Abstract
Today, more than two-thirds of global energy supplies come from fossil fuels. However, dwindling reserve of fossil fuels, precarious global oil market, energy geopolitics, and associated environmental impacts due to fossil fuel burning have compelled almost all the nations of the world to search for clean and renewable alternatives for energy, fuel, and chemicals. Among the renewable energy resources, biomass has been projected as a promising source of future alternative to substitute crude oil-based petro-refinery. Lignin degradation and its conversion into fuel and chemicals is a major unresolved problem in a lignocellulosic biorefinery. Lignin valorization is in nascent stage despite being a high priority in second-generation biofuel programs. Lignin valorization can be done by physical, chemical, physicochemical, and biological routes. This chapter focuses on biological route for bacterial-mediated lignin depolymerization and degradation. The importance of lignin utilization, its synthesis, major sources of its generation, and its structure is discussed. Bacterial strains, enzymes, and pathways involved in lignin degradation have also been elucidated. Furthermore, the role of genomics and proteomics in bacterial lignin degradation has also been discussed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
Lignocellulosic biomass is a carbon-neutral or low-carbon, easy available, and renewable feedstock available for the production of fuel and chemicals. Biomass is currently the single, largest source of renewable energy worldwide, providing 10% (50 EJ) of the global primary energy supply (IEA 2016). Lignocellulosic biomass is composed of cellulose, hemicellulose, and lignin. For the production of second-generation biofuels and chemicals, these components need to be separated into the individual component and must be fully utilized to make the lignocellulosic biorefinery environmentally, socially, and economically feasible (Sims et al. 2010; Singhvi et al. 2014). The lignocellulosic biomass-based research mainly focuses on polysaccharides component of biomass, and lignin is discarded as waste with very limited usage. Therefore, the challenges associated with the success of second-generation biomass-based biorefinery must be addressed (Singhvi et al. 2014). The recent focus on the concept of lignocellulosic biorefinery, i.e., utilization of biomass and byproducts leading to minimal or zero waste generation, can overcome the problems associated with biofuel production at commercial scale. Lignin is very less explored, and the advancement in approaches of lignin valorization into valuable bioproducts is essential for overall economic viability and sustainability of lignocellulosic biorefinery (de Jong et al. 2012; Ragauskas et al. 2014).
Lignin is recalcitrant to degradation due to its complex and heterogeneous structure. Its depolymerization is being done by chemical, thermochemical, and biological processes for conversion into fuel and chemicals. Compared to the chemical processes, the use of microorganism or enzymes for lignin depolymerization is less energy intense, cost effective, eco-friendly, and works at ambient temperature with fewer inhibitors’ generation (Zhu et al. 2017). The discovery of new microbial strains and understanding their enzyme system that is responsible for lignin degradation will help in lignin depolymerization and its conversion into fuel and chemicals (Kumar et al. 2017).
2 Lignocellulosic Biomass and Its Composition
Lignocellulose is the main constituent of plant cell wall and mainly refers to the dry matter of plant biomass. Lignocellulosic biomass is a complex matrix composed of cellulose, hemicellulose, lignin polymers, and a small amount of proteins, extractives, and minerals (Moon et al. 2011; Menon and Rao 2012; Singh et al. 2017). The structure and composition of lignocellulosic biomass are represented in Fig. 4.1. Two-thirds of lignocellulosic biomass are comprised of cellulose and hemicellulose polysaccharides that are made of hexose (C6) and pentose (C5) sugars. These polymers organize themselves into a complex three-dimensional structure. Their organization is nonuniform with a varied composition of cellulose, hemicellulose, and lignin depending on the types of lignocellulosic biomass (Gírio et al. 2010; Scheller and Ulvskov 2010; Menon and Rao 2012). The presence of individual lignocellulosic components in different biomass has been shown in Table 4.1.
Structural organization and composition of lignocellulosic biomass (adapted from Menon and Rao 2012; microbewiki)
2.1 Lignin
The term lignin was coined by a French chemist and botanist Anselme Payen in 1838 and was chemically defined by Schulze in 1885 (Doherty et al. 2011; Lange et al. 2013; Feofilova and Mysyakina 2016). Lignin encrusts the cellulose and hemicellulose components of plant cell wall and provides impermeability, mechanical strength, and rigidity to the plant cell wall. Lignin provides defense barrier and protects the plant against microbial attack. The distribution and concentration of lignin vary in plant cell wall and between different species (Lange et al. 2013). Lignin biosynthesis in plants occurs by phenylpropanoid pathway. Lignin monomer synthesis starts with deamination of phenylalanine to form cinnamic acid through a series of enzymatic reactions catalyzed by various enzymes (Bonawitz and Chapple 2010). The aromatic ring of cinnamic acid undergoes a series of reactions such as hydroxylation, O-methylation, and side chain reduction from acid to alcohol, to form three lignin monomers or monolignols: p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol. After synthesis, the monolignols are transported with the help of ABC transporter to apoplast. The monolignols are polymerized by radical–radical coupling generated by various oxidoreductases to form ether and C-C bonds (Bonawitz and Chapple 2010; Guerriero et al. 2016).
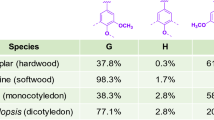
These three phenylpropane units (p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol) after polymerization get incorporated into lignin polymer as p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units (Bonawitz and Chapple 2010; Doherty et al. 2011). Lignin monomer and its polymerization units in lignin polymer are shown in Fig. 4.2. The composition of H, G, and S units differs due to different bonding tendencies in lignin and varies between different species and within tissues of the same plant. The distributions are as follows: angiosperm mostly contains G and S, gymnosperm mainly contains G units, monocot relatively contains more H, coniferous wood mainly contains G followed by Sand H units, deciduous trees contain equal amount of G and S, and grasses contain all three units (Bonawitz and Chapple 2010; Feofilova and Mysyakina 2016).There can be other natural lignin monomers derived from phenolics other than familiar monolignols such as 5-hydroxyconiferyl alcohol, hydroxycinnamate esters, hydroxycinnamaldehydes, dihydrocinnamyl alcohol, hydroxybenzaldehydes, etc. (Ralph et al. 2004).
2.2 Sources of Lignin
Lignocellulosic biomass needs to be pretreated to separate it into its valuable individual components. The basic aim of pretreatment is to improve the biodegradability of cellulose by solubilizing hemicellulose and lignin and cellulosic pulp with reduced crystallinity and lesser degree of polymerization (Harmsen et al. 2010; Menon and Rao 2012; Gomes et al. 2014). Pretreatment methods can be divided into physical, chemical, biological and also their combination to make them more accessible for chemical or enzymatic applications. Selection of pretreatment method depends on feedstock, economic, and environmental impact assessment and biological process to be applied (Doherty et al. 2011; Menon and Rao 2012; Lange et al. 2013). An effective pretreatment method should be cost effective, require less energy input, catalyst recovery, and recycle, avoid inhibitors’ formation, and recovery of lignin and hemicellulose fraction as well as treatment of waste. Chemical pretreatments are most studied processes, and these are generally used in pulp and paper industry. Pretreatment is a prerequisite for feasibility of processes based on plant polysaccharides. Delignification is essential in pulp and paper mill industry and for biofuel production. Pulp and paper and plant polysaccharide-based industries are the major sources of lignin. The major types of lignin generated through different pretreatment processes applied in industries are Kraft lignin, lignosulphonates, soda lignin, and organosolv lignin.
2.3 Lignin Structure and Characterization
The exact structure of natural lignin is still unknown, but with the advancement in analytical technique some linkages commonly found in lignin have been revealed. The linkages are formed between oligomer–monomer, oligomer–oligomer, and monomer–monomer coupling reactions (Ralph et al. 2004). There are various interunit linkages observed in lignin polymer as shown in Fig. 4.3.
Common interunit linkages found in highly complex three dimensional lignin polymer (Longe et al. 2016)
The most abundant coupling is favored at β-carbon of one monolignol with hydroxyl group of the other to form β-O-4-aryl ether linkage. This is the major linkage observed in case of lignin structure. The others are α-aryl ether (α-O-4), phenylcoumaran (β-5), diaryl ether (4-O-5), biphenyl and dibenzodioxocin (5-5), spirodienone (β-1), and pinoresinols (β-β). The interunit linkages present in lignocellulose have been shown in Table 4.2. Coupling at position 4 and 5 is preferentially formed by dilignols and higher oligomers (Lange et al. 2013; Ralph et al. 2004; Chakar and Ragauskas 2004; Lupoi et al. 2015).
3 Lignin Degradation by Bacteria
Lignin is mainly degraded by fungi and bacteria in the natural environment. Fungi, especially white rot fungi, have been studied extensively for degradation of lignin by producing various ligninolytic enzymes (Bugg et al. 2011a; Kumar et al. 2015). The fungal biocatalysts for lignin degradation have not been commercialized till date due to genome complexity and information processing and difficulty in recombinant protein expression. Compared to fungi, the bacterial genome is small and easy to manipulate and recombinant proteins can be easily expressed and produced at a large scale (Bugg et al. 2011a; Kumar et al. 2015). So the focus again has been shifted to identification and characterization of novel bacterial strains and their enzymes responsible for lignin degradation. Recently, various new bacterial strains have been reported with lignin-degrading potential (Priyadarshinee et al. 2016). According to a recent review, 22 actinobacteria, 10 alpha proteobacteria, beta proteobacteria, 11 gamma proteobacteria, one delta proteobacteria, bacteriodes, and archaea, each were reported to be having lignin-degrading genotypes and phenotypes (Tian et al. 2014). Various bacterial enzymes such as β-esterases, DyP-type peroxidase, laccases, and various other oxidative enzymes responsible for lignin degradation have been recently reported (de Gonzalo et al. 2016).
Recently, various bacterial strains have been reported for lignin degradation. Bacterial strains responsible for lignin degradation can be applied in valorization of biomass. Lignin-degrading bacterial strains having cellulose-free xylanase can be an excellent choice for pretreatment. The bacteria responsible for lignin degradation can be found in diverse environments such as soil, digestive system of herbivores, wood-eating insects, effluents from paper industry, sludge, etc. (Brown and Chang 2014; Tian et al. 2014). The bacterial strains recently reported for lignin degradation have been shown in Table 4.3. Advancement in genomics, transcriptomics, and proteomics completely revolutionized the understanding of microbial lignin degradation. Next-generation sequencing (NGS) technology resulted in complete genome sequence of several new microbes that will further enhance our understanding related to lignin degradation (Baldrian and López-Mondéjar 2014; Kameshwar and Qin 2016). The NGS (genomics and transcriptomics) along with proteomics provided various detailed information related to expression of proteins and characterization of new enzymes responsible for lignin utilization (DeAngelis et al. 2013; Lin et al. 2016; Zhu et al. 2017) Discovery of new microbes and further advancement and affordability in these technologies will enhance our knowledge in the near future.
4 Bacterial Peripheral Pathways for Lignin Degradation
Lignin is a complex polymer, and the products obtained after its depolymerization are highly heterogeneous, and therefore various pathways are involved in degradation of lignin. The bacterial enzymes have been characterized for lignin degradation, but degradation of lignin inside the cell is not clearly understood. Thorough understanding of catabolic pathways is very important for biotechnological application of lignin-degrading microbes in lignocellulosic biorefinery. Some prominent pathways have been discussed.
4.1 β-Aryl Ether Degradation Pathway
β-aryl ether linkage is the most predominant linkages (50–70%) in lignin; therefore, cleavage of β-aryl ether bond is crucial for lignin biodegradation. The cleavage of ether bond leads to formation of various industrially important aromatic compounds. Sphingobium sp. SYK-6 has been extensively studied on various lignin model compounds for the degradation of β-aryl ether bond. Lig EFG gene cluster enzymes, lignin peroxidase, and β-aryl-OH elimination followed by decarboxylation, vanillate dehydrogenase, and demethylation mechanism has been reported for β-aryl ether metabolism (Masai et al. 2007; Bugg et al. 2011b). Lig EFG has been discussed in the enzyme section. The degradation of β-aryl ether bond has also been studied in Rhodococcus jostii RHA1, Pseudomonas acidovorans, Pseudomonas putida, Pseudomonas sp. HR199, Novosphingobium, etc. (Masai et al. 2007; Bugg et al. 2011b; Chen and Wan 2017).
4.1.1 Biphenyl Degradation Pathways
Biphenyl linkage is the second most abundant linkage (10%) found in lignin after β-aryl ether. Biphenyl is a major environmental pollutant and affects human health. The degradation pathway has been extensively studied in bacteria. Degradation of biphenyls has been studied in genus Pseudomonas, Ralstonia, Burkholderia, Comamonas, Achromobacter, Rhodococcus, Acinetobacter, and Bacillus. The reaction is initiated by a biphenyl 2, 3-dioxygenase of Rieske nonheme iron oxygenases family. Study on model compounds by S. paucimobilis SYK-6 suggested O-demthylation reaction followed by extradiol ring cleavage by dioxygenase and finally degraded by β-KAP pathway into acetyl-coA and succinyl-CoA (Masai et al. 2007).
4.1.2 Ferulate, Diarylpropane, Phenylcoumarane, and Pinoresinol Catabolic Pathways
Ferulic acid is attached by ester linkage to hemicellulose, and their degradation is carried out by esterases. Ferulate esterases have been identified in several bacteria. There are two types of pathway reported for degradation of ferulate. In one pathway, side chain cleavage occurs to eliminate two carbons from ferulate by two enzymes (a feruloyl-CoA synthetase, feruloyl-CoA hydratase/lyase FerB). The enzymes have been reported in P. putida WCS358, Amycolatopsis sp. HR167, Pseudomonas sp. HR199, Pseudomonas fluorescens, Pseudomonas putida, etc. Another pathway is the release of one carbon by nonoxidative decarboxylation of ferulate side chain. This pathway has been identified in Bacillus sp. BP-7 and Enterobacter sp. Px6-4 (Masai et al. 2007; Bugg et al. 2011b). This degradation of diarylpropane has been studied in Pseudomonas paucimobilis TMY1009. The enzyme responsible for diarylpropane degradation has been characterized while growing this strain on diarylpropane model compounds, but the gene has not been identified. The product obtained is lignostilbene, and the enzyme (lignostilbenedioxygenase) responsible for its degradation into vanillin has been reported (Bugg et al. 2011b). The degradation of phenylcoumarane and pinoresinol has been studied with model compounds in S. paucimobilis SYK-6, but the genes responsible for degradation are still not clear. It was proposed that degradation of these heterocyclic lignin components is initiated by a hydroxylation (Bugg et al. 2011b).
4.2 Central Pathways for Lignin Degradation in Bacteria
4.2.1 Oxidative Cleavage of Aromatic Rings
The process of aromatic ring cleavage is predominantly aerobic, but anaerobic process exists in nature (Fuchs et al. 2011). Funneling pathways for lignin degradation results into formation of derivatives such as vanillic acid, vanillin, syringate, or guaiacol, and these are further converted into few common central intermediates such as protocatechuate (PCA), catechol, and gallic acid. Protocatechuic acid is the most common intermediate formed during funneling pathways of lignin degradation. This central intermediate acts as substrate for ring-cleaving dioxygenases. The pathway for cleavage of aromatic ring can be divided into ortho cleavage and meta cleavage on the basis of position of hydroxyl group and their fission in aromatic ring (Bugg et al. 2011b). Ortho (intradiol) cleavage takes place between two hydroxyl groups catalyzed by Fe3+-dependent dioxygenase, and meta (extradiol) cleavage occurs adjacent to one of the hydroxyl group catalyzed by Fe2+-dependent dioxygenase (Masai et al. 2007; Bugg et al. 2011b). PCA is catalyzed by PCA 2,3-dioxygenase (2,3-PCD), PCA 3,4-dioxygenase (3,4-PCD; intradiol), and PCA 4,5-dioxygenase (4,5-PCD; extradiol). 4,5-PCD pathway has been well studied in S. paucimobilis SYK-6. Meta cleavage of catechol gives acetaldehyde and pyruvate while PCA yields pyruvate. Pathways for oxidative aromatic ring cleavage have been shown in Fig. 4.4. 3,4-PCD is most widely distributed among proteobacteria and actinobacteria, but 4,5-PCD is only found in proteobacteria. The β and γ proteobacteria genus Burkholderia, Pseudomonas, Xanthomonas, Klebsiella, and Ralstonia contains either 3,4-PCD, 4,5-PCD, or both the enzymes (Masai et al. 2007; Bugg et al. 2011b).The pathways for oxidative cleavage of aromatic rings and β-ketoadipate pathway have been shown in Fig. 4.4.
4.2.2 β-Ketoadipate Pathway
β-ketoadipate pathway is a highly conserved metabolic route and has been extensively characterized in Pseudomonas putida, Acinetobacter calcoaceticus, and Agrobacterium tumefaciens (Masai et al. 2007). Ortho cleavage of catechol and PCA leads to β-ketoadipate followed by succinate and acetyl-CoA formation (Fuchs et al. 2011; Abdelaziz et al. 2016). β-KA pathway links the lignin-derived upper funneling pathway with TCA cycle through PCA or catechol intermediates (Harwood and Parales 1996; Pérez-Pantoja et al. 2010). There are several enzymes involved in degradation of PCA and catechol to β-KA, and the enzymes are tightly regulated (Abdelaziz et al. 2016). Among the several lignin-degrading microbes studied so far, ortho or β-KA cleavage pathway was found to be dominant pathway (Bugg et al. 2011b).
5 Bacterial Enzymes Responsible for Funneling Lignin and Its Degradation Intermediates
The major lignin-degrading or modifying enzymes responsible for utilization of lignin or its degradation intermediates are detailed in the following sections.
5.1 DyP-Type Peroxidases
DyP-type peroxidases (DyPs) are heme-containing new class of peroxidase recently identified and are predominantly present in bacteria. This enzyme is also present in fungi, first discovered from Bjerkandera adusta and named so by studying their activity on anthraquinone and azo-dyes (Sugano et al. 2007). Bacterial DyPs have low redox potential than fungal DyPs, but they showed activity toward phenolic as well as nonphenolic compounds. They can perform catalysis by utilizing H2O2 and without H2O2 as oxygenase or hydrolase. The first extracellular lignin-degrading peroxidase from bacteria has been reported in S. viridosporus T7A, and after genomic analysis it was assumed to be DyP (Ramachandra et al. 1988). Several other peroxidases have been reported from this strain responsible for degradation of β-aryl bonds, the most predominant bond in lignin. DyPs were characterized from Rhodococcus jostii RHA1, Pseudomonas fluorescens Pf-5, Amycolatopsis sp. 75iv2ATCC, Bacillus subtilis KCTC2023, Pseudomonas putida MET94, Saccharomonospora viridis DSM 43017, Thermobifida fusca. Their broad substrate range (synthetic dyes, Kraft lignin, lignin model compounds, monophenolic compounds, veratryl alcohol, carotenes, Mn+2) and mechanism of action have also been discussed (de Gonzalo et al. 2016;Priyadarshinee et al. 2016; Bugg et al. 2016). The reaction catalyzed by DyPs has been shown in Fig. 4.5.
Reaction catalyzed by bacterial DyP-type peroxidases. (a) TfuDyP from Thermobifida fusca resulted into dimerization of vanillin. (b) DyPB from Rhodococcus jostii RHA1 formed guaiacol, guaiacol trimmers, and vanillin. (c) TcDyP from Thermomonospora curvata degraded guaiacylglycerol-β-guaiacol into cresol dimers and hydroxylated guaiacol pentamers. (d) BsDyP from Bacillus subtilis KCTC 2023 degraded veratrylglycerol-β-guaiacol ether (adapted from de Gonzalo et al. 2016)
5.2 Laccase (Benzenediol: Oxygen Oxidoreductases; EC 1.10.3.2; AA1)
Laccases are multicopper oxidase that oxidizes various aromatic compounds (phenolics) with reduction of oxygen molecule to water as byproduct. These are extensively distributed among plants, fungi, insects, and bacteria (Riva 2006; Munk et al. 2015). Laccase mostly contains three structural domains, but one or two domains may be lacking in some laccases. Laccase have 4 copper atoms, and these are classified into three groups (type) based on spectral and paramagnetic properties. The type1 (T1) gives blue color (λmax. 600 nm), and it is the main site where oxidation of substrate takes place, Type 2 (T2) contains one copper (EPR active) and Type 3 (T3) contains two copper atoms and together they form trinuclear cluster where oxygen reduction occurs (Bugg et al. 2011b; Munk et al. 2015; Feng et al. 2016). Laccase from fungus has been widely studied and applied but with the advancement in genomics various laccases have been discovered in bacteria (Santhanam et al. 2011; Feng et al. 2016). Streptomyces (S. coelicolor, S. violaceusniger, S. ipomoea CECT 3341, S. ipomoea CECT 3341, S. griseus) has been the most studied bacterial genus for lignin degradation. Various other bacteria such as Pandoraea sp. ISTKB, Bacillus tequilensis SN4, Pantoea ananatis Sd-1, Bacillus pumilus (CotA), Thermus thermophilus HB27, etc., have also been studied (de Gonzalo et al. 2016; Kumar et al. 2018).
Laccase-mediated oxidation of phenolic β-O-4 (most abundant linkage in lignin) and β-1 has been demonstrated on lignin model dimers. Laccase can perform degradation of phenolic as well as nonphenolic substrate in the presence of mediators. Mediators are small molecules that act as an electron carrier, and upon oxidation by laccase they become strong oxidizing intermediates to degrade nonphenolic units and also prevent polymerization of small reactive compounds formed during degradation (Riva 2006; Munk et al. 2015). Mediators may be natural (cinnamic acid, acetosyringone, benzaldehyde, sinapic acid, etc.) or synthetic (ABTS, HBT, TEMPO, etc.). Mechanism of laccase-mediator system and common natural and synthetic mediators are shown in Fig. 4.6.
Reaction mechanism of laccase mediator system and some commonly used synthetic and natural mediators. Lignin is degraded by laccase with the help of mediators. Some of the common synthetic mediators are N-hydroxybenzotriazole (HBT), 2,2,6,6- tetramethylpiperidin-1-yl) oxyl (TEMPO), and 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), and natural mediators are acetosyringone, vanillin, and syringaldehyde
Since laccases do not require H2O2 for substrate oxidation as compared to peroxidases, these enzymes has been used in various industrial applications such as delignification and pretreatment of biomass, wastewater treatment, bioremediation, food and beverages industry, pharmaceuticals and other fine chemicals synthesis, textile dye removal, etc. (Chandra and Chowdhary 2015).
5.3 Glutathione-Dependent β-Etherases
The enzyme glutathione-dependent β-etherases were first discovered and studied in detail in α-proteobacterium Sphingobium SYK- 6. The etherase enzyme system comprising stereospecific Lig DEF was studied in Sphingobium SYK- 6 for degradation of lignin model compounds (Masai et al. 2007). This strain was shown to catalyze the glutathione (thiol group)-mediated cleavage of β-aryl ether linkage in model compounds as shown in Fig. 4.7. The glutathione-dependent cleavage of ether linkages by β-etherase is shown to be enantioselective (de Gonzalo et al. 2016; Feng et al. 2016). Presence of β-etherase in Novosphingobium, Sphingobium SYK-6, Novosphingobium sp. PP1Y, and Thiobacillus denitrificans ATC25259 has been reviewed (de Gonzalo et al. 2016; Feng et al. 2016).
β-etherases catalyzed reaction on lignin model compounds in Sphingobium SYK- 6 (adapted from Bugg et al. 2016)
5.4 Superoxide Dismutases
Superoxidases are generally intracellular enzymes that protect cells from oxidative damage by converting superoxide anions into molecular oxygen and H2O2. Recently two extracellular manganese-dependent superoxide dismutases (MnSOD1 and MnSOD2) have been discovered from Sphingobacterium sp. T2 with lignin-degrading properties (Rashid et al. 2015). These two enzymes are shown to be highly active and can oxidize Organosolv, Kraft lignin, and lignin model substrates into various compounds. The degradation products were obtained from oxidative cleavage of aryl-Cα, Cα-Cβ bond, and O-demethylation activity (Rashid et al. 2015).
5.5 Catalase-Peroxidases
An extracellular catalase-peroxidase was discovered recently in Amycolatopsis sp.75iv2 while growing on lignocellulosic substrate. This is a heme-containing enzyme and showed oxidation of various phenolic model compounds; however, methylated derivatives were not utilized as substrate (Brown et al. 2011). Further research is needed to establish the occurrence of catalase-peroxidases in other strains and their activity to modify polymeric lignin.
5.6 Dehydrogenases
The dehydrogenases have been discovered recently in some bacteria capable of transforming lignin or lignin model compound. The enzyme dehydrogenase from strain SG61-1 L has been characterized for degradation of ether linkages in lignin model compound guaiacylglycerol-guaiacyl ether (GGE) and showed very efficient degradation of all its stereoisomers. Enzyme system Lig DEG (LigD-Cα-dehydrogenase, β-etherase, and glutathione lyase) from Sphingobium sp. SYK6 known for the β-O-4-aryl ether linkage has been expressed in E. coli and has shown to convert complex lignin structure from hardwood and softwood into monomers (Reiter et al. 2013). Dehydrogenases act on toxic aldehyde (generated during degradation) and convert them into their acids that are less toxic to cells and hence play important part in catabolic funneling (Pérez-Pantoja et al. 2010; Abdelaziz et al. 2016).
5.7 O-Demethylases
The enzymes of demethylase system removes methyl group from methoxy-substituted lignin-derived aromatic compounds such as syringate, vanillate, or guaiacol in the presence of cofactors. There are two types of demethylation system. Type one demethylase comprises of an oxygenase (iron-binding site and a Rieske type [2Fe-2S] cluster) and a reductase (a flavin and a [2Fe-2S] redox center) predominant in aerobic microbes such as Pseudomonas and Acinetobacter (Masai et al. 2007; Bugg et al. 2011b; Abdelaziz et al. 2016). Type two demethylase system catalyzes tetrahydrofolate-dependent demethylation of lignin-derived intermediates mainly reported in anaerobic microbes such as Acetobacterium dehalogenans and Acetobacterium woodii. After demethylation, the products converge at a few common intermediates (protocatechuic acid, catechol, or gallic acid), which undergo ring cleavage (intra- or extradiol) and are metabolized further by β-ketoadipate pathway (Masai et al. 2007; Bugg et al. 2011b; Abdelaziz et al. 2016).
5.8 Dioxygenases
Streptomyces sp. SirexAA-E was found to secrete a fusion enzyme SACTE_2871 containing aromatic ring dioxygenase (intradiol) domain and lignin-binding domain while growing on lignocellulosic biomass. This enzyme contains Fe3+ active site and performs oxygen-dependent cleavage of catechol; however, no activity on their methylated derivatives was observed. The secretion is carried out by Tat translocation pathway (Bianchetti et al. 2013). An extradiol dioxygenase was also discovered in the Sphingomonas paucimobilis SYK-6 responsible for lignin degradation. This dioxygenase was identified while studying degradation of biphenyl compound as substrate by this strain (Sonoki et al. 2009). These evidences clearly indicate the role of dioxygenase in lignin degradation.
5.9 Aromatic Alcohol Oxidase (EC 1.1.3.7)
This enzyme belongs to the class oxidoreductases and attacks on the CH–OH group of the donor, with oxygen as acceptor. It generally attacks the primary aromatic alcohol and converts it into aldehyde. It is a monomeric enzyme mostly found in fungi such as Geotrichum candidum, Botrytis cinerea, Pleurotus eryngii, Pleurotus sajor-caju, Pleurotus pulmonarius, Penicillium simplicissimum, Phanerochaete chrysosporium, Brachypsectra fulva, Fusarium solani, Bjerkandera adusta, and Rigidoporus microporus, and one bacterium Sphingobacterium sp. ATM is reported to exhibit this enzyme. Few Pseudomonas sp. under anaerobic conditions also show the presence of veratryl alcohol oxidase analog. This enzyme can efficiently modify or degrade aromatic alcohols produced during the degradation of lignin.
6 Role of Genomics and Proteomics in Understanding Lignin Degradation
Introduction of new molecular techniques in genomics, transcriptomics, and proteomics and advances in instrumental resolution paved the way for improved understanding of lignocellulosic biomass deconstruction by individual microbes and complex microbial communities. The increasingly available genomic data for bacteria and fungi indicate the potential of microbes for biomass degradation across diverse taxa. Comparative analysis of genome gives information regarding their taxonomic classification and possible physiological prospective (Baldrian and López-Mondéjar 2014). Recent development in NGS applied on lignocellulose-degrading fungi, bacteria, and complex community has been reviewed (Kameshwar and Qin 2016). The genome sequence of individual bacterial strain of actinobacteria, α-proteobacteria, β-proteobacteria, and γ-proteobacteria along with their important genomic features responsible for lignin degradation has been reported (Kameshwar and Qin 2016). Improvement in liquid chromatography and mass spectrometry with quantitative proteomics techniques such as isobaric tags for relative and absolute quantitation (iTRAQ) and label-free quantification (LFQ) has provided a solid platform to quantify proteins and their expression studies.
Enzyme production study at different time points can be performed to study the set of proteins expressed at a specified time under different culture conditions (Baldrian and López-Mondéjar 2014; Singh et al. 2017). Novel ligninolytic enzymes and unannotated proteins responsible for degradation can be identified. In recent studies, NGS is complemented with proteomics and metabolomics to get further precise information regarding pattern of bacterial biomass degradation. The genome sequence of some of the recently reported lignin-degrading bacteria are Tolumonas lignilytica, Pandoraea sp. ISTKB, Pseudomonas sp. strain YS-1p, Rhizobium sp. strain YS-1r, and Burkholderia sp. strain LIG30 (Woo et al. 2014; Billings et al. 2015; Prabhakaran et al. 2015; Bao et al. 2015; Kumar et al. 2016). Kumar et al. (2018) recently reported genomics and proteomics for understanding the novel genes, differential expression of the important genes on Kraft lignin and vanillic acid (most common intermediate found during lignin degradation). Novel pathways and enzymes were discovered for phenylacetate and benzoate (Kumar et al. 2018). Lin et al. (2016) used proteomics and genomics approach and engineered Pseudomonas sp. A514 for efficient lignin utilization and bioconversion. The polyhydroxyalkanoate production efficiency reached 73% of bacterial cell dry weight in the engineered strain. Several bacteria like Arthrobacter sp. Rue61a, Amycolatopsis sp. strain ATCC 39116, Novosphingobium sp. Strain MBES04, Cupriavidus basilensisB-8, Halomonas sp. strain KO116, Klebsiella sp. strain BRL6-2, Raoultella ornithinolytica strain S12 have shown to degrade the lignin and its monomers and are validated by whole genome sequencing and followed by identification of enzymes for lignin degradation and its bioconversion (Kameshwar and Qin 2016; Kumar et al. 2016).
7 Conclusion
The addition of value to the lignin generated as waste from plant polysaccharide-based industry will define the success and sustainability of such industries. The biological process is discussed for lignin depolymerization and degradation as it is the most eco-friendly and cost-effective route for its valorization. The pathways for the funneling of heterogeneous lignin derivatives and the enzymes assisting in the lignin utilization were discussed that will further assist in engineering the strains for the enhanced lignin degradation and its valorization. The value addition to lignin will establish the biorefinery concept and also validate the model of circular economy.
References
Abdelaziz OY, Brink DP, Prothmann J et al (2016) Biological valorization of low molecular weight lignin. Biotechnol Adv 34:1318–1346
Baldrian P, López-Mondéjar R (2014) Microbial genomics, transcriptomics and proteomics: new discoveries in decomposition research using complementary methods. Appl Microbiol Biochem 98:1531–1537
Bao W, Zhou Y, Jiang J, Xu Z et al (2015) Complete genome sequence of Raoultella ornithinolytica strain S12, a lignin-degrading bacterium isolated from forest soil. Genome Announc 3:e00104–e00115
Bianchetti CM, Harmann CH, Takasuka TE et al (2013) Fusion of dioxygenase and lignin binding domains in a novel secreted enzyme from cellulolytic Streptomyces sp. SirexAA-E. J Biochem 288:18574–18587
Billings AF, Fortney JL, Hazen TC et al (2015) Genome sequence and description of the anaerobic lignin-degrading bacterium Tolumonas lignolytica sp. Stand Genomic Sci 10:106
Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44:337–363
Brown ME, Chang MC (2014) Exploring bacterial lignin degradation. Curr Opin Chem Biol 19:1–7
Brown ME, Walker M, Nakashige TG et al (2011) Discovery and characterization of heme enzymes from unsequenced bacteria: application to microbial lignin degradation. J Am Chem Soc 133:18006–18009
Bugg TD, Ahmad M, Hardiman EM et al (2011a) The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol 22:394–400
Bugg TD, Ahmad M, Hardiman EM (2011b) Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 28:1883–1896
Bugg TD, Rahmanpour R, Rashid GM (2016) Bacterial enzymes for lignin oxidation and conversion to renewable chemicals. In: Production of biofuels and chemicals from lignin. Springer, Singapore, pp 131–146
Chakar FS, Ragauskas AJ (2004) Review of current and future softwood kraft lignin process chemistry. Ind Crops Prod 20:131–141
Chandra R, Chowdhary P (2015) Properties of bacterial laccases and their application in bioremediation of industrial wastes. Environ Sci Process Impacts 17:326–342
Chen Z, Wan C (2017) Biovalorization strategies for converting lignin in to fuels and chemicals. Renew Sustain Energ Rev 73:610–621
de Gonzalo G, Colpa DI, Habib MH et al (2016) Bacterial enzymes involved in lignin degradation. J Biotechnol 236:110–119
de Jong E, Higson A, Walsh P et al (2012) Bio-based chemicals value added products from biorefineries. IEA Bioen, Task 42 Bioref
DeAngelis KM, Sharma D, Varney R et al (2013) Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1. Front Microbiol 4:280
Doherty WO, Mousavioun P, Fellows CM (2011) Value-adding to cellulosic ethanol: lignin polymers. Ind Crops Prod 33:259–276
Feng J, Jiang J, Yang Z, Su Q et al (2016) Characterization of depolymerized lignin and renewable phenolic compounds from liquefied waste biomass. RSC Adv 6:95698–95707
Feofilova EP, Mysyakina IS (2016) Lignin: chemical structure, biodegradation, and practical application (a review). Appl Microbiol Biochem 52:573–581
Fuchs G, Bol M, Heider J (2011) Microbial degradation of aromatic compounds--from one strategy to four. Nat Rev Microbiol 9:803
Gírio FM, Fonseca C, Carvalheiro F et al (2010) Hemicelluloses for fuel ethanol: a review. Bioresour Technol 101:4775–4800
Gomes FJ, Santos FA, Colodette JL et al (2014) Literature review on biorefinery processes integrated to the pulp industry. Nat Resour 5:419
Guerriero G, Hausman JF, Strauss J et al (2016) Lignocellulosic biomass: biosynthesis, degradation, and industrial utilization. Eng Life Sci 16:1–16
Harmsen PFH, Huijgen W, Bermudez L, et al (2010) Literature review of physical and chemical pretreatment processes for lignocellulosic biomass (No.1184). Wageningen UR Food & Biobased Research
Harwood CS, Parales RE (1996) The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50:553–590
IEA (InternationalEnergyAgency). https://www.iea.org/topics/renewables/subtopics/bioenergy/. Accessed Nov 2016
Kameshwar AKS, Qin W (2016) Lignin degrading fungal enzymes. In: Production of biofuels and chemicals from lignin. Springer, Singapore, pp 81–130
Kumar M, Singh J, Singh MK et al (2015) Investigating the degradation process of kraft lignin by β–proteobacterium, Pandoraea sp. ISTKB. Environ Sci Pollut Res 22:15690–15702
Kumar M, Gazara RK, Verma S et al (2016) Genome sequence of Pandoraea sp. ISTKB, a lignin-degrading betaproteobacterium, isolated from rhizospheric soil. Gen Ann 4:e01240-16
Kumar M, Singhal A, Verma PK et al (2017) Production and characterization of Polyhydroxyalkanoate from Lignin derivatives by Pandoraea sp. ISTKB. ACS Omega 2:9156–9163
Kumar M, Verma S, Gazara RK et al (2018) Genomic and proteomic analysis of lignin degrading and polyhydroxyalkanoates accumulating β–proteobacterium Pandoraea sp. ISTKB Biotechol Biofuel 11:154
Lange H, Decinia S, Crestini C (2013) Oxidative upgrade of lignin – recent routes reviewed. Eur Polym J 49:1151–1173
Lin L, Cheng Y, Pu Y, Sun S et al (2016) Systems biology-guided biodesign of consolidated lignin conversion. Green Chem 18:5536–5547
Longe L, Garnier G, Saito K (2016) Lignin biodegradation with fungi, bacteria and enzymes for producing chemicals and increasing process efficiency. In: Production of biofuels and chemicals from lignin. Springer, Singapore, pp 147–179
Lupoi JS, Singh S, Parthasarathi R et al (2015) Recent innovations in analytical methods for the qualitative and quantitative assessment of lignin. Renew Sustain Energ Rev 49:871–906
Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Bios Biotech Biochem 71:1–15
Menon V, Rao M (2012) Trends in bioconversion of lignocellulose: biofuels, platform chemicals and biorefinery concept. Prog Energy Combust Sci 38:522–550
Moon RJ, Martini A, Nairn J et al (2011) Cellulose nanomaterials review: structure, properties and nanocomposites. Chem Soc Rev 40:3941–3994
Munk L, Sitarz AK, Kalyani DC et al (2015) Can laccases catalyze bond cleavage in lignin? Biotech Adv 33:13–24
Pérez-Pantoja D, González B, Pieper DH (2010) Aerobic degradation of aromatic hydrocarbons. In: Handbook of hydrocarbon and lipid microbiology. Springer, Berlin Heidelberg, pp 799–837
Prabhakaran M, Couger MB, Jackson CA et al (2015) Genome sequences of the lignin degrading Pseudomonas sp. strain YS-1p and Rhizobium sp. strain YS-1r isolated from decaying wood. Gen Ann 3:e00019-15
Priyadarshinee R, Kumar A, Mandal T et al (2016) Unleashing the potential of ligninolytic bacterial contributions towards pulpand paper industry: key challenges and new insights. Environ Sci Pollut Res Int 23:23349–23368
Ragauskas AJ, Beckham GT, Biddy MJ et al (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843
Ralph J, Lundquist K, Brunow G, Lu F et al (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3:29–60
Ramachandra M, Crawford DL, Hertel G (1988) Characterization of an extracellular lignin peroxidase of the lignocellulolytic actinomycete Streptomyces viridosporus. Appl Environ Microbiol 54:3057–3063
Rashid GM, Taylor CR, Liu Y et al (2015) Identification of manganese superoxide dismutase from Sphingobacterium sp.T2 as a novel bacterial enzyme for lignin oxidation. ACS Chem Biol 10:2286–2294
Reiter J, Strittmatter H, Wiemann LO et al (2013) Enzymatic cleavage of lignin β-O-4 aryl ether bonds via net internal hydrogen transfer. Green Chem 15:1373–1381
Riva S (2006) Laccases: blue enzymes for green chemistry. Trends Biotechnol 24:219–226
Santhanam N, Vivanco JM, Decker SR et al (2011) Expression of industrially relevant laccases: prokaryotic style. Trends Biotechnol 29:480–489
Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61:263–289
Sims RE, Mabee W, Saddler JN et al (2010) An overview of second generation biofuel technologies. Bioresour Technol 101:1570–1580
Singh MK, Kumar M, Thakur IS (2017) Proteomic characterization and schizophyllan production by Schizophyllum commune ISTL04 cultured on Leucaena leucocephala wood under submerged fermentation. Bioresour Technol 236:29–36
Singhvi MS, Chaudhari S, Gokhale DV (2014) Lignocellulose processing: a current challenge. RSC Adv 4:8271–8277
Sonoki T, Masai E, Sato K et al (2009) Methoxyl groups of lignin are essential carbon donors in C1 metabolism of Sphingobium sp.SYK-6. J Bas Micro 49(S1):S98–S102
Sugano Y, Muramatsu R, Ichiyanagi A et al (2007) DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase FAMILYASP171 replaces the distal histidine of classical peroxidases. J Biochem 282:36652–36658
Tian JH, Pourcher AM, Bouchez T et al (2014) Occurrence of lignin degradation genotypes and phenotypes among prokaryotes. Appl Microbiol Biotechnol 98:9527–9544
Woo HL, Utturkar S, Klingeman D et al (2014) Draft genome sequence of the lignin-degrading Burkholderia sp. strain LIG30, isolated from wet tropical forest soil. Genome Announc 2:e00637–e00614
Zhu D, Zhang P, Xie C et al (2017) Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnol Biofuels 10:44
Acknowledgment
We would like to express our sincere thanks to Department of Biotechnology, Government of India for providing research grant (BT/IN/Indo-UK/SVP/08/2018-19) through NEWTON BHABHA UK-INDIA project (Project no: 100182-584131) jointly funded by DBT and UKRI.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kumar, M., Morya, R., Gupta, A., Kumar, V., Thakur, I.S. (2021). Bacterial-Mediated Depolymerization and Degradation of Lignin. In: Singh, A., Srivastava, S., Rathore, D., Pant, D. (eds) Environmental Microbiology and Biotechnology. Springer, Singapore. https://doi.org/10.1007/978-981-15-7493-1_4
Download citation
DOI: https://doi.org/10.1007/978-981-15-7493-1_4
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-7492-4
Online ISBN: 978-981-15-7493-1
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)