Abstract
Epilepsy is a well-known neurological disorder which affects moreover 2% of the World’s population. Irregular excessive neuronal activities to the human brain cause epileptic seizures onset. Electroencephalograph (EEG) signals are mostly examined for the detection of epileptic seizure onsets. But an EEG signal consists of a huge amount of complicated information and it is very difficult to analyze it manually. Over the decades, a lot of research has been focused on the development of automated epilepsy diagnosis systems. These systems are dependent on sophisticated feature captureization and classification techniques. The paper aims to present a generalized review and performance comparison of the work reported over a decade in the area of automated epilepsy diagnosis systems that will help future researchers lead a better direction.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Epilepsy is among the commonly existing neurological infirmity and is described as an unexpected change in the usual electrical activities in some parts of the brain or in the entire brain. These abnormal responses in the brain are called epileptic seizures [1]. A normal brain produces low amplitude electrical pulses but if a person is suffering from epilepsy then these pulses are produced in excessive amounts and are not easily controllable. The diagnosis of seizures is not easily predictable and if it is ignored, then it may lead to the death of the patient. According to a recent study, epilepsy affects around 6–10 million people in India, and less than half are treated [2].
Electroencephalogram (EEG) is a method of recording the electrical signals of the brain developed by cortical neurons. As per international standards, 10–20 electrodes are located around the scalp of the person to record the brain signals which are produced by the cerebral cortex nerve cells [3]. Neurologists generally use EEG signals for detection and diagnoses of epileptic seizures. Analyzing the EEG signals visually or manually requires a lot of time and neurological knowledge, making the epilepsy detection process tedious, difficult and may sometimes result in incorrect diagnoses. So, a lot of research has been done to propose computer-based automatic EEG epilepsy detection systems. Such a system must be able to classify the epileptic seizure signal from non-epileptic signals with more accuracy and in less time.
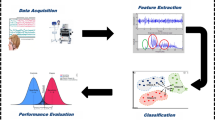
To automatize the seizure detection from an EEG signal, various machine learning approaches have been proposed by various researchers. Machine learning methods are more fast, accurate, and consistent in performance compared to manual labelling [4,5,6,7,8,9]. The machine learning approach is basically based on training a classifier with a repository of existing data consisting of both, epilepsy seizure signals and normal signals. A typical automated diagnosis system for epilepsy detection generally consists of following modules: EEG signal pre-processing, feature extraction and feature selection and finally classification of extracted features as normal, seizure or non-seizure. The performance efficiency of a typical automated epilepsy seizure detection technique is dependent on the type of classifier used, as well as on the features that are inputted to that classifier. Figure 1 presents a typical approach of detecting epileptic seizures for single-channel dataset, for example, the University of Bonn dataset. Similarly, Fig. 2 provides a general framework of epilepsy seizure detection for a multichannel dataset, for example, CHB-MIT dataset. These datasets are discussed in detail in the next section.
The objective of this literature is to present an advanced review of the methods and techniques used for designing these modules. The remaining part of this paper is organized as follows: Sect. 2 focused on the various freely available standard seizure and non-seizure EEG signal datasets. Section 3 briefly summarizes various pre-processing and feature extraction techniques available in the literature. Section 4 studies various classification techniques used in the literature. Finally, the conclusion is given in Sect. 5.
2 EEG Dataset
In literature, many standard datasets for epileptic EEG signals and non-epileptic EEG signals have been utilized by authors for their research work. Online access is available for all such datasets; some are freely accessible and some have paid access. Two well-known datasets are being discussed, in this paper, that have free access online, and these are the University of Bonn dataset and CHB-MIT dataset. The details of these datasets are given below:
-
(i)
University of Bonn dataset
The above-aforementioned EEG dataset was recorded at the University of Bonn, Germany acquired by Andrzejak et al. [10]. This dataset comprises five sets A, B, C, D and E of EEG signal, where every set encompasses 100 single-channel EEG fragments of 23.6 s, which are sampled at a rate of 173.61 Hz, thus giving a total of 4097 samples per set. The spectral bandwidth for all sets ranges between 0.5 and 85 Hz. All the recordings were made using the same 128 channel amplifier system. Set A and B comprises of information gathered from five healthy volunteers with eyes open and closed, separately, utilizing the standardized surface EEG recordings. Sets C, D and E were obtained from five epileptic patients experiencing pre-surgical epilepsy diagnosis. Set C and set D were recorded during epilepsy free interval and set E was recorded during the occurrence of epileptic seizures. More information about this dataset can be gathered from [10]. The authors in papers [4, 6, 12, 13, 16,17,18,19,20] have utilized the University of Bonn dataset for their research work. Table 1 depicts a summary for the University of Bonn dataset and Fig. 3 presents waveforms of typical signals from set A to set E. As can be observed from Fig. 3 the amplitude of set E (ictal with seizure) is much greater than the amplitude of other sets.
-
(ii)
CHB-MIT Dataset
It is also one of the commonly used datasets. This dataset is also referred to as the EEG PhysioNet dataset. It was acquired at Boston Children’s Hospital [11], consisting of multichannel EEG signals that were recorded from 23 paediatric patients (5 males and 18 females) suffering from intractable epilepsy. The paediatric patients were reported to be between age 1.5 and 22 years.
The data was recorded for 23 channels using the international Federation of clinical neurophysiology standard of 10–20 electrode placement. Each channel data was digitized at a sampling rate of 256 Hz with 16-bit resolution. While recording clinical experts manually annotated the beginning and end of each seizure, and classified the EEG signals as epileptic seizures and non-seizures. The acquired dataset was 916 h long and consisted of a total of 198 identified seizures. The shortest duration seizure lasts for 6 s and the longest duration seizure lasts for 752 s with an average seizure duration of 72 s. The authors in papers [7, 8, 27,28,29,30,31] have utilized CHB-MIT dataset for their research work. Table 2 further gives detailed insight into the CHB-MIT dataset. Figure 4 plots a typical multichannel CHB-MIT dataset signal.
3 Pre-processing Techniques and Feature Extraction Methods
Pre-processing is a process of removing unwanted artifacts and noise from raw EEG signal that is collected directly from the scalp. These artifacts/noise may be present either because of blinking eyes, muscle movements, head motion during the record of the data, any problem in the channel/electrode, or any connectivity issue. Pre-processing thus may be utilized not just for artifacts removal but also for improving the Signal to Noise ratio and spatial resolution. Digital filters have been used by most of the authors in the pre-processing step.
In [12] EEG sample data is filtered through a 50 Hz IIR notch filter to eliminate AC power supply noise. In [13] the raw EEG signal is made to pass from a band-pass filter having a lower cut-off frequency of 0.53 Hz and higher cut-off frequency of 40 Hz for the removal of artifacts and noise. In [14] seventh order Chebyshev IIR filter is utilized. In [15] the EEG signal is de-noised by passing it through an FIR low-pass filter designed using Hamming with cut-off frequency at 60 Hz.
For an EEG prediction model to accurately classify between epileptic and non-epileptic signals, feature extraction is a very crucial step. Feature extraction is required because of the non-linear, stochastic and non-stationary behaviour of the EEG signal [6]. A good feature exaction technique must minimize the resources required for accurate classification of the signal as well as the loss of valuable information. The feature extraction process basically converts the raw signal information into a set of mathematical descriptors so that a better understanding of the complex data can be gained. Feature extraction is followed by a feature selection that aims to select the most relevant features and removes redundant features. Although there exist a number of feature selection algorithms in the literature, however Independent Component Analysis (ICA) [14] and Principal Component Analysis (PCA) [18] being the most popular.
The feature extraction techniques used in EEG signals analysis model are either time-based [2,3,4,5,6,7,8,9,10,11,12,13,14, 23], frequency-based [5, 12, 14, 23] or time-frequency based [4, 5, 16,17,18, 20, 21, 23,24,25, 27]. The most popular being the time-frequency analysis since it is able to provide both time and frequency information of the signal simultaneously. Time-domain features are obtained by analyzing the EEG signal as a function of time. Examples of typical features of time-domain include mean, median, standard deviation, kurtosis, amplitude, etc. Likely frequency-domain features are extracted by analyzing EEG data as a function of frequency. Various techniques like Fourier Transform are used for frequency-domain feature extraction. These features are related to spectral information of EEG signals. Examples of such features include spectral flux, power spectrum, spectral centroid, spectral entropy, etc. In the time-frequency domain both time-domain as well as frequency-domain features are extracted simultaneously. Discrete Wavelet Transform (DWT) is the most popular time-frequency domain based feature extraction technique.
In [4] authors have proposed a unique feature extraction technique based on Stockwell-transform (ST). It is a joint time-frequency analysis technique that is applicable to non-stationary signals. Such technique is more noise-robust, able to retain the absolute phase of the individual frequency component and provide more information with less features. After performing S-transform, two features namely, energy and standard deviation are extracted for both seizure and seizure-free EEG signals. In [12] the cross-bispectrum values for three EEG channels are computed to determine various linear (average, maximum and minimum) and non-linear features in EEG data. In [13], for feature extraction EEG signals are sampled twice and plotted as vertical bars. Then a feature called visibility height is computed for the three adjoining neighbours from the top of the referenced bar. Then four various types of entropy are computed to reduce the dimension of data. In [14] after pre-processing the EEG signal, Wavelet Analysis also known as DWT is used for feature extraction. DWT is a time-frequency analysis method used for analyses of non-stationary signals. In this technique the signal is passed through a set of consecutive low-pass and high-pass filters to decompose the signal into set of five sub-bands. Thus, decomposing the initial signal into a set of coefficients that describe the frequency content. But to lower the spectrum usage and time complexities authors in this paper have used only the lowest frequency sub-band for feature extraction. The feature extracted from these identified sub-bands helps in further classification of data. In [15] authors have extracted thirteen features like min amplitude, max amplitude, Entropy, Mean, Median, Standard Deviation, etc. belonging to time, frequency as well as both domains. In [16] also authors have utilized DWT for feature extraction. After DWT decomposition, the sub-bands of EEG signals are discretized using Equal Frequency Discretization (EFD) technique. EFD technique helps to compute the probability density for all sub-band of each EEG segment. Thus, providing the probability densities of both healthy EEG dataset and epilepsy patient EEG dataset. In [17] Local Mean Decomposition (LMD) is utilized that iteratively transforms raw EEG data into a set of product functions (PFs) where a PF is generated by multiplying envelope signal with a frequency modulated signal. This technique has got the advantage of high processing speed along with low computational complexity. From each PF three time-domain features namely maximum amplitude, minimum amplitude, average absolute value, and three frequency-domain features namely maximum power spectral density, skewness, kurtosis are extracted. The authors have further computed the Fractal Dimension (FD), Renyi Entropy (RE) and Hurst exponent for EEG dataset to form a nine-dimensional feature vector. In [18] for feature extraction authors have utilized Daubechies Wavelet order 4 (DB4). Then for the dimensionality reduction, PCA is applied to the extracted data. PCA reduces the data to lower dimensional space by modifying the signal into the new Principal Components (PCs). This is done to reduce the complexity in both space and time. In [19] authors have proposed a Discrete Cosine Transform (DCT) based 3-level multi-rate filter bank that extracts alpha, beta, gamma, delta and theta brain rhythms. Delta brain rhythms are modelled using fractional Brownian motion (fBm) process and the other four brain rhythms namely alpha, beta, gamma and theta are modelled using fractional Gaussian noise (fGn) process. Autoregressive Moving Average (ARMA) model and Hurst exponent (H value) together constitute the extracted feature vector. In [20] Daubechies 4 (DB4) wavelet transform (WT) of the EEG signal is computed to produce wavelet coefficients. Then, phase-space reconstruction (PSR) is computed for the wavelet coefficients of both normal and epileptic EEG signals. Then, in the third step Euclidean Distance (ED), the distance between the origin and the wavelet coefficients plotted in PSR, is calculated. These EDs form the initial feature vector.
4 Classification
An automated epilepsy diagnosis system must be able to accurately classify the EEG signals as epileptic and non-epileptic signal. In this regard many machine learning-based classification algorithms have been proposed in the literature. These algorithms usually train the system using either supervised, unsupervised or semi-supervised learning bypassing the selected features of the relevant class of the training data; and then the trained system performance is evaluated using different parameters for predicting the class of testing data. Although in literature there exists so many machine learning-based classification algorithms that classify the EEG signal as an epileptic class and non-epileptic class but the most popular being Support Vector Machine (SVM) [12, 13, 17, 21, 25, 26], K-nearest neighbour (KNN) [17, 18, 27, 30, 31], Linear Discriminant Analysis (LDA) [17] and Random forest (RF) [8, 13].
SVM is a very commonly used supervised learning-based classifier. It works by a creating hyperplane or set of hyperplanes in a multi-dimensional space that will separate data into various classes linearly. For a two-dimensional space, the hyperplane reduces to a line. The vectors that define the hyperplane are termed as support vector. This algorithm has got good generalization properties but the poor speed of execution. LDA is also one of the most commonly used supervised learning-based pattern reduction and dimensionality reduction algorithms. Like SVM this technique also separates the data into different classes using hyperplanes. In this algorithm, the first separability between different classes is computed which is termed as a between-class variance. Then it calculates within-class variance, i.e. the distance between the mean of the class and the individual samples of the class. Then creates a low-dimensional space that will maximize the between-class variance and minimize the within-class variance. KNN is also a very simple, popular and supervised based learning algorithm. The basic idea behind this is to first make the system learn through training data and as the new instance comes, it is assigned the class that is closest among the k-neighbours. The performance of the algorithm is very much dependent on k value. Greater k value reduces the effect of noise and increases the processing time. Random forest (RF) is a simple, flexible and easy to use machine learning algorithm. RF is also a supervised learning algorithm that works by creating an ensemble of decision trees. As a new instance comes, the class of this instance is decided by travelling from the root node of the tree to its leaf. At each node end, the successor may be chosen if there is a need to split the input space data. For the design of more efficient classifiers, future work can be done in the direction of using various optimization algorithms that have been explored in other applications [32,33,34,35].
The performance of a classifier is very much dependent on the characteristics/features of the data that is being inputted. Thus, if the extracted features are not relevant then the classification algorithm may not be able to accurately identify the class of the data. Also, the performance of a classifier is evaluated by using a number of performance measures/parameters that include classification accuracy, false alarm rate, specificity, sensitivity, latency and Receiving Operating Characteristic (ROC). The performance measures that are being accessed also depends on the dataset that is being used. Authors that have used the University of Bonn dataset have evaluated the accuracy, sensitivity, specificity, confusion matrix, and ROC as the performance parameters, with accuracy and confusion matrix being the most common and widely used performance measure. Table 3 presents the comparison of accuracy level achieved through different classifiers inputting different extracted features. In literature, as briefed in Table 4, it is also found that some researchers have computed the classification accuracy by considering different cases that constitute a different combination of sets (from A to D) with set E from the University of Bonn dataset. For the CHB-MIT dataset apart from performance parameters evaluated for the University of Bonn dataset, two more parameters namely latency and false alarm rate are also evaluated. Table 5 briefs the literature work of different performance parameters evaluated and their experimental values for the CHB-MIT database.
5 Conclusion
This paper presents a review of the automatic epilepsy detection system that is able to classify EEG signals as epileptic or non-epileptic. A brief literature survey of the automated epilepsy detection system with a focus on its modules such as pre-processing, feature extraction and selection, and finally classification is been given. Two most commonly used and freely available datasets, i.e. University of Bonn dataset and CHB-MIT dataset are also discussed. The findings of different authors in terms of different performance parameters have also been summarized and compared. This review will give a clear birds-eye view of the recent advancements and techniques to the upcoming researchers in this field.
References
Pachori RB, Patidar S (2014) Epileptic seizure classification in EEG signals using second-order difference plot of intrinsic mode functions. Comput Methods Programs Biomed 113:494–502
Megiddo I, Colson A, Chisholm D, Dua T, Nandi A, Laxminarayan R (2016) Health and economic benefits of public financing of epilepsy treatment in India: an agent-based simulation model. Epilepsia 57:464–474
Paul Y (2018) Various epileptic seizure detection techniques using biomedical signals: a review. Brain Inform 5
Chatterjee S, Choudhury NR, Bose R (2017) Detection of epileptic seizure and seizure-free EEG signals employing generalised S-transform. IET Sci Meas Technol 11:847–855
Jiang Y, Wu D, Deng Z et al (2017) Seizure classification from EEG signals using transfer learning, semi-supervised learning and TSK fuzzy system. IEEE Trans Neural Syst Rehabil Engg 25(12):2270–2284
Jiang Z, Chung F, Wang S (2019) Recognition of multiclass epileptic EEG signals based on knowledge and label space inductive transfer. IEEE Trans Neural Syst Rehabil Eng 27(4):630–642
Saleem S, Hassan SA, Kamboh AM et al (2018) Dynamic mode decomposition based epileptic seizure detection from scalp EEG. IEEE Access 6:38683–38692
Wu D, Wang Z, Jiang L et al (2019) Automatic epileptic seizures joint detection algorithm based on improved multi-domain feature of cEEG and spike feature of aEEG. IEEE Access 7:41551–41564
Alickovic E, Kevric J, Subasi A (2018) Performance evaluation of empirical mode decomposition, discrete wavelet transform, and wavelet packed decomposition for automated epileptic seizure detection and prediction. Biomed Signal Process Control 39:94–102
Andrzejak RG, Lehnertz K, Mormann F et al (2001) Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: dependence on recording region and brain state. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Top 64(8)
Goldberger AL et al (2000) Physiobank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals. Circulation 101(23):e215–e220
Mahmoodian N, Boese A, Friebe M, Haddadnia J (2019) Epileptic seizure detection using cross-bispectrum of electroencephalogram signal. Seizure 66:4–11
Zeng M, Zhao CY, Meng QH (2019) Detecting seizures from EEG signals using the entropy of visibility heights of hierarchical neighbours. IEEE Access 7:7889–7896
Lasefr Z, Ayyalasomayajula SSVNR, Elleithy K (2017) Epilepsy seizure detection using EEG signals. In: 2017 IEEE 8th annual ubiquitous computing, electronics and mobile communication conference (UEMCON), pp 162–167
Saini J, Dutta M (2018) Epilepsy classification using optimized artificial neural network. Neurol Res 40:982–994
Orhan U, Hekim M, Ozer M, Provaznik I (2011) Epilepsy diagnosis using probability density functions of EEG signals. In: 2011 international symposium on innovations in intelligent systems and applications, pp 626–630
Zhang T, Chen W (2017) LMD based features for the automatic seizure detection of EEG signals using SVM. IEEE Trans Neural Syst Rehabil Eng 25(8):1100–1108
Rahmawati D, Ucn R, Sarno R (2017) Classify epilepsy and normal electroencephalogram (EEG) signal using wavelet transform and K-nearest neighbour. In: 2017 3rd international conference on science in information technology (ICSITech), pp 110–114
Gupta A, Singh P, Karlekar M (2018) A novel signal modeling approach for classification of seizure and seizure-free EEG signals. IEEE Trans Neural Syst Rehabil Eng 26:925–935
Lee SH, Lim JS, Kim JK et al (2014) Classification of normal and epileptic seizure EEG signals using wavelet transform, phase-space reconstruction, and euclidean distance. Comput Methods Programs Biomed 116:10–25
Tiwari AK, Pachori RB, Kanhangad V, Panigrahi BK (2017) Automated diagnosis of epilepsy using key-point-based local binary pattern of EEG signals. IEEE J Biomed Heal Inform 21:888–896
Kaleem M, Guergachi A, Krishnan S (2013) EEG seizure detection and epilepsy diagnosis using a novel variation of empirical mode decomposition. In: 2013 35th annual international conference of the IEEE engineering in medicine and biology society (EMBS), pp 4314–4317
Peker M, Sen B, Delen D (2016) A novel method for automated diagnosis of epilepsy using complex-valued classifiers. IEEE J Biomed Heal Inform 20:108–118
Alsharabi K, Ibrahim S, Djemal R, Alsuwailem A (2016) A DWT-entropy-ANN based architecture for epilepsy diagnosis using EEG signals. In: 2016 2nd international conference on advanced technologies for signal and image processing (ATSIP), pp 288–291
Yuan Y, Xun G, Jia K, Zhang A (2019) A multi-view deep learning framework for EEG seizure detection. IEEE J Biomed Heal Inform 23:83–94
Sharma M, Pachori RB, Rajendra Acharya U (2017) A new approach to characterize epileptic seizures using analytic time-frequency flexible wavelet transform and fractal dimension. Pattern Recogn Lett 94:172–179
Abhijit B, Ram BP (2017) A multivariate approach for patient-specific EEG seizure detection using empirical wavelet transform. IEEE Trans Biomed Eng 64:2003–2015
Fan M, Chou CA (2019) Detecting abnormal pattern of epileptic seizures via temporal synchronization of EEG signals. IEEE Trans Biomed Eng 66:601–608
Samiee K, Kovács P, Gabbouj M (2017) Epileptic seizure detection in long-term EEG records using sparse rational decomposition and local gabor binary patterns feature extraction. Knowl Based Syst 118:228–240
Shanir PPM, Khan KA, Khan YU et al (2018) Automatic seizure detection based on morphological features using one-dimensional local binary pattern on long-term EEG. Clin EEG Neurosci 49:351–362
Bashivan P, Rish I, Yeasin M, Codella N (2019) Learning representations from EEG with deep recurrent-convolutional neural networks. IEEE Int Conf Consum Electron
Rathore NS, Singh VP, Phuc BDH (2019) A modified controller design based on symbiotic organisms search optimization for desalination system. J Water Supply Res Tech Aqua 68(5):337–345
Rathore NS, Singh VP (2019) Whale optimization algorithm based controller design for reverse osmosis desalination plants. Int J Intell Eng Inf 7(1):77–88
Rathore NS, Singh VP, Kumar B (2018) Controller design for DOHA water treatment plant using grey wolf optimization. J Intell Fuzzy Syst 35(5):5329–5336
Singh VP, Prakash T, Rathore NS, Chauhan DPS, Singh SP (2016) Multilevel thresholding with membrane computing inspired TLBO. Int J AI Tools 25(6)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Satyender, Dhull, S.K., Singh, K.K. (2021). A Review on Automatic Epilepsy Detection from EEG Signals. In: Hura, G.S., Singh, A.K., Siong Hoe, L. (eds) Advances in Communication and Computational Technology. ICACCT 2019. Lecture Notes in Electrical Engineering, vol 668. Springer, Singapore. https://doi.org/10.1007/978-981-15-5341-7_110
Download citation
DOI: https://doi.org/10.1007/978-981-15-5341-7_110
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-5340-0
Online ISBN: 978-981-15-5341-7
eBook Packages: EngineeringEngineering (R0)