Abstract
Tuberculosis caused by Mycobacterium tuberculosis (M. tb) with a global burden of 10 million cases in 2019 causes morbidity and mortality if not treated. The constant rise in drug-resistant TB has further aggravated the problems and proves to be the major roadblock in combatting TB. Complete understanding of the pathogen’s physiology and its virulence attributes is essentially required and is important in designing new treatment strategies. M. tb multi-omics strategies are proving to be very useful in gaining insights into the disease. Despite the availability of genomic and transcriptomic data, the pathogenic potential, survival strategies, persistence, immunomodulation, drug resistance mechanisms and the host-pathogen interactions remain poorly understood. Proteomics approaches thus prove to be more informative for studying mycobacteria providing minute details about the true state of the cell under various conditions. Protein profiling of different strains of mycobacteria, clinically relevant, as well as drug-resistant isolates, has tremendously increased our knowledge in the understanding of disease mechanism. Proteomics and bioinformatics approaches have helped greatly in identification and characterization of target proteins which can be exploited for novel therapeutics. This book chapter provides an update on the different proteomics technologies and their application in unravelling TB physiology.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Mycobacterium tuberculosis (M. tb), the aetiological agent for tuberculosis (TB), is ranked as one of the topmost killers among all infectious agents [1]. The major challenges for the management of TB include the rise of multidrug-resistant (MDR) TB, extensively drug-resistant TB (XDR), HIV (human immunodeficiency virus) co-infection and poverty [2]. There is no effective vaccine, and the only vaccine available is the century-old Bacillus Calmette-Guerin (BCG). However, the variable protection of BCG in adults has posed a serious threat to TB elimination programme worldwide [3]. The current therapy comprises first-line drugs, which take 6 to 9 months for completion and have serious side effects [4]. Treatment of MDR/XDR requires much longer duration and includes TB drugs from the second line in addition to first-line drugs, pyrazinamide and a high dose of isoniazid [5]. Despite these treatment regimes, the rise in MDR and XDR cases has created new hurdles for pre-existing drug therapy [6]. Hence, there is an unmet medical need to develop an effective vaccine and improved drugs for TB management.
M. tb is transmitted through inhalation of airborne aerosol bearing the pathogen [7]. Upon entry of M. tb pathogen into the lungs, it infects alveolar macrophages and bypasses the host immune response for its survival and pathogenesis [8]. To further ensure its survival, mycobacteria also dampen anti-mycobacterial defence mechanism utilised by macrophages including autophagy, phagosome acidification and production of reactive oxygen and nitrogen species [9,10,11]. Also, infected alveolar macrophages then secrete chemokines that attract inflammatory cells including neutrophils, macrophages and natural killer cells, further promoting inflammation and formation of multinucleated giant cells called granulomas [12]. These granulomas thus provide a niche for the containment of bacteria and also serve as a reservoir for the spread of the infection.
M. tb-secreted proteins (secretome) play a critical role in subverting the immune response and intracellular growth [13, 14]. Early secreted antigen (ESAT-6), an essential virulence factor of M. tb, is known for regulation of host immune response by inhibiting pro-inflammatory responses, such as interferon (IFN) gamma production [15] and interleukin (IL)-12 production [16]. Furthermore, ESAT -6 stimulates IL-6 production in macrophages [17]. Besides, ESAT-6 also plays an important role in inducing macrophage polarisation and transition into epithelioid macrophages, the major constituent of TB granuloma [18, 19]. It was further demonstrated that M. tb secreted effector Rv1988 localises to the host nucleus and methylate host histone proteins and thus epigenetically modulates macrophage’s anti-mycobacterial functions [20]. These studies cumulatively suggest the critical role of M. tb proteins in the regulation of host functions, and hence, for the understanding of M. tb-associated disease pathology, characterisation of M. tb proteome is warranted [21]. In addition to characterising the role of M. tb proteins in virulence, the M. tb proteome could also be explored for potential antigens that can be utilised as an effective vaccine candidate. In this regard, high-throughput proteome-wide screening of potential M. tb antigens could be helpful in the generation of novel vaccines [22].
Although the genomic makeup of M. tb has been extensively studied, the proteome analysis of M. tb lags due to the complex protocols for M. tb protein isolation and need for sophisticated instrumentation [23]. Also, the proteome of XDR M. tb revealed that more than 30% are hypothetical proteins that have not been characterised to date [24]. The lack of comprehensive exploration of M. tb proteome has further widened the gap in understanding the virulence and pathogenesis of M. tb. Therefore, unravelling the proteomic makeup of M. tb will further be helpful in better understanding of the physiology and virulence of M. tb, which may reveal novel drug targets [23]. Altogether these efforts may help achieve the WHO’s End TB strategy. M. tb H37Rv complete genome of 4.4 Mb consists of around 3924 genes [25]. The identification and characterisation of all the genes are important, but attention should be given to the gene products responsible for virulence and pathogenic attributes. These virulence factors can then be quantitated using proteomics approach amounting for the difference in pathogenicity and drug resistance among lineages.
2 Investigation of Mycobacteria Using Proteomics Approach
Proteomics proves to be an important tool in the identification of novel protein targets which are part of pathogen survival strategies, defence responses by the host and subsequently, the host-pathogen interactions. The upregulation and downregulation of host immune-related proteins and virulence-related proteins of pathogens are indicative of their role in defence or pathogenesis. These upregulation and downregulation are useful in the identification of proteins which may prove to be important as drug targets or development of diagnostic tools representing various stages of the pathogenesis and the level of advancement of the infection. Starting with the identification of any such protein to establishing it as a drug target or a diagnostic marker and to monitor the kinetics of protein contents of different organs in response to infections, we need to proceed with an approach involving the following steps in a sequential pattern:
Identification of novel targets
-
1.
Proteomic analysis
-
2.
Bioinformatic analysis
-
3.
Biochemical and biophysical characterisation
-
4.
Structural determination and functional correlation
2.1 Proteomic Analysis
The whole machinery of a cell (and even acellular living forms) is operated, multiplicated and regulated by the proteins. Thus, the holistic, as well as the individual, study of proteins becomes imperative. Proteomics plays a pivotal role in the identification of novel protein targets which are part of pathogen survival strategies, defence responses by the host and subsequently, the host-pathogen interactions. The differential proteomics investigations output evolved from a large number of proteins laid the foundation of new hypotheses and verification of their functions. Intracellular bacteria have evolved with different mechanisms to interfere with host defence system and are successful as a causative agent of many infectious diseases in human. Particularly, intracellular vacuoles are used as an essential niche by these pathogens to overtake the cellular functions facilitating their replication and survival. M. tb, the causative agent of tuberculosis in human, has not only been able to alter its intraphagosomal fate by blocking phagosome maturation but has also devised strategies to withhold the actions by immune cells and to survive for the long term in the host successfully. Proteomics based on mass spectrometry (MS) and two-dimensional gel electrophoresis (2D-PAGE) followed by western transfer and assisted by N-terminal sequencing (Edman degradation) helps in the identification and quantitative analysis of complex protein mixtures (Fig. 17.1). It is increasingly employed to investigate host-pathogen interactions (Fig. 17.1).
M. tb, the causative agent of tuberculosis, has got much attention from the research community in the near past. Still, it remains one of the leading causes of death and sufferings caused by an infective disease. Today, we can decipher the nucleotide sequence of a prokaryotic genome within hours. However, based on genomic sequence information, it remains unpredictable to characterise the functional properties. Till now, major efforts have focused on the features of the genomic organisation of the tuberculosis pathogen. The genome of more than 10,000 different M. tb strains with varying genotypes and phenotypes has been studied. However, to decipher the causes behind drug resistance and pathogenicity, the application of the whole genome sequencing technology and comparative analysis proves to be limited. The majority of the point mutations that distinguish groups of strains have been found in the promoter regions of the genes and/or regions encoding proteins with a hypothetical function and playing an unknown role in the physiology of mycobacteria. In this context, a functional analysis of the information deciphered in the pathogen genome performed using proteomic testing, including quantitative proteomics, becomes relevant.
The features of the organisation of the cell wall, which is resistant to environmental factors, acids and alkalis, make M. tb a rather complex target for proteomic analysis. This, in its turn, requires the development of unique conditions for protein extraction. The implemented protocols of proteomic analysis of M. tb should also be sufficiently effective, taking into account the complexity of accumulation of a large bacterial mass due to the prolonged culture growth.
2.1.1 Major Gains Using Proteomics in Case of M. tb
Proteomic profiles of virulent H37Rv strains and avirulent strains of M. tb are being used to identify potential candidate for vaccine. Proteomic characterisation of H37Rv suggests a change in DosR regulon protein in hypoxic condition. Comparative proteomic analysis of the proteins of a latent H37Rv strains at the exponential, logarithmic and stationary growth phases was done using the technique of site-specific labelling of cysteine residues (isotope-coded affinity tags, CAT) based on covalent labelling of the cysteine residue in the polypeptide chain by chemically identical but isotopically different reagents. The study showed 193 and 241 proteins presents in exponential and stationary phase, respectively, mostly associated with energy metabolism and protein degradation.

Similarly, proteomic characterisation of Beijing strain shows its association with drug resistance and highly virulent nature. The comparative analysis with H37Rv shows the protein responsible for virulence factors, i.e. Rv0129c, Rv0831c, Rv1096, Rv3117 and Rv3804c, was higher in the Beijing strain than in H37Rv. Proteins Hsp65 (Rv0440), Pst1 (Rv0934) and Rv1886c are low in Beijing strain which helps to avoid host immune response. Furthermore, proteins of the efflux pump Rv0341, Rv2688c and Rv3728 were found only in the Beijing strains. Nowadays, the post-translational modifications (PTMs) of M. tb proteins and their implications are also identified by proteomic analysis like mannosylation decreases the virulence of M. tb. Mostly PTMs are responsible for the regulation of enzymatic activity, interaction with other molecules and lifetime of cellular proteins. Similarly, M. tb antigens are found in surface heparin-binding haemagglutinin that is used for the design of a new vaccine.
2.1.2 For the Proteomic Analysis, the Two-Way Approach Is Followed, Which Is Represented in the Flowchart (Figs. 17.2 and 17.3)
2.1.3 Major Techniques Used in the Proteomic Analysis
2.1.3.1 2D Gel Electrophoresis
This technique is most currently used and done before MS and bioinformatic analysis. It separates proteins according to their isoelectric points and molecular weights, and the major advantage of this technique is to separate differentially post-translationally modified forms of the same protein.
2.1.3.2 Mass Spectroscopy
The principle of MS analysis involves the conversion of the subject molecules to either cations or anions in the ion source, separation according to their mass/charge (m/z) ratios in the mass analyser and subsequent detection. Several configurations of mass spectrometers that combine ES and MALDI with a variety of mass analysers (linear quadrupole mass filter [Q], time-of-flight [ToF], quadrupole ion trap and Fourier transform ion cyclotron resonance [FTICR] instrument) are routinely used.
2.1.3.3 Edman Sequencing
Edman degradation is used to identify the sequence of a protein through labelling and cleaving the peptide without damaging the whole protein in hydrolytic condition. Phenyl isothiocyanate forms phenyl thiocarbamoyl derivative with the N-terminal in less acidic condition form cyclic derivative of PTH. This method can be repeated for the rest of the residues, separating one residue at a time.
2.1.3.4 ELISA (Enzyme-Linked Immunosorbent Assay)
In this technique antigen-antibody is used, it includes an enzyme-labelled antigen or antibody, and enzyme activity is measured calorimetrically. The enzyme activity is measured using a substrate that changes colour when modified by the enzyme. The light absorption of the product formed after substrate addition is measured and converted to numeric values. Depending on the antigen-antibody combination, the assay is called a direct ELISA, indirect ELISA, sandwich ELISA, competitive ELISA, etc.
2.1.3.5 Microarray
Microarray is a high-throughput technique used to identify and quantify a large number of proteins in a short time span. In the case of M. tb, it is effectively used for proteomic analysis of cell lysate, protein constituent of culture media as well as analysis of pleural fluid and other samples of tuberculosis patients.
2.2 Bioinformatic Analysis of Mycobacterial Proteome
Bioinformatic analysis of M. tb proteome has explored the host-pathogen interactions and immunomodulation by mycobacterial infection, protein-drug interactions, epitope-driven vaccine candidate and protein-protein interactions. The complex host-pathogen interactions should be quantified consistently for precision and personalised health care [23]. The structural analysis of the protein involves the determination of primary, secondary, tertiary and quaternary structure. Till now, 4983 crystal structures of M. tb proteins have been submitted in the Protein Data Bank repository which includes proteins (apoprotein, protein-ligand complex, DNA binding proteins, RNA binding proteins, small peptides and post-translationally modified proteins), nucleic acid and carbohydrates. The structural analysis explains the mechanism of action at the molecular levels and provides leads for the development of drug/vaccine. Proteomic analysis can be divided into the following sections.
2.2.1 Database/Tools of Virulence Factors
Virulence factors (VF) are the proteins which are involved in the host-pathogen interactions, the progression of disease and survival inside the host macrophages. There are several online and standalone tools, and databases of VF especially developed for M. tb or bacteria or to different disease-causing pathogens. The Pathosystems Resource Integration Centre (PATRIC) is a repository of integrated omics datasets (genome, transcriptome, protein-protein interactions, the 3D structure of proteins and sequenced data) [26]. The virulence factor database (VFDB) contains the whole genome, sequence, structural and functional data which emphasise on the common and species-or strain-specific VFs. VFDB pipeline is based on comparative pathogenomics and VF analysis. The newly developed VFDB 2019 version has additional pathogenic genera Francisella and Klebsiella [27]. There are other tools which are based on machine learning approaches like VCIMPred, VirulentPred, EffectiveT3 and T4EffPred for the recognition of VFs. VRprofile is a bacterial genome-based server which works on backend database named MobilomeDB that comprises gene cluster loci of bacterial mobile genetic elements which include type III, IV, VI and/or VII secretion apparatuses (T3SSs, T4SSs, T6SSs and/or T7SS), integrative conjugative elements (ICEs), prophages, class I integrons, insertion sequence (IS) elements, pathogenicity islands (PAIs) and antibiotic resistance islands (GIs) (ARIs) [28].
2.2.2 Interactome Analysis of M. tb
TragetTB, MycoPrint, SinCRe-structural interactome computational resource, CHOPIN, Mtb-HID (M. tb-Human Protein-Protein Interaction Map), Prediction of Pathogenic Proteins in Metagenomic Datasets (or MP3) and PHI-base (pathogen-host interactions base) are the tools and databases which explore the host-pathogen interactions and protein-protein interactions based on genome, proteome, sequence, structural and functional annotations. STRING database has been utilised to decipher the protein-protein interactions of M. tb H37Rv. It helped to unravel the possible pathways which lead to drug resistance [29]. TargetTB involves the structural analysis of genome, reactome, interactome, sequence, experimentally validated phenotypic essential data and assessment of the drug on a structural basis by the novel algorithm [30]. MycoPrint and VirulentPred are based on support vector machine (SVM) for the exploration of the interactome of M. tb [31]. MP3 is based on SVM and hidden Markov model for the prediction of pathogenic proteins [32]. Human-M. tb interactions can be analysed by M. tb-HID database, which consists of interactions between five M. tb strains (H37Rv, H37Ra, ATCC, 35801/TMC 107/Erdman, ATCC 35801 and CAS_ NITR204) and the human host. SinCRe interactome has been developed based on protein sequence, domain, functional annotation and tertiary structure which decipher protein-protein and protein-drug interactions and mutation potentially leading to drug resistance [32]. CHOPIN is a web-based interactome which deciphers the drug resistance associated with the mutation based on the structure of the protein (25833954). PHI-base is a database of host-pathogen interactions between several disease-causing agents and host [33]. These host-pathogen, protein-protein and protein-drug interactions which lead to immunomodulation in the host may play an important role in the development of drugs and vaccines.
2.2.3 Structural and Functional Analysis of M. tb Proteome in Drug Discovery
Structural analysis of M. tb proteome involves X-ray & NMR. The functional analysis is involved in the annotation of the protein based on its cellular, biological or molecular function. AgBase-GOanna v.2 is a web-based database which provides information for gene ontology [34]. Drug designing is one of the strategies based on the structural analysis for the development of novel drugs. It involves structure-based and ligand-based drug designing, quantitative structure-activity relationship (QSAR), pharmacophore modelling and virtual high-throughput screening for the scrutinisation of the drug molecule [35]. The potential drug candidates are further analysed by structure-activity relationship (SAR) to determine their specificity, sensitivity and activity [35]. Structural annotation of M. tb has been carried out based on its genome and proteome which has shown its impact on the pathogenesis, virulence, drug discovery, new drug identification and structure-based lead design [36].
2.3 Biochemical and Biophysical Characterisation
The biophysical and biochemical characterisation lays the essential foundation work for structure and functional studies of novel targets, by helping them to crystallise. For example, in 2018, Yang et al. cloned, expressed and purified the transcription factor EF-TU of M. tb, which may be used as a novel drug target. Dynamic light scattering suggested that it is present as monomeric form, and circular dichroism showed it is a well-structured protein. The ITC indicated that it has an intermediate affinity towards GTP and GDP, while ES-MS determined the molecular weight of protein. The structure modelling through docking suggested that they generally bind through H bonds. These experiments helped to understand the chemistry of the protein and their binding properties which also helped to explore the biochemical properties of the protein.
Fluorescence spectrometry helps us in the standardisation of emission/absorption maxima at a particular wavelength by measuring the fluorescence intensity at the particular wavelength. This can also be used for assessing any structural change in the protein as well as any protein-protein or protein-ligand binding interactions, again helpful in drug design.
Western blotting, followed by antibody binding and ELISA, is the most widely used confirmatory technique for biochemical and functional characterisation. The techniques are not only helpful in the quantification of particular protein but are also helpful in drug targeting and diagnostic developments. In drug targeting and diagnostic design, they have immense importance in dosimetry.
2.4 Structural Determination and Functional Correlation
Proteins are involved in many biological pathways as catalysing agent/inhibitors/activators/modifiers by interacting with other macromolecules. Structure determination is a process by which the three-dimensional atomic coordinates and interaction within and other macromolecule are observed using X-ray crystallography, NMR spectroscopy, cryo-electron microscopy and molecular modelling. These noncovalent interactions help to stabilise the 3D structure of the molecule. The specificity and affinity of these interactions are exclusive to biological functions and facilitate many chemical and physical processes. X-ray crystallography is a method to determine the interatomic spacing of most crystalline solids by putting them as a diffraction gradient of X-rays of wavelength 1 Å in order to produce a high-quality structure, while in NMR the applied magnetic field orients atomic nuclei within or against the field. The nuclei absorb EM radiation to fill this energy gap which determines the composition of mixture or product formed during the reaction and also determines the number of hydrogens attached to each carbon. Cryo-EM is a revolutionary technique taking over X-ray crystallography to determine the structure by exposing the flash freeze sample in an electron beam in order to reconstruct the structure of protein and protein complex, and it helps to visualise the protein which is not able to crystallise. This can be well exemplified by the study of Lin et al. 2017 who determined the structure of M. tb RNA polymerase at 3.8 Å through X-ray crystallography which is the target of first-line drug rifampicin which revealed that it inhibits the extension of RNA through stearic occlusion mechanism. Also, non-related RIF compound inhibits the RNA polymerase with no cross-reaction with rifampicin, so if administered together, it will inhibit the growth of M. tb effectively (PDB ID 5UHA). Likewise, CryoEM structure of M. tb 50S and 70S ribosomal subunit determined by Yang et al. in 2019 suggested that the inter-bridge in 70S helps to understand the structural basis of translation in M. tb which led to the development of new drugs (PDB ID 5V93).
3 Differential Proteomic Expression Profiles of H37Rv, H37Ra and BCG and Other Clinical Isolates
The accessibility of genome sequences [37] has facilitated genome-wide comparisons of different mycobacterial species to identify gene mutations, deletions or insertions correlated with its virulence and pathogenic characteristics [38]. Proteomic analyses basically counterpart the genomic data in presenting whichever genes are really expressing and reflect the functional status of the cell under different environment conditions [39]. The availability can expect advancement in the development of novel TB therapeutic measures and vaccine candidates of large-scale differential mycobacterial proteome data.
Proteomic analyses of different mycobacterium species and strains (M. tb H37Rv, H37Ra, BCG) have highlighted the importance of varied gene expression profiles of several proteins involved in survival strategies of the pathogen, emergence of drug resistance, host-pathogen interaction, etc. [40]. Proteome exploration of cellular proteins of M. tb and BCG strains demonstrates 13 proteins unique to M. tb H37Rv and 8 to BCG. These differences in the protein composition between attenuated and virulent strains of M. tb are supportive for the development of novel vaccine candidates and therapeutics [41].
Singh et al. compared proteomes of 12 different pathogenic mycobacterial strains using Insilco tools to investigate the virulence factors of the species and compared with 241 experimentally validated virulence factors of M. tb H37Rv. True, opportunistic and non-pathogenic strains have been found to share 66%, 52% and 34% identity, respectively, with M. tb virulent proteins. Conserved nature of virulent proteins among the genus Mycobacterium points towards their co-option and evolution. Insilco comparative analysis of M. tb with different opportunistic pathogens has shown variable expression and sequence similarities. Proteins belonging to phospholipase, transferase and ESX were observed to be less similar with Mycobacterium indicus pranii (MIP) proteome. Indeed four M. tb proteins from phospholipase family were shown to present in all other pathogenic mycobacterial members but absent in M. leprae. Unique conservation of 14 ESX-related proteins was found in M. bovis and M. marinum. Homologues of M. tb chorismate pyruvate lyase, Fad22, Mmpl17 and lipoprotein Lppx are present in pathogenic mycobacterial strains, thereby highlighting their significance in the virulence and pathogenesis. Two M. tb glycosyl transferases were conserved among M. tb H37Rv, M. bovis, M. leprae and M. marinum. One methyltransferase of M. tb is highly similar to that of M. bovis and M. leprae [42]. The conservation of these transferases emphasises the presumed importance of post-translational modifications in mycobacterial virulence mechanism [43].
Another class of M. tb proteins called PE/PPE/PE_PGRS family is differentially regulated in different strains of mycobacteria. Around 26 PE and 58 PPE proteins of M. tb H37Rv were found to be similar with 7 PE and 21 PPE proteins of MIP, respectively, pointing their importance in the modulation of host immune responses to favour the bacterial survival [42].
In a global protein-protein interaction study by proteome microarray in a yeast expression system, 14 different serum M. tb proteins were found to distinguish active TB patients from recovered individuals. These proteins can, therefore, be used as biomarkers for assessing the treatment outcomes [44]. Besides, proteome microarray revealed the likely regulation of M. tb rhamnose pathway by c-di-GMP (ubiquitous bacterial secondary messenger) [45] and Ser/Thr kinase PknG. This is associated with cell wall synthesis [46]. It shows that M. tb proteome microarray study can be applied to recognise novel molecular targets to combat TB.
The secreted proteins of M. tb that are exported intracellularly to host cells were thought to participate in the phagosomal remodelling and bacterial survival inside phagosomal compartments [47]. A study was conducted to identify expression profiles of culture supernatant proteins of M. tb virulent H37Rv and avirulent H37Ra strains by 2D gel electrophoresis. Protein expression of Rv2346c, Rv2347c, Rv1038c and Rv3620c has been evident in M. tb virulent strain but absent in avirulent bacterium. The location of these protein-coding genes traced in the corresponding area in M. tb H37Ra and multiple mutations was found to be associated with their different expression profiles. The 59th codon CAG coding for glutamine in virulent strain was replaced with a stop codon in H37Ra avirulent strain. This difference in the expression profiles is most likely associated with the attenuating characteristic of avirulent bacterium [40].
Quantitative proteomics studies have highlighted substantial distinctive proteome expression profiles of M. tb H37Rv and M. bovis BCG. The majority of differential expression has been designated particularly to pathways involved in lipid biosynthesis [41, 48] and different growth phases accompanied by nutrient starvation [49, 50]. Furthermore, another study demonstrated the varied proteome profiles of seven clinically significant mycobacterial species from MTBC complex including four M. tb strains (H37RV, LAM, Beijing, CAS), M. avium, M. bovis and M. bovis BCG using LC-MS/MS technique. The objective was to identify relevant phenotypic disparities in different pathogenic strains in terms of immune response generation, virulence and transmission. A total of 3788 unique M. tb proteins out of 4023 theoretical proteomes have been identified. Each of the MTBC members has been identified with an average of 3290 unique proteins, which represent around 82% of their total theoretical proteome. M. avium is represented by 4250 unique proteins that comprised 80% of its theoretical proteome. Although all the classes of proteins were found to be expressed in all strains, the significant quantitative difference was reported between different strains. Relative expression differences in virulence-related proteins have been shown among different strains contributing to bacterial fitness. A total of 989 proteins were similar in four M. tb strains, M. bovis and M. bovis BCG, but they do not share similarity with M. avium. Expression pattern of 168 proteins was uniquely present in M. tb strains only and absented in either of M. bovis BCG, M. bovis or M. avium. These unique proteins can, therefore, be allocated as virulence factors of M. tb that can be explored further. These unique 168 proteins were expressed under different conditions such as starvation, macrophage infection, hypoxia and acidic models that prove the plausible roles of these proteins in conferring an advantage to the bacterium in vivo [51].
PyrB, PyrC, CarA and CarB proteins are present in M. tb pyrimidine biosynthetic operon involved in drug resistance. The Beijing strain of MTBC has been shown to express the highest levels of CarA, PyrB and PyrC. Rv0966 is overexpressed in Latín American Mediterranean (M. tb LAM strain) only. Rv2108, Rv2136c, Rv1002c and Rv2703 are reported to be expressed at higher levels in the Beijing strain. Central Asian strain lineage (CAS strain) of M. tb when compared to other strains specifically upregulates Rv1818c expression. Rv0096, Rv2108, Rv2136c, Rv1002c and Rv2703 have been hypothesised to modulate host functions, and their expression level was enhanced in pathogenic M. tb strains only. Rv2136c is a well-known virulence factor of MTBC encoding undecaprenyl pyrophosphate phosphatase involved in lipid biosynthesis [51, 52]. Rv0833 encoding PE_PGRS33 and Rv3340 encoding O-acetyl homoserine sulfhydrylase MetC are involved in the growth of M. tb and appeared to be highly abundant in M. tb LAM strain, M. tb H37Rv and M. bovis in comparison to BCG and M. avium. Only M. tb LAM and M. avium were shown to have upregulated expression of Rv3621c encoded PPE65. An important toxin protein VapC2 encoded by Rv0301 has reported having the highest expression in the Beijing strain [53].
Within host macrophage, M. tb generally depends on the intracellular setting for the majority of the carbon and iron sources [54, 55]. Thus, in terms of adaption, Rv1346 (Acyl-CoA dehydrogenase MbtN) is profusely expressed in M. tb Beijing strain. Rv3709c encoded aspartate kinase (Ask) was comparatively more abundant in M. tb H37Rv and LAM but almost absent in M. bovis and M. avium. Relatively higher abundance of MetC and aspartate kinase in M. tb LAM strain provides the capacity to synthesise essential amino acids for the bacterium, thereby selectively favouring the bacterial virulence [51]. Rv1346 that encodes for acyl-CoA dehydrogenase, which is an enzyme required for the production of mycobactin (an essential molecule for iron acquisition in infected macrophages), is another important virulence factor for M. tb infection [56].
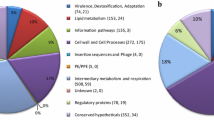
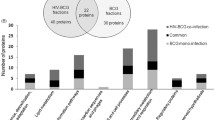
However, another systematic proteomic profiling of two M. tb strains H37Ra and H37Rv along with two clinical isolates BND-433 and JAL-2287 (belonging to CAS lineage) revealed significant insights into the differential protein expression patterns contributing to the differences in drug resistance and virulence capacity. Out of the total 2161 protein groups identified, which covered 54% of M. tb proteome, 257 protein groups were reported to be differentially expressed among different clinical isolates. A total of 13, 12, 13 and 22 proteins groups were found to be specifically expressed in H37Rv, H37Ra, BND and JAL strains, respectively. The majority of proteins expressed from 2161 M. tb proteins were reported to belong cell wall and cell processes (17.2%), intermediary metabolism and respiration (31%) and conserved hypothetical (22.2%), when categorised based on their functional importance [57].
The proteome of JAL strain was found to be significantly distinct in comparison to H37Rv, H37Ra and BND. Cluster gram of identified differentially expressed proteins showed major up- and downregulation of several proteins as compared to other strains. A substantial variation in the regulatory proteins such as transcription factor has been described which has possibly accounted for its intricate regulatory mechanism. Expression profiles of Mce1 and ESX operon proteins in BND and JAL strains, respectively, were very discrete. Proteins expressed from Mce1 operon are expressed in lesser amounts in BND strain [57]. Abrogation of an ESX protein ESAT-6 contributed to the diminished virulence capability of JAL but had minimal effects on other strains. M. tb H37Ra completely lacks ESAT-6 and is considered to be an avirulent strain in nature [58]. An interesting finding pointed the lower levels of Rv2780 encoding l-alanine dehydrogenase in JAL strain, which was the first identified antigen known to be absent in M. bovis BCG vaccine strain [41, 59].
Ninety of the 257 differentially expressed proteins have been identified as enzymes participating in 29 different metabolic cascades. Among those, five belong to membrane metabolism, and the other four belongs to redox metabolism. Both these differentially expressed protein groups are upregulated in JAL strain specifically. Among the 90 differential proteins, 12 have been specified to account in lipid metabolism performing the particular function “beta-oxidation of fatty acids” in all strains (M. tb H37RV, H37Ra, JAL and BND). One and six proteins from this group have been proved to be downregulated in M. tb H37RV and M. tb JAL clinical isolate, respectively. Moreover, this downregulation has not affected the overall lipid metabolism due to the overexpression of other redundant protein in these strains [57].
Altogether, these studies have pointed out a significant role of differential proteome profiles of different mycobacterium strains, which eventually affect the virulence and pathobiology of the pathogen during infection. Study of the distinct protein expression profiles of different strains will aid in better understanding of the biology of the pathogen.
4 Proteomics Studies Delineating the Differences Between Drug-Resistant and Sensitive Strains of M. tb
Tuberculosis caused by M. tb is an important public health concern, and the spike in drug-resistant TB cases has aggravated the issue. As per 2019 WHO report MDR/RR-TB cases accounted for 3.4% and 18% of new and previously treated cases, respectively, and the success rates of treatment of MDR and XDR-TB is just 50% and 34%, respectively [60]. So, to achieve the END TB strategy, a better understanding of the pathogenesis as well as the resistance mechanism of MDR-TB is needed. Mutations in KatG, RpoB, GyrA, GyrB, InhA, PncA, AhpC, EmbB, Rrs, GidB and RpsL account for 36–95% of M. tb-resistant strains. The remaining 5–64% can be accounted for by some other resistance mechanisms like overexpression of porins, efflux pumps [61] or proteins neutralising drugs activity, making proteomics study necessary. Genomics and transcriptomics have provided great insight into TB pathogenesis, but the actual state of the cell can be more informative when it comes to identifying metabolic and physiological characteristics responsible for infection and drug resistance. So proteomic profiling is the new means of unravelling unique features of M. tb strains leading to a different degree of virulence and drug sensitivity.
Proteomics of drug-resistant M. tb has garnered much attention in the last decade with numerous studies giving valuable information about the different drug resistance markers and mechanisms adopted by the bug. Proteomics has been used for exploring the metabolic pathways involved in the action of different anti-TB drugs.
A study comparing proteomes of isoniazid-resistant and sensitive strains revealed five upregulated membrane proteins in resistant strains which are electron transfer flavoprotein FixB (Rv3028c), oxidoreductase (Rv2971), Wag31 (Rv2145c), OpcA (Rv1446c) and RegX3 (Rv0491) [62]. RegX3 belongs to a two-component response regulator enabling mycobacteria to adapt to stress posed by antibiotics. These results did not corroborate with earlier known mechanism of INH resistance, namely, KatG mutations. This points towards the likelihood of these proteins to be exploited as hits for novel therapeutic agents in future.
Another study involving RIF- and INH-resistant isolates found 27 consistently overexpressed proteins, out of which most prominent were Wag31 (Rv2145c), GarA (Rv1827), Rv1437 and Rv2970c [63]. Wag31 presence in both studies makes it an important candidate for further studies. It is earlier known to be a homologue of DivIVA and a substrate for PknA and PknB. GarA (glycogen accumulation regulator protein) plays a role in TCA cycle and metabolism of glutamate and also required for intracellular growth of M. tb inside macrophages. It is also observed that rifampicin resistance occurs because of some different mechanisms other than well-characterised RpoB mutation based on the study showing underexpression of four proteins (FabD, Ino1, PPE60 and EsxK) in rifampicin-treated M. tb [64]. These proteins play a role in cell wall biosynthesis.
Similarly, another study focussed on aminoglycoside (amikacin and kanamycin)-resistant strains employing 2DE coupled with MALDI-TOF/TOF-MS and bioinformatics. Proteins showing increased intensities in resistant isolates using PDQuest advanced software were identified as ferritin (Rv3841), putative short-chain type dehydrogenase/reductase (Rv0148 and Rv3224), bacterioferritin (Rv1876), elongation factor Tu (Rv0685), ATP synthase subunit alpha (Rv1308), alpha-crystallin/HspX (Rv2031c), proteasome subunit alpha (Rv2109c), trigger factor (Rv2462c), 35 kDa hypothetical protein (Rv2744c), transcriptional regulator MoxR1 (Rv1479), dihydrolipoyl dehydrogenase (Rv0462) and universal stress protein (Rv2005c) [65]. Of these, Rv3841, Rv3224, Rv1876 and Rv0685 play an important role in iron metabolism. Acquisition of iron is known to be important due to its role in mycobacterial growth, virulence and dormancy. Rv3841 (ferritin) maintains iron homeostasis in mycobacterial cells and makes it recalcitrant to antibiotics [66].
Streptomycin-resistant M. tb when compared with susceptible strains showed differential expression of 15 proteins which are Rv0350, Rv0440, Rv3846, Rv1860, Rv1636, Rv3418c, Rv1980c, Rv3248c, Rv2140c, Rv1926c Rv0896, Rv3804c, Rv0009, Rv0815c and Rv2334 [67]. Likewise, Rv2896c, Rv2032c, Rv1908c, Rv1827, Rv0635 and Rv0036 were found to be overexpressed in multidrug-resistant strains by a group of researchers. Rv1908c gets activated in phagosomes [68] and is also necessary for growth and persistence in mice, guinea pigs and human peripheral blood monocytes. Rv2896c, Rv2032c, Rv1827, Rv0635 and Rv0036 play a role in intracellular survival.
Proteomic profiling of M. tb exposed to hypoxia revealed a strong induction of DevR/DosR regulon proteins. Since drug-resistant strains mimic dormant cells, total quantification of proteins involved in stress response may also provide some leads to drug resistance mechanisms [69]. The proteome analysis of dormant M. tb compared with reactivated bacteria at different stages of infection revealed the differential and unique expression profiles of around 1871 proteins accounting for 47% of M. tb proteomes. The percentage of proteins identified in different stages of dormancy and reactivation was observed to be very less as compared to that of control. The most significant fluctuations in the expression profiles belong to the proteins involved in the metabolism of cells. Therefore, the proteins that were found to be differentially or uniquely overexpressed in the reactivation stages can serve as promising targets for novel therapeutics and vaccine potential [70]. Mycobacterial Dop protein involved in proteasome-dependent degradation has been upregulated in dormant stages. Degradation of proteasome-mediated Pup-Dop system is important in achieving a dormancy state [70].
Thus, proteomics has helped greatly in deciphering M. tb responses when exposed to drugs providing a peep inside mechanisms of drug action and resistance. Taking all the studies together, it can be implied that differential expression of proteins mostly involved in lipid metabolism, virulence, detoxification and adaptation, cell wall and cell processes, ATP-binding cassette transporters and proteasome function between drug-resistant and sensitive strains shifts the attention from conventional drug resistance mechanisms to novel systems affecting drug efficacy. With the help of these studies, Rv2031c, Rv3692 and Rv0444c are narrowed down as biomarkers for diagnosis of MDR-TB [68].
5 Conclusions
The gain in momentum in proteomics studies over the years proves to be vital for the effective understanding of tuberculosis disease. Advancement in proteomics has made it easier to investigate the pathogen M. tb in more depth, giving insight into the different factors and strategies adopted by the bug for establishing successful infection. This chapter illustrates the role of proteomics in the study of native proteins of M. tb involved in virulence, host-pathogen interaction, immunomodulation and drug resistance. Conventional biomolecule separation techniques like chromatography and gel electrophoresis continue to be exploited to explore M. tb proteome. Advanced MS-based approaches have further helped in refining our knowledge of M. tb pathogenesis. Analysis of differential expression profiles of diverse proteins among various strains of M. tb has provided a comprehensive knowledge of the key players of virulence which includes Fad22, chorismate pyruvate lyase, MMpl17, ESX-related proteins, PE/PPE/PE-PGRS protein, lipoprotein Lppx, etc. Rv1038c, Rv2346c, Rv2347c and Rv3620c were found to be expressed exclusively in M. tb virulent strains. A majority of proteins showing differential expression in H37Ra, H37Rv, BND and JAL strains belong to intermediary metabolism and respiration (31%), cell wall and cell processes (17.2%) and conserved hypothetical groups (22.2%). Likewise, there have been multiple proteomics studies deciphering pathways and markers involved in multidrug and extensive drug resistance. Majorly upregulated proteins in drug-resistant strains includes Rv3028c, Rv2971, Rv2145c, Rv1446c, Rv0491, Rv1827, Rv1437 and Rv2970c, Rv3841, Rv0148, Rv3224, Rv1876, Rv0685, Rv1308, Rv2031c, Rv2109c, Rv2462c, Rv2744c, Rv1479, Rv0462, Rv2005c, Rv0350, Rv0440, Rv3846, Rv1860, Rv1636, Rv3418c, Rv1980c, Rv3248c, Rv2140c, Rv1926c, Rv0896, Rv3804c, Rv0009, Rv0815c and Rv2334, Rv2896c, Rv2032c, Rv1908c, Rv1827, Rv0635 and Rv0036.
To summarise, the current findings pertain to the undoubted significance of differential expression proteins in all arenas of tuberculosis making proteomics studies indispensable for the development of rapid, simple, cost-effective diagnostics, novel therapeutics and efficient vaccine for management of TB.
References
Ukponmwan OE, Rupreht J, Dzoljic M (1986) An analgesic effect of enkephalinase inhibition is modulated by monoamine oxidase-B and REM sleep deprivations. Naunyn Schmiedeberg’s Arch Pharmacol 332(4):376–379
Rachow A, Ivanova O, Wallis R, Charalambous S, Jani I, Bhatt N et al (2019) TB sequel: incidence, pathogenesis and risk factors of long-term medical and social sequelae of pulmonary TB – a study protocol. BMC Pulm Med 19(1):4
Dockrell HM, Smith SG (2017) What have we learnt about BCG vaccination in the last 20 years? Front Immunol 8:1134
Podany AT, Swindells S (2016) Current strategies to treat tuberculosis. F1000Res:5
Tiberi S, Scardigli A, Centis R, D'Ambrosio L, Munoz-Torrico M, Salazar-Lezama MA et al (2017) Classifying new anti-tuberculosis drugs: rationale and future perspectives. Int J Infect Dis 56:181–184
Migliori GB, Tiberi S, Zumla A, Petersen E, Chakaya JM, Wejse C et al (2020) MDR/XDR-TB management of patients and contacts: challenges facing the new decade. The 2020 clinical update by the Global Tuberculosis Network. Int J Infect Dis 92S:S15–S25
Patterson B, Wood R (2019) Is cough really necessary for TB transmission? Tuberculosis (Edinb) 117:31–35
Chai Q, Wang L, Liu CH, Ge B (2020) New insights into the evasion of host innate immunity by Mycobacterium tuberculosis. Cell Mol Immunol 17(9):901–913
Bradfute SB, Castillo EF, Arko-Mensah J, Chauhan S, Jiang S, Mandell M et al (2013) Autophagy as an immune effector against tuberculosis. Curr Opin Microbiol 16(3):355–365
Flannagan RS, Cosio G, Grinstein S (2009) Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol 7(5):355–366
Weiss G, Schaible UE (2015) Macrophage defense mechanisms against intracellular bacteria. Immunol Rev 264(1):182–203
Ehlers S, Schaible UE (2012) The granuloma in tuberculosis: dynamics of a host-pathogen collusion. Front Immunol 3:411
Qiang L, Wang J, Zhang Y, Ge P, Chai Q, Li B et al (2019) Mycobacterium tuberculosis Mce2E suppresses the macrophage innate immune response and promotes epithelial cell proliferation. Cell Mol Immunol 16(4):380–391
Su H, Zhu S, Zhu L, Kong C, Huang Q, Zhang Z et al (2017) Mycobacterium tuberculosis latent antigen Rv2029c from the multistage DNA vaccine A39 drives TH1 responses via TLR-mediated macrophage activation. Front Microbiol 8:2266
Peng H, Wang X, Barnes PF, Tang H, Townsend JC, Samten B (2011) The Mycobacterium tuberculosis early secreted antigenic target of 6 kDa inhibits T cell interferon-gamma production through the p38 mitogen-activated protein kinase pathway. J Biol Chem 286(27):24508–24518
Wang X, Barnes PF, Huang F, Alvarez IB, Neuenschwander PF, Sherman DR et al (2012) Early secreted antigenic target of 6-kDa protein of Mycobacterium tuberculosis primes dendritic cells to stimulate Th17 and inhibit Th1 immune responses. J Immunol 189(6):3092–3103
Jung BG, Wang X, Yi N, Ma J, Turner J, Samten B (2017) Early secreted antigenic target of 6-kDa of Mycobacterium tuberculosis stimulates IL-6 production by macrophages through activation of STAT3. Sci Rep 7:40984
Refai A, Gritli S, Barbouche MR, Essafi M (2018) Mycobacterium tuberculosis virulent factor ESAT-6 drives macrophage differentiation toward the pro-inflammatory M1 phenotype and subsequently switches it to the anti-inflammatory M2 phenotype. Front Cell Infect Microbiol 8:327
Lin J, Jiang Y, Liu D, Dai X, Wang M, Dai Y (2020) Early secreted antigenic target of 6-kDa of Mycobacterium tuberculosis induces transition of macrophages into epithelioid macrophages by downregulating iNOS/NO-mediated H3K27 trimethylation in macrophages. Mol Immunol 117:189–200
Yaseen I, Kaur P, Nandicoori VK, Khosla S (2015) Mycobacteria modulate host epigenetic machinery by Rv1988 methylation of a non-tail arginine of histone H3. Nat Commun 6:8922
Schubert OT, Mouritsen J, Ludwig C, Rost HL, Rosenberger G, Arthur PK et al (2013) The Mtb proteome library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe 13(5):602–612
Kunnath-Velayudhan S, Porcelli SA (2013) Recent advances in defining the immunoproteome of Mycobacterium tuberculosis. Front Immunol 4:335
Bespyatykh JA, Shitikov EA, Ilina EN (2017) Proteomics for the investigation of mycobacteria. Acta Nat 9(1):15–25
Uddin R, Siddiqui QN, Sufian M, Azam SS, Wadood A (2019) Proteome-wide subtractive approach to prioritize a hypothetical protein of XDR-Mycobacterium tuberculosis as potential drug target. Genes Genomics 41(11):1281–1292
Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D et al (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393(6685):537–544
Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL et al (2014) PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42(Database issue):D581–D591
Liu B, Zheng D, Jin Q, Chen L, Yang J (2019) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47(D1):D687–DD92
Li J, Tai C, Deng Z, Zhong W, He Y, Ou HY (2018) VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief Bioinform 19(4):566–574
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M et al (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45(D1):D362–D3D8
Raman K, Yeturu K, Chandra N (2008) targetTB: a target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis. BMC Syst Biol 2:109
Sharma D, Surolia A (2011) Computational tools to study and understand the intricate biology of mycobacteria. Tuberculosis (Edinb) 91(3):273–276
Gupta A, Kapil R, Dhakan DB, Sharma VK (2014) MP3: a software tool for the prediction of pathogenic proteins in genomic and metagenomic data. PLoS One 9(4):e93907
Winnenburg R, Urban M, Beacham A, Baldwin TK, Holland S, Lindeberg M et al (2008) PHI-base update: additions to the pathogen host interaction database. Nucleic Acids Res 36(Database issue):D572–D576
McCarthy FM, Wang N, Magee GB, Nanduri B, Lawrence ML, Camon EB et al (2006) AgBase: a functional genomics resource for agriculture. BMC Genomics 7:229
Macalino SJY, Billones JB, Organo VG, Carrillo MCO (2020) In silico strategies in tuberculosis drug discovery. Molecules 25(3):665
Chandra N, Sandhya S, Anand P (2014) Structural annotation of the Mycobacterium tuberculosis proteome. Microbiol Spectr 2(2)
Cubillos-Ruiz A, Morales J, Zambrano MM (2008) Analysis of the genetic variation in Mycobacterium tuberculosis strains by multiple genome alignments. BMC Res Notes 1:110
ten Bokum AM, Movahedzadeh F, Frita R, Bancroft GJ, Stoker NG (2008) The case for hypervirulence through gene deletion in Mycobacterium tuberculosis. Trends Microbiol 16(9):436–441
Rosenkrands I, King A, Weldingh K, Moniatte M, Moertz E, Andersen P (2000) Towards the proteome of Mycobacterium tuberculosis. Electrophoresis 21(17):3740–3756
He XY, Zhuang YH, Zhang XG, Li GL (2003) Comparative proteome analysis of culture supernatant proteins of Mycobacterium tuberculosis H37Rv and H37Ra. Microbes Infect 5(10):851–856
Jungblut PR, Schaible UE, Mollenkopf HJ, Zimny-Arndt U, Raupach B, Mattow J et al (1999) Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol Microbiol 33(6):1103–1117
Singh Y, Kohli S, Sowpati DT, Rahman SA, Tyagi AK, Hasnain SE (2014) Gene cooption in mycobacteria and search for virulence attributes: comparative proteomic analyses of Mycobacterium tuberculosis, Mycobacterium indicus pranii and other mycobacteria. Int J Med Microbiol 304(5–6):742–748
Wilkinson KA, Newton SM, Stewart GR, Martineau AR, Patel J, Sullivan SM et al (2009) Genetic determination of the effect of post-translational modification on the innate immune response to the 19 kDa lipoprotein of Mycobacterium tuberculosis. BMC Microbiol 9:93
Deng J, Bi L, Zhou L, Guo SJ, Fleming J, Jiang HW et al (2014) Mycobacterium tuberculosis proteome microarray for global studies of protein function and immunogenicity. Cell Rep 9(6):2317–2329
Romling U, Galperin MY, Gomelsky M (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77(1):1–52
Brennan PJ (2003) Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 83(1–3):91–97
Berthet FX, Lagranderie M, Gounon P, Laurent-Winter C, Ensergueix D, Chavarot P et al (1998) Attenuation of virulence by disruption of the Mycobacterium tuberculosis erp gene. Science 282(5389):759–762
Schmidt F, Donahoe S, Hagens K, Mattow J, Schaible UE, Kaufmann SH et al (2004) Complementary analysis of the Mycobacterium tuberculosis proteome by two-dimensional electrophoresis and isotope-coded affinity tag technology. Mol Cell Proteomics 3(1):24–42
Ang KC, Ibrahim P, Gam LH (2014) Analysis of differentially expressed proteins in late-stationary growth phase of Mycobacterium tuberculosis H37Rv. Biotechnol Appl Biochem 61(2):153–164
Albrethsen J, Agner J, Piersma SR, Hojrup P, Pham TV, Weldingh K et al (2013) Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol Cell Proteomics 12(5):1180–1191
Peters JS, Calder B, Gonnelli G, Degroeve S, Rajaonarifara E, Mulder N et al (2016) Identification of quantitative proteomic differences between Mycobacterium tuberculosis lineages with altered virulence. Front Microbiol 7:813
Forrellad MA, Klepp LI, Gioffre A, Sabio y Garcia J, Morbidoni HR, de la Paz Santangelo M et al (2013) Virulence factors of the Mycobacterium tuberculosis complex. Virulence 4(1):3–66
Sala A, Bordes P, Genevaux P (2014) Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 6(3):1002–1020
McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT et al (2000) Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406(6797):735–738
Eisenreich W, Dandekar T, Heesemann J, Goebel W (2010) Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol 8(6):401–412
De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu Y, Barry CE 3rd (2000) The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci USA 97(3):1252–1257
Jhingan GD, Kumari S, Jamwal SV, Kalam H, Arora D, Jain N et al (2016) Comparative proteomic analyses of avirulent, virulent, and clinical strains of Mycobacterium tuberculosis identify strain-specific patterns. J Biol Chem 291(27):14257–14273
Frigui W, Bottai D, Majlessi L, Monot M, Josselin E, Brodin P et al (2008) Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog 4(2):e33
Andersen AB, Andersen P, Ljungqvist L (1992) Structure and function of a 40,000-molecular-weight protein antigen of Mycobacterium tuberculosis. Infect Immun 60(6):2317–2323
Harding E (2020) WHO global progress report on tuberculosis elimination. Lancet Respir Med 8(1):19
Magnet S, Courvalin P, Lambert T (2001) Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother 45(12):3375–3380
Jiang X, Zhang W, Gao F, Huang Y, Lv C, Wang H (2006) Comparison of the proteome of isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis. Microb Drug Resist 12(4):231–238
Singh A, Gopinath K, Sharma P, Bisht D, Sharma P, Singh N et al (2015) Comparative proteomic analysis of sequential isolates of Mycobacterium tuberculosis from a patient with pulmonary tuberculosis turning from drug sensitive to multidrug resistant. Indian J Med Res 141(1):27–45
Meneguello JE, Arita GS, Silva JVO, Ghiraldi-Lopes LD, Caleffi-Ferracioli KR, Siqueira VLD et al (2020) Insight about cell wall remodulation triggered by rifampicin in Mycobacterium tuberculosis. Tuberculosis (Edinb) 101903:120
Sharma D, Kumar B, Lata M, Joshi B, Venkatesan K, Shukla S et al (2015) Comparative proteomic analysis of aminoglycosides resistant and susceptible Mycobacterium tuberculosis clinical isolates for exploring potential drug targets. PLoS One 10(10):e0139414
Pandey R, Rodriguez GM (2012) A ferritin mutant of Mycobacterium tuberculosis is highly susceptible to killing by antibiotics and is unable to establish a chronic infection in mice. Infect Immun 80(10):3650–3659
Sharma D, Bisht D (2017) Secretory proteome analysis of streptomycin-resistant Mycobacterium tuberculosis clinical isolates. SLAS Discov 22(10):1229–1238
Singhal N, Sharma P, Kumar M, Joshi B, Bisht D (2012) Analysis of intracellular expressed proteins of Mycobacterium tuberculosis clinical isolates. Proteome Sci 10(1):14
Sharma D, Bisht D, Khan AU (2018) Potential alternative strategy against drug resistant tuberculosis: a proteomics prospect. Proteomes 6(2):26
Gopinath V, Raghunandanan S, Gomez RL, Jose L, Surendran A, Ramachandran R et al (2015) Profiling the proteome of Mycobacterium tuberculosis during dormancy and reactivation. Mol Cell Proteomics 14(8):2160–2176
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Shariq, M. et al. (2021). Exploration of the Mycobacterial Proteome in the Pathogenesis of TB: A Perspective. In: Hameed, S., Fatima, Z. (eds) Integrated Omics Approaches to Infectious Diseases. Springer, Singapore. https://doi.org/10.1007/978-981-16-0691-5_17
Download citation
DOI: https://doi.org/10.1007/978-981-16-0691-5_17
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-0690-8
Online ISBN: 978-981-16-0691-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)