Abstract
COVID-19, the most destructive global event in 2020, poses gigantic challenges to global medical systems. Meanwhile, the useful concepts and newly-emerging technical terms in medical field generate ambiguity and bring difficulties in extraction, which would set immense obstacles to anti-epidemic actions. To solve this problem, we built a knowledge graph by extracting six kinds of medical entities from titles and abstracts related to COVID-19 in PubMed. Then, we eliminated author name ambiguity and integrated articles’ publication information as well as authors’ affiliation information into the graph. By this way, connections are established between authors, articles, entities and affiliations. Our model which aims at named entity recognition achieved an F1 score of 92.57% on average. This graph not only allows us to seek out hotspots, utilize specific knowledge and transfer research results quickly, but also helps us understand the research development process. It can also aid scholars to focus on specific authors and entities. This method of a knowledge graph is extensible and transplantable, which means it will not be limited to COVID-19 research in the future.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
At the beginning of 2020, COVID-19 has swept the world as a sudden epidemic, disrupting the peace of every family in every country in the world. The spread of COVID-19 is so fast and infectious that it is beyond everyone's imagination. As a result, it has had a catastrophic impact on the world’s population, economy, environment, and education. The severity of the epidemic problem quickly drew a response from scientific researchers in most countries, and academic research on vaccine development, drug research, and disease transmission trend prediction on COVID-19 was quickly launched. Papers from various fields and angles have been included on PubMed.
PubMed is an abstract database developed by the National Center for Biotechnology Information (NCBI) under the National Library of Medicine (NLM). As one of the most influential databases in the biomedical field, PubMed has the advantages of timely update, free access, and high coverage rate. Therefore, we choose PubMed and LitCovid (dataset in PubMed [1, 2]) as our data source.
In this context, a complete and efficient retrieval approach is particularly important. It must meet two requirements: on the one hand, it can enable researchers to quickly obtain research progress in a specific research field, and on the other hand, it also needs to provide a way for researchers to find research partners in the same direction. The powerful information extraction capabilities and intuitive visualization functions of the knowledge graph perfectly meet our needs, so we chose to construct the COVID-19 literature knowledge graph to summarize existing research.
In the research field of bio-entity recognition and knowledge graph, lots of scholars has been fruitful. Song HJ used Word2Vec to complete Bio-NER, and got F1 score of 72.82% [3]. Ling, Luo added attention mechanism on BiLSTM-CRF model to enforce tagging consistency and recognize CHEMDNER corpus and CDR task corpus [4]. Roderic mapped local identifiers to shared global identifiers. He constructed a knowledge graph based on this [5]. Xu trained Bio-BERT model to build a PubMed knowledge graph, and achieved an F1 score of 86.04% [6]. The goal of our study is building a knowledge graph about COVID-19 by extracting valuable information from literatures and integrating multi-source data.
2 Building Methods
2.1 Named Entity Recognition
NER is an important issue in natural language processing and it also plays an important basic role in building a knowledge graph. It can be said that if the problem of NER cannot be resolved reasonably, our follow-up works won’t be possible. Our article uses the BERT-BiLSTM-CRF model to complete the extraction of biological entities in COVID-19 related literature, our process of the model can be shown as Fig. 1.
Bidirectional Encoder Representation from Transformers (BERT) is an Encoder which is based on Bidirectional-Transformer. The Transformer model can be seen as a text sequence architecture depended on the self-attention mechanism. With this transformer, not only could we consider the contextual relationship more clearly and make parallel calculations, but also allow the prediction sequence with no length limit which means we can better capture the semantic features of the context. So, the multi-layer Bidirectional-Transformer in BERT makes the sequence be constrained by the left and right context at the same time. Compared with the ELMo model which was proposed by Matthew E. Peters and others in 2018 [7], Bert can obtain contextual semantic information better.
So, our first step is using the BERT pre-training language model to get the semantic representation of each token. However, the basic BERT is based on common corpus training and cannot be directly applied to our target medical field. It is necessary to fine-tune the existing parameters of the model. We use WordPiece embedding to supplement the missing words, which is an algorithm that decomposes a word into several different units and expresses each unit. The results prove that this method can improve the effect of extracting semantic features of uncommon words.
After getting the vector representation of each token, we input the vector into the BiLSTM model. The structure of basic LSTM can be formalized as follows:
In the formula, \(\sigma\) is the sigmoid activation function, \(x_{t}\) is the input word at the current moment, \(h_{t - 1}\) is the hidden layer state at the previous moment,\( i_{t} ,f_{t} ,o_{t}\) represent the values of the input gate, forget gate, and output gate at time \(t\) respectively. \(W,b\) represent the weight matrix and bias vector, \(\widetilde{{c_{t} }}\) is an intermediate state, and \(h_{t}\) is the output at time \(t\).
BiLSTM uses forward and backward calculations on the basis of LSTM to obtain two different sets of hidden representations and then stitch the vectors to obtain the final hidden representation. The improvement of LSTM allows us to better capture the two-way semantic dependency and master the semantic co-occurrence information of the context more effectively, thereby improving the performance of named entities.
We also set up different tags to predict the type of token, they are BIO (Beginning, inside, out-side), X (subtoken of WordPiece), [CLS] (leading token of sequence), [SEP] (delimiter of a sentence), PAD (padding in sequence). What’s more, the BIO annotation is subdivided into six categories: Gene, Disease, Chemical, Mutation, Species, CellLine. Input the word vector obtained by BERT into BiLSTM and pass through the softmax classification, we can get the probability distribution of each token belonging to different labels.
In order to solve the problem that BiLSTM does not consider the relationship between labeled entity sequences, we introduce Conditional Random Field (CRF) to obtain the globally optimal labeled sequence.
We define matrix \(P\) as the output of the BiLSTM layer, and the size of \(P\) is \(n \times m\), \(n\) is the number of words, \(m\) is the label category. \(P_{i,j}\) represents the probability of the word \(i\) in the sentence belonging to the label \( j\). The probability of the entire prediction sequence \(y = \left\{ {y_{1} ,y_{2} , \cdots ,y_{n} } \right\}\) can be expressed as follows:
Matrix \(A\) is the transition matrix, \(A_{ij}\) represents the probability of transferring from tag \(i\) to tag \(j\).
\(\tilde{y}\) represents the true value of tag, and \(Y_{X}\) represents all possible tag sets. The sequence \(y^{*}\) with the largest overall probability which is output by formula (7), is also the best labeling result obtained after our model training.
2.2 Validation of BERT-BiLSTM-CRF
For the NER model, we need to perform a validity test. All of our data in this article come from PubMed, a website which contains almost all papers in the medical field. The data published on this website has been physically labeled, but the latest published and included articles have not yet labeled information. Therefore, we set 70% of the labeled articles in PubMed as the training data set, 20% as the test set, and 10% as the verification set. The quality of our model is evaluated by the indicators of recall, accuracy and F1 score. In order to verify the effect of the model, we used the unfine-tuned Bert model, Word2Vec, and Att-BiLSTM-CRF to compare and verify the data set. The results are shown in Table 1.
2.3 Author Name Disambiguation
It is common for researchers having the same name or surname, while the names and affiliations of an individual changes over time. Therefore, when constructing a knowledge graph, it is important to disambiguate different authors. So far, the commonly used methods are mainly divided into three categories. The first one is manual disambiguation, searching for the author’s information and comparing the author’s message to make judgments. The advantage of this method is its high accuracy, but it is time-consuming and labor-intensive, which makes it impossible to be applied in huge data sets. The second method is accessing public scholar registration platforms such as ORCID, Google Scholar, and Semantic Scholar to get author’s information. This method can quickly and easily obtain high-precision author identity information, but sometimes the coverage of the research field is limited. The third method is to evaluate the similarity of two same-name authors through algorithms to determine whether they belong to the same author. The acquisition of author’s feature usually depends on the authors’ affiliation information, titles and keywords of the published article, the information of the collaborators, the type of journal, etc. In recent years, with the rapid development of machine learning, the accuracy of such methods has reached a high level.
In our research, we integrate the data and information in Semantic Scholar and Google Scholar to complete the disambiguation and mark the authors. First, we use a two-classifier trained by the Semantic Scholar database to disambiguate each group of authors with the same name, and add the processed authors as increments to the created author dataset. Then use the corresponding author's information obtained in Google Scholar as a supplementary information source. Finally, we correct false disambiguation results manually, while supply the affiliation information of authors not covered.
3 CLKG Construction Process
CLKG is built based on python3.7 and networkx. The output is stored as gpickle. Anyone can get CLKG in https://github.com/spicycock/CLKG. The construction process of CLKG is shown as Fig. 2. Up to the date of writing, we obtained 82365 articles related to COVID-19 on PubMed. First, we use BERT-BiLSTM-CRF model to solve the NER problem and get the entity and its corresponding type from the abstract of each article. In this step, we extract 26,458 entities in total (including 15,437 Disease tags, 3783 Gene tags, 4832 Chemical tags, 316 Mutation tags, 1975 Species tags, and 115 CellLine tags). Then use the method mentioned in 2.3 to extract and disambiguate scholar names, and finally obtain 294655 disambiguated author names. In the third step, construct a knowledge graph based on three types of relationships: entity-entity, author-author, and entity-author. Make a further explanation, we use entity or author as a node. If there is an association between the two nodes, add an undirected edge to connect. In this way, the basic architecture of CLKG can be constructed.
After establishing the basic graph, we integrate the author's affiliation information from Google Scholar into the node information of the graph. At the same time, for each entity-author connection, we added the publication information of the related articles obtained from PubMed, Including journal name, issue time and issue number. By this way, we can expand the information of the knowledge graph to construct CLKG completely.
4 CLKG Visualization
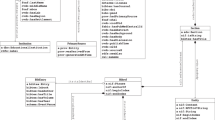
Since CLKG is constructed based on all 82365 documents related to COVID-19, it contains a huge amount of information and the relationship between nodes is also complicated, which means it is difficult to visualize it with general methods. CLKG provides a convenient search interface, allowing us to extract only the relevant fields of interest and reduce the amount of information to the extent that the two-dimensional image can carry. An example of the visualization result is shown in Fig. 3, where the size of the author node is determined by the number of articles published by the author, and the size of the entity node is determined by the number of times mentioned.
Not only could we focus on the overall research status in a certain academical area, but also can conduct more refined searches. On one hand, we can locate a specific well-known scholar to find out what he has said about COVID-19. On the other hand, we can also focus on a specific research entity, listen to other people’s opinions on this issue or look for the future potential partners. We have made two examples to show how it works, Anthony S. Fauci is a well-known infectious disease expert in the United States, who has made great contributions to the control, treatment, and research of the epidemic. We can quickly extract the relevant research content, co-collaborators and other related information from CLKG, and the sub-graph constructed is shown in Fig. 4.
Sinefungin is a medicinal ingredient that is often mentioned in the research on the treatment of COVID-19. As a professional scholar in the medical field, you may be concerned about whether it is proven to be effective. Figure 5 takes sinefungin as an example, and extracts relevant information from CLKG. With Fig. 5, we can clearly see the current status of published research and the clustering relationship among various nodes.
5 Conclusion
As stated at the beginning of this article, COVID-19 is a severe test for each family in every country in the world. We should be one mind to overcome this disaster together. Our article uses all COVID-19 related papers in PubMed as the basis, applies the BERT-BiLSTM-CRF model to solve the key NER problem, disambiguates the researcher with the same name, and finally establishes a comprehensive and complete CLKG based on the relationship across authors and entities. As a knowledge map, CLKG collects and summarizes the research results of the world’s top scientists, pharmacists and other experts on COVID-19 from all over the world and then visualizes the output. Through CLKG, we can not only quickly query the research status, frontier hotspots and research process of COVID-19, but also allows researchers find their academical partners in specific subject areas more quickly and efficiently. This timely and accurate information sharing and sincere cooperation among top scholars will undoubtedly play a key role in overcoming the epidemic, reducing unemployment, restarting the economy and restoring education.
At the same time, CLKG has excellent scalability in both vertical and horizontal directions. Vertically, CLKG can quickly extract the biological entities and add new information to the knowledge map when new literature appears, without complex and time-consuming reconstruction. Horizontally, the CLKG construction method in this article can be easily applied to any field of the same type (such as cancer, heart disease, etc.). Even not only limited to the medical field, global issues such as global warming and environmental pollution can also be extended well.
References
Chen, Q., Allot, A., Lu, Z.: Keep up with the latest coronavirus research. Nature 579(7798), 193 (2020)
Chen, Q., Allot, A., Lu, Z.: LitCovid: an open database of COVID-19 literature. Nucleic Acids Res. 49(D1), D1534–D1540 (2020)
Song, H.-J., Jo, B.-C., Park, C.-Y., Kim, J.-D., Kim, Y.-S.: Comparison of named entity recognition methodologies in biomedical documents. BioMed. Eng. OnLine 17, 158 (2018). https://doi.org/10.1186/s12938-018-0573-6
Luo, L., et al.: An attention-based BiLSTM-CRF approach to document-level chemical named entity recognition. Bioinformatics 34(8), 1381–1388 (2018)
Page, R.D.M.: Ozymandias: a biodiversity knowledge graph. PeerJ 7, e6739 (2019). https://doi.org/10.7717/peerj.6739
Xu, J., et al.: Building a PubMed knowledge graph. Sci. Data 7, 205 (2020)
Devlin, et al.: BERT: Pre-training of deep bidirectional transformers for language understanding (2018).
Yoon, W., et al.: Collaboration of deep neural networks for biomedical named entity recognition. BMC Bioinform. 20(249), 55–65 (2019)
Peters, M.E., Neumann, M., Iyyer, M., et al.: Deep contextualized word representations. In: Proceedings of the Conference of the North American Chapter of the Assocition for Computational Linguistics, pp. 2227–2237 (2018)
Wang, Y., et al.: Named entity recognition in Chinese medical literature using pretraining models. Sci. Program. 2020, 1–9 (2020)
Lee, J., et al.: BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36, 1234–1240 (2019)
Habibi, M., et al.: Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics 33, 37–48 (2017)
Liao, F., Ma, L., Yang, D.: Research on construction method of knowledge graph of US military equipment based on BiLSTM model. In: 2019 International Conference on High Performance Big Data and Intelligent Systems, pp. 146–150. Shenzhen, China (2019)
Hakala, K., Kaewphan, S., Salakoski, T., Ginter, F.: Syntactic analyses and named entity recognition for PubMed and PubMed Central—up-to-the-minute. In: Proceedings of the 15th Workshop on Biomedical Natural Language Processing, pp. 102–107 (2016)
Rossanez, A., dos Reis, J.C., Torres, R.d.S., et al.: KGen: a knowledge graph generator from biomedical scientific literature. BMC Med. Inform. Decis. Mak., 20(Suppl. 4) (2020)
Tosi, M.D.L., dos Reis, J.C.: SciKGraph: a knowledge graph approach to structure a scientific field. J. Inform. 15(1), 101109 (2021)
Berven, A., Christensen, O.A., Moldeklev, S., et al.: A knowledge-graph platform for newsrooms. Comput. Ind. 123, 103321 (2020)
Cho, M., Ha, J., Park, C., et al.: Combinatorial feature embedding based on CNN and LSTM for biomedical named entity recognition. J. Biomed. Inform. 103, 103381 (2020)
Luo, L., Yang, Z., Cao, M., et al.: A neural network-based joint learning approach for biomedical entity and relation extraction from biomedical literature. J. Biomed. Inform. 103, 103384 (2020)
Song, Y., Tian, S., Yu, L.: A method for identifying local drug names in Xinjiang based on BERT-BiLSTM-CRF. Autom. Control. Comput. Sci. 54(3), 179–190 (2020)
Song, M., Kim, E.H.-J., Kim, H.J.: Exploring author name disambiguation on PubMed-scale. J. Informet. 9(4), 924–941 (2015)
Milojević, S.: Accuracy of simple, initials-based methods for author name disambiguation. J. Informet. 7(4), 767–773 (2013)
Veloso, A., Ferreira, A.A., Gonçalves, M.A., et al.: Cost-effective on-demand associative author name disambiguation. Inf. Process. Manage. 48(4), 680–697 (2012)
Ferreira, A.A., Gonçalves, M.A., Almeida, J.M., et al.: A tool for generating synthetic authorship records for evaluating author name disambiguation methods. Inf. Sci. 206, 42–62 (2012)
Schulz, C., Mazloumian, A., Petersen, A.M., Penner, O., Helbing, D.: Exploiting citation networks for large-scale author name disambiguation. EPJ Data Sci. 3(1), 1–14 (2014)
D’Angelo, C.A., van Eck, N.J.: Collecting large-scale publication data at the level of individual researchers: a practical proposal for author name disambiguation. Scientometrics 123(2), 883–907 (2020)
Mu, X., Wang, W., Xu, A.: Incorporating token-level dictionary feature into neural model for named entity recognition. Neurocomputing 375, 43–50 (2020)
Gligic, L., Kormilitzin, A., Goldberg, P., et al.: Named entity recognition in electronic health records using transfer learning bootstrapped Neural Networks. Neural Netw. 121, 132–139 (2020)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Liu, H., Sun, Y., Cao, S. (2022). Building a COVID-19 Literature Knowledge Graph Based on PubMed. In: Su, R., Zhang, YD., Liu, H. (eds) Proceedings of 2021 International Conference on Medical Imaging and Computer-Aided Diagnosis (MICAD 2021). MICAD 2021. Lecture Notes in Electrical Engineering, vol 784. Springer, Singapore. https://doi.org/10.1007/978-981-16-3880-0_38
Download citation
DOI: https://doi.org/10.1007/978-981-16-3880-0_38
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-3879-4
Online ISBN: 978-981-16-3880-0
eBook Packages: EngineeringEngineering (R0)