Abstract
As a complex ecosystem, human gut microbiota have several functions integrated in the host organism through various activities including metabolism, immunity, absorption, etc. One of the human microbiota is viruses, whose composition has not been completely described up till now. Vaious studies proved that the human gut harbors different types of viruses like plant-derived viruses, giant viruses, and bacteriophages. Recently metagenomic methods have allowed to reconstitute entire viral genomes from the genetic material spread in the human gut, opening new avenue on the understanding of the gut virome composition, the importance of gut microbiome, and potential clinical applications. This chapter aims to shed light on the latest evidence on human gut “virome” composition, interaction, its function, and possible future therapeutic applications in human health.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The human microbiome contains communities of commensal, symbiotic, and pathogenic bacteria, archaea, viruses, and small eukaryotes that actively interact with one other and/or with the host to maintain homeostasis (Nicholson et al. 2012). The microbiome acquired at birth is shaped as a result of multiple factors, including the mode of delivery, breastfeeding, solid food, and other environmental exposures (Dominguez-Bello et al. 2010). The human gut microbiome harbors genes that are involved in nutrient synthesis, amino acid metabolism (Lin et al. 2017), carbohydrate metabolism (Rowland et al. 2018), and lipid metabolism (Wang et al. 2016). Other factors such as host genetics (Blekhman et al. 2015), dietary habits (David et al. 2014), lifestyle (Clemente et al. 2015), comorbidities (Nagpal et al. 2018), chemotherapy (Montassier et al. 2015), and antibiotics (Francino 2016) can also disrupt the microbiome balance and may promote disease. The research on microbiome is actively directed to understand the microbial association, function, structure, and trans-kingdom interactions, as well as their cause and effect in the context of health and disease. The techniques used for the isolation and characterization of bacterial communities are relatively well-developed and standardized compared to those of the archaeal, eukaryal, and viral communities, and therefore, most of microbiome studies have been focused on the bacterial component of the human microbiome (Pollock et al. 2018). Nevertheless, characterization of viral communities can be more challenging than bacteria, archaea, and eukaryotes due to the absence of phylogenetically conserved genes. The shotgun metagenomic sequencing is the potential approach to characterize viral communities, a concept known as virome (Santiago-Rodriguez and Hollister 2019). The human virome is defined as a collection of all viruses that are found in human. The human virome comprises eukaryotic and prokaryotic viruses; viruses that cause acute, persistent, or latent infection; and viruses that can integrate themselves into the human genome (Fields et al. 2007). The human intestinal microbiota represents one of the most complex microbial ecosystems that is comprised of bacteria, viruses, fungi, multicellular parasites, and archaea (Moeller et al. 2016), which provide a heterogeneous surface area (>200 m2) for microbial life (Hooper and Macpherson 2010). It is estimated that the human gut contains between 30 and 400 trillion microorganisms from a variety and abundant bacterial hosts that can support a correspondingly rich and varied phage population (Ahmed et al. 2007; Zhang et al. 2014). The DNA and RNA viruses that collectively make up the intestinal virome are at least equivalent in number to the bacterial cells (Reyes et al. 2012), although on gut mucosal surfaces and within the mucus layers they may outnumber bacterial cells by 20:1 (Barr et al. 2013). This viral community encompasses an abundant and diverse collection of viruses that not only infect every domain of life (Eukaryota, Archaea, and Bacteria) but also dominated by viruses that infect and replicate within bacterial cells (bacteriophage or phage) (Reyes et al. 2010). The gut microbial community is now considered to be intimately involved in our health, providing a wide range of beneficial functions such as extraction of additional energy from our diet, shaping the immune systems, and protection from invading pathogens. Additionally, it has emerging roles in modulating mood, behavior, neurocognitive development, and even the aging process (Zoetendal et al. 2006).
1.1 Emerging View of the Human Virome
Until quite lately, human viruses were regarded as pathogens that are able to cause human pandemics and a large variety of illnesses that can cause high mortality rate. Recently, new human-associated viruses, known as human virome, have arisen with the advent of new sequencing technologies that enabled the study of the global viral population (DNA and RNA) in humans (Greninger et al. 2009; Allander 2008). Most of these high-throughput sequencing techniques, however, were conducted using filters with pore sizes varying from 0.2 to 0.45μm, which filter larger viruses, resulting in the human virome’s technical bias. The viral resources and biodiversity in the human body were commonly underestimated under nonpathological conditions. The microbial biodiversity of the human gut is regulated by human-associated viruses (Stern et al. 2012; Marinelli et al. 2012). The very basis of our being, our genome, is influenced by viruses. Reminiscences of human-viral ancestral cohabitation are imprinted in about 100,000 documents for endogenous viral fragments in our genome, comprising about 8% of our genome (Belshaw et al. 2004). Eventually, major physiological roles, such as mammal placental morphogenesis, have been associated with endogenous viral proteins (Mi et al. 2000; Blaise et al. 2003).
Phage-bacteria-human interactome deciphering has only been lately started to appear. The viral metagenomics study of the oral cavity of healthy individuals, reported by Willner et al., showed that phages constitute an essential reservoir for genes of bacterial virulence. These results, therefore, indicated that phages play a dual role in regulating the bacterial population while also contributing through horizontal gene transfer to bacterial pathogenicity and resistance (Willner et al. 2011). Several studies identified the persistent viral scattering from the gastrointestinal tract of dsDNA viruses of the Polyomaviridae family. In healthy children and adults, the polymerase chain reaction (PCR)-based identification of the BK, JC, and SV40 viruses has been reported (Vanchiere et al. 2009). Compared with adults, viral identification was more common in stool samples from infants. These results highlight the hypothesis that the gastrointestinal tract could be a site of persistence of polyomavirus with a potential fecal-oral viral transmission pathway.
In the normal gut viral flora, several RNA viruses, commonly regarded as human pathogens, have also been identified. The existence of many eukaryotic viral families, such as Astroviridae (Gabbay et al. 2005; Méndez-Toss et al. 2004), Caliciviridae (Barreira et al. 2010; Ayukekbong et al. 2011), Picornaviridae, Reoviridae, and Picobirnaviridae, as well as plant viral families, such as Virgaviridae, has been discovered by PCR-based or by metagenomic studies on “healthy” human feces. Reoviridae and Picobirnaviridae are two gastroenteritis-responsible families of dsRNA viruses, but both may occur in healthy humans. For instance, in developing countries, rotaviruses (Reoviridae, genus Rotavirus) are the major cause of death among children below the age of 5. Some genotypes, such as G10P strains, have commonly been correlated in India with asymptomatic neonatal infections (Gómara et al. 2004). The existence of plant viruses in the human gut indicates that, as shown for bacteria, the virome would differ between individuals based on diet (Turnbaugh et al. 2009). Gut virome can also rely on environmental factors, including geography, eating patterns, or ethnic differences, resulting in heterogeneity between individuals. A persistent need for blood transfusions and medical care is reflected by human blood and related products. Blood, however, is also a major viral reservoir, and certain viruses can be pathogenic. Therefore, it has direct implications for public health to identify the viral flora in the blood. A growing body of evidence suggests (Nishizawa et al. 1997) that the blood is not sterile in healthy individuals and can carry several viral organisms. Most of “normal” blood viral flora consists of the most commonly detected ssDNA viruses of the Anelloviridae family of Torque teno viruses (TTVs). TTVs, originally found in a post-transfusion hepatitis patient in Japan, are small viruses of icosahedral symmetry that are non-enveloped and have a high level of genetic diversity. The first genus of Anelloviridae, Alphatorquevirus, currently comprises 29 species of TTV. Globally distributed TTVs are now known to be commensal (Virgin et al. 2009; Breitbart and Rohwer 2005; Biagini et al. 1998). Although replicative forms of TTV DNA have been discovered in peripheral mononuclear blood cells (Okamura et al. 1999), the bone marrow, lung, spleen, and liver have been described with viral loads higher than those in the blood (Okamoto et al. 2001). There have also been studies recording TTV mother-to-child transmission (Bagaglio et al. 2002).
The Parvoviridae family is another ssDNA virus family. The human parvovirus (PARV) was initially detected in the plasma of a person at risk for HIV infection (Jones et al. 2005). However, regular plasma identification of PARV4 and PARV5 has been documented in healthy blood donors as well as symptomatic individuals (Fryer et al. 2007). In blood donors, eukaryotic dsDNA viruses have also been identified. By analyzing blood from 400 donors, Egli et al. confirmed the prevalence of BK and JC polyomaviruses (Egli et al. 2009). Intriguingly, with respect to virus-host interaction and epidemiology, they reported crucial variations between the BK and JC viruses. In addition, lymphotropic polyomavirus and human bocavirus (HBoV) have also been frequently detected in immunocompromised and seemingly healthy individuals in the peripheral blood (Delbue et al. 2010; Bonvicini et al. 2011). Nevertheless, several studies reported the presence of some viral species, as the viral flora, in many parts of the human body such as the respiratory tract, teguments, the nervous system, and the genitourinary tract (Popgeorgiev et al. 2013).
1.2 Human Gut Microbiota Composition
The human intestine is colonized by several microbial strains after birth that fluctuate and change during our life span according to anatomical, dietary, and nutritional status changes (e.g., obese, anorexic, lean nutritional status) and environmental (e.g., climate, familial composition, lifestyle, working place, etc.), pathological (e.g., gastrointestinal and systemic infections), and pharmacological factors (e.g., use of antibiotics, prokinetics, laxatives, and probiotics). The main components of gut microbiota are bacteria, fungi, yeasts, archaea, viruses, and other Eukarya (such as Blastocystis and Amoebozoa) (Mai and Draganov 2009). Bacteria reach more than 1 kg of weight and account for more than 1100 species. Bacteroidetes and Firmicutes are the predominant phyla in adults, followed by Actinobacteria and Proteobacteria (Lozupone et al. 2012). Approximately 1013 bacterial cells and an average of ∼160 distinct species may reside in the adult human alimentary tract (predominantly in the colon), with over 1000 different bacterial species in total associated with the human gut microbiome (Qin et al. 2010). Little information is still known about commensal fungi, archaea, and protozoa. However, some emerging microbiological data on yeast composition and functions have clarified their subsequent clinical use in the modulation of gut microflora. In fact, Saccharomyces boulardii is currently used with significant efficacy over placebo in the treatment of post-infectious and post-antibiotic diarrhea (Dinleyici et al. 2012).
The last three decades of microbiological/clinical research have helped to understand how food, pre−/probiotics, and antibiotics can modulate the intestinal bacteria qualitative/quantitative pattern resulting in different microbial-host functions (Devaraj et al. 2013). The observations of an obese/lean gut microbiota associated with overweight or lean status clarified how microbiota manipulation by diet was possible and how microbiota could be responsible not only for overweight but also for the chronic inflammatory state typical of the metabolic syndrome (Met S) (Geurts et al. 2014). The diet and gut microbiota’s role in obesity pathogenesis is not simply causative as was initially expected. A recent report by Ridaura et al. (2013) showed how co-housing mice with an obese twin’s microbiota and with mice containing the lean co-twin’s microbiota prevented the development of increased body mass and obesity-associated metabolic phenotypes (greater polysaccharide metabolism and protein degradation) in obese cage mates. In a study by Qin et al., the role of diet in gut microbiota modulation showed to be strengthened by the recent metagenome-wide association in type 2 diabetic patients. Indeed, in a diet-associated insulin resistance status, the authors showed that these patients have a peculiar decrease in some butyrate-producing bacteria, an increase in various opportunistic pathogens, and an enrichment of other microbial functions conferring sulfate reduction and oxidative stress resistance (Qin et al. 2012). Based on these results, the possible functions of gut microbiota were quickly related to other organs. The previous association between spontaneous bacterial peritonitis and small bowel bacterial overgrowth in liver cirrhosis (Guarner and Soriano 2005) has led to the understanding of the microbial molecular patterns triggering inflammation and fibrosis in liver diseases such as non-alcoholic fatty liver disease (NAFLD) and its complications, i.e., non-alcoholic steatohepatitis (NASH), liver cirrhosis, and hepatocellular carcinoma (HCC) (Friedman 2013). Actually, gut bacteria seem to interact with the central nervous system (CNS) via the enteric nervous system through the endocannabinoid system. Thus, gut microbiota can affect the neuro-psychiatric state of the host, and also the CNS is able to affect its composition through food intake regulation (Cani et al. 2014).
2 Human Gut Virome Composition: Main Players
The concept of the existence of a “gut virome” has recently discovered (Mai and Draganov 2009), although the presence of viruses as pathogenic organisms in human intestine has been known and documented for more than a century (Lozupone et al. 2012). Recent studies described the temporal dynamics of the human gut virome. It appears that the symbiotic relationships between host and virome develop at a young age, with specific variations occurring during the first 2 years of birth, coinciding with environmental and dietary changes. As a result, individuals on the same diet showed similar gut virome composition (Minot et al. 2013). Norwalk virus, Rotavirus, and enterovirus are the well-known agents of gastroenteritis in man (Lagier et al. 2012). The reason we consider linking these pathogens with the gut virome is that the infection of the gut is responsible for enterocyte and bacterial microflora changes. These pathogens can affect the host in the acute phase of the infection with gastrointestinal complaints such as nausea, vomiting, diarrhea, and weight loss and also in the long-term persistence of symptoms and the possible eliciting of functional gastrointestinal disorders such as functional dyspepsia and post-infectious irritable bowel syndrome (Beatty et al. 2014). Although the viruses are the most numerous (about 1031 viral particles on earth and approximately 108 to 109 per gram of feces) and diverse microbial entities, there are relatively few studies that have focused on the association and function of viruses as part of the human microbiome (Robinson and Pfeiffer 2014). This is due to the challenges encountered in viral isolation, nucleic acid extraction, sequencing, and analysis pipelines (Mukhopadhya et al. 2019). The advent of high-throughput sequencing technologies permitted further insights into the human-microbe complex relationship that revealed the significant associations between microbial ecosystem shifts and disease (termed dysbiosis) and highlighted the diverse and abundant retinue of viruses intimately associated with the human gut microbiome (Breitbart et al. 2003). This human gut virome may be defined as the total population of viruses [or virus-like particles (VLPs)] associated with the underlying gut microbial community. In keeping with the dominance of bacteria in the gut microbiome, the gut virome appears to be predominated by prokaryotic viruses (bacteriophage) (Reyes et al. 2010). Both eukaryotic and prokaryotic viruses share lytic or latent life cycles, which allow different virome/host interactions and promote virus survival and evolution (Virgin 2014). As a result, human eukaryotic viruses can affect host physiology, mainly when chronically infecting particular sites, and virus-derived genetic elements can modify host gene and protein expression once integrated into host chromosomes (Foxman and Iwasaki 2011).
2.1 Eukaryotic Viruses
There are far fewer eukaryotic viruses than bacteriophages in the gut (Lim et al. 2015). According to metagenomic methods, novel enteric eukaryotic viruses were found to be responsible for acute diarrhea in children’s small bowel enteropathy in developing areas of Australia (Scarpellini et al. 2015). These new data were confirmed by quantitative real-time polymerase chain reaction (qRT-PCR), which showed that diarrhea in children contains a higher abundance of viruses, many of them not previously known to be pathogenic (Holtz et al. 2014). Sequencing of eukaryotic viral communities in fecal samples from children has identified Picobirnaviridae, Adenoviridae, Anelloviridae, and Astroviridae family members and species such as bocaviruses, enteroviruses, rotaviruses, and sapoviruses (Minot et al. 2013). Despite being fewer in number, these viruses also have significant effects on human health, both in healthy and immunocompromised subjects, causing acute gastroenteritis, acute enteritis, or colitis (Eckardt and Baumgart 2011). Picobirnaviruses have been found in stool samples from individuals with diarrhea of unknown etiology (Banyai et al. 2003), as well as in healthy subjects (Kapusinszky et al. 2012), leaving their pathogenic capability up for discussion. Among the RNA viruses found in the gut, a prevalence of plant viruses has been introduced in the diet (Minot et al. 2011). In addition, disease-associated viruses such as herpesviruses, polyomaviruses, anelloviruses, adenoviruses, papillomaviruses, polyomaviruses, hepatitis B virus, hepatitis C virus, and human immunodeficiency virus (HIV) are also present in the intestinal viromes of some individuals, indicating that the gastrointestinal (GI) tract contains viruses capable of infecting host cells. As the majority of humans remain asymptomatic, it has been proposed that these pathogenic viruses (pathobionts) have become part of the metagenome of normal individuals, with the majority rarely causing disease and remaining dormant within the host (Hunter 2013). According to experiments in germ-free and antibiotic-treated mice, bacterial microbiome can promote the replication and, in some cases, persistence of enteric viruses (Pfeiffer and Virgin 2016), with the efficient transmission of mouse mammary tumor virus requiring intestinal bacteria (Kane et al. 2011). Thus, the interactions between viruses and bacteria, and other constituents of the intestinal microbiome, are important in influencing the course and outcome of virus infections (Almand et al. 2017).
2.2 Bacteriophages
In recent years, studies of the human gut virome have mainly focused on the analysis of virus-like particles (VLPs) purified from fecal samples and the application of high-throughput metagenomic approaches to characterize these viruses (Reyes et al. 2012). Several studies have provided insights into the diversity and structure of the gut virome, which is likely to reflect the underlying diversity of the bacterial microbiome (Qin et al. 2010). Intestinal bacteriophages (prokaryotic viruses) were recently considered as the main component of the gut virome, accounting for about 90% of its composition (Lozupone et al. 2012). Bacteriophages can be quite literally defined as “viruses of bacteria.” Bacteriophages are commonly known as “bacterial parasites” that inject their genome in their host, integrating with its genetic material (prophage state) and inducing synthesis of other phage particle with bacterial cell lysis (lytic state) (Mokili et al. 2012). Double-stranded DNA phages from the order Caudovirales (Podoviridae, Siphoviridae, and Myoviridae) as well as single-stranded DNA phages from the family Microviridae constituted the bacteriophage component of the gut virome (or phageome) (Minot et al. 2011). The Microviridae family was initially considered as secondary players in the environmental viral community because of their modest genome size. Microviridae are small icosahedral viruses with circular single-stranded DNA genomes, and their members are divided into microviruses (genus Microvirus) and gokushoviruses (subfamily Gokushovirinae) (Wegley et al. 2007). These viruses have been retrieved in bacteria belonging to two genera of the phylum Bacteroidetes: Prevotella and Bacteroides (Roux et al. 2012). The virotypes mostly infect bacteria belonging to the most prevalent phyla within the gut, comprising members of Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Manrique et al. 2017). The adult gut virome may be dominated by just one or a few different virotypes and is characterized by a high degree of stability in terms of its structure over time, with temporal tracking of gut virotypes, revealing the retention of between 80 and 95% of virotypes over a period of 1–2.5 years (Reyes et al. 2012).
Microbial viruses modulate their bacterial hosts directly through affecting their mortality and through horizontal gene transfer and indirectly by reprogramming host metabolism (Dalmasso et al. 2014). The human GI tract contains an estimated 1015 bacteriophages (phages; the phageome) that may represent the richest concentration of biological entities on earth (Dalmasso et al. 2014). Phages can be functionally categorized based on their life cycle after infecting host cells (Weinbauer 2004). Lytic (virulent) phages lyse the infected cells by hijacking the host cell’s replication mechanism to package and produce more phages and lytic enzymes that cause cell lysis to release the newly formed phages (Mukhopadhya et al. 2019). After that, temperate phages incorporate their genetic material into the host cell chromosome as prophages and replicate alongside the host cell (Girons et al. 2000). Recently, analysis of the viral fraction of existing metagenomic studies identified a DNA phage called crAssphage that is highly abundant in the gut microbiome. It has been predicted, based on host co-occurrence profiling, that crAssphage infects Bacteroides species (Guerin et al. 2018). Prokaryotic viruses are known to influence human health by affecting bacterial community structure and function (Reyes et al. 2010). However, the intricate pathways by which this influence is exerted are yet to be fully clarified. Nevertheless, it has been shown that (1) temperate phages are common; (2) bacteriophages vary widely between individual hosts but not within a single subject; and (3) the variety of bacteriophages present increases in adulthood, and the diet affects the composition of phage communities (Minot et al. 2013).
3 Virome Functions Within the Human Gut Microbiome
Up to date, there are few clear data about gut virome functions within the gut microbiota ecosystem, although the life cycle of viruses provides indirect information about their possible roles. The great part of the phages found in the human gut shows atypical “temperate” behavior, which justifies the hypothesis that their composition is quite stable during the host’s life (Lozupone et al. 2012). Furthermore, several authors have used the terms “stability” and “variability” to define phage behavior. The two kinds of these characteristics, belonging to the bacteriophages in the intestine, are linked as in a “virtuous” cycle. In fact, the stability of the viral genome is responsible for that of other microorganisms, such as the bacteria of the gut microbiota (Hofer 2013). This is proved by the fact that gut virome composition mimics the evolution of the infant bacterial microbiota (Palmer et al. 2007). These are common between phages and bacteria and are implicated in bacterial wall adhesion and immunoglobulin receptor synthesis, contributing to maintaining viral-bacterial immune tolerance in the gut. This allows the persistence of bacterial and viral species in the gut, exerting their effects on enterocytes and on the host (Liu et al. 2002). On the other hand, the presence of one intrinsic variability, typical of the few lytic phages found in the intestine, is an interesting characteristic of these viruses, which allowed the generation of new species in a short time frame and allowed them to escape extinction (Hofer 2013). Indeed, among the genes stably conserved during intestinal viral evolution discovered by metagenomics, there are also those involved in energy harvesting such as for carbohydrate transport and degradation (Markine-Goriaynoff et al. 2004). So these properties are common to diet-derived viruses of plants that can modulate human bacterial microbiota/host metabolism (e.g., carbohydrate synthesis/degradation, protein synthesis) (Lagier et al. 2012). According to “Darwin postulate” on animal species survival, the most common genes mapped by the largest part of sampled individuals were that responsible for DNA replication and repair, namely, a feature of “adaptation for survival” (Waller et al. 2014). As a result, gut virome has a significant impact on our gut microbiome and may potentially play a role in human genome maintenance over the generations. Among the genes encoded in cryptic prophages of Escherichia coli, those for resistance to antibiotics and other stress factors have been found (Kumarasamy et al. 2010). These explain the strict interaction between viral and bacterial particles in the intestine, which lead to the classical concept of bacteriophages as “predators” of bacteria. In fact, the transmission of genes between virus and the infected bacteria helps the host to resist oxidative stress and antibiotic use, another proof of the “temperate” lifestyle of the gut virome (Scarpellini et al. 2015).
4 Tools for Human Virome Identification
The absence of validated protocol to study virome might be due to the numerous obstacles in virome study such as viral diversity, host contamination, and lack of common conserved sequence within virus genome (Krishnamurthy and Wang 2017). Plaque assay is a culture-based technique and has been used to study and quantify phages and to detect their host range in environmental samples (Hamdi et al. 2017). However, culture-based approaches are not suitable to study viruses in a complex ecosystem such as human gut (Sutton and Hill 2019). Recently, the development of the metagenomics allowed the study of the natural viruses within complex microbial samples (Ye et al. 2019). Metagenomics is a molecular-based technique of non-culturable organisms used to study environmental samples containing complex of microbes by analyzing their genomes based on function and sequence (Riesenfeld et al. 2004). Most studied viromes are DNA and very low RNA viruses; this is because of the high mutation rate of RNA viruses and lack of standard amplification and bioinformatic tool (Marz et al. 2014).
The main steps in metagenomics include sampling, homogenization, filtration, concentration of the purified viral particle, extraction and amplification of viral nucleic acid, genome sequencing, and, finally, assembly and data analysis (Fig. 4.1) (Kumar et al. 2017; Manoussopoulos and Anastassopoulou 2020). Individualization and combination of various steps were used according to sample source and isolated virus to reach high viral titer and low host cell contamination. Different approaches used in metagenomics are discussed in this section.
4.1 Sampling
The selection of sample type and site is crucial starting in the studying and isolation of virome. Many common clinical samples yielded low abundance viruses and relative noise background from other microbiome and host cells (Haynes and Rohwer 2011; Rascovan et al. 2016).
The most studied human part containing a huge number of natural viruses is the gut and stool sample (Haynes and Rohwer 2011; Popgeorgiev et al. 2013; Rascovan et al. 2016). Other studied biopsies containing viromes include skin, respiratory tract, genitourinary tract, nervous cells, and blood (Popgeorgiev et al. 2013). Collected samples should be homogenized with isotonic buffer and then purified immediately or kept at −80 °C to prevent the change of the ratio between different present microbes (Shkoporov et al. 2018). Fecal material stored at −80 °C was found to be very stable for several years and reproducible to virus particles (Reyes et al. 2010).
4.2 Genome Purification and Concentration
Viral DNA represents about 2–5% of total microbial DNA found in human (Reyes et al. 2010). For that, it is very challenging to choose the most suitable technique or combination of techniques for the purification of high titer of viral genome and isolate all types of viruses, while decreasing the contamination of other microbial and host genome (Vibin et al. 2018). Purification and concentration of samples containing high density of virus-like particles (VLPs) are achieved by resuspending the sample in an osmotically neutral buffer. After that, separate viral small particle from other large contaminated particles by filtration and/or centrifugation. The used protocol is greatly affecting the viral purified rate and purity (Castro-Mejía et al. 2015). Furthermore, the study of active and silent virome can be achieved by using both extraction methods of VLPs and total nucleic acid isolation (TNAI) (Garmaeva et al. 2019).
4.2.1 Tangential Flow Filtration (TFF)
This technique is used to recover and concentrate the VLPs from large sample volume and low viral density using a 0.2-μm filter followed by concentration using an ultra centrifugal filter. However, this tool renders the isolation of large virus particles and thus decreases the yield of isolated viruses (Castro-Mejía et al. 2015). For that, the use of a bigger pore size filter (0.45μm) was recommended to get high viral DNA titer, but at the same time this elevates the contamination with host particles (Hoyles et al. 2014). TFF is one of the most common methods used for purification of viruses from bacteria and host cells (Castro-Mejía et al. 2015).
4.2.2 Cesium Chloride (CsCl) Density Gradient Ultracentrifugation
This method is characterized by removing the host DNA contamination to produce highly pure viral isolate. This technique has several drawbacks, including the fact that it is not repeatable, labor-intensive, and biassed in isolating a specific type of virus based on density range (Callanan et al. 2018; Shkoporov et al. 2018). Furthermore, attention was needed while using CsCl in quantitative virome studies (Kleiner et al. 2015).
4.2.3 Precipitation with Polyethylene Glycol
This method is used when large sample volume is present. The polyethylene glycol (PEG) was added to the sample followed by centrifugation, filtration, and dialysis. Precipitation approach is very useful and more effective than TFF in virome extraction (Castro-Mejía et al. 2015).
4.2.4 Chloroform and Nuclease Treatment
This method favors small circular genome than large linear one and is more successful with DNA viruses. Chloroform disrupts the lipid layer of a bacterial cell; then the free bacterial genome is removed by the addition of DNase and RNase to decrease sample contamination (McLaughlin et al. 2006). However, chloroform alters the stability of some enveloped and non-enveloped viruses (Conceição-Neto et al. 2015).
4.2.5 Flow-Cytometry-Based Methods
Flow cytometric analysis is used for counting the stained phage particles (Brown et al. 2015) or to fractionate the phage from mixed microbial sample such as human fecal sample according to its size and fluorescence intensity using fluorescence-activated cell sorting (FACS) (Džunková et al. 2015). The main concept of flow cytometry is using a fluorescent dye such as SYBR Green II for labeling of the VLPs and then separation (Roux et al. 2016). This method decreases significantly the host and bacterial contamination and is an added value to neglect the amplification step of genome before sequencing (Shkoporov and Hill 2019). The main limitation here is the low sensitivity in virus detection, thereby leading to great loss of many viruses within the sample (Warwick-Dugdale et al. 2019).
4.3 Extraction and Amplification of VLP-Derived DNA
The nucleic acid of viruses was extracted after purification step/s, but with an abundance level under the limit needed for sequencing. To overcome low viral genome abundance, numerous amplification methods are used. Viruses with DNA genome are the most prescribed gut virome in humans. In RNA viruses, the RNA genome is converted to cDNA by reverse-transcription PCR (RT-PCR) with random primers followed by PCR amplification (Emerson et al. 2018).
4.3.1 Multiple Displacement Amplification (MDA)
MDA is a fast and sensitive isothermal approach; it amplifies DNA viral genome before sequencing using Phi29 polymerase and random hexamers (Angly et al. 2006). MDA amplifies large amount of the whole viral genome (WVG) of both single-stranded DNA (ssDNA) and dsDNA. The limitation is the overamplification of small circular ssDNA viruses but low presentation of high GC content viruses (Kim et al. 2008). This method favors small circular genome than large linear one and is more successful with DNA viruses (Chen et al. 2014a; Roux et al. 2016).
4.3.2 Linker Amplified Shotgun Library (LASL)
LASL is used to amplify DNA based on PCR. The viral DNA template is ligated with dsDNA linker and then amplified using Vent DNA polymerase, ligated again into vector for cloning and finally electroporated. This approach overcomes the problem of bactericidal genes within viral DNA and the use of modified nucleotides. However, the main disadvantages of using PCR are as follows: only double-stranded genome can be amplified, and large DNA concentration is required, which leads to mixed amplification templates (Breitbart et al. 2003; Hindiyeh et al. 2019; Reyes et al. 2012).
4.3.3 Random Amplified Shotgun Library (RASL)
RASL method, like LASL, is based on the use of thermal cycler but with the use of random primers to amplify DNA in randomly amplified shotgun libraries. This procedure is rapid, very useful to amplify nanograms of DNA with non-culturable viruses, and suitable for shotgun sequencing (Rohwer et al. 2001).
4.3.4 Adaptase-Linker Amplification (A-LA)
A-LA is one of the most used genome amplification tools for both ssDNA and dsDNA templates. This method involves the use of adaptase before the linker amplification (Roux et al. 2016).
4.3.5 Virus Discovery cDNA-AFLP (VIDISCA) Technique
This method is a cDNA amplification approach that doesn’t require prior genome sequencing (de Vries et al. 2011; van der Heijden et al. 2012). ViSeq, for example, is one of these amplification techniques which is sensitive and give quantitative results that uses adapter-specific primers to document the viral entire genome in humans (Cotten et al. 2014).
4.3.6 Fully Automated Virus Extraction
MagNA Pure 96 and eMAG are automated, sensitive, and specific viral genome extraction tools (Hindiyeh et al. 2019).
4.4 Sequencing Strategies
Next-generation sequencing method enables the sequencing and identification of unculturable novel viruses. This technique represents the basic step in metagenomic studies (Reyes et al. 2012).
4.4.1 Pyrosequencing
This technique is based on the detection of the released pyrophosphate by pyrosequencing. This yields a long genome length of 400 to 500 nucleotides each run (de Vries et al. 2011).
4.4.2 Illumina Sequencing
Based on the emitted fluorescent sequence unique to each base, Illumina sequencing simultaneously identifies the DNA bases. The HiSeq2000 platform (Xie et al. 2016) and the MiSeq deep sequencing platform (Cotten et al. 2014) are two examples of Illumina sequencing technique.
4.5 Quality Control
Quality control measurements should be applied to overcome the limitations of different approaches and the absence of valid protocol to study virome. The main boundaries during the study of human viromes are the contamination with host genome, abundance of viral genome, and detection of prophages (Sutton and Hill 2019). For that, the quality of sequenced sample, based on the mentioned obstacles, should be checked before the assembly and annotation.
4.5.1 Viral Quantification
4.5.1.1 Epifluorescence Microscopy
It is a rapid, simple, and reproducible tool to count the number of viruses. The sample was stained by SYBR Gold after filtration; then the slides were examined under the microscope, images were captured, and finally, the viral titers were calculated (Patel et al. 2007; Thurber et al. 2009).
4.5.1.2 Transmission Electron Microscope (TEM)
Tem is an old but labor-intensive method. the samples are fixed and stained using a negative staining technique, and then electron micrographs are taken to estimate the number of virus particles (Hoyles et al. 2014).
4.5.2 Prophage Identification Applications
Many automated and computational applications are present to predict prophages in host genome, and their main approach is to detect a sequence similar to known sequence of viral genome. Phage-finder (Fouts 2006), other program was developed based on the detection of prophages by analyzing the difference of dinucleotide relative abundance (Srividhya et al. 2007), Prophinder (Lima-Mendez et al. 2008), PHAge Search Tool (Zhou et al. 2011), PhiSpy application that identifies de novo or known virome based on similarity and composition analysis (Akhter et al. 2012), and VirSorter that can detect prophages in complete and fragmented (meta)genomic databases (Roux et al. 2015).
4.6 Computational Approaches for Characterizing Sequenced Viromes
Bioinformatics is the most used method to analyze the produced data. The choice of annotation or assembly software had a great impact on virome analysis outcome more than sequence technology used (Sutton et al. 2019). It could be divided into similarity-dependent and similarity-independent approaches (Haynes and Rohwer 2011). The main advantages of approaches are as follows: they are fast and simple and no special reagents are needed. Furthermore, they are able to distinguish between novel and known viruses by the conserved region on the genome (Delwart 2007).
4.6.1 Similarity-Dependent or Reference-Based Approach
This is the original and most used method to analyze the sequence data of microbiome by database search for finding segment similarity. The limitations of this tool in virome study are that they are not applicable in studying de novo viruses and the high diversity of viral genome makes the virus not similar to any known sequence (Brister et al. 2015). For similarity search, BLAST is the most used tool, which is based on the sequence of nucleic acid and amino acid (McGinnis and Madden 2004). In addition, microarray hybridization pattern is used to characterize new viral nucleic acids by comparing the shared sequences (Urisman et al. 2005).
4.6.2 Similarity-Independent or De Novo Approaches
In this approach, there is no need for database search, while more focus on the viral dark matter is applied. Phage Communities from Contig Spectrum (PHACCS) is a program for the detection and modeling of de novo viral diversity by using the spectrum of contig from the sequenced data (Angly et al. 2005). Numerous programs are used in the assembly of human virome, while some approaches are used to diagnose particular taxa based on GC % in the genome or dinucleotide combination rate (Sutton et al. 2019; Willner et al. 2009). SPAdes (meta) software (Nurk et al. 2017) showed the best accuracy and genome recovery (Sutton et al. 2019). However, the main limitation of de novo approaches is the finding of the lysogenic or silent virome from host genome (Delwart 2007).
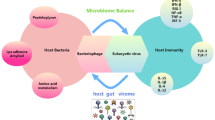
5 Virome-Associated Dysbiosis
Imbalances in the makeup of gut microbiome, also termed dysbiosis, are now increasingly linked with a wide spectrum of diseases and disorders (both gut associated and those relating to extra-intestinal organ systems). These range from inflammatory bowel diseases, cancer, to metabolic disorders, obesity, and even autism and Alzheimer’s (Lynch and Pedersen 2016). Emphasis is now being placed on delineating whether dysbiosis of the microbiome is a cause or consequence of some of these diseases and how manipulation of the gut microbiome may aid prophylaxis, diagnosis, or treatment (Sommer et al. 2017). The large part of the gut microbiota is composed of viruses and contracts both prokaryotic and eukaryotic cells to form the gut virome (Santiago-Rodriguez and Hollister 2019). As the role of the virome in the gut microbial community has been started to be uncovered, evidences have highlighted that this viral community also reflects the driving diversity and functionality co-evolution of host and microbe within the gut (Koskella and Brockhurst 2014). Recognition of the potential of phage to drive ecological functioning and evolutionary change (Koskella and Lively 2009) has understandably ignited interest in investigating the role of these prokaryotic viruses within the human gut virome and as part of the human gut microbiome. The concept of dysbiosis and the impacts of such perturbations on human health have begun to be considered from the perspective of the virome or phageome. There is a growing consensus that this concept should also be extended to the phage component of the gut ecosystem (Dalmasso et al. 2014). Although translational animal models are used to understand cause-effect relationships, virome analyses of the human gut are providing evidence on the potential role(s) of viruses in maintaining homeostasis or promoting disease (Fig. 4.2).
5.1 Type 1 Diabetes (T1D)
Type 1 diabetes (T1D) is a proinflammatory disease that targets beta cells of the pancreatic islet, resulting in a loss of these cells. In this disease, the genetic and environmental factors are incorporated, including complex genetic elements, patient exposures, and the gut microbiome. Viral infections and broader gut dysbioses have been identified as contributing factors or potential causes of T1D. However, human studies have not yet identified microbial functional or compositional triggers that are predictive of T1D or islet autoimmunity. T1D can be also known as the presence of antibodies against beta-cell autoantigens including insulin, zinc transporter 8, and islet antigen 2 (Morahan 2012). Any changes in the lifestyle of the person or changes in the dietary habits are known to alter the gut microbiome, suggesting that altered gut microbiome composition might be associated with T1D progression (Needell and Zipris 2016). In comparison to T1D patients, bacteria such Prevotella, Faecalibacterium, Eubacterium, Fusobacterium, Anaerostipes, and Subdoligranulum have been found to be common in healthy controls (Brown et al. 2011). In contrast, Lactobacillus, Lactococcus, Bifidobacterium, and Streptococcus were more abundant in T1D subjects. Researchers found that viruses have been implicated with T1D by triggering human T1D but the relationship between infections and progression of disease has not been established (Zipris 2008; Ghazarian et al. 2012; de Beeck and Eizirik 2016). Previous reports have demonstrated that viruses and virus-specific antibodies can be found frequently in a person with recent diabetes onset compared to a healthy person (King et al. 1983; Dotta et al. 2007). The viruses with reported implication in human diabetes are mumps virus, cytomegalovirus, rotavirus, rubella virus, Epstein-Barr virus, and varicella zoster virus (Zipris 2009). Enterovirus and Coxsackie B virus play a particular role in triggering the destruction of beta cell (Jaeckel et al. 2002; Jun and Yoon 2003). A virus infection was observed to harm the pancreatic islets of Langerhans in T1D murine models, suggesting a link between the virus and T1D model (Filippi and Herrath 2008). In 1993, an increase in diabetes rate was observed in young children notably after 2 years of released measles epidemic, suggesting that the increased T1D rate might have been associated to the measles outbreak (Lipman et al. 2002). To date, it is unknown how viral infections lead to T1D. It might be that the viral infection leads to diabetes due to a number of mechanisms, including molecular mimicry, bystander activation of T cells, beta-cell damage resulting in autoantigen release and activation of autoreactive T cells, and the induction of stress pathways in beta cells (Ghazarian et al. 2012) (Fig. 4.2).
5.2 Type 2 Diabetes Mellitus (T2DM)
Type 2 diabetes mellitus (T2DM) is a metabolic disorder distinguished by a high blood glucose level due to insulin resistance (IS) (Karim et al. 2014). T2D progression is associated with many factors, especially both genetic and environmental factors, in particular dietary habits, and most recently has been associated with change in microbiome composition (Larsen et al. 2010). Continuation studies identified several Clostridium spp., as well as Bacteroides intestinalis, Akkermansia muciniphila, and E. coli being enriched in subjects with T2D (Qin et al. 2012). Based on these data, an association was found between specific members of the human gut microbiome and T2D. Additionally, an association was observed between T2D and those viruses belonging to the Siphoviridae family, including Lactobacillus, Pseudomonas, and Staphylococcus (temperate and some are strictly lytic) (Ma et al. 2018). Viral infection is one of the important causes of diabetes, liver damage, neurological disorders, and several other diseases worldwide. The most important viruses related to the T2DM are HSV, hepatitis viruses, West Nile virus (WNV), influenza viruses (H1N1, H5N1 serotypes), picornavirus, cytomegalovirus (CMV), enterovirus, and Borna disease virus (BDV) (Cadranel et al. 2008; Carter 2010). Herpes simplex virus type 1 is a member of herpesvirus family. The HSV genome is large, double stranded, containing 74 genes, and encased within an icosahedral capsid protein, which is covered by an envelope (Mettenleiter et al. 2006; McGeoch et al. 2006). Recent clinical and epidemiological studies have reported association between HSV infection and T2DM (Cadranel et al. 2008; Ott 1999; Kroner 2009). Chronic inflammation is closely and early involved in the pathogenesis of T2DM (Pickup 2004; Helmersson et al. 2004). Cytomegalovirus (CMV) belongs to Herpesviridae family, and it is extremely a cause of human infections (Sweet 1999; Koichi et al. 2007; Ryan and Ray 2004). Human CMV infects ~40% of the world population, especially elderly people, without causing any specific symptoms (Offermanns and Rosenthal 2008). Reports have showed that CMV harms cells in the pancreas and causes T1D and T2D. Due to T1D, the immune system becomes weak which favors more susceptibility to infection with CMV, and chances of acquiring T2DM become 12 times higher (Rachael 2012; Sarah 2012). Enteroviruses are members of the Picornaviridae family and cause infections to humans and other mammals (Li et al. 2005). A study involving a group of patients diagnosed with T2DM were compared to others without T2DM explored the link between enterovirus infection and diabetes. It was observed that 40% of the pancreas of people with T2DM contained the enteroviral protein. The presence of enterovirus in the pancreas of people diagnosed with T2DM was three times higher than that in those without it (Richardson et al. 2009) (Fig. 4.2).
5.3 Inflammatory Bowel Disease (IBD)
Inflammatory bowel disease (IBD) is an immune-mediated disease and also defined as a chronic illness of childhood that causes inflammation to the gastrointestinal tract and includes Crohn’s disease (CD) and ulcerative colitis (UC). IBD requires medication in some cases, and in extreme cases, surgery is required. Current research suggests that several factors including genetic, environmental, autoimmune, dietary, and microbial play a role in the pathogenesis of IBD (Adamiak et al. 2013; Eszter Muller et al. 2014). Bacteria, fungi, and viruses are enteric microbiome purported that play a role in the pathogenesis of IBD (Wang et al. 2015). Bacteria can differ based on the kind of IBD (CD versus UC) and the operation has been performed (Halfvarson et al. 2017). In general, these bacteria include Faecalibacterium prausnitzii, Roseburia spp., Bifidobacterium spp., and Lactobacillus spp., as well as E. coli, Oscillospira, and unclassified Ruminococcaceae (Morgan et al. 2012; Celiberto et al. 2018). Various studies have investigated the association between the enteric virome and IBD. Early studies to estimate that link showed an increased abundance of phages infecting bacterial orders in subjects with IBD, including Alteromonadales, Clostridiales, and Clostridium acetobutylicum, and also elevated viruses from the Retroviridae family (Pérez-Brocal et al. 2015). Another study looked at the gut virome of the mucosa of a murine model of colitis and found that the quantity of viruses from the Caudovirales order had increased, but the variety had decreased (Duerkop et al. 2018). Interestingly, phages infecting enterobacteria were safely more represented in mice with colitis. Increased abundance and decreased diversity of phages are in agreement with a reduced number of phage-related functions related with UC (Duerkop et al. 2018). These results are totally opening the possibility of examining therapeutics to target the virome in IBD subtypes. An interesting study reported by Norman et al. detected disease-specific changes. The gut virome was less presented in both CD and UC. The primary difference in the IBD-associated virome was the increased abundance of Caudovirales phages on a taxonomic level; however, the exact viruses that may perform such change were different in CD when compared to UC (Norman et al. 2015). All data obtained by Norman et al. was recently reanalyzed by Clooney et al. The results confirmed IBD-specific changes in the virome, loss of the “core phageome,” and the stimulated development of phages in patients with CD (Clooney et al. 2019). Moreover, the changes in the structure of virome reflected shifts in bacterial structure. Indeed, both virome and bacteriome alterations were more obvious in patients with CD when compared to those with UC, which reflect the disease severity. Hence, integrating both bacteriome and virome assessment offers higher classification power between healthy and unhealthy states in IBD (Clooney et al. 2019) (Fig. 4.2).
5.4 Cancer
Pioneering research studies that linked microorganisms and cancer have led to enormous implications for public health. Helicobacter pylori and its relation to gastric cancer helped to classify this bacterium as a class I carcinogen (Santiago-Rodriguez and Hollister 2019). Hepatitis B virus, hepatitis C virus, human papillomavirus (HPV), HIV-1, Epstein-Barr virus (EBV), and human T-cell lymphotropic virus type 1 (HTLV-1) are viruses linked with different types of cancer that are also classified as class I carcinogens (Chen et al. 2014b). Some of these viruses are known as a part of the human virome due to their implications with cancer. However, certain viruses, including HPV, may be known as a part of the human virome as those are considered to be low and high risk, and those with unknown pathogenicity have been identified with no symptoms (Santiago-Rodriguez and Hollister 2019). Furthermore, only a small percentage of those subjects infected with high-risk HPV develop cancer (Van Dyne et al. 2018). Other viruses can also cause cancer including human polyomavirus (Prado et al. 2018). Knowledge regarding eukaryotic viruses and disease might lead to understand the relationship between disease and gut microbiome. This knowledge provides key information regarding the mechanisms during which viral infection could cause various cancer types, including chronic inflammation, immunodeficiency, and virus oncogenes. Furthermore, a growing number of research investigations have linked microorganisms and certain bacteria members to several malignancies, including melanoma (Matson et al. 2018; Routy et al. 2018; Gopalakrishnan et al. 2018), non-Hodgkin’s lymphoma (Montassier et al. 2015), cervical cancer (Lam et al. 2018), acute lymphoblastic leukemia (ALL) (Chua et al. 2017), and colorectal cancer (CRC). CRC is characterized by decreased bacterial variety in feces and mucosal samples (Wong et al. 2017). This decreased diversity is related to the absence of bacteria that may be implicated in preserving a healthy state and the detection of taxa associated with CRC and tumorigenesis, including Desulfovibrio spp., Bilophila wadsworthia, Fusobacterium nucleatum, Parvimona, Alistipes, and E. coli (Rubinstein et al. 2013; Veziant et al. 2016; Maisonneuve et al. 2017; Tilg et al. 2018). These bacteria are being considered as potential candidates for therapeutic approaches (Matson et al. 2018; Routy et al. 2018; Gopalakrishnan et al. 2018) and potential diagnostic. Although the eukaryotic viral infection of the enteric virome has been associated with CRC to a lower extent, phage communities have the potential to be the most effective in this context (Hannigan et al. 2018). Like bacteria, viral infection has been identified in CRC, mostly being bacteriophages from the Siphoviridae and Myoviridae families (Hannigan et al. 2018). Another study that did not isolate viruses specifically found >20 viral genera that distinguish between subjects with CRC and healthy controls (Nakatsu et al. 2018). Orthobunyavirus, Tunavirus, Phikzvirus, Betabaculovirus, and Zindervirus are eukaryotic viruses that were remarkably more represented in subjects with CRC (Nakatsu et al. 2018). Other viruses such as Fromanvirus seemed to be represented only in the healthy cohort. Also, the most abundant in subjects with CRC include the phageome, Streptococcus phage SpSL1, Streptococcus phage 5093, Streptococcus phage K13, Enterobacteria phage HK544, and Vibrio phage pYD38-A (Nakatsu et al. 2018). Alterations in the gut virome represent the opportunity to elucidate bacterial-viral-host interactions in promoting CRC (Fig. 4.2).
6 Communication Between Enteric Virome and Human Gut Probiotics: Implication on Gut Health
It is well described that gut microbiomes have a duty in the modulation of the immune system, keeping balance and preventing infections (Domínguez-Díaz et al. 2019). The prokaryotes, eukaryotes, archaea, and viruses (Laforest-Lapointe and Arrieta 2018; Mukhopadhya et al. 2019) living coexist in the intestinal lumen that is an optimal environment for microbial community interactions (Hillman et al. 2017). Hundreds or thousands of bacteriophage-bacteria pairs could interact at any time, making this hard to study (Sausset et al. 2020). The very recent study unveiling these interactions was reported by Marbouty et al. (2020) who used meta3C proximity ligation to analyze the phage-bacteria interaction in healthy human guts. In this study, they identified 6651 unique host-phage relationships. Half of the detected phages seemed to be lysogenic phages, and one-fourth represented potentially active phages (Marbouty et al. 2020). This outcome matches with previous reports that stated that the predominant state of a bacteriophage in the gut is the lysogenic state instead of the lytic state (Anthenelli et al. 2020).
At the moment, there is a sharp gap in interpreting the gut virome and gut prokaryote interaction, because the current models are not enough to translate this complex relation. However, some predictions have been done in this direction (Beller and Matthijnssens 2019).
6.1 Probiotics and Bacteriophages
Probiotics are “live organisms that, when administered in adequate amounts, confer a health benefit to the host.” Although traditionally probiotics, mainly Lactobacillus and Bifidobacterium, have been obtained from the gut and fermented foods, the next generation of probiotics (NGP) are based on commensal bacteria (Martín and Langella 2019). These commensal bacteria are colonizing human mucosal surfaces (Khan et al. 2019). The enteric virome might also be provided the health benefits of probiotics (Łusiak-Szelachowska et al. 2017), and they probably have a stabilizing role in the gut ecosystem (Draper et al. 2018).
The virome can interact with bacterial probiotics. For example, treatment with lytic and lysogenic bacteriophages increases the abundance of Lactobacillus and Bifidobacterium in mice (Bao et al. 2018). Also, the reduction of Lactococcus in Parkinson’s disease patients is associated with a higher abundance of Lactococcus-lytic phages in comparison to healthy controls (Tetz et al. 2018).
Lactobacillus reuteri is a probiotic that produces antibiotics and can shape the commensal microbiota in the gut (Mu et al. 2018). Nearly all human strains of these bacteria contain active prophages. Studies on an L. reuteri harboring two active prophages from the Siphoviridae family showed that prophages reduce the fitness of the bacteria during gastrointestinal transit. The phages are induced in the distal intestinal tract rather than in small intestinal regions in an SOS-dependent manner. The phage production provides a competitive advantage by killing a competitor strain (Oh et al. 2019).
6.2 Strategies of Interactions Between Prokaryotes and Viruses in the Gut
Bacteriophage predation and lysogenic conversion play an important role in the regulation of bacterial biomass and microbial diversity (Shkoporov and Hill 2019). However, in the gut, there is no observation of biomass control from phages, and the virus-to-microbe ratio remains low (Shkoporov and Hill 2019). This might be explained by the recently proposed “piggyback-the-winner” strategy. This strategy proposes that phages take advantage of the high microbial abundance and growth rates of their hosts by remaining integrated in them (as a prophage) (Anthenelli et al. 2020; Guo et al. 2020; Silveira and Rohwer 2016). This strategy, observed in the gut and mucosal surfaces, is different to the models explaining the interaction and co-evolution between virus and hosts in other ecosystems, like the ocean where the phage/bacteria ratio is 10:1 (Maurice 2019). These strategies are as follows: first, “the arms race” model - in which the host acquires a mutation that makes it resistant to the virus, but then the virus acquires a mutation that allows it to reinfect the new resistant population (Avrani et al. 2012). Second, the “kill-the-winner” model in which the growth of the most active bacteria population is controlled by a virus and, therefore, there is an increase in the diversity of microbial communities (Maslov and Sneppen 2017; Winter et al. 2010).
Guo et al. (2020) proposed that the interactions between phages and hosts follow the piggyback-the-winner strategy (Guo et al. 2020). Research comparing the gut virome of stunted and non-stunted children demonstrated that non-stunted children had more temperate phages than stunted children. In the latter, microbial interactions could be following the “kill-the-winner” strategy (Mirzaei et al. 2020). An alteration in the abundance of lytic phages in comparison to temperate phages could lead to the development of diseases such as Parkinson’s disease (Tetz et al. 2018).
6.3 Role of Lytic Phages in the Gut
Bacteriophages might control and regulate the abundance of bacteria in the gut (Bao et al. 2018), but little is known about the dynamics of phage predation in the human gastrointestinal tract (Hsu et al. 2018). Phages in the gut, apart from having an impact (knockdown) on the population of susceptible bacteria, might also have an impact on other microbial populations after a cascade effect caused by inter-bacterial interactions (Hsu et al. 2018). Although Hsu et al. (2018)) worked with phages with a narrow host range, recent studies pointed out that in the ecosystems, there are more bacteriophages with a broad range of hosts than previously thought (de Jonge et al. 2019). In relation to this, Marbouty et al. (2020) found out that most gut phages are specific to their host but nearly one-third of the identified phages showed contact with more than one MAG (Marbouty et al. 2020).
To understand the bacteriophage-host interaction, it is necessary to know what the host target of a bacteriophage. To predict this, several computation analyses have been used such as sequence homology, CRISPR spacers, occurrence profiles (Edwards et al. 2016), and abundance profiles (Stern et al. 2012). Recent research has found out that the abundance of phages is related to the abundance of susceptible bacteria (Oh et al. 2019). Programs such as VirHostMatcher (Ahlgren et al. 2017) and WIsH (Galiez et al. 2017) have shown high accuracy with the predictions.
6.4 Horizontal Gene Transfer Between Bacteriophages and Bacteria
The viruses that inhabit the gut are involved in horizontal gene transfer to prokaryotes (Mukhopadhya et al. 2019). This mechanism is determinant to shape complex ecosystems, including the gut, and acquire important genes (Callier 2019; Sutton and Hill 2019). The dynamics of how phage-mediated HGT occurs and regulates the evolution of bacteria in this ecosystem have been recently studied by Frazão et al. (2019)). Their experiment in a mice’s gut showed that in the presence of a resident E. coli, the colonization of an invading E. coli can be successful if it adapts via HGT followed by mutation. The invader E. coli showed a rapid evolution, and it acquired two new genomic regions from the resident E. coli. They corresponded to two complete prophage regions, named KingRac and Nef. Further studies to characterize the process of inductions of each prophage responsible for the HGT revealed that the prophages of resident and invader E. coli can form active phage particles. The lysogenic invaders acquired phage-killing potential and a metabolic advantage related to the uptake of carbon sources (Frazão et al. 2019).
The HGT of antibiotic resistance (AR) genes in the gut is of great interest; however, how it works remains unknown (Kent et al. 2020). Górska et al. (2018) suggested that phages could be involved in the HGT of AR genes. Their pilot research has suggested that the abundance of phages carrying antibiotic resistance genes increases in patients that are under antibiotic treatment. However, further research is needed to know the insights of the integration of the phage in the bacterial genome (Górska et al. 2018). Other genes acquired by horizontal gene transfer are the Shiga genes that codify for the main virulence factors of some E. coli strains (Krüger and Lucchesi 2015) and the cholera toxin that was transferred to Vibrio cholerae (Faruque and Mekalanos 2012).
6.5 Pathogenic Interactions
There is evidence that enteric virome can cause significant diseases and could be utilizied in the gut to promote infection. Apparently, infection with enteric viruses is affected in the absence of gut microbiota because viral-bacteria mixed infections are worse than viral infections alone. How bacteria could facilitate the viral co-infection of cells is an active area of research and has been dug mainly in poliovirus and norovirus (Berger and Mainou 2018; Huang 2020).
Poliovirus is a non-enveloped ssRNA virus, which was a major cause of paralysis until 1950. It has been shown that polysaccharides of bacterial surfaces containing N-acetylglucosamine, like LPS, could bind to poliovirus and stabilize the particles by preventing the premature release of RNA. Also, LPS helps in the attachment of poliovirus to the host cells (Robinson et al. 2014). Bacteria can, as well, enhance the genetic recombination between two different viruses that helps them to increase the viral fitness and drive adaptation (Erickson et al. 2018). Human norovirus (NoV) causes viral gastroenteritis. The presence of commensal bacteria allows for efficient infection with that virus (Baldridge et al. 2015). It has been described that NoV binds to histo-blood group antigens (HBGAs), expressed by several enteric bacteria such as Enterobacter cloacae. This moiety is differing between bacterial strains (Almand et al. 2019).
6.6 Gaps in Knowledge
The processes by which bacteriophages regulate the microbial community is multifactorial (Maronek et al. 2020). Current research is being carried out in phage-bacteria interactions seeking to find out what conditions of the human gut can trigger the lytic stage of prophages and how that influences the community. It is known that if phages are released from bacteria, then the competition between them to infect new hosts would increase. Also, the death of many bacteria would set free a niche for many other bacteria to occupy (that could be pathogenic or commensal), and this gives place to new interactions (Maurice 2019; Mirzaei and Maurice 2017). Finally, it has been recently suggested that the interactions between phages and bacteria could be age dependent (Mirzaei et al. 2020). Understanding and learning about these interactions will lead to the potential use of the enteric virome in therapy (Altamura et al. 2020).
7 Human Virome Therapeutic Implications and Future Directions
Bacteriophages are the most abundant members of the microbiota and have the ability to shape microbial communities (Garmaeva et al. 2019). It was suggested that they play a role in dysbiosis (Lin and Lin 2019), which is the decline in the diversity of the gut microbiome (DuPont et al. 2020). This microbial imbalance, alongside genetics, immune responses, and functional microbial activity, is associated with many gut diseases (Kostic et al. 2014). The study of the participation of the enteric virome in the development of intestinal diseases could lead to the development of a new diagnostic biomarker or antiviral drugs (Ansari et al. 2020). Also, bacteriophage-based therapies could emerge as a particular treatment option for microbiota-related diseases (Gogokhia et al. 2019).
7.1 Importance of the Enteric Virome in Fecal Microbial Transplantation
FMT (fecal microbial transplantation) is the administration of fecal slurry from a donor to the intestinal tract of a recipient, aiming to restore microbiota diversity and composition and provide a health benefit. This treatment has been proven to be effective against Clostridium difficile infection (CDI) (Gupta et al. 2016) with cure rates up to 90% (Basson et al. 2020).
The virome participation in the effectiveness of FMT treatment has been poorly studied, but there are signs that bacteriophages might have a role in the FMT outcomes (Broecker et al. 2017). For example, Ott et al. (2017)) showed that sterile fecal filtrate transfer (without living organisms) is effectively eliminating the symptoms of CDI. These results suggested that bacteriophages could be taking part in the FMT results (Ott et al. 2017). Zuo et al. (2018) further studied the differences in the phageome community between CDI patients and healthy controls and their importance in FMT. They found that CDI patients have more abundance but less diversity, evenness, and richness of Caudovirales compared to healthy controls. When the Caudovirales richness of the donor was higher than that of the recipient, the CDI patients achieved a positive response for FMT. Unlike the FMT non-responders, the FMT responders after the treatment showed more donor-derived Caudovirales contigs present in larger fractions in their enteric virome (Zuo et al. 2018). Recent research pointed out that the virome of the recipient after FMT ends up being very similar to the donor and that this can last up to 12 months (Draper et al. 2018). These might indicate that phages could be used as disease treatment (Manrique et al. 2017).
7.2 Phage Therapy
Bacteriophages can be used as biocontrol agents for specific hosts (Bao et al. 2018). Based on that, phage therapy is reemerging due to the rising antibiotic resistance problem. However, it is restricted for its use in humans because they can interact with our immune system and evolve inside the body. Additionally, they could influence the composition of the gut microbiome (Divya Ganeshan and Hosseinidoust 2019). Several studies have been done to show their effectiveness and specificity. For example, Hu et al. (2018) did an in vitro study with SalmoFresh™, which is a mixture of six strictly lytic phages of Salmonella (Zhang et al. 2019). They revealed that this phage cocktail can be used to target that bacteria in a very specific way. In comparison to the antibiotic azithromycin, phage treatment did not cause perturbation in non-targeted microbial communities (Hu et al. 2018).
Llanos-Chea et al. (2019)) evaluated the effect of the bacteriophage Φ2457T, which is a Shigella flexneri 2457 T-specific bacteriophage, in a human intestinal organoid-derived epithelial monolayer model. Their results showed that Φ2457T efficiently killed Shigella flexneri 2457 T in a highly specific manner (Llanos-Chea et al. 2019).
Dissanayake et al. (2019)) used “Foodborne Outbreak Pill” (FOP) (a combination of E. coli O157:H7, Salmonella spp., and L. monocytogenes-targeting lytic bacteriophages) to combat pathogenic E. coli in mice. FOP was demonstrated to be effective against E. coli O157:H7. They found out that FOP was highly specific and was better to maintain the natural richness and diversity of the gut microbiome in comparison to the control treated with ampicillin (Dissanayake et al. 2019). These results were consistent with those of Cieplak et al. (2018)) who used E. coli specific bacteriophages against E. coli DSM 1058 (Cieplak et al. 2018).
Adherent-invasive E. coli (AIEC) may be correlated with sustaining inflammation in the preexisting inflammatory mucosa (Lee et al. 2019) and with the exacerbation of intestinal inflammation (Delmas et al. 2019). Regular treatments with AIEC-bacteriophages isolated from the gut microbiome can reduce and control the outgrowth of targeted bacteria in the intestine and protect from AIEC and invasive bacteria-exacerbated colorectal cancer (Gogokhia et al. 2019). An important challenge to sort out when orally consuming bacteriophages would be to protect the phages from low pH due to the gastric acids. Therefore, encapsulating them with resistant material would make the treatment more effective (Vinner et al. 2019).
7.3 Enteric Virome Associated with Gastrointestinal Diseases
7.3.1 Inflammatory Bowel Disease (IBD)
Inflammatory bowel disease (IBD) is a convoluted, microbiome-driven immunological inflammatory disorder. There are two subtypes of IBD: Crohn’s disease (CD) and ulcerative colitis (UC) (Panaccione 2013). UC and CD patients have different bacteriophage communities in comparison to healthy donors. The order of Caudovirales increased in UC and CD patients. Also, it has been suggested that the virome is specific for patients with UC and for patients with CD (Norman et al. 2015). As a therapy, FMT has proven to be not as effective as with CD (Basson et al. 2020). However, mixing donor products has been a way to expand the potential value of FMT when treating chronic diseases (DuPont et al. 2020).
Since the disease phenotypes in adult and pediatric-onset IBD are different, a study in children alone was necessary. Research in kids has shown that minor patterns in gut virome differentiate IBD patients and healthy donors. Among the differences they found was that Caudovirales were more abundant in CD (Crohn’s disease) and UC (ulcerative colitis) patients than in healthy controls. It was also found that the richness of Microviridae is higher in controls than in CD patients (Fernandes et al. 2019). A study of virome transference in FMT between three pediatric UC patients and a healthy donor confirmed that the transference of viruses is mainly associated with temperate-phage replication, specially associated with the group Siphoviridae. None of them can replicate in human cells (Chehoud et al. 2016).
7.3.2 Celiac Disease Autoimmunity (CDA)
Celiac disease autoimmunity (CDA) is an autoimmune disease that is triggered mainly by genetics and dietary gluten. Recent reports indicated that microbiome disturbance could be behind celiac disease pathogenesis. However, the role or the virome participation in CDA is debatable (Akobeng et al. 2020). Recently, a study with children carrying a genotype for increased risk of celiac disease suggested that Enterovirus A and Enterovirus B are significantly associated with CDA. This research, using longitudinal birth control analysis, also discarded the possibility that adenovirus is involved with CDA (Kahrs et al. 2019). Also, it seems that there is a cumulative effect of enterovirus and high gluten intake for the development of CDA (Lindfors et al. 2020). On the other hand, previous studies have pointed out that the infection with reovirus could trigger CDA (Bouziat et al. 2017).
7.3.3 Enteric Virome Implication in Obesity and Diabetes
Obesity and type 2 diabetes are also related to gut microbiota dysbiosis (Maruvada et al. 2017; Ridaura et al. 2013). Fecal virome transplantation (FVT) – where sterile filtered donor feces that contain enteric viruses but not bacteria are transferred – has proven to reduce weight gain and normalize blood glucose parameters in mice (Rasmussen et al. 2020). In the case of type 1 diabetes (TID), differences between the viromes of patients with TID and controls have also been reported. Zhao et al. (2017)) found that the gut viromes of patients were less diverse than those of healthy controls. For example, healthy patients were richer in the eukaryotic virus Circoviridae and the bacteriophages Microviridae, Myoviridae, and Podoviridae (Zhao et al. 2017). Park and Zhao (2018) found differences in the virome population preceding initial signs of T1D (Park and Zhao 2018).
7.3.4 Enteric Virome Implication in Parkinson’s Disease
Enteric virome has been recently linked to Parkinson’s disease (PD) as a possible factor for the development of this neurodegenerative disorder. In PD patients, there is a significant reduction of Lactococcus and Lactobacillus. Lactococcus is involved in the production of microbiota-derived neurochemicals such as dopamine and with gut permeability. Their decrease could be involved with the triggering of PD. Tetz et al. (2018) showed that the decline of Lactococcus was not accompanied by the decline of their phages. The parallel decrease would have been normal in case the bacteriophages were integrated as prophage. They correlated that to the higher abundance of lactococcal lytic phages, especially the ones belonging to c-2-like and group 936, in PD patients. This finding could be useful to use the virome composition as a diagnostic tool or target for therapeutic intervention (Santos et al. 2019; Tetz et al. 2018).
7.4 Eukaryotic Viruses and Their Implications in Gastrointestinal Diseases
Apart from bacteriophages, the gut virome has eukaryotic viruses. Although they are considered as pathogens, they can live innocuously in the healthy human intestine (Mukhopadhya et al. 2019). The differences in the eukaryotic virome between patients and healthy controls have been reported. For example, Norman et al. (2015) found more presence of sequences from the eukaryotic virus Anellovirus in IBD patients (Norman et al. 2015). Also, Ansari et al. (2020) reported fewer abundance and diversity of members of Megavirales (Ansari et al. 2020). According to Conceição-Neto et al. (2018), who studied the implication of eukaryotic viruses in FMT treatment, healthy donors have a lower richness of eukaryotic virome than UC patients. Surprisingly, UC patients that responded to the FMT treatment already presented lower viral richness than non-responders. This suggests that eukaryotic virome richness could be important in UC and might lead to an increase in treatment success (Conceição-Neto et al. 2018).
7.5 Challenges and Future Directions
There are several challenges that need to be overcome to understand the role of the virome in human health. First is the inefficiency to identify all viruses due to the lack of a universal viral sequence. Second, the detection of DNA viruses has an advantage over RNA viruses because of inadequate sampling strategies that currently focus only on sequencing DNA, leaving aside an important source of diversity. Third is the absence of culture systems to propagate components of the virome and perform the Koch’s postulate to know if the virome plays a causative role. Fourth is the need to change from in vitro systems to experimental animal infection models (Wang 2020). There are some gaps in knowledge for using FMT in IBD, such as identifying the optimal route, dose, and frequency of FMT, as well as what makes an optimum donor. Further studies to understand the potential contribution of the virome to the efficacy of the FMT in IBD are needed (Yalchin et al. 2019). Also, even though FMT has proven to be effective, there is a need to develop targeted microbiome therapies (Russell et al. 2018). These will come out as more information is generated on the mechanisms and diversity of human microbiome.
References
Adamiak T, Walkiewicz-Jedrzejczak D, Fish D et al (2013) Incidence, clinical characteristics, and natural history of pediatric IBD in Wisconsin: a population-based epidemiological study. Inflamm Bowel Dis 19:1218–1223
Ahlgren NA, Ren J, Lu YY, Fuhrman JA, Sun F (2017) Alignment-free d_2^* oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res 45(1):39–53
Ahmed S, Macfarlane GT, Fite A, McBain AJ, Gilbert P, Macfarlane S (2007) Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol 73:7435–7442. https://doi.org/10.1128/AEM.01143-07
Akhter S, Aziz RK, Edwards RA (2012) Nucleic Acids Res 40:1–13
Akobeng AK, Singh P, Kumar M, Al Khodor S (2020) Role of the gut microbiota in the pathogenesis of coeliac disease and potential therapeutic implications. Eur J Nutr 59(8):3369–3390. https://doi.org/10.1007/s00394-020-02324-y
Allander T (2008) Human bocavirus. J Clin Virol 41(1):29–33
Almand EA, Moore MD, Jaykus LA (2017) Virus bacteria interactions: an emerging topic in human infection. Viruses 9:58
Almand EA, Moore MD, Jaykus L-A (2019) Characterization of human norovirus binding to gut-associated bacterial ligands. BMC Res Notes 12(1):607
Altamura F, Maurice CF, Castagner B (2020) Drugging the gut microbiota: toward rational modulation of bacterial composition in the gut. Curr Opin Chem Biol 56:10–15
Angly F, Rodriguez-Brito B, Bangor D, McNairnie P, Breitbart M, Salamon P, Felts B, Nulton J, Mahaffy J, Rohwer F (2005) PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinform 6:1–9
Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F (2006) PLoS Biol 4:2121–2131
Ansari MH, Ebrahimi M, Fattahi MR, Gardner MG, Safarpour AR, Faghihi MA, Lankarani KB (2020) Viral metagenomic analysis of fecal samples reveals an enteric virome signature in irritable bowel syndrome. BMC Microbiol 20(1):123
Anthenelli M, Jasien E, Edwards R, Bailey B, Felts B, Katira P, Nulton J, Salamon P, Rohwer F, Silveira CB, Luque A (2020) Phage and bacteria diversification through a prophage acquisition ratchet, p 2020.04.08.028340. https://doi.org/10.1101/2020.04.08.028340
Avrani S, Schwartz DA, Lindell D (2012) Virus-host swinging party in the oceans: incorporating biological complexity into paradigms of antagonistic coexistence. Mob Genet Elem 2(2):88–95
Ayukekbong J, Lindh M, Nenonen N, Tah F, Nkuo-Akenji T, Bergström T (2011) Enteric viruses in healthy children in Cameroon: viral load and genotyping of norovirus strains. J Med Virol 83(12):2135–2142
Bagaglio S, Sitia G, Prati D, Cella D, Hasson H, Novati R, Lazzarin A, Morsica G (2002) Mother-to-child transmission of TT virus: sequence analysis of non-coding region of TT virus in infected mother-infant pairs. Arch Virol 147(4):803–812
Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, Diamond MS, Ivanova Y, Artyomov M, Virgin HW (2015) Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 347(6219):266–269
Banyai K, Jakab F, Reuter G et al (2003) Sequence heterogeneity among human picobirnaviruses detected in a gastroenteritis outbreak. Arch Virol 148(12):2281–2291
Bao H-D, Pang M, Olaniran A, Zhang X-H, Zhang H, Zhou Y, Sun L-C, Schmidt S, Wang R (2018) Alterations in the diversity and composition of mice gut microbiota by lytic or temperate gut phage treatment. Appl Microbiol Biotechnol 102(23):10219–10230
Barr JJ, Auro R, Furlan M et al (2013) Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A 110:10771–10776
Barreira DMPG, Ferreira MSR, Fumian TM, Checon R, de Sadovsky ADI, Leite JPG, Miagostovich MP, Spano LC (2010) Viral load and genotypes of noroviruses in symptomatic and asymptomatic children in southeastern Brazil. J Clin Virol 47(1):60–64
Basson AR, Zhou Y, Seo B, Rodriguez-Palacios A, Cominelli F (2020) Autologous fecal microbiota transplantation for the treatment of inflammatory bowel disease. Transl Res 226:1–11. https://doi.org/10.1016/j.trsl.2020.05.008
Beatty JK, Bhargava A, Buret AG (2014) Post-infectious irritable bowel syndrome: mechanistic insights into chronic disturbances following enteric infection. World J Gastroenterol 20:3976–3985
Beller L, Matthijnssens J (2019) What is (not) known about the dynamics of the human gut virome in health and disease. Curr Opin Virol 37:52–57
Belshaw R, Pereira V, Katzourakis A, Talbot G, Pačes J, Burt A, Tristem M (2004) Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci 101(14):4894–4899
Berger AK, Mainou BA (2018) Interactions between enteric Bacteria and eukaryotic viruses impact the outcome of infection. Viruses 10(1):19. https://doi.org/10.3390/v10010019
Biagini P, Gallian P, Cantaloube JF, De Micco P, de Lamballerie X (1998) Presence of TT virus in French blood donors and intravenous drug users. J Hepatol 29(4):684–685
Blaise S, de Parseval N, Bénit L, Heidmann T (2003) Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci 100(22):13013–13018
Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, Spector TD, Keinan A, Ley RE, Gevers D et al (2015) Host genetic variation impacts microbiome composition across human body sites. Genome Biol 16:191
Bonvicini F, Manaresi E, Gentilomi GA, Di Furio F, Zerbini M, Musiani M, Gallinella G (2011) Evidence of human bocavirus viremia in healthy blood donors. Diagn Microbiol Infect Dis 71(4):460–462
Bouziat R, Hinterleitner R, Brown JJ, Stencel-Baerenwald JE, Ikizler M, Mayassi T, Meisel M, Kim SM, Discepolo V, Pruijssers AJ, Ernest JD, Iskarpatyoti JA, Costes LMM, Lawrence I, Palanski BA, Varma M, Zurenski MA, Khomandiak S, McAllister N et al (2017) Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 356(6333):44–50
Breitbart M, Rohwer F (2005) Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 39(5):729–736
Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F (2003) J Bacteriol 185:6220–6223
Brister JR, Ako-Adjei D, Bao Y, Blinkova O (2015) Nucleic Acids Res 43:571–577
Broecker F, Russo G, Klumpp J, Moelling K (2017) Stable core virome despite variable microbiome after fecal transfer. Gut Microbe 8(3):214–220
Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M et al (2011) Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One 6:e25792
Brown MR, Camézuli S, Davenport RJ, Petelenz-Kurdziel E, Øvreås L, Curtis TP (2015) Flow cytometric quantification of viruses in activated sludge. Water Res 68:414–422
Cadranel JF, Di Martino V, Lambrey G et al (2008) Prevalence of hepatitis C infection and risk factors in hospitalized diabetic patients: results of a cross-sectional study. Eur J Gastroenterol Hepatol 20:829–836
Callanan J, Stockdale SR, Shkoporov A, Draper LA, Ross RP, Hill C (2018) Viruses 10:1–17
Callier V (2019) Core concept: gene transfers from bacteria and viruses may be shaping complex organisms. Proc Natl Acad Sci U S A 116(28):13714–13716
Cani PD, Geurts L, Matamoros S et al (2014) Glucose metabolism: focus on gut micro-biota, the endocannabinoid system and beyond. Diabete Metab 40:246–257
Carter CJ (2010) Familial and late-onset Alzheimer’s disease: evidence for an auto-immune component triggered by viral, microbial and allergen antigens with homology to beta-amyloid and APP mutant peptides. Nat Preced 1:4662
Castro-Mejía JL, Muhammed MK, Kot W, Neve H, Franz CMAP, Hansen LH, Vogensen FK, Nielsen DS (2015) Optimizing protocols for extraction of bacteriophages prior to metagenomic analyses of phage communities in the human gut. Microbiome 3:64
Celiberto LS, Graef FA, Healey GR, Bosman ES, Jacobson K, Sly LM, Vallance BA (2018) Inflammatory bowel disease and immunonutrition: novel therapeutic approaches through modulation of diet and the gut microbiome. Immunology 155:36–52
Chehoud C, Dryga A, Hwang Y, Nagy-Szakal D, Hollister EB, Luna RA, Versalovic J, Kellermayer R, Bushman FD (2016) Transfer of viral communities between human individuals during fecal microbiota transplantation. MBio 7(2):e00322
Chen Y, Williams V, Filippova M, Filippov V, Duerksen-Hughes P (2014a) Viral carcinogenesis: factors inducing DNA damage and virus integration. Cancers (Basel) 6:2155–2186
Chen M, Song P, Zou D, Hu X, Zhao S, Gao S, Ling F (2014b) PLoS One 9:1–12
Chua LL, Rajasuriar R, Azanan MS, Abdullah NK, Tang MS, Lee SC, Woo YL, Lim YAL, Arian H, Loke P (2017) Reduced microbial diversity in adult survivors of childhood acute lymphoblastic leukemia and microbial associations with increased immune activation. Microbiome 5:35
Cieplak T, Soffer N, Sulakvelidze A, Nielsen DS (2018) A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbe 9(5):391–399
Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó et al (2015) The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183
Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, Ryan FJ, Draper LA, Plevy SE, Ross RP et al (2019) Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 26:764–778
Conceição-Neto N, Zeller M, Lefrère H, De Bruyn P, Beller L, Deboutte W, Yinda CK, Lavigne R, Maes P, Van Ranst M, Heylen E, Matthijnssens J (2015) Sci Rep 5:1–14
Conceição-Neto N, Deboutte W, Dierckx T, Machiels K, Wang J, Yinda KC, Maes P, Van Ranst M, Joossens M, Raes J, Vermeire S, Matthijnssens J (2018) Low eukaryotic viral richness is associated with faecal microbiota transplantation success in patients with UC [Review of Low eukaryotic viral richness is associated with faecal microbiota transplantation success in patients with UC]. Gut 67(8):1558–1559
Cotten M, Oude Munnink B, Canuti M, Deijs M, Watson SJ, Kellam P, Van Der Hoek L (2014) PLoS One 9:e93269
Dalmasso M, Hill C, Ross RP (2014) Exploiting gut bacteriophages for human health. Trends Microbiol 22:399–405
David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA et al (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563
De Beeck AO, Eizirik DL (2016) Viral infections in type 1 diabetes mellitus—why the [beta] cells? Nat Rev Endocrinol 12(5):263–273. https://doi.org/10.1038/nrendo.30
de Jonge PA, Nobrega FL, Brouns SJJ, Dutilh BE (2019) Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol 27(1):51–63
de Vries M, Deijs M, Canuti M, van Schaik BDC, Faria NR, van de Garde MDB, Jachimowski LCM, Jebbink MF, Jakobs M, Luyf ACM, Coenjaerts FEJ, Claas ECJ, Molenkamp R, Koekkoek SM, Lammens C, Leus F, Goossens H, Ieven M, Baas F, van der Hoek L (2011) A sensitive assay for virus discovery in respiratory clinical samples. PLoS One 6
Delbue S, Tremolada S, Elia F, Carloni C, Amico S, Tavazzi E, Marchioni E, Novati S, Maserati R, Ferrante P (2010) Lymphotropic polyomavirus is detected in peripheral blood from immunocompromised and healthy subjects. J Clin Virol 47(2):156–160
Delmas J, Gibold L, Faïs T, Batista S, Leremboure M, Sinel C, Vazeille E, Cattoir V, Buisson A, Barnich N, Dalmasso G, Bonnet R (2019) Metabolic adaptation of adherent-invasive Escherichia coli to exposure to bile salts. Sci Rep 9(1):2175
Delwart E (2007) Viral metagenomics. Rev Med Virol 17:115–131
Devaraj S, Hemarajata P, Versalovic J (2013) The human gut microbiome andbody metabolism: implications for obesity and diabetes. Clin Chem 59:617–628
Dinleyici EC, Eren M, Ozen M et al (2012) Effectiveness and safety of Saccharomyces boulardii for acute infectious diarrhea. Expert Opin Biol Ther 12:395–410
Dissanayake U, Ukhanova M, Moye ZD, Sulakvelidze A, Mai V (2019) Bacteriophages reduce pathogenic Escherichia coli counts in mice without distorting gut microbiota. Front Microbiol 10:1984
Divya Ganeshan S, Hosseinidoust Z (2019) Phage therapy with a focus on the human microbiota. Antibiotics (Basel, Switzerland) 8:131. https://doi.org/10.3390/antibiotics8030131
Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107:11971–11975
Domínguez-Díaz C, García-Orozco A, Riera-Leal A, Padilla-Arellano JR, Fafutis-Morris M (2019) Microbiota and its role on viral evasion: is it with us or against us? Front Cell Infect Microbiol 9:256
Dotta F, Censini S, van Halteren AGS, Marselli L, Masini M, Dionisi S et al (2007) Coxsackie B4 virus infection of beta cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc Natl Acad Sci U S A 104(12):5115–5120
Draper LA, Ryan FJ, Smith MK, Jalanka J, Mattila E, Arkkila PA, Ross RP, Satokari R, Hill C (2018) Long-term colonisation with donor bacteriophages following successful faecal microbial transplantation. Microbiome 6(1):220
Duerkop BA, Kleiner M, Paez-Espino D, ZhuW BB, Hassell B, Winter SE, Kyrpides NC, Hooper LV (2018) Murine colitis reveals a disease associated bacteriophage community. Nat Microbiol 3:11023–11031
DuPont HL, Jiang Z-D, DuPont AW, Utay NS (2020) Abnormal intestinal microbiome in medical disorders and potential reversibility by fecal microbiota transplantation. Dig Dis Sci 65(3):741–756
Džunková M, D'Auria G, Moya A (2015) Direct sequencing of human gut virome fractions obtained by flow cytometry. Front Microbiol 6:955
Eckardt AJ, Baumgart DC (2011) Viral gastroenteritis in adults. Recent Pat Antiinfect Drug Discov 6(1):54–63
Edwards RA, McNair K, Faust K, Raes J, Dutilh BE (2016) Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol Rev 40(2):258–272
Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, Gosert R, Hirsch HH (2009) Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis 199(6):837–846
Emerson JB, Roux S, Brum JR, Bolduc B, Woodcroft BJ, Jang HB, Singleton CM, Solden LM, Naas AE, Boyd JA, Hodgkins SB, Wilson RM, Trubl G, Li C, Frolking S, Pope PB, Wrighton KC, Crill PM, Chanton JP, Saleska SR, Tyson GW, Rich VI, Sullivan MB (2018) Nat. Microbiology 3:870–880
Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK (2018) Bacteria facilitate enteric virus co-infection of mammalian cells and promote genetic recombination. Cell Host Microbe 23(1):77–88.e5
Eszter Muller K, Laszlo Lakatos P, Papp M et al (2014) Incidence and Paris classification of pediatric inflammatory bowel disease. Gastroenterol Res Pract 2014:904307
Faruque SM, Mekalanos JJ (2012) Phage-bacterial interactions in the evolution of toxigenic Vibrio cholerae. Virulence 3(7):556–565
Fernandes MA, Verstraete SG, Phan TG, Deng X, Stekol E, LaMere B, Lynch SV, Heyman MB, Delwart E (2019) Enteric virome and bacterial microbiota in children with ulcerative colitis and Crohn disease. J Pediatr Gastroenterol Nutr 68(1):30–36
Fields BN, Knipe DM, Howley PM (2007) Fields’ virology, 5th edn. Lippincott-Williams, Philadelphia, PA
Filippi CM, Herrath MG (2008) von viral trigger for type 1 diabetes. Diabetes 57:2863–2871
Fouts DE (2006) Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res 34:5839–5851
Foxman EF, Iwasaki A (2011) Genome-virome interactions: examining the role of common viral infections in complex disease. Nat Rev Microbiol 9(4):254–264
Francino MP (2016) Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol 6:1543
Frazão N, Sousa A, Lässig M, Gordo I (2019) Horizontal gene transfer overrides mutation in Escherichia coli colonizing the mammalian gut. Proc Natl Acad Sci U S A 116(36):17906–17915
Friedman SL (2013) Liver fibrosis in 2012: convergent pathways that cause hepatic fibrosis in NASH. Nat Rev Gastroenterol Hepatol 10:71–72
Fryer JF, Delwart E, Hecht FM, Bernardin F, Jones MS, Shah N, Baylis SA (2007) Frequent detection of the parvoviruses, PARV4 and PARV5, in plasma from blood donors and symptomatic individuals. Transfusion 47(6):1054–1061
Gabbay YB, da Luz CRN, Costa IV, Cavalcante-Pepino EL, Sousa MS, Oliveira KK, Wanzeller ALM, Mascarenhas JD, Leite JPG, Linhares AC (2005) Prevalence and genetic diversity of astroviruses in children with and without diarrhea in São Luís, Maranhão, Brazil. Mem Inst Oswaldo Cruz 100(7):709–714
Galiez C, Siebert M, Enault F, Vincent J, Söding J (2017) WIsH: who is the host? Predicting prokaryotic hosts from metagenomic phage contigs. Bioinformatics 33(19):3113–3114
Garmaeva S, Sinha T, Kurilshikov A, Fu J, Wijmenga C, Zhernakova A (2019) BMC Biol 17:1–14
Geurts L, Neyrinck AM, Delzenne NM et al (2014) Gut microbiota controls adipose tissue expansion, gut barrier and glucose metabolism: novel insights into molecular targets and interventions using prebiotics. Benef Microb 5:3–17
Ghazarian L, Diana J, Simoni Y, Beaudoin L, Lehuen A (2012) Prevention or acceleration of type 1 diabetes by viruses. Cell Mol Life Sci 70(2):239–255. https://doi.org/10.1007/s00018-012-1042-1
Girons IS, Bourhy P, Ottone C et al (2000) The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: construction of an L. biflexa– Escherichia coli shuttle vector. J Bacteriol 182:5700–5705
Gogokhia L, Buhrke K, Bell R, Hoffman B, Brown DG, Hanke-Gogokhia C, Ajami NJ, Wong MC, Ghazaryan A, Valentine JF, Porter N, Martens E, O’Connell R, Jacob V, Scherl E, Crawford C, Stephens WZ, Casjens SR, Longman RS, Round JL (2019) Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe 25(2):285–299.e8
Gómara MI, Kang G, Mammen A, Jana AK, Abraham M, Desselberger U, Brown D, Gray J (2004) Characterization of G10P [11] rotaviruses causing acute gastroenteritis in neonates and infants in Vellore, India. J Clin Microbiol 42(6):2541–2547
Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Homan K, Wei SC et al (2018) Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359:97–103
Górska A, Peter S, Willmann M, Autenrieth I, Schlaberg R, Huson DH (2018) Dynamics of the human gut phageome during antibiotic treatment. Comput Biol Chem 74:420–427
Greninger AL, Runckel C, Chiu CY, Haggerty T, Parsonnet J, Ganem D, DeRisi JL (2009) The complete genome of klassevirus–a novel picornavirus in pediatric stool. Virol J 6(1):82
Guarner C, Soriano G (2005) Bacterial translocation and its consequences in patients with cirrhosis. Eur J Gastroenterol Hepatol 17(1):27–31
Guerin E, Shkoporov A, Stockdale S et al (2018) Biology and taxonomy of crAss-like bacteriophages, the most abundant virus in the human gut. Cell Host Microbe 24:653–664
Guo M, Liu G, Chen J, Ma J, Lin J, Fu Y, Fan G, Lee SM-Y, Zhang L (2020) Dynamics of bacteriophages in gut of giant pandas reveal a potential regulation of dietary intake on bacteriophage composition. Sci Total Environ 734:139424
Gupta S, Allen-Vercoe E, Petrof EO (2016) Fecal microbiota transplantation: in perspective. Therap Adv Gastroenterol 9(2):229–239
Halfvarson J, Brislawn CJ, Lamendella R, Vázquez-Baeza Y, Walters WA, Bramer LM, D’Amato M, Bonfiglio F, McDonald D, Gonzalez A et al (2017) Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2:17004
Hamdi S, Rousseau GM, Labrie SJ, Tremblay DM, Kourda RS, Slama KB, Moineau S (2017) Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci Rep 7:40349
Hannigan GD, Duhaime MB, Run MT, Koumpouras CC, Schloss PD (2018) Diagnostic potential and interactive dynamics of the colorectal cancer virome. MBio 9
Haynes M, Rohwer F (2011) The human virome. Springer, New York, NY, pp 63–77
Helmersson J, Vessby B, Larsson A, Basu S (2004) Association of type 2 diabetes with cyclooxygenase-mediated inflammation and oxidative stress in an elderly population. Circulation 109:1729–1734
Hillman ET, Lu H, Yao T, Nakatsu CH (2017) Microbial ecology along the gastrointestinal tract. Microb Environ 32(4):300–313
Hindiyeh M, Mor O, Pando R, Mannasse B, Kabat A, Assraf-Zarfati H, Mendelson E, Sofer D, Mandelboim M (2019) Comparison of the new fully automated extraction platform eMAG to the MagNA PURE 96 and the well-established easyMAG for detection of common human respiratory viruses. PLoS One 14:e0211079
Hofer U (2013) Variation in the gut virome. Nat Rev Microbiol 11(9):596
Holtz LR, Cao S, Zhao G et al (2014) Geographic variation in the eukaryotic virome of human diarrhea. Virology 468–470:556–564
Hooper LV, Macpherson AJ (2010) Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10(3):159–169
Hoyles L, McCartney AL, Neve H, Gibson GR, Sanderson JD, Heller KJ, van Sinderen D (2014) Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res Microbiol 165:803–812
Hsu BB, Gibson TE, Yeliseyev V, Liu Q, Bry L, Silver PA, Gerber GK (2018) Bacteriophages dynamically modulate the gut microbiota and metabolome, p 454579. https://doi.org/10.1101/454579
Hu YOO, Hugerth LW, Bengtsson C, Alisjahbana A, Seifert M, Kamal A, Sjöling Å, Midtvedt T, Norin E, Du J, Engstrand L (2018) Bacteriophages synergize with the gut microbial community to combat Salmonella. mSystems 3(5):e00119. https://doi.org/10.1128/mSystems.00119-18
Huang PN (2020) Microbiota and enteric viruses infection. Med Microecol 3:100006
Hunter P (2013) The secret garden’s gardeners: research increasingly appreciates the crucial role of gut viruses for human health and disease. EMBO Rep 14:683–685
Jaeckel E, Manns M, von Herrath M (2002) Viruses and diabetes. Ann N Y Acad Sci 958:7–25
Jones MS, Kapoor A, Lukashov VV, Simmonds P, Hecht F, Delwart E (2005) New DNA viruses identified in patients with acute viral infection syndrome. J Virol 79(13):8230–8236
Jun HS, Yoon JW (2003) A new look at viruses in type 1 diabetes. Diabetes Metab Res Rev 19(1):8–31
Kahrs CR, Chuda K, Tapia G, Stene LC, Mårild K, Rasmussen T, Rønningen KS, Lundin KEA, Kramna L, Cinek O, Størdal K (2019) Enterovirus as trigger of coeliac disease: nested case-control study within prospective birth cohort. BMJ 364:l231
Kane M, Case LK, Kopaskie K et al (2011) Successful transmission of a retrovirus depends on the commensal microbiota. Science 334:245–249
Kapusinszky B, Minor P, Delwart E (2012) Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol 50(11):3427–3434
Karim S, Mirza Z, Kamal MA, Abuzenadah AM, Azhar EI, Al-Qahtani MH, Sohrab SS (2014) An association of virus infection with type 2 diabetes and Alzheimer’s disease. CNS Neurol Disord Drug Targets 13(3):429–439. https://doi.org/10.2174/18715273113126660164
Kent AG, Vill AC, Shi Q, Satlin MJ, Brito IL (2020) Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial hi-C. Nat Commun 11(1):4379
Khan R, Petersen FC, Shekhar S (2019) Commensal bacteria: an emerging player in defense against respiratory pathogens. Front Immunol 10:1203
Kim KH, Chang HW, Do Nam Y, Roh SW, Kim MS, Sung Y, Jeon CO, Oh HM, Bae JW (2008) Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl Environ Microbiol 74:5975–5985
King ML, Shaikh A, Bidwell D, Voller A, Banatvala JE (1983) Coxsackie- B-virus-specific IgM responses in children with insulin-dependent (juvenile-onset; type I) diabetes mellitus. Lancet 1(8339):1397–1399
Kleiner M, Hooper LV, Duerkop BA (2015) Evaluation of methods to purify virus-like particles for metagenomic sequencing of intestinal viromes. BMC Genomics 16:1–15
Koichi Y, Arvin AM, Gabriella C et al (2007) Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge
Koskella B, Brockhurst MA (2014) Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev 38(5):916–931. https://doi.org/10.1111/1574-6976.12072
Koskella B, Lively CM (2009) Evidence for negative frequency‐dependent selection during experimental coevolution of a freshwater snail and a sterilizing trematode. Evol Int J Organ Evol 63(9):2213–2221. https://doi.org/10.1111/j.1558-5646.2009.00711.x
Kostic AD, Xavier RJ, Gevers D (2014) The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146(6):1489–1499. https://doi.org/10.1053/j.gastro.2014.02.009
Krishnamurthy SR, Wang D (2017) Origins and challenges of viral dark matter. Virus Res 239:136–142
Kroner Z (2009) The relationship between Alzheimer’s disease and diabetes: type 3 diabetes? Altern Med Rev 14:373–379
Krüger A, Lucchesi PMA (2015) Shiga toxins and stx phages: highly diverse entities. Microbiology 161(Pt 3):451–462
Kumar A, Murthy S, Kapoor A (2017) Evolution of selective-sequencing approaches for virus discovery and virome analysis. Virus Res 239:172–179
Kumarasamy KK, Toleman MA, Walsh TR et al (2010) Emergence of a new antibi-otic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602
Laforest-Lapointe I, Arrieta M-C (2018) Microbial eukaryotes: a missing link in gut microbiome studies. mSystems 3(2):e00201. https://doi.org/10.1128/mSystems.00201-17
Lagier JC, Million M, Hugon P et al (2012) Human gut microbiota: repertoire and variations. Front Cell Infect Microbiol 2:136
Lam KC, Vyshenska D, Hu J, Rodrigues RR, Nilsen A, Zielke RA, Brown NS, Aarnes E-K, Sikora AE, Shulzhenko N et al (2018) Transkingdom network reveals bacterial players associated with cervical cancer gene expression program. Peer J 6:e5590
Larsen N, Vogensen FK, van Den Berg FWJ, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M (2010) Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5:e9085
Lee JG, Han DS, Jo SV, Lee AR, Park CH, Eun CS, Lee Y (2019) Characteristics and pathogenic role of adherent-invasive Escherichia coli in inflammatory bowel disease: potential impact on clinical outcomes. PLoS One 14(4):e0216165
Li L, He Y, Yang H (2005) Genetic characteristics of human Enterovirus 71 and Coxsackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J Clin Microbiol 43(8):3835–3839
Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, Holtz LR (2015) Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 21(10):1228–1234
Lima-Mendez G, Van Helden J, Toussaint A, Leplae R (2008) Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics 24:863–865
Lin DM, Lin HC (2019) A theoretical model of temperate phages as mediators of gut microbiome dysbiosis. F1000Research 8. https://doi.org/10.12688/f1000research.18480.1
Lin R, Liu W, Piao M, Zhu H (2017) A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids 49:2083–2090
Lindfors K, Lin J, Lee H-S, Hyöty H, Nykter M, Kurppa K, Liu E, Koletzko S, Rewers M, Hagopian W, Toppari J, Ziegler A-G, Akolkar B, Krischer JP, Petrosino JF, Lloyd RE, Agardh D, TEDDY Study Group (2020) Metagenomics of the faecal virome indicate a cumulative effect of enterovirus and gluten amount on the risk of coeliac disease autoimmunity in genetically at risk children: the TEDDY study. Gut 69(8):1416–1422
Lipman TH, Chang Y, Murphy KM (2002) The epidemiology of type 1 diabetes in children in Philadelphia 1990–1994: evidence of an epidemic. Diabetes Care 25(11):1969–1975
Liu M, Deora R, Doulatov SR et al (2002) Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295:2091–2094
Llanos-Chea A, Citorik RJ, Nickerson KP, Ingano L, Serena G, Senger S, Lu TK, Fasano A, Faherty CS (2019) Bacteriophage therapy testing against Shigella flexneri in a novel human intestinal organoid-derived infection model. J Pediatr Gastroenterol Nutr 68(4):509–516
Lozupone CA, Stombaugh JI, Gordon JI et al (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230
Łusiak-Szelachowska M, Weber-Dąbrowska B, Jończyk-Matysiak E, Wojciechowska R, Górski A (2017) Bacteriophages in the gastrointestinal tract and their implications. Gut Pathog 9:44
Lynch SV, Pedersen O (2016) The human intestinal microbiome in health and disease. N Engl J Med 375(24):2369–2379. https://doi.org/10.1056/NEJMra1600266
Ma Y, You X, Mai G, Tokuyasu T, Liu C (2018) A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 6:24
Mai V, Draganov PV (2009) Recent advances and remaining gaps in our knowledge of associations between gut microbiota and human health. World J Gastroenterol 15:81–85
Maisonneuve C, Irrazabal T, Martin A, Girardin SE, Philpott DJ (2017) The impact of the gut microbiome on colorectal cancer. Annu Rev Cancer Biol 2:229–249
Manoussopoulos YN, Anastassopoulou CG (2020) Virome: the prodigious little cousin of the family. In: Microbiomics, pp 53–73
Manrique P, Dills M, Young M (2017) The human gut phage community and its implications for health and disease. Viruses 9:141. https://doi.org/10.3390/v9060141
Marbouty M, Thierry A, Koszul R (2020) Phages—bacteria interactions network of the healthy human gut. https://doi.org/10.1101/2020.05.13.093716
Marinelli LJ, Fitz-Gibbon S, Hayes C, Bowman C, Inkeles M, Loncaric A, Russell DA, Jacobs-Sera D, Cokus S, Pellegrini M, Kim J (2012) Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. MBio 3(5):e00279
Markine-Goriaynoff N, Gillet L, Van Etten JL et al (2004) Glycosyltransferases encoded by viruses. J Gen Virol 85:2741–2754
Maronek M, Link R, Ambro L, Gardlik R (2020) Phages and their role in gastrointestinal disease: focus on inflammatory bowel disease. Cell 9(4):1013. https://doi.org/10.3390/cells9041013
Martín R, Langella P (2019) Emerging health concepts in the probiotics field: streamlining the definitions. Front Microbiol 10:1047
Maruvada P, Leone V, Kaplan LM, Chang EB (2017) The human microbiome and obesity: moving beyond associations. Cell Host Microbe 22(5):589–599
Marz M, Beerenwinkel N, Drosten C, Fricke M, Frishman D, Hofacker IL, Hoffmann D, Middendorf M, Rattei T, Stadler PF, Töpfer A (2014) Bioinformatics 30:1793–1799
Maslov S, Sneppen K (2017) Population cycles and species diversity in dynamic kill-the-winner model of microbial ecosystems. Sci Rep 7:39642
Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF (2018) The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359:104–108
Maurice CF (2019) Considering the other half of the gut microbiome: bacteriophages. mSystems 4(3). https://doi.org/10.1128/mSystems.00102-19
McGeoch DJ, Rixon FJ, Davison AJ (2006) Topics in herpesvirus genomics and evolution. Virus Res 117(1):90–104
McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32:20–25
McLaughlin MR, Balaa MF, Sims J, King R (2006) Isolation of Salmonella bacteriophages from swine effluent lagoons. J Environ Qual 35(2):522–528
Méndez-Toss M, Griffin DD, Calva J, Contreras JF, Puerto FI, Mota F, Guiscafré H, Cedillo R, Munoz O, Herrera I, López S (2004) Prevalence and genetic diversity of human astroviruses in Mexican children with symptomatic and asymptomatic infections. J Clin Microbiol 42(1):151–157
Mettenleiter TC, Klupp BG, Granzow H (2006) Herpesvirus assembly: a tale of two membranes. Curr Opin Microbiol 9(4):423–429
Mi S, Lee X, Li XP, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403(6771):785–789
Minot S, Sinha R, Chen J et al (2011) The human gut virome: interindividual variation and dynamic response to diet. Genome Res 21(10):1616–1625
Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD (2013) Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A 110(30):12450–12455
Mirzaei MK, Maurice CF (2017) Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol 15(7):397–408
Mirzaei MK, Khan MAA, Ghosh P, Taranu ZE, Taguer M, Ru J, Chowdhury R, Kabir MM, Deng L, Mondal D, Maurice CF (2020) Bacteriophages isolated from stunted children can regulate gut bacterial communities in an age-specific manner. Cell Host Microbe 27(2):199–212.e5
Moeller AH, Caro-Quintero A, Mjungu D et al (2016) Cospeciation of gut microbiota with hominids. Science 353:380–382
Mokili JL, Rohwer F, Dutilh BE (2012) Metagenomics and future perspectives in virus discovery. Curr Opin Virol 2:63–77
Montassier E, Gastinne T, Vangay P, Al-Ghalith GA, Bruley Des Varannes S, Massart S, Moreau P, Potel G, de La Cochetière MF, Batard E et al (2015) Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment Pharmacol Ther 42:515–528
Morahan G (2012) Insights into type 1 diabetes provided by genetic analyses. Curr Opin Endocrinol Diabetes Obes 19:263–270
Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB et al (2012) Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13:R79
Mu Q, Tavella VJ, Luo XM (2018) Role of Lactobacillus reuteri in human health and diseases. Front Microbiol 9:757
Mukhopadhya I, Segal JP, Carding SR, Hart AL, Hold GL (2019) The gut virome: the “missing link” between gut bacteria and host immunity? Therap Adv Gastroenterol 12:1756284819836620
Nagpal R, Newman TM, Wang S, Jain S, Lovato JF, Yadav H (2018) Obesity-linked gut microbiome Dysbiosis associated with derangements in gut permeability and intestinal cellular homeostasis independent of diet. J Diabetes Res 2018:1–9
Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, Li X, Szeto CH, Sugimura N, Lam TYT et al (2018) Alterations in enteric Virome are associated with colorectal Cancer and survival outcomes. Gastroenterology 155:529–541.e5
Needell JC, Zipris D (2016) The role of the intestinal microbiome in type 1 diabetes pathogenesis. Curr Diab Rep 16:89
Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S (2012) Host-gut metabolic interactions. Science 336:1262–1268
Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M (1997) A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun 241(1):92–97
Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P et al (2015) Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160:447–460
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834
Offermanns S, Rosenthal W (2008) Encyclopedia of molecular pharmacology, 2nd edn. Springer, Berlin, pp 437–438
Oh J-H, Lin XB, Zhang S, Tollenaar SL, Özçam M, Dunphy C, Walter J, van Pijkeren J-P (2019) Prophages in Lactobacillus reuteri are associated with fitness trade-offs but can increase competitiveness in the gut ecosystem. Appl Environ Microbiol 86(1):e01922. https://doi.org/10.1128/AEM.01922-19
Okamoto H, Nishizawa T, Takahashi M, Asabe S, Tsuda F, Yoshikawa A (2001) Heterogeneous distribution of TT virus of distinct genotypes in multiple tissues from infected humans. Virology 288(2):358–368
Okamura A, Yoshioka M, Kubota M, Kikuta H, Ishiko H, Kobayashi K (1999) Detection of a novel DNA virus (TTV) sequence in peripheral blood mononuclear cells. J Med Virol 58(2):174–177
Ott A (1999) Diabetes mellitus and the risk of dementia: the Rotterdam study. Neurology 53:1937–1942
Ott SJ, Waetzig GH, Rehman A, Moltzau-Anderson J, Bharti R, Grasis JA, Cassidy L, Tholey A, Fickenscher H, Seegert D, Rosenstiel P, Schreiber S (2017) Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 152(4):799–811.e7
Palmer C, Bik EM, Di Giulio DB et al (2007) Development of the human infant intestinal microbiota. PLoS Biol 5:e177
Panaccione R (2013) Mechanisms of inflammatory bowel disease. Gastroenterol Hepatol 9(8):529–532
Park A, Zhao G (2018) Mining the virome for insights into type 1 diabetes. DNA Cell Biol 37(5):422–425
Patel A, Noble RT, Steele JA, Schwalbach MS, Hewson I, Fuhrman JA (2007) Nat Protoc 2:269–276
Pérez-Brocal V, García-López R, Nos P, Beltrán B, Moret I, Moya A (2015) Metagenomic analysis of Crohn’s disease patients identifies changes in the virome and microbiome related to disease status and therapy, and detects potential interactions and biomarkers. Inflamm Bowel Dis 21:2515–2532
Pfeiffer JK, Virgin HW (2016) Viral immunity: transkingdom control of viral infection and immunity in the mammalian intestine. Science 351. https://doi.org/10.1126/science.aad5872
Pickup JC (2004) Inflammation and activated innate immunity in the pathogenesis of type 2 diabetes. Diabetes Care 27:813–823
Pollock J, Glendinning L, Wisedchanwet T, Watson M (2018) The madness of microbiome: attempting to find consensus “best practice” for 16S microbiome studies. Appl Environ Microbiol 84:e02627
Popgeorgiev N, Temmam S, Raoult D, Desnues C (2013) Describing the silent human virome with an emphasis on giant viruses. Intervirology 56(6):395–412
Prado J, Monezi T, Amorim A, Lino V, Paladino A, Boccardo E (2018) Human polyomaviruses and cancer: an overview. Clinics 73:e558s
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. https://doi.org/10.1038/nature08821
Qin J, Li Y, Cai Z et al (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60
Rachael R (2012) Type 2 diabetes linked to common virus. In: My health news daily, p 1
Rascovan N, Duraisamy R, Desnues C (2016) Metagenomics and the human virome in asymptomatic individuals. Annu Rev Microbiol 70:125–141
Rasmussen TS, Mentzel CMJ, Kot W, Castro-Mejía JL, Zuffa S, Swann JR, Hansen LH, Vogensen FK, Hansen AK, Nielsen DS (2020) Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut 69(12):2122–2130. https://doi.org/10.1136/gutjnl-2019-320005
Reyes A, Haynes M, Hanson N, Angly FE, Andrew C, Rohwer F, Gordon JI (2010) Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–338
Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI (2012) Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol 10:607–617. https://doi.org/10.1038/nrmicro2853
Richardson SJ, Willcox A, Bone AJ (2009) The prevalence of enteroviral capsid protein vp1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia 52(6):1143–1151
Ridaura VK, Faith JJ, Rey FE et al (2013) Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214
Riesenfeld CS, Schloss PD, Handelsman J (2004) Annu Rev Genet 38:525–552
Robinson CM, Pfeiffer JK (2014) Viruses and the microbiota. Annu Rev Virol 1:55–69
Robinson CM, Jesudhasan PR, Pfeiffer JK (2014) Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 15(1):36–46
Rohwer F, Seguritan V, Choi DH, Segall AM, Azam F (2001) Production of shotgun libraries using random amplification. Biotechniques 31:108–118
Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP et al (2018) Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359:91–97
Roux S, Krupovic M, Poulet A et al (2012) Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One 7:e40418
Roux S, Enault F, Hurwitz BL, Sullivan MB (2015) VirSorter: mining viral signal from microbial genomic data. Peer J 2015:1–20
Roux S, Solonenko NE, Dang VT, Poulos BT, Schwenck SM, Goldsmith DB, Coleman ML, Breitbart M, Sullivan MB (2016) Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. Peer J 4:e2777
Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K (2018) Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr 57:1–24
Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW (2013) Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/_-catenin signaling via its fad a Adhesin. Cell Host Microbe 14:195–206
Russell L, Monaghan T, Kao D (2018) The need to move away from fecal transplant towards targeted, refined microbiome therapy [Review of The need to move away from fecal transplant towards targeted, refined microbiome therapy]. J Thorac Dis 10(10):E755–E757
Ryan KJ, Ray CG (2004) Sherris medical microbiology, 4th edn. McGraw Hill, New York, pp 566–569
Santiago-Rodriguez TM, Hollister EB (2019) Human Virome and disease: high-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut. Viruses 11(7):656. https://doi.org/10.3390/v11070656
Santos SF, de Oliveira HL, Yamada ES, Neves BC, Pereira A Jr (2019) The gut and Parkinson’s disease-a bidirectional pathway. Front Neurol 10:574
Sarah S (2012) Could a common virus Lead to type 2 diabetes in seniors? In: My health news daily, p 1
Sausset R, Petit MA, Gaboriau-Routhiau V, De Paepe M (2020) New insights into intestinal phages. Mucosal Immunol 13(2):205–215. https://doi.org/10.1038/s41385-019-0250-5
Scarpellini E, Ianirob G, Attili F, Bassanellid C, De Santisd A, Gasbarrinib A (2015) The human gut microbiota and virome: potential therapeutic implications. Dig Liver Dis 47:1007–1012
Shkoporov AN, Hill C (2019) Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 25(2):195–209
Shkoporov AN, Ryan FJ, Draper LA, Forde A, Stockdale SR, Daly KM, McDonnell SA, Nolan JA, Sutton TDS, Dalmasso M, McCann A, Ross RP, Hill C (2018) Microbiome 6:68
Silveira CB, Rohwer FL (2016) Piggyback-the-winner in host-associated microbial communities. NPJ Biofilms Microbiomes 2:16010
Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P (2017) The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol 15(10):630–638. https://doi.org/10.1038/nrmicro.2017.58
Srividhya KV, Alaguraj V, Poornima G, Kumar D, Singh GP, Raghavenderan L, Mohan Katta AVSK, Mehta P, Krishnaswamy S (2007) Identification of prophages in bacterial genomes by dinucleotide relative abundance difference. PLoS One 2:e1193
Stern A, Mick E, Tirosh I, Sagy O, Sorek R (2012) CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res 22(10):1985–1994
Sutton TDS, Hill C (2019) Gut bacteriophage: current understanding and challenges. Front Endocrinol 10:784
Sutton TDS, Clooney AG, Ryan FJ, Ross RP, Hill C (2019) Choice of assembly software has a critical impact on virome characterisation. Microbiome 7:1–15
Sweet C (1999) The pathogenicity of cytomegalovirus. FEMS Microbiol Rev 23(4):457–482
Tetz G, Brown SM, Hao Y, Tetz V (2018) Parkinson’s disease and bacteriophages as its overlooked contributors. Sci Rep 8(1):10812
Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F (2009) Laboratory procedures to generate viral metagenomes. Nat Protoc 4:470–483
Tilg H, Adolph TE, Gerner RR, Moschen AR (2018) The intestinal microbiota in colorectal cancer. Cancer Cell 33:954–964
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M (2009) A core gut microbiome in obese and lean twins. Nature 457(7228):480–484
Urisman A, Fischer KF, Chiu CY, Kistler AL, Beck S, Wang D, DeRisi JL (2005) E-Predict: a computational strategy for species identification based on observed DNA microarray hybridization patterns. Genome Biol 6:R78
van der Heijden M, de Vries M, van Steenbeek FG, Favier RP, Deijs M, Brinkhof B, Rothuizen J, van der Hoek L, Penning LC (2012) Sequence-independent VIDISCA-454 technique to discover new viruses in canine livers. J Virol Methods 185:152–155
Van Dyne EA, Henley SJ, Saraiya M, Thomas CC, Markowitz LE, Benard VB (2018) Trends in human papillomavirus–associated cancers—United States, 1999–2015. MMWR Morb Mortal Wkly Rep 67:918–924
Vanchiere JA, Abudayyeh S, Copeland CM, Lu LB, Graham DY, Butel JS (2009) Polyomavirus shedding in the stool of healthy adults. J Clin Microbiol 47(8):2388–2391
Veziant J, Gagnière J, Jouberton E, Bonnin V, Sauvanet P, Pezet D, Barnich N, Miot-Noirault E, Bonnet M (2016) Association of colorectal cancer with pathogenic Escherichia coli: focus on mechanisms using optical imaging. World J Clin Oncol 7:293
Vibin J, Chamings A, Collier F, Klaassen M, Nelson TM, Alexandersen S (2018) Metagenomics detection and characterisation of viruses in faecal samples from Australian wild birds. Sci Rep 8:1–23
Vinner GK, Richards K, Leppanen M, Sagona AP, Malik DJ (2019) Microencapsulation of enteric bacteriophages in a pH-responsive solid oral dosage formulation using a scalable membrane emulsification process. Pharmaceutics 11(9):475. https://doi.org/10.3390/pharmaceutics11090475
Virgin HW (2014) The virome in mammalian physiology and disease. Cell 157(1):142–150
Virgin HW, Wherry EJ, Ahmed R (2009) Redefining chronic viral infection. Cell 138(1):30–50
Waller AS, Yamada T, Kristensen DM et al (2014) Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J 8:1391–1402
Wang D (2020) 5 challenges in understanding the role of the virome in health and disease. PLoS Pathog 16(3):e1008318
Wang W, Jovel J, Halloran B et al (2015) Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory bowel diseases reveals interplay of viruses and bacteria. Inflamm Bowel Dis 21:1419–1427
Wang Z, Koonen D, Hofker M, Fu J (2016) Gut microbiome and lipid metabolism: from associations to mechanisms. Curr Opin Lipidol 27:216–224
Warwick-Dugdale J, Solonenko N, Moore K, Chittick L, Gregory AC, Allen MJ, Sullivan MB, Temperton B (2019) Long-read viral metagenomics captures abundant and microdiverse viral populations and their niche-defining genomic islands. Peer J 7:e6800
Wegley L, Edwards R, Rodriguez-Brito B et al (2007) Metagenomic analysis of the microbial community associated with the coral porites astreoides. Environ Microbiol 9:2707–2719
Weinbauer MG (2004) Ecology of prokaryotic viruses. FEMS Microbiol Rev 28:127–181
Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F (2009) Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One 4:e7370
Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, Relman DA, Angly FE, McDole T, Mariella RP, Rohwer F, Haynes M (2011) Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci 108(supplement 1):4547–4553
Winter C, Bouvier T, Weinbauer MG, Thingstad TF (2010) Trade-offs between competition and defense specialists among unicellular planktonic organisms: the “killing the winner” hypothesis revisited. Microbiol Mol Biol Rev 74(1):42–57
Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, XuW XX, Kwong TNY, Tsoi H, Wu WKK et al (2017) Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology 153:1621–1633
Xie H, Guo R, Zhong H, Feng Q, Lan Z, Qin B, Ward KJ, Jackson MA, Xia Y, Chen X, Chen B, Xia H, Xu C, Li F, Xu X, Al-Aama JY, Yang H, Wang J, Kristiansen K, Wang J, Steves CJ, Bell JT, Li J, Spector TD, Jia H (2016) Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst 3:572–584.e3
Yalchin M, Segal JP, Mullish BH, Quraishi MN, Iqbal TH, Marchesi JR, Hart AL (2019) Gaps in knowledge and future directions for the use of faecal microbiota transplant in the treatment of inflammatory bowel disease. Therap Adv Gastroenterol 12:1756284819891038
Ye SH, Siddle KJ, Park DJ, Sabeti PC (2019) Benchmarking metagenomics tools for taxonomic classification. Cell 178:779–794
Zhang Z, Geng J, Tang X, Fan H, Xu J, Wen X et al (2014) Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISMEJ 8:881–893. https://doi.org/10.1038/ismej.2013.185
Zhang X, Niu YD, Nan Y, Stanford K, Holley R, McAllister T, Narváez-Bravo C (2019) SalmoFreshTM effectiveness in controlling Salmonella on romaine lettuce, mung bean sprouts and seeds. Int J Food Microbiol 305:108250
Zhao G, Vatanen T, Droit L, Park A, Kostic AD, Poon TW, Vlamakis H, Siljander H, Härkönen T, Hämäläinen A-M, Peet A, Tillmann V, Ilonen J, Wang D, Knip M, Xavier RJ, Virgin HW (2017) Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci U S A 114(30):E6166–E6175
Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS (2011) Nucleic Acids Res 39:347–352
Zipris D (2008) Innate immunity and its role in type 1 diabetes. Curr Opin Endocrinol Diabetes Obes 15(4):326–331
Zipris D (2009) Epidemiology of type 1 diabetes and what animal models teach us about the role of viruses in disease mechanisms. Clin Immunol 131(1):11–23
Zoetendal EG, Vaughan EE, de Vos WM (2006) A microbial world within us. Mol Microbiol 59:1639–1650. https://doi.org/10.1111/j.1365-2958.2006.05056.x
Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W, Ching JYL, Chan PKS, Chan MCW, Wu JCY, Chan FKL, Yu J, Sung JJY, Ng SC (2018) Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 67(4):634–643
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
AboNahas, H.H. et al. (2022). Virome and Microbiome Interaction and Their Implication in the Human Gut Health. In: Sayyed, R.Z., Khan, M. (eds) Microbiome-Gut-Brain Axis. Springer, Singapore. https://doi.org/10.1007/978-981-16-1626-6_4
Download citation
DOI: https://doi.org/10.1007/978-981-16-1626-6_4
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-1625-9
Online ISBN: 978-981-16-1626-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)