitrogen. Ammonium is the most reduced inorganic form of nitrogen and preferred source of nitrogen for cyanobacteria. When present in the environment, a decrease in the abundance of nitrogen assimilatory enzymes and a reduced expression of nitrogen transport systems leads to a process referred as global nitrogen control (Esteves-Ferreira et al. 2018). Ammonium indirectly represses the expression of nif genes by blocking the transcription of NtcA. In natural environments, ammonium is generally present at low concentrations; therefore, specific permeases namely Amt1, Amt2, and Amt3 are required for efficient cellular uptake of ammonium (Esteves-Ferreira et al. 2018). It has been identified that Amt1 is the main permease for ammonium uptake in Synechocystis (Montesinos et al. 1998).
Nitrate and nitrite are the most frequent sources of nitrogen for cyanobacteria. In order to be assimilated by cyanobacteria, nitrate is reduced to ammonium via two sequential reactions catalyzed by enzymes nitrate reductase and nitrite reductase. The reductions of nitrate to nitrite and nitrite to ammonium are Fd-dependent and energetically costly (Flores and Herrero 2005). Nitrate uptake and nitrate reductase system are not found in heterocyst. Nitrate and nitrite are actively transported by the ABC-type NrtABCD transporter in freshwater cyanobacterial strains (Maeda et al. 2015). However, it has been reported that nitrate utilization by cyanobacteria in saline environments may be mediated by NapA (NrtP) rather than NrtABCD transporters. The genes for NrtABCD transporter (nrtA, nrtB, nrtC, and nrtD) are commonly present in the nirA operon (i.e., nirA-nrtABCD-narB). The nirA and narB genes encode the enzymes Fd-nitrite reductase (NirA) and Fd-nitrate reductase (NarB), respectively, which lead to the formation of ammonium. In Synechocystis, nirA has been found to be separated from nrtABCD-narB (Ohashi et al. 2011). Certain marine and saline water cyanobacterial strains have nitrite transporter of the formate/nitrite transporter (FNT) family, and the cyanate ABC-type transporter which transport nitrite with a much lower affinity than for cyanate (Maeda and Omata 2009; Maeda et al. 2015). A transporter, encoded by the gene nrtP, additionally displays high affinity for nitrate and nitrite and was distinguished in the genome of cyanobacterial strains from freshwater and marine conditions (Sakamoto et al. 1999; Bird and Wyman 2003; Maeda et al. 2015).
Many cyanobacteria have shown to import urea at concentrations as low as 0.1–0.6 mM (Mitamura et al. 2000). But before assimilation, urea needs to be hydrolyzed to ammonium and CO2 catalyzed by a Ni2+-dependent urease. The urease is typically a constitutive enzyme which is not regulated by nitrogen-containing compounds (Ludwig and Bryant 2012). However, a low urease action has been noted in some cyanobacterial strains in presence of ammonium (Singh 1992). These cyanobacteria, such as Synechocystis, have high-affinity urea ABC-type transporter responsible for urea uptake at concentrations lower than 1 mM (Esteves-Ferreira et al. 2018). The urea ABC-type transporter is encoded by urt genes urtA, urtB, urtC, urtD, and urtE. These genes are normally organized in an operon, although in Synechocystis they have been found to spread along the chromosome (Valladares et al. 2002).
12.5 Ammonium Incorporation into Carbon Skeletons
In cyanobacteria, ammonium is incorporated into carbon skeletons mainly through the glutamine synthetase/glutamine oxoglutarate aminotransferase (GS/GOGAT) cycle. GS catalyzes the ATP-dependent incorporation of ammonium into glutamate to form glutamine. In the following reaction, GOGAT (glutamate synthase) catalyzes the transfer of amide group from glutamine to 2-oxoglutarate (2-OG) to form two molecules of glutamate. Subsequently, aminotransferases can transfer the amino group from glutamate to other carbon skeletons to form other amino acids (Esteves-Ferreira et al. 2018).
In cyanobacteria, there is only one GS (GSI) which is encoded by the gene glnA. GSI activity is negatively regulated in presence of ammonium by protein-protein interaction of two inactivating factors (i.e., IF7 and IF17). In contrast, under nitrogen deficiency or in the presence of nitrogen sources other than ammonium, glnA expression is up-regulated (Esteves-Ferreira et al. 2018). Interestingly, in some cyanobacterial strains such as Synechocystis, Synechococcus, and Gloeocapsa sp. PCC 7428, a second type of GS encoded by glnN, referred as GS type III (GSIII) has also been observed (Reyes and Florencio 1994). It has been observed that glnN transcription is more sensitive to ammonium and nitrate than glnA, and maximal GSIII activity can reach at most 24% of GSI activity in cells under nitrogen starvation (Reyes and Florencio 1994). In Pseudanabaena sp. strain PCC 6903, only the GSIII has been found to be responsible for all ammonium assimilation. This indicates that the presence of an additional glutamine synthetase (GSIII) in some cyanobacterial strains indicates its possible role in increasing the efficiency of ammonium assimilation under nitrogen deficiency.
GOGAT (glutamine 2-oxoglutarate aminotransferase) is a constitutive expressed enzyme. Two different forms of GOGAT have been described in photosynthetic organisms, one is a Fd-GOGAT encoded by glsF (gltS), and, second is a complex of NADH-GOGAT encoded by gltB and gltD gltB (large subunit) and gltD (small subunit) (Muro-Pastor and Florencio 2003). All cyanobacteria encompass Fd-GOGAT (Muro-Pastor et al. 2005) though NADH-GOGAT has been also identified in Synechocystis and Plectonema boryanum (Wang et al. 2004). Although, both the forms of this enzyme are simultaneously active in these strains, but Fd-GOGAT is found to be more active and has a more prominent role in ammonium assimilation and growth.
Alternatively, the glutamate dehydrogenase (GDH) can catalyze the formation of glutamate directly from 2-oxoglutarate and ammonium in an NADPH-dependent reaction. However, since GDH has higher Km (and consequently low affinity) for ammonium than GSI, hence, this enzyme is not likely to be intricated in main ammonium assimilation in cyanobacteria. It has been suggested that this enzyme is involved in the removal of excess cellular ammonium, so as to guard the proton gradient within the thylakoid and periplasmic spaces (Chávez et al. 1999).
12.6 Cyanobacteria as Biofertilizer
Due to ability of cyanobacteria to fix the atmospheric nitrogen, they are conventionally used as biofertilizers in agriculture field. Their importance as biofertilizer has increased mainly to avoid usage of synthetic fertilizers. Cyanobacteria not only increase the nitrogen content in the soil, but also improve the soil structure via released polysaccharides, increase the soil aeration by their filamentous structure, improve soil health by releasing growth hormones, decrease the soil salinity, and improve water holding capacity of the soil (Kuraganti et al. 2020). Because of their ecofriendly organic nature and economic feasibility, cyanobacteria are commonly used as biofertilizer mainly in the paddy field. Generally, Anabaena, Nostoc, Aulosira, and Tolypothrix are used as biofertilizer. However, use of Azolla-Anabaena symbiotic N2-fixing complex has also been reported and it is found to have this symbiotic complex as more advantageous than free-living cyanobacteria from agronomic point of view.
12.7 Conclusions
Nitrogen-fixing capability of cyanobacteria has made this group of organisms very important for the research purpose. Cyanobacteria require two interdependent cell types for growth, i.e., vegetative cells for oxygenic photosynthesis and heterocyst for nitrogen fixation. Heterocyst differentiation process involves many proteins like NtcA, HetR, HetP, HetZ, PatS, PatX, PatC, PatD, PatN, PknH, HepA, HepK, etc. The first sensing signal for induction of heterocyst formation is synthesis of 2-oxoglutarate which is a part of Kreb cycle as alpha ketoglutarate. Increase in concentration of 2-oxoglutarate triggers the synthesis of NtcA protein. NtcA further activates the other genes involved in heterocyst differentiation. Development of heterocyst and their spatial distribution both are equally important. Proteins of het gene family are involved in differentiation and proteins of Pat gene family regulate distribution pattern.
In this chapter, it is discussed that most commonly used nitrogen sources by cyanobacteria are nitrate, ammonium, urea, and dinitrogen, but ammonium is most preferred nitrogen source. In nitrogen fixation, dinitrogen is reduced to ammonia in multiple electron transfer reactions requiring 16 ATPs per N2 fixed. In nitrogen assimilation by cyanobacteria, nitrate is reduced to ammonium via two sequential reactions catalyzed by enzymes nitrate reductase and nitrite reductase. Nitrate and nitrite are actively transported by the ABC-type NrtABCD transporter in freshwater cyanobacterial strains, and in saline environments may be mediated by NapA (NrtP). Urea is hydrolyzed to ammonium and CO2 by a Ni2+-dependent urease. The urea ABC-type transporter is encoded by urt genes.
In cyanobacteria, ammonium is combined into carbon skeletons mostly through the GS/GOGAT cycle. GS is encoded by the gene glnA. Two diverse forms of GOGAT have been described in photosynthetic organisms, a Fd-dependent GOGAT (glsF) and NADH-dependent GOGAT (gltB and gltD). Alternatively, GDH catalyzes the synthesis of glutamate from 2-oxoglutarate and ammonium in an NADPH-dependent reaction, albeit with low affinity.
Studies involving nitrogen metabolism led to understanding of molecular mechanism of nitrogen fixation on exposure to various nitrogen sources. Better understanding of the mechanisms involved in nitrogen metabolism would increase the biotechnological potential of these organisms (Zehr and Capone 2020).
References
Bergman B, Sandh G, Lin S, Larsson J, Carpenter EJ (2013) Trichodesmium–a widespread marine cyanobacterium with unusual nitrogen fixation properties. FEMS Microbiol Rev 37(3):286–302
Berman-Frank I, Lundgren P, Falkowski P (2003) Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res Microbiol 154(3):157–164
Bird C, Wyman M (2003) Nitrate/nitrite assimilation system of the marine picoplanktonic cyanobacterium Synechococcus sp. strain WH 8103: effect of nitrogen source and availability on gene expression. Appl Environ Microbiol 69(12):7009–7018
Chávez S, Lucena JM, Reyes JC, Florencio FJ, Candau P (1999) The presence of glutamate dehydrogenase is a selective advantage for the cyanobacterium Synechocystis sp. strain PCC 6803 under nonexponential growth conditions. J Bacteriol 181(3):808–813
Esteves-Ferreira AA, Cavalcanti JHF, Vaz MGMV, Alvarenga LV, Nunes-Nesi A, Araújo WL (2017) Cyanobacterial nitrogenases: phylogenetic diversity, regulation and functional predictions. Gen Mol Biol 40(1):261–275
Esteves-Ferreira AA, Inaba M, Fort A, Araújo WL, Sulpice R (2018) Nitrogen metabolism in cyanobacteria: metabolic and molecular control, growth consequences and biotechnological applications. Crit Rev Microbiol 44(5):541–560
Fay P (1992) Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Mol Biol Rev 56(2):340–373
Flores E, Herrero A (1994) Assimilatory nitrogen metabolism and its regulation. In: The molecular biology of cyanobacteria. Springer, Dordrecht, pp 487–517
Flores E, Herrero A (2005) Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem Soc Trans 33(1):164–167
Forchhammer K, Lüddecke J (2016) Sensory properties of the PII signalling protein family. The FEBS J 283(3):425–437
Forchhammer K, Selim KA (2020) Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol Rev 44(1):33–53
Harish SK (2020) Molecular circuit of heterocyst differentiation in cyanobacteria. J Basic Microbiol. https://doi.org/10.1002/jobm.202000266
Herrero A, Stavans J, Flores E (2016) The multicellular nature of filamentous heterocyst-forming cyanobacteria. FEMS Microbiol Rev 40(6):831–854
Huang X, Dong Y, Zhao J (2004) HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. PNAS USA 101(14):4848–4853
Huergo LF, Dixon R (2015) The emergence of 2-oxoglutarate as a master regulator metabolite. Microbiol Mol Biol Rev 79(4):419–435
Kuraganti G, Edla S, Pallaval VB (2020) Cyanobacteria as biofertilizers: current research, commercial aspects, and future challenges. In: Yadav A, Rastegari A, Yadav N, Kour D (eds) Advances in plant microbiome and sustainable agriculture. Microorganisms for sustainability, vol 20. Springer, Singapore. https://doi.org/10.1007/978-981-15-3204-7_11
Lee RE (2018) Phycology. Cambridge University Press, New York
Ludwig M, Bryant DA (2012) Acclimation of the global transcriptome of the cyanobacterium Synechococcus sp. strain PCC 7002 to nutrient limitations and different nitrogen sources. Front Microbiol 3:145
Maeda SI, Murakami A, Ito H, Tanaka A, Omata T (2015) Functional characterization of the FNT family nitrite transporter of marine picocyanobacteria. Life 5(1):432–446
Maeda SI, Omata T (2009) Nitrite transport activity of the ABC-type cyanate transporter of the cyanobacterium Synechococcus elongatus. J Bacteriol 191(10):3265–3272
McRose DL, Zhang X, Kraepiel AM, Morel FM (2017) Diversity and activity of alternative nitrogenases in sequenced genomes and coastal environments. Front Microbiol 8:267
Mitamura O, Kawashima M, Maeda H (2000) Urea degradation by picophytoplankton in the euphotic zone of Lake Biwa. Limnology 1(1):19–26
Montesinos ML, Muro-Pastor AM, Herrero A, Flores E (1998) Ammonium/methylammonium permeases of a cyanobacterium. Identification and analysis of three nitrogen-regulated amt genes in Synechocystis sp. PCC 6803. J Biol Chem 273(47):31463–31470
Muro-Pastor MI, Florencio FJ (2003) Regulation of ammonium assimilation in cyanobacteria. Plant Physiol Biochem 41(6–7):595–603
Muro-Pastor AM, Hess WR (2020) Regulatory RNA at the crossroads of carbon and nitrogen metabolism in photosynthetic cyanobacteria. BBA-Gene Regul Mech 1863(1):194477
Muro-Pastor MI, Reyes JC, Florencio FJ (2001) Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J Biol Chem 276(41):38320–38328
Muro-Pastor MI, Reyes JC, Florencio FJ (2005) Ammonium assimilation in cyanobacteria. Photosyn Res 83(2):135–150
Ohashi Y, Shi W, Takatani N, Aichi M, Maeda SI, Watanabe S, Omata T (2011) Regulation of nitrate assimilation in cyanobacteria. J Exp Bot 62(4):1411–1424
Reyes JC, Florencio FJ (1994) A new type of glutamine synthetase in cyanobacteria: the protein encoded by the glnN gene supports nitrogen assimilation in Synechocystis sp. strain PCC 6803. J Bacteriol 176(5):1260–1267
Rodriguez IB, Ho TY (2014) Diel nitrogen fixation pattern of Trichodesmium: the interactive control of light and Ni. Sci Rep 4:4445
Sakamoto T, Inoue-Sakamoto K, Bryant DA (1999) A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol 181(23):7363–7372
Schirrmeister BE, de Vos JM, Antonelli A, Bagheri HC (2013) Evolution of multicellularity coincided with increased diversification of cyanobacteria and the great oxidation event. PNAS USA 110(5):1791–1796
Schrautemeier B, Neveling U, Schmitz S (1995) Distinct and differently regulated Mo- dependent nitrogen-fixing systems evolved for heterocysts and vegetative cells of Anabaena variabilis ATCC 29413: characterization of the fdxH1/2 gene regions as part of the nif1/2 gene clusters. Mol Microbiol 18(2):357–369
Singh S (1992) Regulation of urease cellular levels in the cyanobacteria Anacystis nidulans and Nostoc muscorum. Biochem Physiol Pflanz 188(1):33–38
Thiel T (1996) Isolation and characterization of the VnfEN genes of the cyanobacterium Anabaena variabilis. J Bacteriol 178(15):4493–4499
Thiel T, Pratte BS (2014) Regulation of three nitrogenase gene clusters in the cyanobacterium Anabaena variabilis ATCC 29413. Life 4(4):944–967
Thiel T, Lyons EM, Erker JC (1997) Characterization of genes for a second Mo-dependent nitrogenase in the cyanobacterium Anabaena variabilis. J Bacteriol 179(16):5222–5225
Thorsteinsson MV, Bevan DR, Ebel RE, Weber RE, Potts M (1996) Spectroscopical and functional characterization of the hemoglobin of Nostoc commune UTEX 584 (cyanobacteria). BBA Protein Struct Mol Enzym 1292(1):133–139
Valladares A, Montesinos ML, Herrero A, Flores E (2002) An ABC type, high affinity urea permease identified in cyanobacteria. Mol Microbiol 43(3):703–715
Wang HL, Postier BL, Burnap RL (2004) Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J Biol Chem 279(7):5739–5751
Zehr JP, Capone DG (2020) Changing perspectives in marine nitrogen fixation. Science 368(6492):eaay9514. https://doi.org/10.1126/science.aay9514
Zhang CC, Zhou CZ, Burnap RL, Peng L (2018) Carbon/nitrogen metabolic balance: lessons from cyanobacteria. Trends Plant Sci 23(12):1116–1130
Acknowledgments
MK acknowledges financial assistance from University Grants Commission (UGC), New Delhi in a form of JRF-NET (NTA Ref. No. 191620001766). Contribution of VS to this study was financially supported by UGC, New Delhi, India in the form of BSR meritorious fellowship [F.25-a/2014-15(BSR)/7-125/2007(BSR)]. Financial support from Department of Science and Technology, New Delhi for laboratory infrastructure is also acknowledged (SERB File Number: EEQ/2020/000011). We are thankful to the Head, Department of Botany for providing necessary facility.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Seth, K. et al. (2021). Nitrogen Metabolism in Cyanobacteria. In: Rastogi, R.P. (eds) Ecophysiology and Biochemistry of Cyanobacteria. Springer, Singapore. https://doi.org/10.1007/978-981-16-4873-1_12
Download citation
DOI: https://doi.org/10.1007/978-981-16-4873-1_12
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-4872-4
Online ISBN: 978-981-16-4873-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)