Abstract
Autophagy is a process that is involved in the bulk recycling of cellular materials to replenish cellular demands during crisis. However, in cancer cells, the process of autophagy is dual-faced. Generally, during the initial stages of tumorigenesis, autophagy inhibits proliferation, but when the cancer cells face extreme nutrient deprivation and other metabolic stress then autophagy protects them to metastasize. Advances in clinical findings have identified many targets involved in the autophagic pathway of cancer. This led to the development of trials using protective autophagy inhibitor and lethal autophagy inducer in order to trigger cell death dependent on autophagy. Interestingly, the existence of solely autophagy-dependent death of the cancer cells is yet to be established in patients. Nevertheless, the present concept of autophagy-dependent cell death is highly debatable from the point of therapy. Specificity and efficacy issues of autophagy-dependent cell death remain largely uncertain. This brings us to focus on the controversies and lacunas in the understanding of cell death and autophagy. The tumor microenvironment (TME) is regarded as a bed of metabolic cascade that ensures proper refueling of the nutrient-deprived core facing extremes of pH, starvation, hypoxia, immunogenic intrusion, and therapeutic insults. The novelty of this chapter lies in its comprehensive outlook to highlight how autophagy modulation alters TME and its significance in rewiring the metabolism.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Introduction
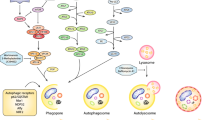
Tumors achieve their metabolic rewiring due to the fueling from their immediate microenvironment that makes them dynamic pseudoorgans (Lyssiotis and Kimmelman 2017). Tumor microenvironment (TME) represents an ecosystem of cells of heterogeneous lineage which is mainly comprised of cancer cells, stromal cells, interstitial fluids, immune cells, cytokines, chemokines, tumor-associated macrophages (TAM), cancer-associated fibroblasts (CAFs), and endothelial cells. TME provides the tumor a niche that makes them metabolically independent and breaks free off the homeostatic control of the normal cells thereby making them behave like a self-regulating organ(oid) like structure (Fig. 5.1). In order to understand the metabolic rewiring in a tumor, it becomes imperative to understand the phenomena of autophagy by virtue of which it can beat all stress response.
Exploring the arm that fuels the tumor microenvironment: ecosystem of TME comprises of heterogeneous cellular components from extracellular matrix (ECM) comprising cancer-associated fibroblasts, stromal cells, to various immune cells (T, NK, B cells, macrophages), and fats. TME creates a shield-like structure that is impervious to nutrient uptake, drug bioavailability, or access to growth factors, resulting in massive autophagy increase in the core where there is high stress due to starvation and hypoxia, accumulation of metabolic wastes, thereby resulting in the production of therapy-resistant cancer cells. This figure was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com
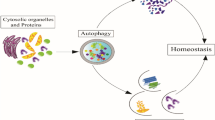
The term autophagy comes from the Greek meaning for “self-eating” which encompasses a highly evolutionarily conserved catabolic process digesting the cell’s own machinery to bypass a stress condition. Generally, the mechanism of autophagy remains conserved from yeast to higher mammals and involves the development of a crescent-shaped double-membrane structure called phagophore or isolation membrane, sequestering damaged part of the stressed cell en masse and gives rise to a ring-shaped “autophagosome”. This autophagosome fuses with a lysosome to give rise to “autolysosome” where the acidic milieu degrades the cellular cargo into recyclable metabolites which are absorbed into the cytosolic pool for further cellular nourishment. Till now 41 Atg (Autophagy-related) genes have been identified to regulate the process of autophagy, and most of the genes have been identified using yeast as a model organism. This phenomenon remains holistically conserved across different eukaryotes.
5.2 Fundamentals of Autophagy and its Duality in TME
Briefly speaking, autophagy can be categorized broadly into three types –macroautophagy, microautophagy, and chaperone-mediated autophagy. In this chapter, we will be dealing with macroautophagy which will be mentioned henceforth simply as autophagy. Before we open the topic about the relevance of autophagy in the tumor microenvironment, let us understand how the mechanism of autophagy function and why there is so much debate regarding this. Stress due to various stimuli like a response to therapy, increasing population of cancer cells, hypoxic tumor core region, alteration of pH, temperature, nutrient starvation, and pressure from infiltrating immune cells signal envisages inactivation of mTOR (mammalian Target Of Rapamycin) triggering the de novo synthesis of a double-layered isolation membrane that leads to nucleation of the initiation step of autophagy. Stress evokes a natural autophagic response in a cell through the class III phosphatidylinositol-3-kinase, hVps34 (human vesicular protein sorting 34) complex. Binding of Beclin1 with hVps34 is involved in rapid catalysis of PI3P (phosphatidylinositol-3-phosphate) production. PI3P, Beclin1, and ATG14 mostly remains tethered together as a specialized pre-autophagic structure called omagesome (named as such due to the Ω shaped structure) giving rise to the initial development of autophagosome. The presence of PI3P phosphatases like PTEN, JUMPY plays an important regulatory signal in the development of budding autophagic vesicle structure (Mukhopadhyay et al. 2016).
In the mammalian system, the initiation of autophagy is quite complex and involves the ULK1/2 complex which comprises FIP200, ATG 13, and ATG101 proteins. After initiation, the second major phase of autophagy is elongation, which is taken care of by the two ubiquitination-like systems LC3 and ATG5-ATG12 processing which gives rise to a mature double-membrane autophagosome structure. Finally, the autophagosome docks and fuses with the lysosome thereby forming autolysosome which help to inundate the cytoplasm with nutrients. Laura et al. showed that survivability of the disseminated solitary dormant breast cancer and metastatic tumor recurrence rely on triggering autophagy-mediated through ATG7 (Vera-Ramirez et al. 2018). The topic of autophagy is a highly complicated process with different and diverse nature at every turn of the tumorigenesis process. Any sort of stress implicated in a living cell provides an impetus to evoke an autophagy response. However, the dynamics of autophagy is not operated by a simple “on/off” switch in context to a cell that is undergoing tumorigenesis to develop into a cancer cell (Fig. 5.2).
Schematic representation of the dynamics of autophagy. Healthy cells exhibiting normal homeostasis undergoes cytoprotective autophagy during stress, however, when this continues for a protracted period beyond a point of no return then it becomes lethal and causes autophagy-dependent cell death. A cell undergoing tumorigenesis face extreme metabolic inhibition by cytoprotective autophagy, however, once the malignancy sets in then it serves as a promoter of tumor proliferation. A characteristic duality of autophagy which is different from the normal cellular autophagy is defined as oncophagy and the status of its progression according to the stages of the tumor is shown
Initially, autophagy behaves in a cytoprotective manner to reduce the stress and tumorigenic consequences in a cell. But once the benign tumor metastasizes to full-blown malignancy this cytoprotective mechanism shifts into a lethal phenotype by supporting and/or fueling the growing tumor cells to ensure their rapid proliferation and hijacks the metabolic fueling from the body.
Work in this field deserves special attention because of the janus role of autophagy in cancer. In a healthy cell, the cytoprotective role of autophagy prevents the buildup of excess stress-mediated damage and tumorigenesis. Protracted stress-mediated autophagy in these cells commits the cell to cross the “autophagic point of no return” an elusive concept that commits the cell to crossover an irreversible autophagy barricade that pushes the cytoprotective phenomena to a lethal makeshift (as elaborated in Fig. 5.2, Loos et al. 2011) and undergoes autophagy-dependent cell death (type II programmed cell death). Therefore, for a cancer cell, it becomes crucial to understand the autophagic point of no return to target them without making them therapeutically latent to pose risks of cancer recurrence. Autophagic modulation of TME helps the cancer cells strive for austere conditions like hypoxia, extreme acidosis, and accumulation of biological wastes in the interstitial spaces along with shielding the infiltrating chemotherapy.
Emerging research has focused on the vital role of TME that is capable of modulating autophagy and vice-versa. The immediate TME faces a rush of a diverse cellular niche. How the tumor environment can modulate the autophagy quotient during therapy to act against or in favor of the clinician is a hot field of research in the present time and will be discussed in detail in the later sections. There exists a lot of lacunae between medical research and patient therapy. This work tries to highlight these points with a focus on the major paradoxes and paradigms of the molecular mechanism at work behind the autophagic dynamics that modulate the TME inside a cancer cell.
5.3 Autophagic Regulation of Different Metabolites in TME
5.3.1 Autophagic Contributions of Glycolytic Intermediates
Tumor cells become attuned to dysregulated glucose metabolism leading to disease recurrence and cancer cell survival benefit under acidic pH stress by glycolysis to mediate autophagic stress through the PI3K/AKT/mTOR pathway that prolongs cancer cell survival by resisting apoptosis (Lue et al. 2017; Wojtkowiak et al. 2012). Low glucose in TME triggers energy sensor AMP-activated protein kinase (AMPK) mediated autophagy induction which becomes responsible for the degradation of damaged or long-lived components to replenish nutrients to compensate the stress (Williams et al. 2009). On the other hand, high glucose TME accelerated the SREBP1-autophagy axis to play a crucial role in pancreatic ductal adenocarcinoma (PDAC) progression (Zhou et al. 2019). Karsli-Uzunbas et al. showed that conditionally deleted ATG7 is expendable for short-term survival, but help to avoid the lethal hypoglycemic condition, cachexia during starvation highlighting a new role of autophagy in glucose homeostasis, and lung tumor maintenance (Karsli-Uzunbas et al. 2014). Chemotherapeutic insults in autophagy inhibited condition under glucose restricted condition led to the upregulation of period circadian clock 2 (PER2), a tumor suppressor protein in the colorectal cancer cell (Schroll et al. 2017). Interestingly, GPx1 degradation caused by glucose deprivation triggered ROS/AMPK signaling mediated autophagy in PDAC. Also, GPx1 may modulate glycolysis inhibition in pancreatic cancer under the glucose-starved situation while GPx1 overexpression and autophagy inhibition resulted in caspase-dependent apoptosis (Meng et al. 2018).
Upregulation of another glycolytic enzyme hexokinase (HKII) was found to be involved in escalating autophagy in the hypoxic TME of tongue squamous cell carcinoma (Chen et al. 2018). In a stressful TME, miRNA-7 inhibits autophagy by upregulation of LKB1-AMPK-mTOR signaling, thereby diminishing the supply of intracellular glucose pool available to fuel glycolysis (Gu et al. 2017). Glucose shortage mediated metabolic catastrophe resulted in autophagy-dependent BCR/Abl protein degradation in chronic myeloid leukemia cells, which are resistant to tyrosine kinase inhibitor-based therapy (Bono et al. 2016). βIII-Tubulin was known to decrease the reliance of cells on glycolytic metabolism, priming them to cope with low glucose stress in TME and protect from the endoplasmic reticulum (ER) by co-immunoprecipitating with GRP78 in non-small cell lung cancer (NSCLC) (Parker et al. 2016). GRP78 expression in limited glucose condition, led to dysregulation of pyruvate kinase M2 (PKM2) that triggered mitochondrial pyruvate dehydrogenase A (PDHA) and B (PDHB), causing a metabolic transition from glycolysis to the TCA cycle (Li et al. 2015). Glucose limitation in TME led to GRP78 mediated autophagic activation that led to degradation of IKKβ, which instigated inactivation of the NF–κB pathway and consequently altered the expression of PKM2, GLUT1, and HIF-1α.
Another glycolytic metabolite lactate helps the viability of glucose deprived melanoma TME in hypoxic conditions by repressing autophagy (Matsuo et al. 2019). In this regard tumor suppressor gene ANKDD1A reduces the half-life of HIF1α through an increase of FIH1; downregulation of glucose uptake and lactate production impedes autophagy along with triggering apoptosis in hypoxic glioblastoma multiforme (GBM) TME (Feng et al. 2019). Hypoxic TME of PDAC is involved in autophagy-mediated degradation of stromal lumican involved in regulating cancer progression (Sarcar et al. 2019).
Acidification of hypoxic and glucose deprived tumor cores by exogenous lactate supplementation is reported to prevent cell death by inhibiting autophagy in B16 melanoma cells (Matsuo and Sadzuka 2018). On the other hand, depletion of MCT4 on NK cells executes a compensatory metabolic rewiring by inducing autophagy to minimize the acidic extracellular breast tumor microenvironment that is involved with the export of lactate (Long et al. 2018). Following the phenomenon of reverse Warburg effect, cancer cells utilize lactate as an active metabolite and shuttle to TME to inflict metabolic reprogramming. In this regard, 2-methoxyestradiol, can inverse l-lactate-induced metabolic reprogramming in osteosarcoma 143B tumor cells (Gorska-Ponikowska et al. 2017). Phosphoproteomics screening established that mitochondrial Akt-PDK1 signaling alters tumor metabolism toward glycolysis by inhibiting autophagy, apoptosis, and support proliferation of hypoxic tumors (Chae et al. 2016).
5.3.2 Autophagy Contributions of Amino Acids
In this section, we briefly report about the major contributions of amino acids that are under the autophagic regulatory control network in TME. Among the amino acids, the glutamine plays a crucial role to fuel the tricarboxylic acid (TCA) cycle and maintain the pool size of antioxidants and ammonia.
Glutaminolysis-derived ammonia diffuses into the interstitial space of TME and is involved in regulating anti autophagy protein TIGAR (Mariño and Kroemer 2010) in MCF7 cells. Coculturing tamoxifen-resistant cancer cell with stromal fibroblasts drives catabolism of tumor stroma along with the anabolic activity of tumor cell (Albanese et al. 2011). Witkiewicz et al. showed that screening of lethal breast TME supported “Autophagic Tumor Stroma Model of Cancer Metabolism” and identified that loss of stromal caveolin-1 status in breast cancers leads to autophagy-mediated recycling of nutrients like lactate, ketones, and glutamine to feed anabolic events in cancer that led to metastasis and poor clinical prognosis (Witkiewicz et al. 2011). Interestingly, from the perspective of autophagy, it was reported that SNAT7 is a critical primary lysosomal glutamine exporter essential for the growth and proliferation of cancer (Verdon et al. 2017). Besides this, the relevance of arginine in TME was elucidated by work from Beth Levine’s lab where it was shown that autophagic impairment leads to the secretion of arginase and subsequent degradation of arginine was recognized as a metabolic vulnerability in cancer (Poillet-Perez et al. 2018).
On the other hand, preconditioning of primary human renal proximal tubular epithelial cells without tryptophan led to enhanced survivability in hypoxic conditions by triggering autophagy (Eleftheriadis et al. 2017). Exemplary findings established that the autophagic modulation in pancreatic stellate cells controls alanine secretion which feeds the PDAC in an austere tumor microenvironment to promote growth (Sousa et al. 2016).
5.3.3 Autophagy Contributions of Lipids
Vital research is being carried out to understand how the TME is being inundated with lipid molecules which in turn like a vicious cycle promote more fat-derived energy to fuel the tumor cell. Wen et al. co-cultured adipocytes with colon cancer cells to establish the mechanism by virtue of which adipocyte-derived free fatty acids are released to the cancer cells, thereby inducing autophagy as a result of AMPK activation (Wen et al. 2017). SIRT3 mediated autophagy was reported to adipocyte differentiation, and lipophagy mediated through increased activity of pyruvate dehydrogenase under high salt conditions which may result in an influx of lipid droplets in TME (Wei et al. 2019; Zhang et al. 2020).
5.4 Autophagic Regulation of Immune System in TME
An ecosystem in a TME comprises a plethora of immunomodulatory signals secreted by the cancer cells, CAFs, stromal cells, fibroblasts, macrophages, and T cells that modify each other to define the metabolic plasticity of the tumor. Tumor-associated macrophages (TAMs) create an environment conducive to facilitate the progression of the tumor. Tumor cell-released autophagosomes (TRAPs) stimulate IL-10-producing B cells and inhibit neutrophil production to repress antitumor immunogenic signals. Moreover, within TME, TRAPs are also responsible for converting TAMs into immunosuppressive M2 like macrophage secreting PD-L1, IL10. Inhibition of autophagy by Beclin1 silencing led to a decrease of TRAPs and enhanced T cell activation (Wen et al. 2018). The rush of antitumor response combined with translocation of calreticulin (CRT), extracellular release of ATP, danger-associated molecular patterns (DAMPs), high mobility group box 1 (HMGB1), and stimulation of type I interferon (IFN) led to a form of cell death which is regarded as immunogenic cell death (ICD). Targeting ICD by a combination of autophagy and chemotherapy is a potential approach to improve the prognosis of cancer patients with a long-term immune memory response to protect against the possible chance of tumor recurrence (Wang et al. 2018). For instance, Sigma1 utilizes autophagy to eradicate the functional PD-L1 from the cell surface to regulate the tumor immune microenvironment (Maher et al. 2018).
There is debate over the topic of mechanism that autophagy utilizes to trigger anticancer immunity while there are reports that show autophagy disarm anticancer immunity mediated by cytotoxic T cells and natural killer (NK) cells (Li et al. 2017). Autophagy-dependent regulation of TGF-β in myeloid cells leads to M2 macrophage accumulation in TME and is involved in controlling metastasis along with epithelial-mesenchymal transition (EMT) of tumor cells (Jinushi et al. 2017). The motility of aggressive metastatic cancer cell in a TME cannot be attributed due to a single factor, rather a hypoxic tumor core due to limited vascularization creating a hypoxic region which gives rise to mesenchymal-like carcinoma cells that exhibit high EMT and acquire stem cell-like propensity (Pietilä et al. 2016). Hypoxia-induced autophagy eliminates pro-apoptotic NK-derived serine protease GZMB/granzyme B, thereby blocking NK-mediated target cell apoptosis in breast tumor (Viry et al. 2014). Interestingly, chemotherapy dependent p53 activation helps in granzyme B and NK cells mediated breast tumor killing through induction of autophagy (Chollat-Namy et al. 2019).
Recent work confirmed that hypoxic TME activates autophagy as well as suppresses the immune surveillance of melanoma by NK cells through modulation of Cx43-mediated intercellular communications (Tittarelli et al. 2015). Although autophagy can be expendable for chemotherapy-induced cell death, but it is critical for its immunogenicity (Michaud et al. 2011). Tumor cells with functional autophagy responded to chemotherapy by attracting dendritic cells and T lymphocytes into TME. Inhibition of autophagy suppressed the discharge of ATP from dying cancel cell. Besides it will be interesting to figure out how autophagy can be used as a tool in a framework where cancer immunoediting integrates the immune system’s dual host-protective and tumor-promoting roles (Schreiber et al. 2011).
On the other hand inhibition of autophagy, as clinically identified by loss of LC3, HMGB1 staining followed by characteristic immune infiltration, and metastasis-free survival was identified to be a driver of tumor progression due to their adverse role in anticancer immunosurveillance (Ladoire et al. 2016). On a similar note, Atg5-mutant KRas (G12D)-driven lung cancer was reduced by depletion of CD25+ Treg cells thereby demonstrating that autophagy accelerates tumor progression; however, it represses early oncogenesis, highlighting a role between autophagy and regulatory T cell controlled anticancer immunity (Rao et al. 2014).
Gene profiling studies in early tumors of FIP200 conditional knockout mice saw T cells infiltration and CXCL10 secretion in TME (Wei et al. 2011). Interestingly, proinflammatory cytokine IL-1β secretion via an unconventional export pathway depending on Atg5 and involving Golgi reassembly stacking protein (GRASP) paralogues, GRASP55 (GORASP2) and Rab8a (Dupont et al. 2011). Moreover, elimination of LC3B and Beclin 1 was showed to accompany an increase in ROS, along with heightened stimulation of caspase-1 and secretion of interleukin 1β (IL-1β) and IL-18 (Nakahira et al. 2011). Autophagy blockade led to M2 to M1 repolarization of TAM to increased sensitivity of laryngeal cancer cells to cisplatin in mice (Guo et al. 2019).
Autophagic involvement in TME often presents a puzzle in that the infiltrating lymphocytes with a TME are dysfunctional in situ, however, they exhibit stem cell-like properties with massive metastatic potential. It was reported that increased abundance of extracellular potassium restricts T cell effector activity by restraining nutrient uptake, thus evoking autophagy and decline of histone acetylation at effector and exhaustion loci. This in turn produces CD8+ T cells with enhanced in vivo persistence, multipotency, and tumor clearance (Vodnala et al. 2019).
Besides this, the nonconventional role of autophagy was recently highlighted by Cunha et al., where they showed antitumor effects of LC3-associated phagocytosis (LAP) machinery impairment require tumor-infiltrating T cells, dependent upon STING and type I interferon response. Furthermore, they also showed that autophagy induction by myeloid cells in TME suppress T lymphocytes by effecting LAP (Cunha et al. 2018).
5.5 Concluding Remarks
The most important aspect of the knowledge of autophagic relevance holds a clue to modify the TME to determine the route to sensitization of a tumor (Mukhopadhyay et al. 2016). The conventional therapeutic regime in clinical trials focusing on autophagy, makes use of CQ/HCQ-based autophagy inhibition followed by different drugs like cisplatin, paclitaxel, FOLFOX, or radiation. Considering the duality of autophagy (Panda et al. 2015) and the intersecting cross-talking pathway shared with apoptosis (Mukhopadhyay et al. 2014) it becomes important to understand the autophagic point of no return.
But the biggest challenge in this regard of utilizing autophagy inhibition/induction-based medicine is that we still do not have any standard drug for clinical use that is extremely potent and specific towards tumor cells. Most of the drugs result in a holistic modulation of the tumor and all the neighboring cells so it becomes difficult for the effector killer cells or the therapy to intrude the thick TME and reach the core of the highly aggressive cancer cell that have already achieved high autophagic potential as an anti-therapeutic defense response. Subsequent to this effect it becomes a very big problem to estimate the accurate dose of therapy. It remains a clinical dilemma, to hit the right target without inducing side effects (Yang and Klionsky 2020). Moreover, there is also variation in stage-specific autophagy level that adds complexity to its dual character. So, it becomes imperative to search for any cancer stage-specific genetic event that can be assured to be a good modulatory marker of TME. An in-depth investigation revealing the molecular mechanism of the potential clinical drug on the interplay of TME’s autophagy and its metabolic signatures should be carried out to deliver a coherent approach for tackling this Achilles’ heel of cancer.
References
Albanese C, Machado FS, Tanowitz HB (2011) Glutamine and the tumor microenvironment: understanding the mechanisms that fuel cancer progression. Cancer Biol Ther 12(12):1098–1100. https://doi.org/10.4161/cbt.12.12.18856. Epub 2011 Dec 15
Bono S, Lulli M, D’Agostino VG, Di Gesualdo F, Loffredo R, Cipolleschi MG, Provenzani A, Rovida E, Dello Sbarba P (2016) Different BCR/Abl protein suppression patterns as a converging trait of chronic myeloid leukemia cell adaptation to energy restriction. Oncotarget 7(51):84810–84825. https://doi.org/10.18632/oncotarget.13319
Chae YC, Vaira V, Caino MC, Tang HY, Seo JH, Kossenkov AV, Ottobrini L, Martelli C, Lucignani G, Bertolini I, Locatelli M, Bryant KG, Ghosh JC, Lisanti S, Ku B, Bosari S, Languino LR, Speicher DW, Altieri DC (2016) Mitochondrial Akt regulation of hypoxic tumor reprogramming. Cancer Cell 30(2):257–272. https://doi.org/10.1016/j.ccell.2016.07.004
Chen G, Zhang Y, Liang J, Li W, Zhu Y, Zhang M, Wang C, Hou J (2018) Deregulation of hexokinase II is associated with glycolysis, autophagy, and the epithelial-mesenchymal transition in tongue squamous cell carcinoma under hypoxia. Biomed Res Int 2018:8480762. https://doi.org/10.1155/2018/8480762. eCollection 2018
Chollat-Namy M, Ben Safta-Saadoun T, Haferssas D, Meurice G, Chouaib S, Thiery J (2019) The pharmalogical reactivation of p53 function improves breast tumor cell lysis by granzyme B and NK cells through induction of autophagy. Cell Death Dis 10(10):695. https://doi.org/10.1038/s41419-019-1950-1
Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, Quarato G, Boada-Romero E, Kalkavan H, Johnson MDL, Natarajan S, Turnis ME, Finkelstein D, Opferman JT, Gawad C, Green DR (2018) LC3-associated phagocytosis in myeloid cells promotes tumor immune tolerance. Cell 175(2):429–441.e16. https://doi.org/10.1016/j.cell.2018.08.061. Epub 2018 Sep 20
Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V (2011) Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J 30(23):4701–4711. https://doi.org/10.1038/emboj.2011.398
Eleftheriadis T, Pissas G, Sounidaki M, Antoniadis N, Antoniadi G, Liakopoulos V, Stefanidis I (2017) Preconditioning of primary human renal proximal tubular epithelial cells without tryptophan increases survival under hypoxia by inducing autophagy. Int Urol Nephrol 49(7):1297–1307. https://doi.org/10.1007/s11255-017-1596-9. Epub 2017 Apr 17
Feng J, Zhang Y, She X, Sun Y, Fan L, Ren X, Fu H, Liu C, Li P, Zhao C, Liu Q, Liu Q, Li G, Wu M (2019) Hypermethylated gene ANKDD1A is a candidate tumor suppressor that interacts with FIH1 and decreases HIF1α stability to inhibit cell autophagy in the glioblastoma multiforme hypoxia microenvironment. Oncogene 38(1):103–119. https://doi.org/10.1038/s41388-018-0423-9. Epub 2018 Aug 6
Gorska-Ponikowska M, Kuban-Jankowska A, Daca A, Nussberger S (2017) 2-Methoxyestradiol reverses the pro-carcinogenic effect of l-lactate in osteosarcoma 143B cells. Cancer Genomics Proteomics 14(6):483–493. Review
Gu DN, Jiang MJ, Mei Z, Dai JJ, Dai CY, Fang C, Huang Q, Tian L (2017) microRNA-7 impairs autophagy-derived pools of glucose to suppress pancreatic cancer progression. Cancer Lett 400:69–78. https://doi.org/10.1016/j.canlet.2017.04.020. Epub 2017 Apr 25
Guo Y, Feng Y, Cui X, Wang Q, Pan X (2019) Autophagy inhibition induces the repolarisation of tumour-associated macrophages and enhances chemosensitivity of laryngeal cancer cells to cisplatin in mice. Cancer Immunol Immunother 68(12):1909–1920. https://doi.org/10.1007/s00262-019-02415-8. Epub 2019 Oct 22
Jinushi M, Morita T, Xu Z, Kinoshita I, Dosaka-Akita H, Yagita H, Kawakami Y (2017) Autophagy-dependent regulation of tumor metastasis by myeloid cells. PLoS One 12(7):e0179357. https://doi.org/10.1371/journal.pone.0179357. eCollection 2017
Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, White E (2014) Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4(8):914–927. https://doi.org/10.1158/2159-8290.CD-14-0363. Epub 2014 May 29
Ladoire S, Enot D, Senovilla L, Ghiringhelli F, Poirier-Colame V, Chaba K, Semeraro M, Chaix M, Penault-Llorca F, Arnould L, Poillot ML, Arveux P, Delaloge S, Andre F, Zitvogel L, Kroemer G (2016) The presence of LC3B puncta and HMGB1 expression in malignant cells correlate with the immune infiltrate in breast cancer. Autophagy 12(5):864–875. https://doi.org/10.1080/15548627.2016.1154244. Epub 2016 Mar 16
Li Z, Wang Y, Newton IP, Zhang L, Ji P, Li Z (2015) GRP78 is implicated in the modulation of tumor aerobic glycolysis by promoting autophagic degradation of KKβ. Cell Signal 27(6):1237–1245. https://doi.org/10.1016/j.cellsig.2015.02.030. Epub 2015 Mar 5
Li YY, Feun LG, Thongkum A, Tu CH, Chen SM, Wangpaichitr M, Wu C, Kuo MT, Savaraj N (2017) Autophagic mechanism in anti-cancer immunity: its pros and cons for cancer therapy. Int J Mol Sci 18(6):E1297. Review. https://doi.org/10.3390/ijms18061297
Long Y, Gao Z, Hu X, Xiang F, Wu Z, Zhang J, Han X, Yin L, Qin J, Lan L, Yin F, Wang Y (2018) Downregulation of MCT4 for lactate exchange promotes the cytotoxicity of NK cells in breast carcinoma. Cancer Med 7(9):4690–4700. https://doi.org/10.1002/cam4.1713. Epub 2018 Jul 26
Loos B, Genade S, Ellis B, Lochner A, Engelbrecht AM (2011) At the core of survival: autophagy delays the onset of both apoptotic and necrotic cell death in a model of ischemic cell injury. Exp Cell Res 317(10):1437–1453. https://doi.org/10.1016/j.yexcr.2011.03.011. Epub 2011 Mar 21
Lue HW, Podolak J, Kolahi K, Cheng L, Rao S, Garg D, Xue CH, Rantala JK, Tyner JW, Thornburg KL, Martinez-Acevedo A, Liu JJ, Amling CL, Truillet C, Louie SM, Anderson KE, Evans MJ, O'Donnell VB, Nomura DK, Drake JM, Ritz A, Thomas GV (2017) Metabolic reprogramming ensures cancer cell survival despite oncogenic signaling blockade. Genes Dev 31(20):2067–2084. https://doi.org/10.1101/gad.305292.117
Lyssiotis CA, Kimmelman AC (2017) Metabolic interactions in the tumor microenvironment. Trends Cell Biol 27(11):863–875. Review. https://doi.org/10.1016/j.tcb.2017.06.003. Epub 2017 Jul 19
Maher CM, Thomas JD, Haas DA, Longen CG, Oyer HM, Tong JY, Kim FJ (2018) Small-molecule Sigma1 modulator induces autophagic degradation of PD-L1. Mol Cancer Res 16(2):243–255. https://doi.org/10.1158/1541-7786.MCR-17-0166. Epub 2017 Nov 8
Mariño G, Kroemer G (2010) Ammonia: a diffusible factor released by proliferating cells that induces autophagy. Sci Signal 3(124):pe19. Review. https://doi.org/10.1126/scisignal.3124pe19
Matsuo T, Sadzuka Y (2018) Extracellular acidification by lactic acid suppresses glucose deprivation-induced cell death and autophagy in B16 melanoma cells. Biochem Biophys Res Commun 496(4):1357–1361. https://doi.org/10.1016/j.bbrc.2018.02.022. Epub 2018 Feb 5
Matsuo T, Daishaku S, Sadzuka Y (2019) Lactic acid promotes cell survival by blocking autophagy of b16f10 mouse melanoma cells under glucose deprivation and hypoxic conditions. Biol Pharm Bull 42(5):837–839. https://doi.org/10.1248/bpb.b18-00919
Meng Q, Xu J, Liang C, Liu J, Hua J, Zhang Y, Ni Q, Shi S, Yu X (2018) GPx1 is involved in the induction of protective autophagy in pancreatic cancer cells in response to glucose deprivation. Cell Death Dis 9(12):1187. https://doi.org/10.1038/s41419-018-1244-z
Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, Rello-Varona S, Tailler M, Menger L, Vacchelli E, Galluzzi L, Ghiringhelli F, di Virgilio F, Zitvogel L, Kroemer G (2011) Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 334(6062):1573–1577. https://doi.org/10.1126/science.1208347
Mukhopadhyay S, Panda PK, Sinha N, Das DN, Bhutia SK (2014) Autophagy and apoptosis: where do they meet? Apoptosis 19(4):555–566. Review. https://doi.org/10.1007/s10495-014-0967-2
Mukhopadhyay S, Sinha N, Das DN, Panda PK, Naik PP, Bhutia SK (2016) Clinical relevance of autophagic therapy in cancer: investigating the current trends, challenges, and future prospects. Crit Rev Clin Lab Sci 53(4):228–252. Review. https://doi.org/10.3109/10408363.2015.1135103. Epub 2016 Feb 16
Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM (2011) Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 12(3):222–230. https://doi.org/10.1038/ni.1980. Epub 2010 Dec 12
Panda PK, Mukhopadhyay S, Das DN, Sinha N, Naik PP, Bhutia SK (2015) Mechanism of autophagic regulation in carcinogenesis and cancer therapeutics. Semin Cell Dev Biol 39:43–55. Review. https://doi.org/10.1016/j.semcdb.2015.02.013. Epub 2015 Feb 25
Parker AL, Turner N, McCarroll JA, Kavallaris M (2016) βIII-Tubulin alters glucose metabolism and stress response signaling to promote cell survival and proliferation in glucose-starved non-small cell lung cancer cells. Carcinogenesis 37(8):787–798. https://doi.org/10.1093/carcin/bgw058
Pietilä M, Ivaska J, Mani SA (2016) Whom to blame for metastasis, the epithelial-mesenchymal transition or the tumor microenvironment? Cancer Lett 380(1):359–368. Review. https://doi.org/10.1016/j.canlet.2015.12.033. Epub 2016 Jan 11
Poillet-Perez L, Xie X, Zhan L, Yang Y, Sharp DW, Hu ZS, Su X, Maganti A, Jiang C, Lu W, Zheng H, Bosenberg MW, Mehnert JM, Guo JY, Lattime E, Rabinowitz JD, White E (2018) Autophagy maintains tumour growth through circulating arginine. Nature 563(7732):569–573. https://doi.org/10.1038/s41586-018-0697-7. Epub 2018 Nov 14
Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, Sykacek P, Frank L, Schramek D, Komnenovic V, Sigl V, Aumayr K, Schmauss G, Fellner N, Handschuh S, Glösmann M, Pasierbek P, Schlederer M, Resch GP, Ma Y, Yang H, Popper H, Kenner L, Kroemer G, Penninger JM (2014) A dual role for autophagy in a murine model of lung cancer. Nat Commun 5:3056. https://doi.org/10.1038/ncomms4056
Sarcar B, Li X, Fleming JB (2019) Hypoxia-induced autophagy degrades stromal lumican into tumor microenvironment of pancreatic ductal adenocarcinoma: a mini-review. J Cancer Treat Diagn 3(1):22–27. https://doi.org/10.29245/2578-2967/2019/1.1165. Epub 2019 Feb 22
Schreiber RD, Old LJ, Smyth MJ (2011) Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331(6024):1565–1570. Review. https://doi.org/10.1126/science.1203486
Schroll MM, LaBonia GJ, Ludwig KR, Hummon AB (2017) Glucose restriction combined with autophagy inhibition and chemotherapy in hct 116 spheroids decreases cell clonogenicity and viability regulated by tumor suppressor genes. J Proteome Res 16(8):3009–3018. https://doi.org/10.1021/acs.jproteome.7b00293. Epub 2017 Jul 3
Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, Asara JM, Evans RM, Cantley LC, Lyssiotis CA, Kimmelman AC (2016) Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536(7617):479–483; 540(7631):150. https://doi.org/10.1038/nature19084. Epub 2016 Aug 10
Tittarelli A, Janji B, Van Moer K, Noman MZ, Chouaib S (2015) The selective degradation of synaptic Connexin 43 protein by hypoxia-induced autophagy impairs natural killer cell-mediated tumor cell killing. J Biol Chem 290(39):23670–23679. https://doi.org/10.1074/jbc.M115.651547. Epub 2015 Jul 28
Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE (2018) Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun 9(1):1944. https://doi.org/10.1038/s41467-018-04070-6
Verdon Q, Boonen M, Ribes C, Jadot M, Gasnier B, Sagné C (2017) SNAT7 is the primary lysosomal glutamine exporter required for extracellular protein-dependent growth of cancer cells. Proc Natl Acad Sci U S A 114(18):E3602–E3611. https://doi.org/10.1073/pnas.1617066114. Epub 2017 Apr 17
Viry E, Baginska J, Berchem G, Noman MZ, Medves S, Chouaib S, Janji B (2014) Autophagic degradation of GZMB/granzyme B: a new mechanism of hypoxic tumor cell escape from natural killer cell-mediated lysis. Autophagy 10(1):173–175. https://doi.org/10.4161/auto.26924. Epub 2013 Nov 15
Vodnala SK, Eil R, Kishton RJ, Sukumar M, Yamamoto TN, Ha NH, Lee PH, Shin M, Patel SJ, Yu Z, Palmer DC, Kruhlak MJ, Liu X, Locasale JW, Huang J, Roychoudhuri R, Finkel T, Klebanoff CA, Restifo NP (2019) T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 363(6434):eaau0135. https://doi.org/10.1126/science.aau0135
Wang YJ, Fletcher R, Yu J, Zhang L (2018) Immunogenic effects of chemotherapy-induced tumor cell death. Genes Dis 5(3):194–203. Review. https://doi.org/10.1016/j.gendis.2018.05.003. eCollection 2018 Sep
Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL (2011) Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev 25(14):1510–1527. https://doi.org/10.1101/gad.2051011
Wei T, Huang G, Liu P, Gao J, Huang C, Sun M, Shen W (2019) Sirtuin 3-mediated pyruvate dehydrogenase activity determines brown adipocytes phenotype under high-salt conditions. Cell Death Dis 10(8):614. https://doi.org/10.1038/s41419-019-1834-4
Wen YA, Xing X, Harris JW, Zaytseva YY, Mitov MI, Napier DL, Weiss HL, Mark Evers B, Gao T (2017) Adipocytes activate mitochondrial fatty acid oxidation and autophagy to promote tumor growth in colon cancer. Cell Death Dis 8(2):e2593. https://doi.org/10.1038/cddis.2017.21
Wen ZF, Liu H, Gao R, Zhou M, Ma J, Zhang Y, Zhao J, Chen Y, Zhang T, Huang F, Pan N, Zhang J, Fox BA, Hu HM, Wang LX (2018) Tumor cell-released autophagosomes (TRAPs) promote immunosuppression through induction of M2-like macrophages with increased expression of PD-L1. J Immunother Cancer 6(1):151. https://doi.org/10.1186/s40425-018-0452-5
Williams T, Forsberg LJ, Viollet B, Brenman JE (2009) Basal autophagy induction without AMP-activated protein kinase under low glucose conditions. Autophagy 5(8):1155–1165. Epub 2009 Nov 16
Witkiewicz AK, Kline J, Queenan M, Brody JR, Tsirigos A, Bilal E, Pavlides S, Ertel A, Sotgia F, Lisanti MP (2011) Molecular profiling of a lethal tumor microenvironment, as defined by stromal caveolin-1 status in breast cancers. Cell Cycle 10(11):1794–1809. Epub 2011 Jun 1
Wojtkowiak JW, Rothberg JM, Kumar V, Schramm KJ, Haller E, Proemsey JB, Lloyd MC, Sloane BF, Gillies RJ (2012) Chronic autophagy is a cellular adaptation to tumor acidic pH microenvironments. Cancer Res 72(16):3938–3947. https://doi.org/10.1158/0008-5472.CAN-11-3881. Epub 2012 Jun 19
Yang Y, Klionsky DJ (2020) Autophagy and disease: unanswered questions. Cell Death Differ 27:858. Review. https://doi.org/10.1038/s41418-019-0480-9 [Epub ahead of print]
Zhang T, Liu J, Tong Q, Lin L (2020) SIRT3 Acts as a Positive Autophagy Regulator to Promote Lipid Mobilization in Adipocytes via Activating AMPK. Int J Mol Sci 21(2):372. https://doi.org/10.3390/ijms21020372
Zhou C, Qian W, Li J, Ma J, Chen X, Jiang Z, Cheng L, Duan W, Wang Z, Wu Z, Ma Q, Li X (2019) High glucose microenvironment accelerates tumor growth via SREBP1-autophagy axis in pancreatic cancer. J Exp Clin Cancer Res 38(1):302. https://doi.org/10.1186/s13046-019-1288-7
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Mukhopadhyay, S. (2020). Exploring the Metabolic Implications of Autophagy Modulation in Tumor Microenvironment. In: Bhutia, S.K. (eds) Autophagy in tumor and tumor microenvironment . Springer, Singapore. https://doi.org/10.1007/978-981-15-6930-2_5
Download citation
DOI: https://doi.org/10.1007/978-981-15-6930-2_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-6929-6
Online ISBN: 978-981-15-6930-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)