Abstract
The hydrating water molecules play a crucial role in enhancing the structural stability, protein folding, and competition with the ligand for its occupancy at the binding site. This chapter describes the importance of hydrating water molecules in biomolecular processes explained via thermodynamic approaches. The focus is on understanding the change of magnitude of hydrating water molecules present at the surface of the protein and their shift to the bulk water and protein environment. Due to significant inhomogeneity in water density, polarity, and mobility assisted by intramolecular interactions such as hydrophobic, electrostatic forces, and hydrogen-bonding network, the thermodynamic information are hard to obtain through experiments. Computational elucidation of water displacements by ligand molecules is characterized by the entropic and enthalpic compensation where the inhibition of mobility of the water molecule and solvent environment sets a limit on the entropic contribution and ligand reactivity. Most importantly, computational simulations can provide a better understanding of the most intriguing aspects of hydration water molecules. Water is an influential determinant for protein–protein interactions, protein–nucleic acid interactions, and other enzymes-assisted or cofactor-assisted interactions that are driven by hydrophilic forces, whereas the hydrophobic effects facilitate the binding energy of the ligand.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Protein hydration
- Enthalpy and entropy compensation
- Hydrophilic and hydrophobic
- Water displacement and thermodynamics
8.1 Introduction
Drug designer uses proteins as a therapeutic option that becomes challenging without knowing the nature and the conformational characteristics of the specific protein (Lagasse et al. 2017). Proteins are represented by four forms of structures (primary structure, secondary structure, tertiary structure, and quaternary structure). There are 20 naturally occurring amino acids, grouped into ten polar and ten nonpolar amino acids based on their side chain features (Kangueane and Nilofer 2018). Analytical methods such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and electron microscopy are used to determine the structural stability of the proteins. Protein stability can be disrupted by various factors such as external stress factors including temperature, pH, removal of water, presence of hydrophobic surfaces, metal ions, etc. (Thomas 2020; Tripathi 2013). Water plays a crucial role in protein folding and actively participates in the long range of water-mediated contacts through the hydrogen-bonded network. Similarly, adding water molecules will enhance protein folding and drug design strategies with high specificity (Rhee et al. 2004). Each water molecule acts as a magnet comprising three atoms (two hydrogen atoms that have a positive charge and one oxygen atom that has a negative charge). The oxygen atom of the nucleus (with eight positively charged protons) in the water molecule attracts electrons strongly than the hydrogen nuclei (with only one positively charged proton). The different part of the water molecule has a separate charge named as a polarity in which a particular part of the molecule is positively charged, and another part of the molecule is negatively charged.
Water molecules play an essential role in the stability, folding, dynamics, and function of the biomolecules (Levy and Onuchic 2004). The hydration forces determine the stabilization and packing of the protein structure. These hydrating water molecules involved in the formation of hydrogen bond networks. Hydration of water molecules close to the protein surface that exhibits dynamical properties from those of bulk are crucial for stabilizing the folded proteins (Bizzarri and Cannistraro 2002). Hydrating water fluctuations can affect the protein function, and disturb the protein dynamics factor, having consequent changes in the protein motions. A solvent is an active participant in energy associated with a protein (Ansari et al. 1992; Fenimore et al. 2002). Hydrophobic interactions contribute to protein stability and are very important for making protein stable and biologically active conformation (Zapadka et al. 2017). The energetics of protein folding and binding involves significant trade-offs between losses of protein–water interactions.
Nowadays, thermodynamic signatures have become popular and are considered as the most important criteria for the selection of potential drug molecules (Ladbury et al. 2010). The only limitation from the thermodynamic signatures is their unpredictable motions in the local water structure. Due to the thermodynamic nature of the binding, water molecules have a dominant effect. Specific interactions were observed in water molecules for the promotion of protein–ligand, but the thermodynamic signals are not involved in the proximity of the ligand (Spyrakis et al. 2017). Water not only interacts with the protein surface, but it will also have interplay with the protein backbone and side chains (Levy and Onuchic 2004). Mutations affect the placement of the water molecules in the protein structures and disturb the main interactions of the water network that leads to destabilization (Covalt et al. 2001). Interactions observed in water-mediated network exhibit entropy-enthalpy compensations, which has a minor effect on affinity and significant impact on the thermodynamic signatures. The contributions of solvent–solvent interactions play a vital role in molecular recognition processes. Therefore, experimentally predicted binding enthalpy value is extracted from isothermal titration calorimetry and have a mix of several positive and negative thermodynamic signatures which strongly restrain the solvent changes (Ruhmann et al. 2015).
The major limitation and accuracy of molecular simulations depend on the relevant biological systems and the core force fields. A biological system consists of large molecules with several internal degrees of freedom and solvent molecules. Sampling of all conformations is a complicated process and inorder to overcome these issues, we can calculate the free energy perturbation (FEP) simulations, which give differences in estimated free energy between closely related systems, e.g., small incremental changes in ligands (Manzoni and Ryde 2018). The selection of appropriate force field is very challenging in the case of nonavailable experimental evidence.
The water in the hydrogen bond is neither weak nor too strong. Each water molecule forms two hydrogen bonds and are covalently attached to the oxygen of a water molecule (492.21 kJ mol−1) (Boyarkin et al. 2013). To predict the location of a particular water molecule from the same crystal structure, different methods and their applied force fields can give huge different estimates on the position and conformation. A recent study has investigated the relationship between water models and the computed free energies in protein folding (Anandakrishnan et al. 2019). Computed protein folding free energy landscape obtained from water–water interactions seems to have the most significant discrepancies due to the long range of electrostatic interactions and not because of Van der Waals interactions (Kuffel 2017).
This chapter will review the identification of the location of water molecules and its thermodynamics properties (ΔHsolv and ΔSsolv), which are very important for protein stability and the drug discovery process. Protein hydrations are essential for the determination of protein 3D structures (Wüthrich et al. 1992). Most of the water molecules interact with the protein, and the resulting hydration structure is an integral part of the proteins. Due to the amino acid substitution on the protein surface, slight changes are observed in protein stability and hydration. There are various tools available to predict the hydration sites, and it will be explained in the subsequent section, which may help in defining the solvent sites through different computational techniques applied on an explicitly solvated binding pocket (Mondal et al. 2014). This extensive thermodynamics study will give a significant insight on molecular design and drug discovery.
8.2 Water Molecules in Proteins
Water molecules present in the protein or between the proteins are linked by hydrogen bonds similar to the interface between a ligand and protein. When compared with the protein, protein–ligand complexes (one or two) with the presence of water has a higher B-factor (Lu et al. 2007). The term “buried” defines that a molecular entity’s solvent-accessible surface area is ≤5%, and the B-factor of such buried water holding three or four bonds is lower than the structural average of the protein because of their location in different secondary structure elements. Significantly, low B-factor with buried water is conserved in the crystal structures of evolutionarily linked proteins (Takano et al. 2003).
Interestingly, the B-factor relation also holds vice versa: Protein atoms show lower B-factor when hydrogen bond is formed to buried water molecules exclusively, rather than hydrogen bonding to another protein atom. Similarly, the protein backbone amide nitrogen can form only one H-bond, while the carbonyl oxygen can form two simultaneously. Mostly, amino acid side chains buried in the protein core are hydrophobic, so these water molecules forms hydrogen bonds to the protein backbone. Hence, the hydrophobic forces are involved in the denaturation of the proteins and their temperature dependency (Van Dijk et al. 2015).
After the hydrophobic folding process, water uses protein to satisfy their requirement of hydrogen bond formation. Consequently, backbone hydrogen bonds formed with the water molecules and not with other protein residues are localized in regions that are neither involved in helical structure nor pleated sheet conformation (Finkelstein and Ptitysyn 2016). Waters are mostly observed in α-helices than β-sheets and often found in coil regions. The difference between α-helices and β-sheets for hosting water molecules is not because of the number of cavities or hydrophobicity but it is likely due to secondary structure flexibility and thus residence times (Gromiha 2000).
Buried polar side chains are flexible or evolutionarily optimized enough to find other protein atoms to bond and do not rely on water molecules. Protein–ligand interface consists of charged and polar amino acids such as Arg and Glu, responsible for the main hydrated side chains. Commonly hydrophobic residues such as Val, Leu, Ile, Phe, and Met have an interior buried polar side chain exposed to solvent (Malleshappa Gowder et al. 2014). Lys is a charged amino acid, frequently hydrated in rigid protein–ligand interfaces, but when it has a large cavity, obtain rotameric states (Perutz et al. 1965). Such characteristics are mostly found in disordered crystal structures that impart this disorder on nearby water molecules. The amino acids Tyr and Trp constituting aromatic moieties are more hydrating in nature.
In apo-protein crystal structures, among all 20 amino acids, Gly has the lowest hydration propensity because of its extensive flexibility and small size. This situation is reversed in the rigid interfaces of protein–ligand complexes, where it has the highest main chain hydration propensity comparable to the Ser side chain as its backbone is accessible than the backbone of other amino acids (Pappas et al. 2012). On the other hand, the amide nitrogen of Pro does not form hydrogen bonds. Yet, it is frequently observed to be located close to buried water molecule due to its rigidness, B-factor will decrease. Interestingly, Pro in protein–ligand systems has the lowest hydration propensity when compared to other side chains (Rose and Wolfenden 1993). Gly and Pro behave inversely as Pro is a nonpolar amino acid that gets displaced upon ligand binding.
There is no cutoff kept for buried water molecules present in protein surface, but the overwhelming bulk is found at less than 3 Å, or about one protein atom depth. Nearly 60% of buried waters are “alone,” 20% is as a cluster of two molecules, 8% as a cluster of three molecules, and so on. Each additional water molecule beyond the first forms about 1–1.5 hydrogen bonds to the protein, while the remaining interacts with the other water molecules (Maurer and Oostenbrink 2019). Generally, waters present in the core region will enhance the protein stability with the hydrating polar atoms (Ball 2008).
8.2.1 Protein Hydration: An Essential Factor for Various Biological Processes
As mentioned, water or protein hydration is vital to form and maintain the 3D structural and functional aspects of protein. The combined use of experimental (X-ray, neutron scattering and diffraction, NMR, terahertz spectroscopy) and computational (Molecular simulations) techniques have confirmed the essentiality of water in protein stability and consequent processes (Bellissent-Funel et al. 2016). Layers of hydrating water molecules determine the protein structure resolution and reliability. Proteins solved in a resolution of about 1.5–1.6 Å have a continuum of hydration layers with a mono-layer covering >1.5 moles H2O mol−1 amino acid residue.
8.2.2 Role of Water Molecules in Protein Stability
Proteins are essential for biological functions and are polymers of alpha-amino-acids. Under normal pH, temperature, and ionic strength conditions, the native structure of protein assumes an ideal 3D fold. The proteins hold different functional groups in their side chains (amide, carboxylic, hydroxyl, thiol, aromatic). This specific native protein structure catalyzes and regulates reactions, transport substrates, code, and transcribes genetic information. It has been extensively appreciated that water molecules are essential for maintaining the structure, stability, dynamics, and function of biomolecules. In reality, the absence of water molecules may lead to a lack of functional activity. However, it is found in recent years that water has been prominently treated as an integral component of biomolecular systems. Mixtures of experimental and computational studies acknowledge the active role of water in biomolecule structure, stability, and dynamics. Molecular dynamics (MD) simulations can balance the knowledge gained through experiments and explain the biomolecule and solvent as well as the time dependency of their dynamics (Karplus and McCammon 2002). The importance of water in structure, stability, and dynamics of proteins and nucleic acids is discussed below, besides the underlying molecular recognitions are also addressed.
8.2.3 Hydrating Water Around the Protein Surface
Water molecules interact with the surface groups by reorientation of both themselves and protein. In contrast, the other water molecules link this association to the rest of the bulk in an orderly manner to remain in active form (Zhong et al. 2011). The movement of the surface water molecules is due to the hydrogen bonding, and the dielectric constant is constrained, which is imposed by the protein (Seyedi and Matyushov 2018). The surface of the protein contains the electron acceptor groups than the donor, along with the excluded-volume effects, which is the cause for unoccupied hydrogen bonds in the region of the neighboring water molecules. The number of hydration sites in a protein depends on two factors, namely (a) conformational variability and (b) the freedom that the water has to hydrate the protein (Parsegian 2002). Protein conformations requiring greater hydration are facilitated by the compactness of waters containing many weak, delicate, or twisted hydrogen bonds, whereas the other form of conformations requires low-density water containing many strong intramolecular hydrogen bonds. The waters around the surface region are held captive by the basic amino acids, and those waters, when exposed to the bulk solvent through the groups present in that region, paves the way to greater flexibility and chain movement (Bandyopadhyay et al. 2005a, b). The core hydration layers around the protein surface are affected by the chemical composition of the amino acids and molecular interactions. Conversely, hydrating water molecules are slower in the presence of hydrophobic residues than the hydrophilic residues.
Adding water molecules can ease the structure prediction through the knowledge-based potential to an established Hamiltonian. The study shows that water not only induces protein folding and binding but also actively participates via water-mediated contacts. Water present at the surface of biological macromolecules defines a layer, which has been termed “biological water.” The closeness of the “biological water” at the protein surface distinctly exhibits dynamical properties from that of the bulk, also its residence times in the sub-nanosecond range.
8.2.4 Protein Folding–Hydrogen Bond Network–Water Clusters–Domain Motions
The hydrating water molecules influence the reactions and interactions of coenzymes and cofactors. Despite changes in the ionic strength, there seems to be a less impact on the protein charge distribution extended by the effects of hydrating waters (Virtanen et al. 2014). Second, the most important biological phenomena “protein folding” depends on the drive of the hydrophobic groups, which is usually clustered away from the protein surface and is controlled by the interactions with polar based residues and cooperative hydration (Huggins 2016). Also, the protein folding is characterized by the decrease in the β-sheet linking hydrogen bonds (Novak and Grdadolnik 2017). For a protein to be biologically active, it should form a 2D hydrogen-bonded network spanning the protein surface connecting all the surface hydrogen-bonded water clusters (Oleinikova et al. 2005; Smolin et al. 2005). Changes in hydration sites are necessary to account for the collective domain motions, with water molecules corresponding to structural transitions through movements of the protein cavity observed through alteration in the formation of hydrogen bonds. Besides this, interior protein motions are combined through the involvement of hydrogen-bonded water bound in the protein structure that facilitate through the distribution of information between the functionally important regions of the protein.
8.2.5 Electrostatic Forces and Buried Waters
Intramolecular forces contribute significantly to protein structure and stability, and the forces are more influential in α-helices rather than β-sheets (Nakasako 2004). Water molecules can help to bridge different peptide links by catalyzing the interaction between the carbonyl oxygen atoms and amide protons, forming more interactions that stabilize protein–ligand and protein–protein interfaces (Cao and Bowie 2014). Internal water molecules enable the folding of proteins and are cast away from the hydrophobic central core (Ikura et al. 2004). MD simulations will facilitate the computation of buried waters, and multi-water bridges (Cheung et al. 2002), and these buried waters form structurally important hydrogen-bonded linkages. Usually, the first hydration shell around the protein surface has high proton transfer, well-resolved hydration sites, and organized hydrogen bond interactions with large net dipole fields (Pradhan et al. 2019). Hydrogen bonds formed by these hydration waters around the surface have longer lifetimes than the bulk water (Yokomizo et al. 2005). The organization of the waters present in the nearby proteins expands its electrostatic visibility to visiting ligands (Chakraborty et al. 2007). Moreover, the protein folding nature can be determined by the electrostatic effect of the water directed towards specific hydrogen bonding, which extends into the bulk from the surface of hydrogen-bonding to the amino acid side chains (Hildebrandt et al. 2007).
8.2.6 Role of Thermodynamics in Protein Stability
Temperature and pressure play a significant role in protein unfolding when the temperature of the system increases, the entropy also increases, and proteins adapt themselves in folded form. The major role of hydration on protein folding can be studied by MD simulation, umbrella sampling, coarse-grained models, and knowledge-based structure prediction models. These calculations showed that despite the hydrophobic disruption in the protein chain, the water molecules enrich the folding process by forming intermediates with the protein backbone. Entropy is considered being the most critical parameter in understanding the stability of the proteins. Thermodynamic studies help to predict the protein conformational changes under environmental conditions and thermodynamic parameters useful in analyzing protein stability. The thermodynamic properties (enthalpy, entropy, and free energy) always have a standard state while it changes during folding and unfolding of proteins, which aids in understanding the protein stability (Gummadi 2003).
Entropy is a mathematical concept that describes the distribution of energy within a system. Free energy is a thermodynamic function that relates enthalpy and entropy to spontaneity and equilibrium constants. Hence there are theoretical approaches such as WaterMap (Sect. 8.3.10) that will help in locating the hydration sites and thermodynamic properties of water molecules. This facilitates solvate protein-binding sites that offer rich physical insights into the properties of the pocket and the hydrophobic forces driving the binding of small molecules.
8.2.7 Thermodynamics Properties in the Stability of Molecular Interactions
Water is an essential part of enzymes, which penetrates and determines how molecules move, bind to each other, and solvate charges. For example, the proton transfer takes place inside of proteins carried out by the Grotthuss mechanism, and it requires a group of hydrogen bonds involving internal water molecules and protein amino acid residues. Consecutively, water molecules can assist the enzymes catalytic reactions by behaving as a temporary proton donor/acceptor. Hence, to understand the function, it is essential to know about the location of internal water molecules available in the protein structures. To find the location of the internal water molecules, it is best to observe while they are forming hydrogen bonds, but due to its dynamic nature, the crystallographic prediction of the hydrating water molecules becomes less feasible. Only crystal structures with high resolution can be able to capture a few water molecules with certainty per protein (Kubinyi 2001).
Non-covalent interactions play a significant role in deducing the structure stability, and specific Thermodynamic and Spectroscopic studies are used to identify and compute the non-covalent interactions. Due to the enthalpic and entropic contribution, there is a difference in free energy between native and unfolded states of a protein. Numerous forces bring small and differing contributions to protein stability. Few recent experimental techniques can elucidate the structure and denatured states of the protein. The ignorance of the structural details of unfolded proteins makes it difficult to analyze all data quantitatively. Electrostatic interactions, particularly the salt bridges and cation interactions, also the Van der Waals interactions, play essential roles in defining the stability of proteins. The hydrophobic effect, hydrogen bonds, and water molecules are important contributors to protein structure and stability.
Frank and Evans explained the low solubility of nonpolar variety in the water at the molecular level. In the late 1950s, Kauzmann introduces the concept of “hydrophobicity,” which explains the protein folding complexity (Baldwin 2014). Water is very fundamental in protein folding because of hydrophobic attractions. Water form clusters around nonpolar groups (hydrophobic hydration), which lead to slight decrease in entropy of the system, and when they are released from the hydrophobic surfaces, there is a gain in entropy contributing to protein stability. The hydrophobic “collapse” of the protein is necessarily accompanied and explored by hydrogen bond formation between favorable functional groups (Fernández et al. 2003). For biological phenomena like protein folding water molecules are essential to define the “hydrophobic interactions.” Hydrogen bonds will have a positive contribution towards protein stabilization and balance between entropy and enthalpy terms. Regardless of the little involvement build towards protein stability via hydrogen bonds, if an intermolecular hydrogen bond in a protein is broken or deleted without the possibility of forming a compensating hydrogen bond to solvent, the protein will lose its structure and get destabilized.
Hydrogen bonding is nothing but dipole-dipole interaction between molecules, e.g., the hydrogen bond between water molecules; O atom is attracted to an H atom in the second molecule. The strength of a hydrogen bond is around 4–50 kJ mol−1. This is, however, not necessarily the amount of energy that the hydrogen bond contributes to the stabilization of a folded protein. In the unfolded state, potential hydrogen-bonding partners in the polypeptide chain are satisfied by hydrogen bonds to water. When the protein folds, the hydrogen bonds are broken, some are replaced by intra-protein hydrogen bonds and the entropy of the solvent increases. The balance between the entropy and enthalpy terms is close, but hydrogen bonds contribute positively to protein stability. Despite the small contribution made to protein stability by hydrogen bonds, if an intermolecular hydrogen bond in a protein is broken or deleted without the possibility of forming a compensating H-bond to solvent, the protein will be destabilized and can lose its structure (Ragone 2001).
Apart from conferring stability to protein’s structure, hydrating waters take part in most of the protein–protein (Lehmkuhler et al. 2017), protein–DNA ((a) Rodier et al. 2005 (b) Lo Conte et al. 1999), and protein–ligand ((a) Lu et al. 2007 (b) Panigrahi and Desiraju 2007; Reddy et al. 2001) interactions and aid in the molecular recognition and both the binding thermodynamics and kinetics (Bodnarchuk 2016) properties. Enthalpy contribution is characterized by its mobility in the displacement process, and complete arrest sets a limit on its involvement (Maurer and Oostenbrink 2019). Usually, protein–protein and protein–DNA binding surfaces have hydrophilic residues, and their interaction happens with the help of water molecules. The comparative parameters between the binding cavity and the ligand are hydrophobic. The binding energy attributes to changes in the free energy when two hydrophobic surfaces not in contact can be seen in hydrophobic effects (Chaplin 2008).
8.2.8 Displacement of Water Molecules in the Protein Cavity and Its Associated Thermodynamics
The kinase inhibitors have a specific recognition mechanism following the water occupancy in the binding pocket (Maurer and Oostenbrink 2019). Attributes of water, such as its smaller size, polarity, conformational flexibility, interaction strength, and directions, directly contribute to elasticity and reversibility. The main force for binding relies on the displacement of the unstable waters, and the dependent energetic gain through favorable relative binding energy is feasible through molecular rearrangement of the hydration water molecules (Snyder et al. 2014; Levinson and Boxer 2014; Lim et al. 2012; Jana and Bandyopadhyay 2012).
The thermodynamics concept of ligand binding is influenced by the relationship between enthalpy and entropy contributions of the binding event. Protein–ligand interactions involve attractive forces and hydration effects. Accurately positioned polar groups will make a way to specific interactions (Hydrogen bonds, salt-bridges, polar-polar interactions, and non-classical interactions) such as whole mediated halogen bonding results in enthalpy gain. To use this enthalpy, binding partners should be in optimal orientation, since the binding energy is highly sensitive to both the distance and the angle of the interacting atoms. The feature of interactions and the associated binding thermodynamics profile impact selectivity against off-targets (Tarcsay and Keserű 2015). Enthalpically optimized compounds possess carefully positioned ligand-binding site atom pairs to achieve the desired gain in binding enthalpy. Due to the improper orientation of the ligand, the desired protein interactions cannot yield the enthalpic contribution to binding free energy. In contrast, entropically optimized compounds have less positional constraints, and desolvation of the polar moieties can result in entropy gain due to the lower dependence from the binding environment. The compounds have, therefore, a higher propensity to form attractive interactions with off-targets. Through, the analysis of the thermodynamic properties, the structural and functional characteristics of the protein can be easily elucidated by the occupancy of high-energy hydration sites. Somewhere high-energy hydration sites are mostly localized near hydrophilic protein motifs (Olsson et al. 2008). Furthermore, there was no significant correlation between the hydration site-free energy and the solvent-accessible surface area of the site. Besides, the distribution of high-energy hydration sites on the protein surface can identify the location of binding sites and that binding sites of druggable targets have a higher density of thermodynamically unstable hydration sites.
The water molecules observed in crystal structures are less stable on average than bulk water due to the high degree of spatial localization resulting in loss of entropy. These findings must help to a better understanding of the water characteristics at the surface of proteins and lead to insights into the structure-based drug design efforts. Nonpolar ligand groups assist displacement of waters from the binding site due to the decrease in the interference of the previously bound water with the protein’s internal hydrogen-bonding and improved bulk hydrogen-bonding (Snyder et al. 2014).
8.3 Solvent Mapping Tools
In this section, we will discuss a few commercial and freely available tools and software that can predict the water hydration sites. The list of open-source software is listed in Table 8.1.
8.3.1 WatAA
This tool is newly developed which is known as atlas of amino acid hydration in proteins (Černy et al. 2017) that helps in exploring the synergies between data mining and ab-initio calculations using Turbo-Mole v6.4 program (Ahlrichs et al. 1989) with the DFT-D/RI-TPSS/TZVP method and calculating the interaction energies (Eint) of biomolecular fragments; the calculations were compared with the computationally intensive benchmark CCSD (T)/CBS method (Jurecka et al. 2007). The available data in the atlas was taken from two sources, such as experimental data and ab-initio quantum mechanics calculations. Validation of water replacement using electron density maps in crystallographic refinement helps in locating water molecules mediating protein–ligand interactions and MD simulations. The quantum mechanics calculations and validation can be performed by optimization and stabilizing the water position of each hydration site, by providing hydrogen atom positions and quantifying the interaction energy. A non-redundant set of 2818 high-resolution protein crystal structures are collected from protein data bank (PDB) and classified according to the secondary structure and X1 rotameric state. Data mining analysis provides the yielded statistical data on each amino acid residue hydration site. Hydration sites are positioned near local energy minima, and interaction energies were calculated that helps in assessing the water molecule hydration sites individually. Fourier averaging was also performed for the water densities and displacement. The displacement term is the spatial distance between the position of the crystal-derived water molecule and its optimized position (Schneider et al. 1993).
8.3.2 WATCLUST
This tool helps in identifying the specific and freely available water sites (WS). MD simulation (explicit water) can be performed using the VMD program. By using trajectory files, the WATCLUST plug-in can be used by the option “Extensions > Analysis > WATCLUST.” This plug-in helps in determining the WS and subgroup of residues where the WS will be calculated. The program computes: (a) water finding probability (WEP), (b) R90, (c) WS-protein mean interaction energy (<Eωρ>), (d) WS water mean interaction energy with respect to bulk (ΔΕint), and finally (e) excess rotational (Sr) and translational (St) entropies (Lopez et al. 2015).
This method is mostly applicable for the identification of WS in protein–ligand binding sites, presence of hydrophilic hot-spots in the protein–protein interface and exclusively aids in the identification of water structures, waters in the ion channels, and arrangement of catalytic waters. The disadvantage of this method is that it cannot be applied to the regions highly hydrophobic.
8.3.3 SZMAP (Solvent-Zap-Mapping)
SZMAP and GamePlan, developed by OpenEye scientific available at Santa Fe (2013). The gamePlan is used for analyzing the water sites; further running of SZMAP provides the coordinates of the protein–ligand binding site and analyzing the results, respectively (Grant et al. 2001). SZMAP is a semi-explicit solvent mapping (Tanger and Pitzer 1989; Rashin and Bukatin 1991) approach that uses the semi-continuum model to study the thermodynamics properties of water in the protein–ligand system using both explicit and implicit solvent modeling methods and address the displacement of the water-binding site which either increases or decreases the binding affinity. Specific water orientations were analyzed using classical statistical mechanics and sampling. SZMAP calculates ΔG, ΔH, and TΔS, which differentiate the explicit probe and the continuum water. A positive ΔG indicates that the continuum model estimates the cost of displacing water at that position. This will help in comparing the water probe to the ligand atom that displaces it. The difference in free energy between standard and uncharged water is negative, where standard water is more favored and positive. SZMAP can calculate stabilization energies from the neutral difference values as reaction energy:
where bulk-solvent is defined as 0 kcal mol−1.
SZMAP predicts very low B-factors where water molecule has less entropy and is more buried. Therefore, the crystallographic waters occupy hydrophilic positions on an SZMAP n_ddG contour map. Higher burial terms (near one) had lower B-factors. In addition, it could predict water conservation across a series of protein–ligand complexes and also predict optimal directions for deriving ligands during lead optimization. The SZMAP neutral probe entropy difference (n_dTdS) is a single physics-based descriptor that provides an accurate prediction of water conservation in active sites of protein structures using a simple threshold-value model.
8.3.4 3D-RISM
3D reference interaction site model (3D-RISM) is developed by a chemical group and works on a genetic algorithm; besides, the local minima problem was neglected by the desirability function. For detecting the potential water sites, 3D-RISM uses a double filter procedure and the minimum threshold value for the density distribution. Gridpoint was set as default g(r) > −5, followed by spatial constraint applied in the center and radius of the grid. The density distribution from the 3D-RISM calculation is transformed to a population function by using the equation P(T) = ρbulk Vvoxel g(r⃗), where ρbulk is the density of the bulk solvent, Vvoxel is the volume of one voxel in the grid, and g(r⃗) is the density function. Following this, each water site is detected in the first phase of the program. Followed by scoring was defined as the weighted ratio of the number of incompatible water sites. Hence this tool will be helpful in prediction and calculation of solvent density distribution. Besides this, GAsol is capable of finding the network of water molecules that best fits a particular 3D-RISM density distribution rapidly and accurately (Fusani et al. 2018).
8.3.5 ProBiS H2O
ProBiS H2O is a solvent mapping tool, which helps in the prediction of conserved water sites available in the protein data bank (PDB) by using a local structure alignment algorithm. It is an innovative approach, uses the existing experimental structures, and helps in the prediction of conserved water sites. This program is the first tool to perform local superimposition for the prediction of conserved water using DroP algorithm with Cambridge Structural Database (CSD) (Colin et al. 2016), a small molecule crystal database used for defining the water molecules interactions using geometric criteria, and Acqua Alta algorithm used to reproduce the water molecules interactions (Rossato et al. 2011). The density-based spatial clustering of applications with noise (3D-DBSCAN) algorithm was implemented in the scikit-learn machine learning Python library and clusters were defined as dense regions defined as the ε-neighborhood of a data point (p) with n or more data points (q). A dense region comprising a cluster is then calculated as:
ε-neighborhood of an object contains at least n objects pε-neighborhood
Nε (p):{q\d(p,q) < ε} (ε-neighborhood with objects (p,q) within a radius ε from an object)
Setting of ε to 0.9, this equal to a sphere with a radius of 0.9 Å. Then n parameter is increased iteratively from n = 1, indicating random water molecules, to n = N,where N is the number of superimposed chains of similar proteins.
The repetitively clustering calculation was done with an increase in the density until no more clusters are recognized. The ProBiS H2O plug-in is useful in finding the conserved and water networks in protein and designing the drug molecule and plays a role in protein-structure stability (Jukic et al. 2017).
8.3.6 WaterFLAP
A new approach developed by molecular discovery for predicting binding site water molecules. It uses a continuum solvent method and Grid inhomogeneous solvation theory (GIST) Molecular Interaction Fields (MIFs). Water scoring was done using two new GRID fields: the combined hydrophobicity and lipophilicity (CRY) field for combined hydrophobicity and lipophilicity, entropic (ENTR) field to estimate the entropic character of a particular water molecule and OH2 water enthalpy. In addition to this, water enthalpy prediction and various properties like structural, displaceable, and bulkiness can be assessed. GRID-binding sites in terms of their MIFs enable straight forward structure-based design. Water network creation can also be done using WaterFLAP; the water is placed on the most favorable positions (Baroni et al. 2007).
8.3.7 WAP
A web-based package used to calculate geometrical parameters between water molecules present in the protein and nucleic acid structure (Shanthi et al. 2002). There are two ways of implementing the program: (a) structure available from the PDB. This tool will provide information about the unit cell parameters, symmetry, and space group to the users. Distances are set to 2.5 Å (minimum) and 3.6 Å (maximum), followed by an angle set to default 0. (b) 3D atomic coordinates from the client machine. Here, the user has to provide input PDB files from the client machine. It displays the chains, metal ions, and inhibitor information. Protein angle and distance were calculated. This package gives information about all side chain/backbone polar atoms, protein, water, nitrogen, oxygen atoms.
8.3.8 Consolv
Consolv uses knowledgebase prediction, hybrid K nearest neighbors’ classifier, and genetic algorithm for the identification of hydration sites (free and ligand-bound water molecules) in the free environment; the water molecule’s crystallographic temperature factor, the number of hydrogen bonds between the water molecules and protein, density, and hydrophilicity of the neighboring protein atoms. The ability to predict water-mediated and polar interactions have an essential role in protein structure and function. A training set of 13 non-homologous proteins was used in the identification of conserved and displaced water molecules in the active site with 75% accuracy. Water molecules within 3.6 Å of protein surface atoms capable of making Van der Waal’s contacts or hydrogen bonds to atoms in the protein are considered being first-shell waters (De Beer et al. 2010; Raymer et al. 1997).
8.3.9 WaterScore
WaterScore is a method developed by researchers (Garcia-Sosa and Mancera 2003) for identifying between bound and displaceable water molecules. The structural properties of water molecules can be performed using multivariate logistic statistical analysis. WaterScore uses multivariate logistic models (Glonek and McCullagh 1995) and develops structural features of water molecules present in the empty binding site of a protein. There is a chance of observing the same water molecules (bound) after ligand binding. In WaterScore, the generated model will be considered based on the B-factor, the solvent-contact surface area, the total hydrogen bond energy, and the number of protein atomic contacts. This consistent approach gives an idea that the addition of water molecules towards the binding site of protein transversely differs from the various biomolecular applications. Models will be verified by checking the water molecules that are placed in bounded form or displaced in the binding site by using a better resolution of 3D structure. Secondly, in the structure-based drug design, the methodology adopts sorting, analyzing, and including the selection of water molecules in the protein structures. These criteria will help in the prediction of more accurate binding mode and energies for designing ligands. This tool also has several applications extended to de novo drug design and molecular docking and plays a significant role in ligand binding.
8.3.10 WaterMap
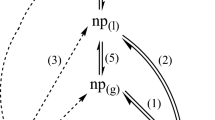
WaterMap software is developed by Schrödinger, based on inhomogeneous solvation theory (Lazaridis 1998). It shows the hydration site around the ligand-binding site. It can be used for enzymes such as GPCRs, bromodomains, nucleic acids, and protein–protein interfaces. Salient features of WaterMap (WaterMap, Schrödinger, LLC, New York, NY, 2020) includes identification of the water sites and thermodynamic properties present in the protein-binding sites and detailed examination of the thermodynamics binding comprising the free energy changes resulting from displacing water molecules in the active site (Fig. 8.1). It is represented by a sphere where there is a region of space, and the water molecules tend to aggregate. Therefore, each hydration site has several associated thermodynamic properties. WaterMap calculation is a multistep process; the first step is “simulation,” where the system is simulated for 2 ns (300 K) with full explicit solvent and heavy restraints were applied. The second step is “Clustering,” the position of water molecule will be clustered based on the clustering algorithm and water density, which can be measured for each position. The highest water location represents the hydration sites. The third stage is calculating the thermodynamic property of each water molecule. The calculation corresponds to the average of excess enthalpy, entropy, and free energy. Negative ΔH value of hydration site results in stronger interaction with the nearby water molecules and proteins in solution (e.g., charged group). Similarly, a positive ΔH value of hydration site represents weaker interactions with the adjacent water molecules and proteins in solution (e.g., near a hydrophobic residue). The hydration site (number) will give you the extent of volume measure. Meanwhile, hydration sites energies result in the enclosure and hydrophobic/hydrophilic balance of binding sites (Fig. 8.2). The net water transfer energy from the binding site to bulk, and from bulk to the binding site, contributes a broad ligand binding free energy. We majorly focus on a tiny (drug-sized) cluster of binding sites with highly unstable hydration sites. There are four following choices for designing a ligand:
-
(a)
Displace: Hydration site has both ΔG and ΔH >> 0 kcal mol-1, generally favorable to displace (hydrophobic regions).
-
(b)
Replace: Hydration sites with ΔH << 0 kcal mol−1 but with ΔG ≳ 0 kcal mol−1 are candidates for replacement.
-
(c)
Interact: Highly conserved hydration sites, forming bridging waters.
-
(d)
Avoid: When ΔG << 0 kcal mol−1, water molecules are highly stable, such cases it could be easier to avoid.
Schematic representation of the thermodynamic decomposition of ligand/protein binding adapted from WaterMap Schrödinger (WaterMap 2020)
8.3.11 Quantitative WaterMap
WaterMap scoring considers only the ΔG. The contribution of each hydration site from the WaterMap is summarized using the simple equation.
where O(a,s) measures the overlap of the atom “a” with site “s.” The water scoring equation was taken from WaterMap Schrödinger (Garcia-Sosa and Mancera 2003).
WM/MM scoring calculation is similar to MM-GBSA calculation implemented in Prime (Prime, Schrödinger, LLC, New York, NY, 2019), which uses the advanced VSGB solvation models and optimized potential for liquid simulations (OPLS) 2.1 force field. For WaterMap simulations, the binding site was defined by the position of a co-crystal ligand in the protein and ligand site was used to describe the volume for the calculation of hydration sites that was carried out by thermodynamic analysis (Biswal et al. 2020). The Holo-WaterMap Scoring is relatively simple; integrating the continuous WaterMap within the vicinity of the ligand gives an estimate of the ligand effect on the solvent. Druggability, activity, and selectivity analyses will all be used for the ligand-binding site. WaterMap is used for more quantitative ligand scoring and can be calculated using the equation mentioned above (Eq. 8.1).
8.3.12 Druggability and Selectivity Using WaterMap
Druggability is a term used in drug discovery where the biological target binds with a drug molecule (binding pocket) (Kwon 2012). The concept of druggability is restricted to a small molecule, but it also extended to biomedical products, e.g., monoclonal antibodies (Stockwell and Roark 2011). It is estimated that 10–15% of human proteins are disease linked and druggable, 1–25% of disease-modifying are likely to be druggable. There are several tools available for predicting the nature of druggability (Stockwell 2011).
There is no single approach that will deliver a good target on demand; potential starting points can emerge from methods, namely genome sequencing studies, genomic screens, phenotypic screening, and existing drugs (Lansdowne 2018). WaterMap tool can be used to find the druggability of the compound, and the hydration site thermodynamics of a binding site also provides similar information. Protein structures, which are constant and high energy, are distributed on hydration sites besides being helpful in the identification of ligand-binding sites. Druggable cavities have unstable water molecules, and hydration sites provide the measure of the volume, whereas the energies of hydration site evidence the hydrophobic/hydrophilic balance of a binding site. A binding site with drug size small cluster comprises highly unstable hydration sites (Beuming et al. 2012). Usually, the drug-sized molecule contains the significant binding from occupying the binding site. The binding sites with unstable hydration sites are likely to be shallow or polar to bind a drug-like molecule. The binding site contains many stable water molecules and mostly hydrophobic in character. There are several methods, namely WaterMap, SZMAP, and WaterFLAP, for druggability prediction. Kohlmann et al. have applied MM-GB/SA and WaterMap methods for affinity calculation.
8.3.13 DOWSER++
DOWSER uses a semi-empirical modification program for protein hydration based on average energy from MD simulation (Morozenko and Stuchebrukhov 2016). The position of the water molecules will be predicted and compared with the experimental data. A cutoff of −10 kcal mol−1 is used to determine how much water is occupied in the interior cavities before MD simulations. The script WaterDock helps to discover the number of potential water locations, which we further analyze with the methods implemented in the Dowser code previous version. Thus, the main reason for using WaterDock is to locate internal cavities that are unnoticed by the Dowser program. With the help of the PDB file, internal cavities present in the protein with a water probe of 1 Å and water molecules will be filled based on the minimized energies. Specific model (water-protein interaction) will be used for evaluating the water energies. This method will predict the water molecules present in the X-ray structure (Carugo and Bordo 1999; Zhang and Hermans 1996; Morozenko et al. 2014).
8.3.14 WaterDock
WaterDock is an algorithm that uses the freely available AutoDock Vina tool (Trott and Olson 2010) to predict the location of ordered water molecules in ligand-binding sites to very high accuracy (Sridhar et al. 2017). WaterDock was confirmed against high-resolution crystal structures, neutron diffraction data, and MD simulations approach. For justification, a set of proteins (14 structures of OppA structure) with high-resolution X-ray structures were used, and it predicts 88% of “consensus” water sites with a mean error of 0.78 Å using Acqua Alta method (Rossato et al. 2011). WaterDock is accurate and predicts 97% of the ordered water molecules, with a standard of 1 false-positive per structure. Ligand functional groups will be identified first, and their hydration sites were done based on semi-empirical function. “Favorable” protein–water interactions can also be identified. Two probabilistic water molecules are developed to predict WaterDock predictions by using data mining, heuristic, and machine learning techniques. The individual hydration of functional groups was first calculated from MD simulations of ligands (Ross et al. 2012).
8.3.15 WATsite: Hydration Site Prediction Program with PyMOL Interface
A graphical user interface (GUI) developed with inbuilt PyMOL plug-in free of charge for calculating the thermodynamic properties and hydration sites with the help of enthalpy and entropy of the water molecule. MD simulation can be performed, followed by hydration site identification and free energy estimations. This tool will solvate the proteins with explicit water molecules and performs MD simulations, where the fluctuations of water molecules in the protein-binding site will be analyzed. The clustering algorithm and quality threshold (QT) (Glenn 2001) were applied for the prediction of locations of water molecules, which is followed by hydration sites that are used for constructing the pharmacophore models and enrichment analysis. Therefore, if a ligand restores the water present in the hydration site, it can be expected to increase the binding free energy. The “occupancy” term denotes the probability of a water molecule was observed in the hydration site using MD simulations approach. This tool will help in the predicted hydration sites and estimate the desolvation free energies occupied by the replacing water molecules present in the protein–ligand binding site within the user-specified distance (default value 1 Å). The main aim of this tool is to estimate the desolvation free energy of water molecules present in the protein-binding site. Once the ligand site is fixed, the water molecules are retained for simulation. The energy minimization was performed using the steepest descent algorithm with the periodic boundary conditions. The binding free energy for protein includes other important contributions, such as the direct protein–ligand interaction energy and desolvation energy of the ligand.
The desolvation free energy of each hydration site is determined by analyzing the enthalpy and entropy of the water molecules inside a hydration site.
where ΔHhs and ΔShs denote the enthalpic and entropic changes of water molecules transferred from the bulk solvent into the hydration site of the protein cavity.
where Ehs term denotes the interaction energy of a water molecule in the hydration site, based on the average sum of van der Waals and electrostatic interactions between each water molecule inside a given hydration site with the protein and all the other water molecules. Ebulk is the interaction energy of a water molecule with its surrounding environment in the bulk solvent.
Assuming no alteration at the moment apart of the partition function on transferring a water molecule from the bulk solvent into the protein cavity, ΔShs can be estimated by
where C° is the concentration of pure water (1 molecule/29.9 Å3), R is the gas constant, and pext (q) is the external mode probability density function of the water molecules’ translational and rotational motions during the MD simulation (Hu and Lill 2014).
8.4 Selectivity
There is an essential role in the binding site of hydration in the ligand selectivity, as analyzed for kinase target in the earlier study (Robinson et al. 2010). Displacement of potential water molecules site has a more significant impact on affinity and selectivity. Longer chains in kinases determine the thermodynamically unstable hydration sites (Knegtel and Robinson 2011), resulting in binding affinity gain, and a slight change in the mobility of the hydration site towards the ligand corresponds to the selectivity of the kinases. Energetics and hydration sites will be useful to analyze the selectivity differences.
8.5 Limitations
The hydration site analysis is carried out based on the protein rigidness. Restraints will be challenging for hydration sites, where the number and strength of hydrogen bonds are affected by the variability of hydrogen-bonding partners. Similarly, inappropriate binding site analysis will cause flexibility in proteins. Restricted binding sites might have difficulties to be solvated as the buried sites cannot be reached without conformational rearrangements. WaterMap will compare all the energies for each hydration sites using thermodynamic approaches. It does not provide absolute binding free energies, but offer only relative free energy changes upon water displacement. The estimation for relative contributions of individual hydration sites to the macroscopic thermodynamic property is formulated as a sum of water enthalpy and entropy from MD using different theoretical approaches. WaterMap provides a comparative ranking of similar ligands and finds the linking between the binding site desolvation and affinity. Any interpretation of the structure–activity relationship (SAR) is only meaningful if the binding is primarily due to hydrophobic and not electrostatic interactions. Therefore, WaterMap is not a replacement for scoring functions or advanced binding affinity estimations.
8.6 Conclusions
Water molecules are essential for protein–ligand interactions and the study of the thermodynamic properties during binding. The chapter covered the significance of hydration sites in conferring stability, influencing protein folding, and key biomolecular interactions. Vital intramolecular forces such as hydrophobic, hydrophilic, electrostatic, and hydrogen bond network aid to establish the interaction of the first-order hydrating water molecules to bulk and the protein environment. Likewise, thermodynamic signatures such as the entropy and enthalpic contributions about the mobility of the water and the influence sought by the environment are contributing factors for water displacements. Computational approaches like prediction tools have been discussed here. In the recent era, hydration site analysis is an essential criterion for structure-based drug design projects for further exploration of the binding site profiling will highlight the fundamental interactions. This chapter depicts the importance of hydration sites in the SAR or analogs around the lead compound might help in designing lead molecules and optimization. In structure-based drug design, potency and ADMET properties are important factors to water interactions; this helps in understanding the molecular detection of protein–ligand complexes. There is a significant importance using computational tools to realize a lot about water role and signifying opportunities in structure-based design. We summarized concepts and critical applications towards hydration sites, which are useful in drug design projects.
References
Abel R, Young T, Farid R, Berne BJ, Friesner RA (2008) Role of the active-site solvent in the thermodynamics of factor Xa ligand binding. J Am Chem Soc 130:2817–2831
Ahlrichs R, Bar M, Haser M, Horn H, Kolmel C (1989) Electronic structure calculations on workstation computers: the program system turbomole. Chem Phys Lett 162(3):165–169
Anandakrishnan R, Izadi S, Onufriev AV (2019) Why computed protein folding landscapes are sensitive to the water model. J Chem Theory Comput 15(1):625–636
Ansari A, Jones CM, Henry ER, Hofrichter J, Eaton WA (1992) The role of solvent viscosity in the dynamics of protein conformational changes. Science 256(5065):1796–1798
Baldwin RL (2014) Dynamic hydration shell restores Kauzmann’s 1959 explanation of how the hydrophobic factor drives protein folding. Proc Natl Acad Sci U S A 111(36):13052–13056
Ball P (2008) Water as an active constituent in cell biology. Chem Rev 108(1):74–108
Bandyopadhyay S, Chakraborty S, Bagchi B (2005a) Secondary structure sensitivity of hydrogen bond lifetime dynamics in the protein hydration layer. J Am Chem Soc 127(47):16660–16667
Bandyopadhyay S, Chakraborty S, Balasubramanian S, Bagchi B (2005b) Sensitivity of polar solvation dynamics to the secondary structures of aqueous proteins and the role of surface exposure of the probe. J Am Chem Soc 127(11):4071–4075
Baroni M, Cruciani G, Sciabola S, Perruccio F, Mason JS (2007) A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for ligands and proteins (FLAP): theory and application. J Chem Inf Model 47(2):279–294
Bellissent-Funel MC, Hassanali A, Havenith M, Henchman R, Pohl P, Sterpone F, van der Spoel D, Xu Y, Garcia AE (2016) Water determines the structure and dynamics of proteins. Chem Rev 116(13):7673–7697
Beuming T, Che Y, Abel R, Kim B, Shanmugasundaram V, Sherman W (2012) Thermodynamic analysis of water molecules at the surface of proteins and applications to binding site prediction and characterization. Proteins 80(3):871–883
Biswal J, Jayaprakash P, Suresh Kumar R, Venkatraman G, Poopandi S, Rangasamy R, Jeyaraman J (2020) Identification of Pak1 inhibitors using water thermodynamic analysis. J Biomol Struct Dyn 38(1):13–31
Bizzarri AR, Cannistraro S (2002) Molecular dynamics of water at the protein-solvent interface. J Phys Chem B 106(26):6617–6633
Bodnarchuk MS (2016) Water, water, everywhere... It’s time to stop and think. Drug Discov Today 7:1139–1146
Boyarkin OV, Kosheleva MA, Aseev O, Maksyutenko P, Rizzo TR, Zobov NF, Lodi L, Tennyson T, Polyansky OL (2013) Accurate bond dissociation energy of water determined by triple-resonance vibrational spectroscopy and ab initio calculation. Chem Phys Lett 568–569:14–20
Cao Z, Bowie JU (2014) An energetic scale for equilibrium H/D fractionation factors illuminates hydrogen bond free energies in proteins. Protein Sci 23(5):566–575
Carugo O, Bordo D (1999) How many water molecules can be detected by protein crystallography? Acta Crystallogr D Biol Crystallogr 55(Pt 2):479–483
Černy J, Schneider B, Biedermannová L (2017) WatAA: atlas of protein hydration. Exploring synergies between data mining and ab initio calculations. Phys Chem Chem Phys 19(26):17094–17102
Chakraborty S, Sinha SK, Bandyopadhyay S (2007) Low-frequency vibrational spectrum of water in the hydration layer of a protein: a molecular dynamics simulation study. J Phys Chem B 111(48):13626–13631
Chaplin MF (2008) Water in biological recognition processes. In: Begley TP (ed) Wiley Encyclopedia of chemical biology. Wiley, New York, pp 1–8
Cheung MS, Garcia AE, Onuchic JN (2002) Protein folding mediated by solvation: water expulsion and formation of the hydrophobic core occur after the structural collapse. Proc Natl Acad Sci U S A 99(2):685–690
Colin RG, Lan JB, Matthew PL, Suzanna CW (2016) The Cambridge structural database. Acta Crystallor B Struct Sci Cryst Eng Mater 72(2):171–179
Covalt JC Jr, Roy M, Jennings PA (2001) Core and surface mutations affect folding kinetics, stability and cooperativity in IL-1 beta: does alteration in buried water play a role? J Mol Biol 307(2):657–669
De Beer SB, Vermeulen NP, Oostenbrink C (2010) The role of water molecules in computational drug design. Curr Top Med Chem 1:55–66
Fenimore PW, Frauenfelder H, McMahon BH, Parak FG (2002) Slaving: solvent fluctuations dominate protein dynamics and functions. Proc Natl Acad Sci U S A 99(25):16047–16051
Fernández A, Kardos J, Goto Y (2003) Protein folding: could hydrophobic collapse be coupled with hydrogen-bond formation? FEBS Lett 536(1–3):187–192
Finkelstein A, Ptitysyn O (2016) Protein physics: a course of lectures, 2nd edn. Elsevier, Academic
Fusani L, Wall I, Palmer D, Cortes A (2018) Optimal water networks in protein cavities with GAsol and 3D-RISM. Bioinformatics 34(11):1947–1948
Garcia-Sosa AT, Mancera RL, Dean PM (2003) WaterScore: a novel method for distinguishing between bound and displaceable water molecules in the crystal structure of the binding site of protein-ligand complexes. J Mol Model 9(3):172–182
Glenn F (2001) A comprehensive overview of basic clustering algorithms. Comp Sci 2:1–37
Glonek GFV, McCullagh P (1995) Multivariate logistic models. J R Stat Soc 57(3):533–546
Grant JA, Pickup BT, Nicholls A (2001) A smooth permittivity function for poisson boltzmann solvation methods. J Comput Chem 22(6):608–640
Gromiha MM (2000) Protein bioinformatics: from sequence to function, 1st edn. Elsevier, Academic
Gummadi SN (2003) What is the role of thermodynamics on protein stability? Biotechnol Bioprocess Eng 8:9–18
Hildebrandt A, Blossey R, Rjasanow S, Kohlbacher O, Lenhof HP (2007) Electrostatic potentials of proteins in water: a structured continuum approach. Bioinformatics 23(2):e99–e103
Hu B, Lill MA (2014) WATsite: hydration site prediction program with PyMOL interface. J Comput Chem 35(16):1255–1260
Huggins DJ (2016) Studying the role of cooperative hydration in stabilizing folded protein states. J Struct Biol 196(3):394–406
Ikura T, Urakubo Y, Ito N (2004) Water-mediated interaction at a protein- protein interface. Chem Phys 307(2–3):111–119
Jana M, Bandyopadhyay S (2012) Conformational flexibility of a protein-carbohydrate complex and the structure and ordering of surrounding water. Phys Chem Chem Phys 14(18):6628–6638
Jukic M, Konc J, Gobec S, Janezic D (2017) Identification of conserved water sites in protein structures for drug design. J Chem Inf Model 57(12):3094–3103
Jurecka P, Cerny J, Hobza P, Salahub DR (2007) Density functional theory augmented with an empirical dispersion term. Interaction energies and geometries of 80 noncovalent complexes compared with ab initio quantum mechanics calculations. J Comput Chem 28(2):555–569
Kangueane P, Nilofer C (2018) Protein-protein and domain-domain interactions, 1st edn. Springer, New York
Karplus M, McCammon JA (2002) Molecular dynamics simulations of biomolecules. Nat Struct Biol 9(9):646–652
Knegtel RM, Robinson DD (2011) A role for hydration in interleukin-2 inducible T cell kinase (Itk) selectivity. Mol Inform 30(11–12):950–959
Kubinyi H (2001) Hydrogen bonding: the last mystery in drug design. Pharmacokinetic optimization in drug research, 1st edn. Wiley, New York
Kuffel A (2017) How water mediates the long-range interactions between remote protein molecules. Phys Chem Chem Phys 19(7):5441–5448
Kwon B (2012) Chemical biologist targets ‘undruggable’ proteins linked to cancer in quest for new cures. Brent Stockwell interview Medical Xpress. Accessed 17 May 2012
Ladbury JE, Klebe G, Freire E (2010) Adding calorimetric data to decision making in lead discovery: a hot tip. Nat Rev Drug Discov 9(1):23–27
Lagasse HA, Alexaki A, Simhadri VL, Katagiri NH, Jankowski W, Sauna ZE, Kimchi-Sarfaty C (2017) Recent advances in (therapeutic protein) drug development. F1000Res 1:6–113
Lansdowne LE (2018). Target identification & validation in drug discovery. https://www.technologynetworks.com/drug-discovery/articles/target-identification-validation-in-drug-discovery-312290. Accessed 20 Mar 2018
Lazaridis T (1998) Inhomogeneous fluid approach to solvation thermodynamics. 1. Theory Chemistry 102(18):3531–3541
Lehmkuhler F, Forov Y, Elbers M, Steinke I, Sahle CJ, Weis C, Tsuji N, Itou M, Sakurai Y, Poulain A, Sternemann C (2017) Temperature dependence of the hydrogen bond network in trimethylamine N-oxide and guanidine hydrochloride-water solutions. Phys Chem Chem Phys 19(41):28470–28475
Levinson NM, Boxer SG (2014) A conserved water-mediated hydrogen bond network defines bosutinib’s kinase selectivity. Nat Chem Biol 10(2):127–132
Levy Y, Onuchic JN (2004) Water and proteins: a love-hate relationship. Proc Natl Acad Sci U S A 101(10):3325–3326
Lim VI, Curran JF, Garber MB (2012) Hydration shells of molecules in molecular association: a mechanism for biomolecular recognition. J Theor Biol 301:42–48
Lo Conte L, Chothia C, Janin J (1999) The atomic structure of protein-protein recognition sites. J Mol Biol 285(5):2177–2198
Lopez ED, Arcon JP, Gauto DF, Petruk AA, Modenutti CP, Dumas VG, Marti MA, Turjanski AG (2015) WATCLUST: a tool for improving the design of drugs based on protein-water interactions. Bioinformatics 31(22):3697–3699
Lu Y, Wang R, Yang CY, Wang S (2007) Analysis of ligand-bound water molecules in high-resolution crystal structures of protein-ligand complexes. J Chem Inf Model 47(2):668–675
Malleshappa Gowder S, Chatterjee J, Chaudhuri T, Paul K (2014) Prediction and analysis of surface hydrophobic residues in tertiary structure of proteins. Sci World J 2014:971258
Manzoni F, Ryde U (2018) Assessing the stability of free-energy perturbation calculations by performing variations in the method. J Comput Aided Mol Des 32(4):529–536
Maurer M, Oostenbrink C (2019) Water in protein hydration and ligand recognition. J Mol Recognit 32(12):e2810
Mondal J, Friesner RA, Berne BJ (2014) Role of desolvation in thermodynamics and kinetics of ligand binding to a kinase. J Chem Theory Comput 10(12):5696–5705
Morozenko A, Stuchebrukhov AA (2016) Dowser++, a new method of hydrating protein structures. Proteins 84(10):1347–1357
Morozenko A, Leontyev IV, Stuchebrukhov AA (2014) Dipole moment and binding energy of water in proteins from crystallographic analysis. J Chem Theory Comput 10(10):4618–4623
Nakasako M (2004) Water-protein interactions from high-resolution protein crystallography. Philos R Soc Lond B: Biol Sci 359(1448):1191–1204
Novak U, Grdadolnik J (2017) The hydration of concanavalin a studied by infrared spectroscopy. J Mol Struct 1135:138–143
Oleinikova A, Smolin N, Brovchenko I, Geiger A, Winter R (2005) Formation of spanning water networks on protein surfaces via 2D percolation transition. J Phys Chem B 109(5):1988–1998
Olsson TS, Williams MA, Pitt WR, Ladbury JE (2008) The thermodynamics of protein-ligand interaction and solvation: insights for ligand design. J Mol Biol 384(4):1002–1017
Panigrahi SK, Desiraju GR (2007) Strong and weak hydrogen bonds in the protein-ligand interface. Proteins 67(1):128–141
Pappas CG, Tzakos AG, Gerothanassis IP (2012) On the hydration state of amino acids and their derivatives at different ionization states: a comparative multinuclear NMR and crystallographic investigation. J Amino Acids 2012:565404
Parsegian VA (2002) Protein-water interactions. Int Rev Cytol 215:1–31
Perutz MF, Kendrew JC, Watson HC (1965) Structure and function of haemoglobin: II some relations between polypeptide chain configuration and amino acid sequence. J Mol Biol 13(3):669–678
Pradhan MR, Nguyen MN, Kannan S, Fox SJ, Kwoh CK, Lane DP, Verma CS (2019) Characterization of hydration properties in structural ensembles of biomolecules. J Chem Inf Model 59(7):3316–3329
Ragone R (2001) Hydrogen-bonding classes in proteins and their contribution to the unfolding reaction. Protein Sci 10:2075–2082
Rashin AA, Bukatin MA (1991) Continuum based calculations of hydration entropies and the hydrophobic effect. J Comput Chem 95(8):2942–2944
Raymer ML, Sanschagrin PC, Punch WF, Venkataraman S, Goodman ED, Kuhn LA (1997) Predicting conserved water-mediated and polar ligand interactions in proteins using a K-nearest-neighbors genetic algorithm. J Mol Biol 265(4):445–464
Reddy CK, Das A, Jayaram B (2001) Do water molecules mediate protein-DNA recognition? J Mol Biol 314(3):619–632
Rhee YM, Sorin EJ, Jayachandran G, Lindahl E, Pande VS (2004) Simulations of the role of water in the protein-folding mechanism. Proc Natl Acad Sci U S A 101(17):6456–6461
Robinson DD, Sherman W, Farid R (2010) Understanding kinase selectivity through energetic analysis of binding site waters. ChemMedChem 5(4):618–627
Rodier F, Bahadur RP, Chakrabarti P, Janin J (2005) Hydration of protein-protein interfaces. Proteins 60(1):36–45
Rose GD, Wolfenden R (1993) Hydrogen bonding, hydrophobicity, packing, and protein folding. Annu Rev Biophys Biomol Struct 22:381–415
Ross GA, Morris GM, Biggin PC (2012) Rapid and accurate prediction and scoring of water molecules in protein binding sites. PLoS One 7(3):e32036
Rossato G, Ernst B, Vedani A, Smiesko M (2011) Acqua Alta: a directional approach to the solvation of ligand-protein complexes. J Chem Inf Model 51(8):1867–1881
Ruhmann E, Betz M, Fricke M, Heine A, Schafer M, Klebe G (2015) Thermodynamic signatures of fragment binding: validation of direct versus displacement ITC titrations. Biochim Biophys Acta 1850(4):647–656
Santa Fe NM (2013) SZMAP 1.5.0.2: OpenEye Scientific Software
Schneider B, Cohen DM, Schleifer L, Srinivasan AR, Olson WK, Berman HM (1993) A systematic method for studying the spatial distribution of water molecules around nucleic acid bases. Biophys J 65(6):2291–2303
Schrödinger Release (2019) Prime, Schrödinger. LLC, New York, NY
Schrödinger Release (2020) 2020–1: WaterMap, Schrödinger. LLC, New York, NY
Seyedi S, Matyushov D (2018) Dipolar susceptibility of protein hydration shells. Chem Phys Lett 713:210–214
Shanthi V, Rajesh CK, Jayalakshmi J, Vijay VG, Sekar K (2002) WAP: water analysis package—a web-based package to calculate geometrical parameters between water oxygen and protein atoms. J Appl Crystallogr 36(1):167–168
Smolin N, Oleinikova A, Brovchenko I, Geiger A, Winter R (2005) Properties of spanning water networks at protein surfaces. J Phys Chem B 109(21):10995–11005
Snyder PW, Lockett MR, Moustakas DT, Whitesides GM (2014) Is it the shape of the cavity, or the shape of the water in the cavity? Eur Phys J Spec Top 223:853–891
Spyrakis F, Ahmed MH, Bayden AS, Cozzini P, Mozzarelli A, Kellogg GE (2017) The roles of water in the protein matrix: a largely untapped resource for drug discovery. J Med Chem 60(16):6781–6827
Sridhar A, Ross GA, Biggin PC (2017) Waterdock 2.0: water placement prediction for Holo-structures with a pymol plugin. PLoS One 12(2):e0172743
Stockwell B (2011) Outsmarting Cancer. A biologist talks about what makes disease-causing proteins so difficult to target with drugs. Sci Am 305(4):20
Stockwell B, Roark B (2011) The quest for the cure: the science and stories behind the next generation of medicines. Columbia University Press, New York
Takano K, Yamagata Y, Yutani K (2003) Buried water molecules contribute to the conformational stability of a protein. Protein Eng 16(1):5–9
Tanger JC, Pitzer KS (1989) Calculation of the thermodynamic properties of aqueous electrolytes to 1000.degree.C and 5000 bar from a semi continuum model for ion hydration. J Phys Chem 93(12):4941–4951
Tarcsay Á, Keserű GM (2015) Is there a link between selectivity and binding thermodynamics profiles? Drug Discov Today 1:86–94
Thomas L (2020) “Protein Structure Determination”. News-Medical. https://www.news-medical.net/life-sciences/Protein-StructureDetermination.aspx. Accessed 20 Mar 2020
Tripathi T (2013) Calculation of thermodynamic parameters of protein unfolding using far-ultraviolet circular dichroism. J Proteins Proteomics 4(2):85–91
Trott O, Olson AJ (2010) AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Van Dijk E, Hoogeveen A, Abeln S (2015) The hydrophobic temperature dependence of amino acids directly calculated from protein structures. PLoS Comput Biol 11(5):e1004277
Virtanen JJ, Sosnick TR, Freed KF (2014) Ionic strength independence of charge distributions in solvation of biomolecules. J Chem Phys 141(22):22D503
Wüthrich K, Otting G, Liepinsh E (1992) Protein hydration in aqueous solution. Faraday Discuss 93:35–45
Yokomizo T, Nakasako M, Yamazaki T, Shindo H, Higo J (2005) Hydrogen-bond patterns in the hydration structure of a protein. Chem Phys Lett 401(4–6):332–336
Zapadka KL, Becher FJ, Gomes Dos Santos AL, Jackson SE (2017) Factors affecting the physical stability (aggregation) of peptide therapeutics. Interf Focus 7(6):20170030
Zhang L, Hermans J (1996) Hydrophilicity of cavities in proteins. Proteins 24(4):433–438
Zhong D, Pal SK, Zewali AH (2011) Biological water: a critique. Chem Phys Lett 503(1–3):1–11
Acknowledgement
JJ thank the DST INDO-TAIWAN (GITA/DST/TWN/P-86/2019 dated: 04/03/2020), Board of Research in Nuclear Sciences (BRNS) (35/14/ 02/2018 BRNS/35009), Indian Council for Medical Research (ICMR) (No.BIC/12(07)/2015), DST-Science and Engineering Research Board (SERB) (No.EMR/2016/000498), UGC Research Award (No. F. 30-32/2016(SA-II) dt 18.04.2016), DST- Fund for Improvement of S&T Infrastructure in Universities & Higher Educational Institutions (FIST) (SR/FST/LSI-667/2016) (C), DST-Promotion of University Research and Scientific Excellence (PURSE) (No. SR/PURSE Phase 2/38 (G), 2017 and MHRD-RUSA 2.0, New Delhi (F.24-51/2014-U, Policy (TNMulti-Gen), Dept. of Edn. Govt. of India, Dt.09.10.2018). JB is grateful to the UGC OBC National Fellowship (F./2015-16/NFO-2015-17-OBC-PON-29027)) A special word of gratitude to Mr. Raghu Rangasamy, Vice-President, and the Application Scientists of Schrodinger, India, for their timely help and suggestions.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Biswal, J., Jayaprakash, P., Rangaswamy, R., Jeyakanthan, J. (2020). Synergistic Effects of Hydration Sites in Protein Stability: A Theoretical Water Thermodynamics Approach. In: Singh, D., Tripathi, T. (eds) Frontiers in Protein Structure, Function, and Dynamics. Springer, Singapore. https://doi.org/10.1007/978-981-15-5530-5_8
Download citation
DOI: https://doi.org/10.1007/978-981-15-5530-5_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-5529-9
Online ISBN: 978-981-15-5530-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)