Abstract
Glycoside hydrolases are group of enzymes belonging to class 3 enzymes as per classification by IUBMB. They specifically break down glycosidic bonds of complex polysaccharides and are generally named upon the substrates on which they act (e.g. lactase acting on lactose; chitinase acting on chitin, sucrase acting on sucrose, etc). Present chapter introduces glycoside hydrolases, its identification and occurrence in nature, highlighting the diverse existence of these enzymes. It covers a detailed report on classification and available three-dimensional structures of the enzymes of the group. Glycoside hydrolases have been classified under 166 families which are grouped in 16 different superfamilies and three added clans. The chapter gives a complete documentation of all available data on glycoside hydrolases, which will be beneficial for researchers working in the domain.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Glycoside hydrolases (Glycosidase or glycosyl hydrolases) are specific catalyst that breaks down glycosidic bonds of complex polysaccharides, such as cellulose, hemicelluloses, starch etc. These enzymes with their specific functions are called cellulase, amylases, xylanases, arabinases and includes several others. In addition to degradation of plant polysaccharides they also function in anti bacterial defense mechanism (lysozymes), in normal cellular functioning (such as biosynthesis of N-linked glycoprotein through mannosidases), pathogenesis through viral neuraminidase (Bourne and Henrisatt 2001; Henrisatt and Davies 1997)
Occurrence and Importance
Present in almost all domains of life, glycoside hydrolases are found both as intracellular and extracellular enzymes in prokaryotes and are majorly involved in hydrolysis of glycoside molecules, nutrient acquisition, regulation of expression of operon, post translational modification, lysosomal storage in higher organisms, biosynthesis and degradation of glycogen. In prokaryotes and lower eukaryotes they are present intracellular as well as secreted as extracellular enzymes. In higher organisms they are found within endoplasmic reticulum and golgi apparatus (processing of N-linked glycoproteins); in lysosome (degradation of carbohydrate structure) in intestinal tract and in saliva as carbohydrate degraders (amylase), in gut as glycosylphosphatidyl anchored enzymes on endothelial cells as enzyme lactase (degradation of milk sugar lactose), enzyme O-GlcNAcase (removal of N-acetylglucosamine groups from cytoplasmic and nuclear located serine and threonine residues)
Mechanisms of Glycoside Hydrolases
Basis their mechanism of action, Glycoside hydrolases are broadly classified as inverting glycoside hydrolases and retaining glycoside hydrolases.
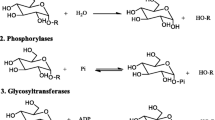
Inverting enzymes utilize two enzymic residues (generally carboxylate), one act as acid and other as base. Figure 1 shows mechanism of action of a β-glucosidase.
Retaining glycoside hydrolases conduct hydrolysis through two main mechanisms, with each step resulting in inversion. Two residues involved are generally enzyme accepted carboxylates; of which one acts as a nucleophile and the other as acid/base. First step involves attack of nucleophile to the anomeric center forming a glycosyl enzyme intermediate (acidic assistance provided by acid carboxylate), followed by hydrolysis of glycosyl-enzyme transitional state through nucleophilic water (assisted by deprotonated acidic carboxylate, that acts as a base). This process results in net retention of stereochemistry. Mechanism has been illustrated as example for hen egg white lysozyme (Fig. 2) (Vocadlo et al. 2001).
Enzyme mechanisms that carried out hydrolysis with retention of stereochemistry occurs through a substrate bound nucleophilic residue, rather than being directly attached to the enzyme. Such mechanism can be seen for certain N-acetylhexosaminidases (Fig. 3), where an acetamido group present on the enzyme can participate with neighboring group and forms and intermediate oxazoline or oxazolinium ion following two steps mechanism of distinct inversions making net retention of configuration.
Applications of Glycoside Hydrolases
Glycoside hydrolases are one of the major catalysts of ensuing generation owing to its varied applications in biorefining processes. These includes hydrolysis of plant materials (cellulases, xylanases) for production of value added components, applications in food industries (invertase for production of invert sugars; amylase for production of maltodextrins), usage in paper and pulp industry, in detergent manufacturing (cellualses for washing of cotton fabrics for maintenance of fabric colour by removing microfibre) (Linares-Pastén et al. 2014).
These enzymes are used as synthetic catalysts (performing reverse hydrolysis/transglycosylation); reversing equilibrium and enabling the retaining glycoside hydrolase to catalyze transfer of glycosyl moiety from an activated glycoside to an acceptor alcohol. Glycosynthases (formed from retaining glycoside hydrolases by site directed mutagenesis of enzymic nucleophile to some less nucleophilic group like alanine/glycine) are enzymes catalyzing high yield of glycosides from activated glycosyl donors like gycosyl fluorides. Thioglycoligases also are mutant glycoside hydrolases, formed by site directed mutagenesis of the acid base residue of retaining glycoside hydrolases and catalyze condensation of activated glycosides and thiol comprising acceptors.
Glycoside hydrolases show usefulness in matrix polysaccharide within extracellular polymeric substances (EPS) of microbial biofilm (Fleming and Rumbaugh 2017). Degrading microbial biofilm, increases antibiotic efficacy, potentiating host immune function (Fleming et al. 2017).
Inhibitors if Glycoside Hydrolases
There are various natural and synthetic compounds that have been reported to act as inhibitors of glycoside hydrolases. This includes naturally occurring nitrogen containing sugar shaped heterocycles (deoxynojirimycin, swainsonine, australine, castanospermine) working as natural templates for developing modified inhibitors (e.g. isofagomine, deoxygalactonojirimycin, unsaturated compounds such as PUGNAc). Glycoside hydrolase inhibitors finding clinical usage includes anti-diabetic drugs (acarbose and miglitol) and antiviral drugs (oseltamivir and zanamivir). Few proteins have also been identified as Glycoside hydrolase inhibitors.
Classification of Glycoside Hydrolases
According to enzyme nomenclature by International Union of Biochemistry and Molecular Biology (IUBMB), Glycoside hydrolases are classified into EC 3.2.1 as enzymes catalyzing the hydrolysis of O- or S-glycosides (Sinnott 1990). They are also classified based on stereochemical outcome of hydrolysis reaction (retaining or inverting enyzmes); on exo (non-reducing end) and endo (middle of molecule) acting, on sequence and structure based classification. Sequence based classification has suggested more than 150 different families of glycoside hydrolases (Henrissat et al. 1995; Henrissat and Davies 1995; Bairoch 1999), these are available on CAZy Carbohydrate-Active Enzymes web site, supported by CAZypedia [Cazy family Glycoside hydrolase; Cazypedia; Henrissat and Coutinho 1999]. Sequence based classification significantly helps in prediction of mechanism of action of enzyme, active site residues and possible substrates. Enzymes further classified as clans of related structure based on three dimensional structural similarities obtained from available sequences (Naumoff 2006, 2011).
Glycoside hydrolases (O-Glycosyl hydrolases) EC 3.2.1.X catalyzing hydrolysis of glycosidic bond are classified based on sequence similarity to be most reliable and led to the definition of 128 families and 14 clans (based on folds of proteins) of the same (Henrissat et al. 1995; Henrissat and Davies 1995; Henrissat and Bairoch 1996). This sequence based classification is available on the CAZy (Carbohydrate- Active Enzymes) (Cantarel et al. 2009). Glycoside hydrolases are also classified based on their localization in cell (secreted, monotopic, peripheral, lysosomal, located in eukaryotic plasma, inner membrane of Gram positive bacteria).
Following section in the chapter will deal with the major features and mode of action of all glycoside hydrolases classified. One prominent known structure from each family is depicted in Fig. 4a-r5.
Glycoside Hydrolase Family 1
Enzymes of family 1 follow the IUBMB EC 3.2.1 nomenclature and catylzes hydrolysis of glycosidic bond between two carbohydrate molecules or a carbohydrate and non carbohydrate molecule (Lombard et al. 2014; Cazypedia.org and Cazypedia Consortium). Major enzymes of Glycoside Hydrolase family 1 are; beta-glucosidase (EC 3.2.1.21); beta-galactosidase (EC 3.2.1.23); 6-phospho-beta-galactosidase (EC 3.2.1.85); 6-phospho-beta glucosidase (EC 3.2.1.86); lactase-phlorizin hydrolase (EC 3.2.1.62), lactase (EC 3.2.1.108); beta mannosidase (EC 3.2.1.25); myrosinase (EC 3.2.1.147). Figure depicts general structure of Glycoside hydrolase family 1. According to the PROSITE documentation (https://prosite.expasy.org/cgi-bin/prosite/prosite-search-ac?PDOC00495#description) (Henrissat 1991a, b; Gonzalez-Candelas et al. 1990; El Hassouni et al. 1992) classifies β-glucosidases (EC 3.2.1.21); β-Galactosidases (EC 3.2.1.23); 6-phospho- β-Galactosidases (EC 3.2.1.85); β-Glucosidases (EC 3.2.1.86); plant myroniases (EC 3.2.1.147; e g. synigrinases or thioglucosidases); Mammalian lactase-phlorizin hydrolase (LPH; EC 3.2.1108/EC 3.2.1.62) in family 1 of Glycoside hydrolases. Conserved regions are central glutamic acid residues, acting as nucleophile during glycosidic bond cleavage. This is marked as signature pattern for this group of enzymes with another along with a conserved region containing glutamic acid found at their N-terminal extremity (Withers et al. 1990). This group belongs to the TIM Barrel glycoside hydrolase superfamily contains the range of enzymes belonging to group that posses a TIM barrel fold merging clans GH-A, GH-D, GH_H and GH-K. It contains 57 families and 259,156 domains and was built by A Bateman (Naumov and Karreras 2009). All members of family 1 glycoside hydrolases for which localization is known are typically restricted to plasma membrane and endoplasmic reticulum membrane (Davies and Henrissat 1995; Henrissat et al. 1995) (Fig. 4a).
Glycoside Hydrolase Family 2
Glycoside hydrolases family 2 enzymes comprises enzymes with β-galactosidases (EC 3.2.1.23); β-mannosidases (EC 3.2.1.25), β-glucuronidase (EC 3.2.1.31). Glutamic acid residue is the general acid/base catalyst at active sites of these enzymes (Gebler et al. 1992). The catalytic domain of β-galactosidases contains TIM barrel core surrounded beta domains, with sugar binding domain forming jelly–roll fold, containing immunoglobulin like beta-sandwich domain (Jacobson et al. 1994). General structure of glycoside hydrolase family 2 enzymes is elaborated as its complete picture, sugar binding domain and TIM Barrel domain as depicted in Fig. 4b–d respectively.
Sugar binding domain belongs to the galactose binding domain like superfamily, the clan that contains 70 families and 124,105 domains. This superfamily has prominent sandwich domains with a jelly roll topology and is mainly involved in carbohydrate recognition. They share very little sequence similarity and weak sequence motif with conserved bulge (possibly helps in bending of beta sheets) in the C-terminal beta sheet, enabling curvature of sheet that forms a sugar binding site (Murzin and Bateman 1998).
Glycoside Hydrolase Family 3
Glycoside hydrolase family 3 enzymes are two domain globular proteins N-glycosylated at 3 sites (Varghese et al. 1999). This family comprises of β-glucosidases (EC 3.2.1.21); β-xylosidase (EC 3.2.1.37), N-acetyl β-glucoseaminidase (EC 3.2.1.52), Glucan β-1,3-glucosidase (EC 3.2.1.58), cellodextrinase (EC 3.2.1.74); exo 1,3-1,4-glucanase (EC 3.2.1.). Crystal structures for N-terminal and C-terminal domain of family 3 enzymes are available (Fig. 4e). Conserved region of these enzymes have been centered on a conserved aspartic acid residue depicted in β-glucosidase A3 in Aspergillus wentii (Bause and Legler 1980).
Glycoside Hydrolase Family 4
Major enzyme activities of this group comprise 6-phospho-β-glucosidase (EC 3.2.1.86); 6-phospho-α-glucosidase (EC 3.2.1.122), α-galactosidase (EC 3.2.1.22); of these 6-phospho-α-glucosidase requires both NAD(H) and divalent metal (Mn2+, Fe2+, Co2+, or Ni2+) for activity (Thompson et al. 1998). Figure shows structure of 6-phospho-β-glucosidase from Thermotoga maritime in tetragonal form with manganese, NAD+ and glucose-6-phosphate (GH F-4), belonging to FAD/NAD(P)-binding Rossmann fold Superfamily (CL0063) (Fig. 4f).
These are redox enzymes containing a catalytic domain (provides substrate specificty) and Rossmann-fold domain (contains alpha-beta folds with central beta-sheet surrounded by 5 alpha-helices in order 654123; binds to NAD+), where NAD+ reversibly binds to hydride ion (lost/gained during redox process). Inter sheet crossover of the stands in the sheet form the NAD+ binding site (Bashton and Chothia 2002) and in some distantly related Rossmann NAD+ is replaced by FAD. Clan has been buit by RD Finn.
Structure of GH F-4 C terminal domain belongs to LDH C-terminal domain-like superfamily (CL 0341). This superfamily includes the C-terminal domain of lactate/malate dehydrogenase as well as the C-terminal domain of the GH F-4.
Glycoside Hydrolase Family 5
They are membrane localized enzymes, basically involved in polysaccharides like cellulose and xylan and have been reported to be produced from fungi as well bacteria. This family comprises of enzymes namely endoglucanase (EC 3.2.1.4); beta-mannanase (EC 3.2.1.78); exo-1,3-glucanase (EC 3.2.1.58); endo-1,6-glucanase (EC 3.2.1.75); xylanase (EC 3.2.1.8); endoglycoceramidase (EC 3.2.1.123). Active site includes conserved glutamic acid residue (Henrissat 1991a; Py et al. 1991). Relationship between thermal stability and structural rigidity of members of GH F-5 enzymes have been reported after being studied through molecular dynamics (Bedieyan et al. 2012). It belongs to TIM Barrel Glycoside hydrolase Superfamily (CL0058) (Fig. 4g).
Glycoside Hydrolase Family 6
They were formerly known as Cellulase family B including endoglucanses (EC 3.2.1.4) and cellobiohydrolases (EC 3.2.1.91). They are alpha-beta proteins (Fig. 4h) having similar folds as that seen in triose phosphate isomerase. This was studied in 3D structure of cellobiohydrolase II (CBHII) from the fungus Trichoderma reesei, having active site located at C-terminal end of a parallel beta barrel present in an enclosed tunnel. Probable catalytic residues are two aspartic acid residues (positioned in center of tunnel) (Rouvinen et al. 1990).
Glycoside Hydrolase Family 7
Known activities of enzymes (formerly known as cellulase family C) classified under this group are endoglucanase (EC 3.2.1.4) and cellobiohydrolase (EC 3.2.1.91, end-acting cellobiohydrolase (EC 3.2.1.176); chitosanase (EC 3.2.1.132); endo-β-1,3-1,4-glucanase (EC 3.2.1.73) from eukaryotes catalyzing glycosidic bond using double-displacement mechanism. This mechanism helps in net retention of the conformation at the anomeric carbon. Endoglucanases cleaves the beta-1,4 linkages of cellulose and CBH cleaves off cellobiose disaccharide units from the reducing end of the chain. Their catalytic region is generally bound to cellulose binding domain (CBD) via a proline and/or hydroxy-amino acids rich linker region. In type I exoglucanases, the CBD domain is found at the C-terminal extremes (this domain forms a hairpin loop structure stabilized by 2 disulphide bridges). Active site contains glutamic acid residue as catalytic nucleophile/base as well as proton donor and catalysis progress through retaining mechanism (Divne et al. 1994; Mackenzie et al. 1998; Ducros et al. 2003; Klarskov et al. 1997; Sulzenbacher et al. 1997; Viladot et al. 1998).
GH F-7 enzymes belong to Concanavalin-like lectin/glucanase superfamily (contains 49 families and 153254 domains) (Bateman..). Conserved protein domain of endoglucanase and cellobiohydrolase are grouped under O-glycosyl hydrolases domain family (cd07999) contains a total of 95 families which includes enzymes of glycoside hydrolase family 7 as well.
Beta-jellyroll folded framework, containing two antiparallel beta-sheet packed face-to-face forming a curved beta-sandwich is the predominant three dimensional feature (Fig. 4i). These sandwich structures are extended along both the ends through several loops, resulting in an elongated assembly of ~50 Å, with a substrate binding structure running perpendicular to the β-strands of the inner, concave β-sheet. Some loops have short alpha-helical segments present at the periphery. Endoglucanases specifically contains open substrate binding domain, while cellobiohydrolases have additional elongated loops that bend around the active site giving it a closed tunnel form. Some key studies include, boat confirmation required prior to hydrolysis (Sulzenbacher et al. 1997), presence of discrete glycoside binding subsites (Divne et al. 1998); product ejection mechanism during hydrolysis of cellulose also suggesting prior release of cellobiose products (Ubhayasekera et al. 2005); flexibility of sugar binding within tunnel of cellobiohydrolase (Parkkinen et al. 2008). Numerous commercial enzyme of this group is also available produced through Megazymes (Company).
Glycoside Hydrolase Family 8
These enzymes are of prokaryotic/eukaryotic origin belonging to clan GH-M of six hairpin glycosidase superfamily (CL0059). Enzymes classified under this superfamily share common structure composed of six helical hairpins and are chitosanase (EC 3.2.1.132), cellulase (EC 3.2.1.4), licheninase (EC 3.2.1.73), endo-1,4-β-xylanase (EC 3.2.1.8) and reducing-end-xylose releasing exo-oligoxylanase (EC 3.2.1.156). GH F-8 enzymes cleaves β-1,4 linkages in β-1,4 glucans, xylans (or xylooligosaccharides), chitosans, and lichenans (1,3-1,4-β-D-glucan). They are majorly endo acting with few having exo-activity. They were classified by hydrophobic cluster analysis, and was previously known as “Cellulase Family D” (Henrissat et al. 1989; Gilkes et al. 1991). These enzymes have inverting mechanism of action (Fierobe et al. 1993; Petersen et al. 2009). General acid (proton donor to the leaving group; located at N-terminal end of α4 helix), base of GH8a (proton acceptor from the nucleophilic water) and base of GH8b subfamily was first identified in CelA from C. thermocellum as Glu95 (Alzari et al. 1996; Adachi et al. 2004), CelA from C. thermocellum as Asp278 (Alzari et al. 1996) and chitosanase from Bacillus sp. K17 as Glu309 (based on its crystal structure and by making E309Q mutant) (Adachi et al. 2004) respectively. They are divided in three subfamilies based on positions of general base. Subfamily ‘a’ has Aspartate at the N-terminal end of α8 helix (cellulases, xylanases); ‘b’ has Asparagine, and the general base is a Glu residue located in a long loop inserted between α7 and α8 helices (chitinosases, lichenisases, cellulases). They have (α/α)6 fold like Glycoside Hydrolase Family 48. Atomic (0.94 Å) resolution structure of CelA in complex with substrate (PDB ID 1kwf) and has been determined (Guérin et al. 2002; Honda and Kitaoka 2006; Béguin et al. 1985) (Fig. 4j).
Glycoside Hydrolase Family 9
These enzymes were earlier known as cellulase family E. Enzymes include endoglucanases (EC 3.2.1.4), cellobiohydrolases (EC 3.2.1.91) (exoglucanases), or xylanases (EC 3.2.1.8) (Beguin, 1990; Gilkes et al. 1991). Enzymes from diverse group of organisms (fungi, bacteria, amaebozoa, invertebrate metazoan, ferns, mosses, gymnosperms and angiopserms) are reported to belong to this family. They belong to six-hairpin glycosidase super family and major localized to plasma membrane (Fig. 4k).
Glycoside Hydrolase Family 10
Enzymes of this family were formerly known as cellulase family F and comprise of endoglucanases (EC 3.2.1.4), cellobiohydrolases (EC 3.2.1.91) (exoglucanases), or xylanases (EC 3.2.1.8) activities. All family 10 xylanases hydrolyze the glycosidic bond in a double-displacement ‘retaining’ mechanism using two catalytic acidic residues, where one residue acts a nucleophile (base) and the other acts as a general acid/base (Fig. 4l). They undergo retaining type catalytic mechanism, with conserved glutamic acid residues acting as catalytic nucleophile/base and catalytic proton donor. Three dimensional structure has prominent (β/α)8 barrels.
Glycoside Hydrolase Family 11
This family was earlier known as cellulase family G (Henrissat and Bairoch 1996) and is monspecific, consisting of xylanases. They belong to Concanavalin-like lectin/glucanase superfamily (CL0004). Major enzymes are xylanases from Aspergillus awamori, Bacillus circulans, Clostridium stercorarium, Fibrobactor succinogenes etc. Domains folds into a jelly roll shaped, and anti-parallel β strands bending almost 90 °C to produce a substrate binding grooves are characteristics of this family. Three dimensional structure has two catalytic Glu residues facing each other from opposite sides of the groove and catalysis is carried through double-displacement mechanism, with one Glu residue acting as a general acid/base catalyst and the other as a nucleophile. Two catalytically active conserved regions are centrally located on glutamic acid residues (studied in Bacillus pumilis, Lombard et al. 2014) (Fig. 4m).
Glycoside Hydrolase Family12
Family 12 comprises of enzymes Concanavalin-like lectin/glucanase superfamily (CL0004). These include endoglucanase (EC 3.2.1.4), xyloglucan hydrolase (EC 3.2.1.151), β-1,3-1,4-glucanase (EC 3.2.1.73) and xyloglucan endotransglycosylase (EC 3.2.1.207). These enzymes were formerly known as cellulase family H. Three dimensional structures has β-jelly roll and glutamic acid residues acts as catalytic site nucleophile/base and proton donor (Fig. 4n).
Glycoside Hydrolase Family 13
These are maltogenic alpha-amylase, catalyzing hydrolysis of (1-4)-alpha-D-glucosidic linkages in polysaccharides that helps in removal of successive alpha-maltose residues from the non-reducing ends of chains, during conversion of starch to maltose. Other enzymes in this family include neopullulanase, which hydrolyses pullulan to panose, and cyclomaltodextrinase, which hydrolyses cyclodextrins. They belong to TIM Barrel glycosyl hydrolase family (CL0058) (Fig. 4o,p).
Glycoside Hydrolase Family 14
Enzymes of this family have β-amylase (EC 3.2.1.2) activities. They belong to TIM Barrel glycoside hydrolase superfamily (CL0058) containing catalytic glutamic acid residues and have three highly conserved regions (Mikami et al. 1988; Friedberg and Rhodes 1988). First on N-terminal (contains catalytic aspartic acid, Nitta et al. 1989) and second conserved domain is centrally located (catalytic glutamic acid, Totsuka et al. 1994) (Fig. 4q). Various three dimensional structures of enzymes belonging to this family have been studied; like soyabean beta-amylase with an inhibitor, alpha-dextrin etc (Table 1).
Glycoside Hydrolase Family 15
Major enzymes of family 15 are glucoamylase (EC 3.2.1.3; catalyze release of D-glucose from the non-reducing end of starch and other oligo and polysaccharaides); alpha-glucosidase (EC 3.2.1.20) and glucodextranase (EC 3.2.1.70). Glucoamylases have three closely clustered acidic conserved residues at catalytic site (Sierks et al. 1990 and Ohnishi et al. 1992) and 3-D structure have been reported to belong to mainly alpha-class and contains 19 helices and 9 strands (Aleshin et al. 1994). They belong to Six- hairpin glycosidase superfamily (Fig. 4r).
Glycoside Hydrolase Family 16
Enzymes of family 16 belong to Concanavalin-like lectin/glucanase superfamily (CL0004) and are group with functionally heterogenous members, like lichenase (EC 3.2.1.73); xyloglucan xyloglucosyltransferase (EC 2.4.1.207); agarase (EC 3.2.1.81); kappa-carrageenase (EC 3.2.1.83); endo-beta-1,3-glucanase (EC 3.2.1.39); endo-beta-1,3-1,4-glucanase (EC 3.2.1.6); endo-beta-galactosidase (EC 3.2.1.103). All these enzymes have a common ancestor and have diverged significantly in their primary sequences (Fig. 4s).
Catalytic domain have this family has sandwich-like β-jelly roll fold formed by two anti-parallel beta sheet that are closely packed. All enzymes of GH F-16 feature a common catalytic motif E-[ILV]-D-[IVAF]-[VILMF](0,1)-E. The two glutamic acid residues in the conserved motif act as nucleophile and base in the catalytic reaction and aspartic acid residue maintains relative position of catalytic amino acids (Allouch et al. 2003; Michel et al. 2001) (Fig. 5).
Active site of glycoside hydrolase family 19 papaya chitinase (PDB: 3cql). Conserved residues are shown surrounding the catalytic acid, Glu67. A GlcNAc2 unit binding in the -1 and +1 subsites is shown in narrow stick representation and an arrow indicates the position of the glycosidic oxygen (Eijsink et al. 2010)
Glycoside Hydrolase Family 17
This family comprise of enzymes endo-1,3-beta-glucosidase (EC 3.2.1.39); lichenase (EC 3.2.1.73); exo-1,3-glucanase (EC 3.2.1.58) and have been reported only in eukaryotes namely plants and fungi. Central section has the conserved region, containing tryptophan residue, which is involved in interaction with the glucan substrate (Ori et al. 1990). It also contains a glutamate residue acting as nucleophile in the catalytic reaction (Varghese et al. 1994) (Fig. 4t).
Glycoside Hydrolase Family 18
Chitinase class II group, including chitinase and chitodextrinase are enzymes belonging to this family, falling under TIM-barrel Glycoside hydrolase superfamily (CL0058) and located on endoplasmic reticulum membrane. They majorly catalyze hydrolysis of chitin oligosaccharides. This family also includes chitinase like protein that binds to chitin but does not hydrolyse it, and also varied glycoproteins (from mammals, cartilage and oviduct specific) (Fig. 4u).
Glycoside Hydrolase Family 19
Like GH F-18, this family also has enzymes only with chitinase activity [EC 3.2.1.14], catalyzing hydrolysis of beta-1,4-N-acetyl-D-glucosamine linkages in chitin polymers (Flach et al. 1992). These enzymes also known as class IA/I and IB/II are of plant origin functioning as defense tool against fungal and insect pathogen. Catalytic domain of these enzymes consists of 220-230 residues (Henrissat 1991b) of which class IA/I has an extra N-terminal chitin domain. There are two highly conserved regions on this enzyme, of which one is located N-terminal section (containing cysteines involved in disulphide bond formation). They belong to Lysozyme like superfamily having structurally invariant core consisting of two-helices and a three stranded beta sheet that forms substrate binding and catalytic cleft (Monzingo et al. 1996) (Fig. 4v).
Glycoside Hydrolase Family 20
Beta-hexosaminidase (EC 3.2.1.52); lacto-N-biosidase (EC 3.2.1.140) are major enzymes of this family belonging to TIM-Barrel glycosyl hydrolase superfamily (CL0058). Catalytic nucleophile/base of these enzymes are carbonyl oxygen of the C-2 acetamide group. Beta-hexosaminidase degrade GM2 gangliosides by hydrolysing the terminal non-reducing N-acetyl-D-hexoamine residues. This enzyme is found in three forms; hexosaminidase A is a trimer, with one alpha, one beta-A and one beta-B chain; hexosaminidase B is a tetramer of two beta-A and two beta-B chains; and hexosaminidase S is a homodimer of alpha chains (Fig. 4w,x).
Glycoside Hydrolase Family 21
There is no entry of enzyme been classified under family 21 of glycoside hydrolases as per source of cazypedia.org and a detailed study of enzymes to be classified under this family is required.
Glycoside Hydrolase Family 22
Lysozyme type C (EC 3.2.1.17) lysozyme type I (EC 3.2.1.17) and alpha-lactalbumins are enzymes classified as GH Family 22. Specifically based on activities they belong to Lysozyme like superfamilies and the catalytic nucleophile/base for the same is aspartate and/or carbonyl oxygen of the C-2 acetamido group. Five different classes of lysozymes are known; namely C (chicken) type, G (goose) type, Phage type, fungal and bacterial and all these exhibit minor sequence similarities. Though being functionally divergent, primary sequence and structure of lysozyme type C and alpha-lactalbumin are very similar, reflecting their common origin (Nitta and Sugai 1989) where approximately 40% of residues and 4 disulphide bonds are conserved. All lactalbumins and few lysozymes have the property of binding to calcium while catalysis (Nitta et al. 1987; Stuart et al. 1986) in order to obtain a stable structure. Catalytic site of these enzymes have three aspartic acid residues (Fig. 4y).
Glycoside Hydrolase Family 23
Lysozyme type G (EC 3.2.1.17) and chitinase (EC 3.2.1.14) are activities of enzymes of glycoside hydrolase family 23. This family also include peptidoglycan lyase (EC 4.2.2.n1) also known as peptidoglycan lytic transglycosylase. Glutamic acid is the catalytic proton donor, mechanism and catalytic nucleophile/base is not known. Enhanced and elaborate studies are still under progress (Fig. 4z).
Glycoside Hydrolase Family 24
Lysozymes (EC 3.2.1.17) are the activity reported in this group and enzymes belong to Lysozyme like superfamily. Lambda phage lysozyme and Escherichia coli endolysin (Weaver and Matthews 1987) are included in this family. These enzymes have inverting mechanism of action and glutamic acid acts as a catalytic proton donor. Three dimensional structure of the protein has both alpha helices and beta sheets (Fig. 4a1).
Glycoside Hydrolase Family 25
Family 25 also comprises of enzymes with lysozyme activity (EC 3.2.1.17; cell wall lytic enzymes) (Henrissat 1991b; Croux and García 1991), with retaining mode of catalytic mechanism where aspartic acid is catalytic nucleophile or base and glutamic acid is the proton donor. DxE is a conserved active site motif, helping in catalysis by formation of oxazoline intermediate. Aspartate residue initially protonates the leaving facilitating its departure and subsequently acts as a general base to activate the hydrolytic water molecule; and the glutamic residue stabilized or deprotonated the oxazoline N- atom. Aspartic acid at sixth position might be involved in catalysis with the glutamic acid at DxE motif, provided the hydrolysis has taken place via net inversion of anomeric configuration, with the aspartic acid residue acting as general base activating the nucleophilic water molecule and glutamic acid of DxE motif acting as general acid and protonating the departing oxygen atom in a concerted fashion as the bonds cleaves (Rau et al. 2001; Korczynska et al. 2010; Hermoso et al. 2003; Al-Riyami et al. 2016). Three dimensional structure consists of (β/α)5 (β)3 TIM-like domain and thus they classified under TIM Barrel glycoside hydrolase super family. Majorly studied enzymes include lysozymes (lysine) from Streptococcus pneumoniae bacteriophages of the Cp family, lysozyme (endolysin) from Lactococcus delbrueckii phage mv1, autolytic lysozyme from Clostridium acetobutylicum, lysozyme M1 from Streptomyces globisporus and N,O-diacetylmuramidase (lysozyme ch) from the fungus Chalaropsis (Fig. 4b1).
Glycoside Hydrolase Family 26
Enzymes in this group has activities of mannanase (EC 3.2.1.78) and β-1,3-xylanase (EC 3.2.1.32) of which main enzyme is Mannan endo-1,4-beta-mannosidase catalyzing specific hydrolysis of mannan and galactomannan and showing very little hydrolysis towards 1,4-beta-D-linkages in mannans, galacto-mannans, glucomannans and galactoglucomannans (Braithwaite et al. 1995) (Fig. 4c1).
Glycoside Hydrolase Family 27
Alpha-galactosidases (Melibiase) (EC 3.2.1.22) (Dey and Pridham 1972) α-N-acetylgalactosaminidase (EC 3.2.1.49); isomalto-dextranase (EC 3.2.1.94); β-L-arabinopyranosidase (EC 3.2.1.88); galactan: galactan galactosyltransferase (EC 2.4.1.-), are major activities of GH Family 27 enzymes, catalyzing hydrolysis of melibiose to glucose and galatose. Together with members of family 11 and 36, these enzymes form clan GH-D (alpha-galactosidase; alpha-N-acetylgalactosaminidases, and isomaltodextranases); belonging to TIM Barrel glycoside hydrolase superfamily. NAD and magnesium acts as cofactors for few enzymes of prokaryotic origin. Mode of action for these enzymes is retaining type, where aspartic acid is catalytic nucleophile and base as well as proton donor. These enzymes find mechanistic commonality with family GH36 demonstrated by Comfort et al. 2007 (Fig. 4d1).
Glycoside Hydrolase Family 28
Activities of enzymes belonging to this group includes polygalacturonase (EC 3.2.1.15); α-L-rhamnosidase (EC 3.2.1.40); exo-polygalacturonase (EC 3.2.1.67); exo-polygalacturonosidase (EC 3.2.1.82); rhamnogalacturonase (EC 3.2.1.171); rhamnogalacturonan α-1,2-galacturonohydrolase (EC 3.2.1.173); xylogalacturonan hydrolase (EC 3.2.1.-); all these enzymes have inverting mechanism for catalysis, with aspartic acid residue as catalytic nucleophile/base and catalytic proton donor. Three dimensional structures have prominent beta- helices. These enzymes belong to pectate-lyase like beta-helix Superfamily. Main feature of this superfamily is presence of right handed beta helix similar to first found in pectate lyase (Jenkins et al. 1998). Polygalatorunoase/pectinase catalyzes hydrolysis of 1,4-alpha-D-galactosiduronic linkages in pectate/galacturonans (Ruttkowski et al. 1990). Exo-poly-alpha-D-galacturonosidase (EC 3.2.1.82) (exoPG) hydrolyzes peptic acid from the non-reducing end, releasing digalacturonate (He and Collmer 1990) (Fig. 4e1).
Glycoside Hydrolase Family 29
Alpha-L-fucosidase (EC 3.2.1.51) and α-1,3/1,4-L-fucosidase (EC 3.2.1.111) are major activities of this family, belonging to the TIM barrel glycoside hydrolase superfamily, these emzymes have retaining mechanism of action and has hexamer structure with (β/α)8 domain observed in their three dimensional structures. Aspartic acid and glutamic acid acts as catalytic nucleophile base and proton donor, respectively (Fisher and Aronson 1989; Sulzenbacher et al. 2004). Alpha-L-fucosidase catalyze hydrolysis of alpha-1,6-linked fucose joined to the reducing-end N-acetylglucosamine (glycoproteins) (Fig. 4f1).
Glycoside Hydrolase Family 30
Endo-β-1,4-xylanase (EC 3.2.1.8); β-glucosidase (3.2.1.21); β-glucuronidase (EC 3.2.1.31); β-xylosidase (EC 3.2.1.37); β-fucosidase (EC 3.2.1.38); glucosylceramidase (EC 3.2.1.45); β-1,6-glucanase (EC 3.2.1.75); glucuronoarabinoxylan endo-β-1,4-xylanase (EC 3.2.1.136); endo-β-1,6-galactanase (EC:3.2.1.164); [reducing end] β-xylosidase (EC 3.2.1.-) are major activities of enzymes belonging to glycoside hydrolase family 30. These include enzymes of mammalian origin (Dinur et al. 1986). They follow retaining type mechanism of action, with glutamic acid as catalytic nucleophile/base as well as proton donor. Three dimensional structure has (β/α)8 domains (Fig. 4g1).
Glycoside Hydrolase Family 31
Alpha-glucosidase (EC 3.2.1.20); α-galactosidase (EC 3.2.1.22); α-mannosidase (EC 3.2.1.24); α-1,3-glucosidase (EC 3.2.1.84); sucrase-isomaltase (EC 3.2.1.48) (EC 3.2.1.10); α-xylosidase (EC 3.2.1.177); α-glucan lyase (EC 4.2.2.13); isomaltosyltransferase (EC 2.4.1.-); oligosaccharide α-1,4-glucosyltransferase (EC 2.4.1.161); sulfoquinovosidase (EC 3.2.1.-) are activities of enzymes of family 31 of glycoside hydrolases, with retaining mechanism of action and aspartic acid residues as nucleophile/base as well proton donor, these enzymes have been reported to be of archaea, eukaryotes, prokaryotes (bacteria) origin (Kinsella et al. 1991; Naim et al. 1991; Hermans et al. 1991). These enzymes belong to TIM barrel glycoside hydrolase superfamily clan-D (Fig. 4h1).
Glycoside Hydrolase Family 32
Invertase (EC 3.2.1.26); endo-inulinase (EC 3.2.1.7); β-2,6-fructan 6-levanbiohydrolase (EC 3.2.1.64); endo-levanase (EC 3.2.1.65); exo-inulinase (EC 3.2.1.80); fructan β-(2,1)-fructosidase/1-exohydrolase (EC 3.2.1.153); fructan β-(2,6)-fructosidase/6-exohydrolase (EC 3.2.1.154); sucrose:sucrose 1-fructosyltransferase (EC 2.4.1.99); fructan:fructan 1-fructosyltransferase (EC 2.4.1.100); sucrose:fructan 6-fructosyltransferase (EC 2.4.1.10); fructan:fructan 6G-fructosyltransferase (EC 2.4.1.243); levan fructosyltransferase (EC 2.4.1.-); [retaining] sucrose:sucrose 6-fructosyltransferase (6-SST) (EC 2.4.1.-); cycloinulo-oligosaccharide fructanotransferase (EC 2.4.1.-) forms major activities of enzymes of glycoside hydrolase family 32, with retaining mechanisms of action and fivefold β-propeller in their 3-D structure (Alberto et al. 2004), has aspartic acid and glutamic acid residue at their active site that serves as nucleophile/base and proton donor respectively. They belong to clan GH-J of beta-fructosidase/furanosidase superfamily (composed of glycoside hydrolase enzymes having five-bladed beta-propeller fold, built by J Mistry) (Hettwer et al. 1998; Nurizzo et al. 2002a) (Fig. 4i1, j1).
Glycoside Hydrolase Family 33
Sialidase or neuraminidase (EC 3.2.1.18); trans-sialidase (EC 2.4.1.-); anhydrosialidase (EC 4.2.2.15); Kdo hydrolase (EC 3.2.1.-); 2-keto-3-deoxynononic acid hydrolase/KDNase (EC 3.2.1.-) are enzyme activities of family 33.ave. They catalyze through retaining mode of activity and have sixfold β-propeller domain as found in 3-D structure. Tyrosine and glutamic acid acts as catalytic nucleophile/base (Rothe et al. 1991; Luo et al. 1998) (Fig. 4k1).
Glycoside Hydrolase Family 34
Sialidase or neuraminidase (EC 3.2.1.18) are activities of enzymes of glycoside hydrolase family 34, with retaining catalytic mechanism, tyrosine and glutamic acid serves as nucelophile/base, these enzymes have sixfold-beta propellers. They belong to Sialidase superfamily (CL0434). Neuraminidases cleave the terminal sialic acid (negatively charged sugars associated with proteins and lipids portion of the lipoproteins) residues from carbohydrate chains in glycoproteins. These enzymes have been extensively studied and have been widely reported to act as endo and exo-enzymes (Kim et al. 2013) (Fig. 4l1).
Glycoside Hydrolase Family 35
Beta-galactosidase (EC 3.2.1.23); exo-β-glucosaminidase (EC 3.2.1.165); exo-β-1,4-galactanase (EC 3.2.1.-); β-1,3-galactosidase (EC 3.2.1.-), are major enzymes of this group, with retaining mode of action they belong to clan GH-A, where glutamic acid residue acts as catalytic nucleophile/base and proton donor and the three dimensional structure of these enzymes have (β/α)8 barrels. They are reported to be produced from various sources including mammals, fungi, plants and bacteria (Fig. 4m1).
Glycoside Hydrolase Family 36
Alpha-galactosidase (EC 3.2.1.22); α-N-acetylgalactosaminidase (EC 3.2.1.49); stachyose synthase (EC 2.4.1.67); raffinose synthase (EC 2.4.1.82) are prominent activities of the family with retaining mechanism of catalysis, they belong to GH-D, containing (β/α)8 motif. Aspartic acid residues in the conserved domain work as nucleophile/base and proton donor. They show similarity in catalytic mechanism as glycoside hydrolase family 27 (Fig. 4n1).
Hydrolysis of melibiose into galactose and glucose is done by alpha-galactosidase, prokaryotic enzyme requires NAD and magnesium as co-factor. Glycoside hydrolase family 36 have been subdivided into 11 families GH36A to GH36K (Dey and Pridham 1972; Aslanidis et al. 1989; Wang et al. 1990; Peterbauer et al. 2002; Naumoff 2011).
Glycoside Hydrolase Family 37
Alpha,α-trehalase (EC 3.2.1.28) is the activity of the enzymes of this family, with inverting mode of catalytic mechanism and conserved glutamic and aspartic acid acting as nucleophile/base an proton donor they belong clan GH-G, of six-hairpin glycosidase superfamily. The three dimensional structure of same contains (α/α)6 (Kopp et al. 1993). Trehalases catalyses degradation of disaccharide α,α-trehalose yielding two-glucose subunits (Fig. 4o1).
Glycoside Hydrolase Family 38
Alpha-mannosidase (EC 3.2.1.24); mannosyl-oligosaccharide α-1,2-mannosidase (EC 3.2.1.113); mannosyl-oligosaccharide α-1,3-1,6-mannosidase (EC 3.2.1.114); α-2-O-mannosylglycerate hydrolase (EC 3.2.1.170); mannosyl-oligosaccharide α-1,3-mannosidase (EC 3.2.1.-) are major activities of enzymes of the group with a prominent (β/α)7 domain in their three-dimensional structure. Aspartic acid residue at the catalytic site acts as nucelophile/base and these enzymes show retaining mode of catalytic mechanism. They carry catalysis of N-linked carbohydrates released during glycoprotein turnover, catalyzing hydrolysis of terminal, non-reducing alpha-D mannose residues in α-D-mannosides and are localized in plasma membrane and golgi membrane. N-terminal domain of these enzymes belongs to glycoside hydrolase/deacetylase superfamily (CL0158) and C-terminal domain is categorized in Galactose Mutarotase-like superfamily (CL0103). Both the clans were built by A Bateman. Clan CL0158 contains diverse carbohydrate catalysing enzymes including hydrolases and deacetylases (Heikinheimo et al. 2003). Clan CL0103 is group of enzymes composed of beta-sandwich acting on sugar substrate and can be observed in any location (eg., domain 5 of beta-galatosidase, central domain of copper amine oxidase; c-terminal of chondroitinase and hyaluronate lyase, N terminal of maltose phosphorylase and galactose mutarotase) (Thoden and Holden 2002) (Fig. 4p1, q1, r1).
Glycoside Hydrolase Family 39
Alpha-L-iduronidase (EC 3.2.1.76); β-xylosidase (EC 3.2.1.37), are activities of the family. These enzymes belong to TIM-barrel glycoside hydrolase superfamily (CL0058) with prominent (β/α)8 motif in its three dimensional structure. They follow retaining type catalytic mechanism with conserved glutamic acid residue working as nucleophile/base and proton donor (Henrissat and Bairoch 1996; Henrissat et al. 1995) (Fig. 4s1).
Glycoside Hydrolase Family 40
This family does not have specified studied enzymes classified under its group. No data available through CAZypedia.
Glycoside Hydrolase Family 41
This family does not have specified studied enzymes classified under its group. No data available through CAZypedia.
Glycoside Hydrolase Family 42
Beta-galactosidase (EC 3.2.1.23); α-L-arabinopyranosidase (EC 3.2.1.-) are activities of enzymes of this family. They undergo retaining type catalytic mechanism with conserved glutamic acid residue acting as nucleophile/base and proton donor. Three dimensional structure has (β/α)8 barrels. They share domains with three superfamilies (beta-galactosidase, beta-galactosidase trimerisation domain and beta-galactosidase C-terminal domain belongs to TIM-barrel glycoside hydrolase, Class I glutamine amidotransferase and glycoside hydrolase domain superfamily respectively). These enzymes catalyze hydrolysis of terminal and non-reducing terminal beta-D-galactosidase residues (Shimizu et al. 1995). Class I glutamine amidotransferase superfamily contains glutaminase enzymes and also the members of DJ-1/PfpI family (peptidases, catalytic triad Cys-His-Glu). This clan was built by A Bateman. Class I glutamine amidotransferase superfamily also has Cys-His-Glu triad, but differs from that of peptidases PfpI. Glycoside hydrolase domain superfamily includes the C-terminal domain of sugar-lytic enymes and was also developed by A Bateman (Fig. 4t1, u1, v1).
Glycoside Hydrolase Family 43
This family consists of numerous enzymes namely with activities β-xylosidase (EC 3.2.1.37); α-L-arabinofuranosidase (EC 3.2.1.55); xylanase (EC 3.2.1.8); α-1,2-L-arabinofuranosidase (EC 3.2.1.-); exo-α-1,5-L-arabinofuranosidase (EC 3.2.1.-); [inverting] exo-α-1,5-L-arabinanase (EC 3.2.1.-); β-1,3-xylosidase (EC 3.2.1.-); [inverting] exo-α-1,5-L-arabinanase (EC 3.2.1.-); [inverting] endo-α-1,5-L-arabinanase (EC 3.2.1.99); exo-β-1,3-galactanase (EC 3.2.1.145); β-D-galactofuranosidase (EC 3.2.1.146). They exhibit inverting mode of catalytic mechanism with aspartic acid and glutamic acid as catalytic nucleophile/base and proton donor respectively. Fivefold β-propellers are prominent feature of three-dimensional structure with long V-shaped groove forming a single extended substrate binding surface across the face of propeller. These enzymes belong to beta-fructosidase superfamily and have membrane localization (Nurizzo et al. 2002b) (Fig. 4w1).
Glycoside Hydrolase Family 44
Endoglucanase (EC 3.2.1.4) and xyloglucanase (EC 3.2.1.151) forms major activities of this family with retaining mechanism of catalytic activity and glutamic acid at conserved domain acting as nucleophile/base and proton donor. They belong to TIM barrel glycoside hydrolase superfamily (CL0058) and has (β/α)8 domain in its three-dimensional structure (Kitago et al. 2007; Ariza et al. 2011). This family was formerly known as cellulase family J (Fig. 4x1).
Glycoside Hydrolase Family 45
Endoglucanase (EC 3.2.1.4); xyloglucan-specific endo-β-1,4-glucanase/endo-xyloglucanase (EC 3.2.1.151); endo-β-1,4-mannanase (EC 3.2.1.78) are prominent activities of enzymes of this group and were formerly known to classified under cellulase family K. They exhibit inverting mode of catalytic reaction, with conserved aspartic acid residues (conserved N-terminal domain with several cysteines involved in forming disulphide bonds, referred as signature sequence) working as nucelophile/base and proton donor (Davies et al. 1993). Enzymes of this family belong to Double Psi beta barrel glucanase superfamily (CL0199). This clan represents barwin like barrels and was developed by A Bateman (Mizuguchi et al. 1999; Castillo et al. 1999). Examples of enzymes of this group includes endoglucanase 5 from Humicola insolens, Trichoderma reesei¸ endoglucanase K from Fusarium oxysporum etc (Fig. 4y1).
Glycoside Hydrolase Family 46
Chitosanases (EC 3.2.1.132) is activity of enzymes of this group, they belong to lysozyme like superfamilies (CL0037) and have inverting mode of chitinase activity, with aspartic acid and glutamic acid at probable catalytic conserved site acting as nucleophile/base and proton donor, respectively. Chitosanase catalyze the endohydrolysis of beta 1,4-linkages between N-acetyl-D-glucosamine and D-glucosamine residues in a partly acetylated chitosan (Fig. 4z1).
Glycoside Hydrolase Family 47
Alpha-mannosidase (EC 3.2.1.113) is activity of enzymes found in this family and these protein have (α/α)7 domain in their three dimensional structure with conserved glutamic acid acting as catalytic proton donor. Alpha-mannosidase catalyzes maturation of Asn-linked oligosaccharides (Lal et al. 1994). Hydrolysis of terminal 1,2-linked alpha-D-mannose residues into oligo-mannose oligosaccharide man(9)(glcnac)(2) is calcium dependent process and mannose residues get trimmed to produce man(8)glcnac(2) followed by man(5)(glcnac)(2) structures. They are prominently located in endoplasmic reticulum membrane, golgi membrane and cell membrane (Fig. 4a2).
Glycoside Hydrolase Family 48
Reducing end-acting cellobiohydrolase (EC 3.2.1.176); endo-β-1,4-glucanase (EC 3.2.1.4); chitinase (EC 3.2.1.14) are major enzymes of the group of the family (formerly known as cellulase family L). Conserved glutamic acid residues act as catalytic proton donor with the enzyme exhibiting inverting catalytic mechanism. Three dimensional structure has (α/α)6 barrels, these enzymes belong to six-hairpin glycosidase superfamily (CL0059) (Te’o et al. 1995) (Fig. 4b2).
Glycoside Hydrolase Family 49
Dextranase (EC 3.2.1.11); isopullulanase (EC 3.2.1.57); dextran 1,6-α-isomaltotriosidase (EC 3.2.1.95); sulfated arabinan endo-1,4-β-L-arabinanase (EC 3.2.1.-) and catalyze hydrolysis of alpha-1,6-glycosidic bonds in dextran polymers, through catalytic mechanism with conserved aspartic acid residues acting as catalytic nucleophile/base and proton donor. They belong to Pectate lyase-like beta helix with prominent beta helix seen in three dimensional structures (Fig. 4c2).
Glycoside Hydrolase Family 50
Beta-agarase (EC 3.2.1.81) is the activity reported for this group. These enzymes are less explored and probably follow retaining mode of activity with glutamic acid residues as nucleophile/base and proton donor. They belong to clan GH-A of Tim barrel glycoside hydrolase superfamily. Few bacterial enzymes of the group are reported (Fig. 4d2).
Glycoside Hydrolase Family 51
Endoglucanase (EC 3.2.1.4); endo-β-1,4-xylanase (EC 3.2.1.8); β-xylosidase (EC 3.2.1.37); α-L-arabinofuranosidase (EC 3.2.1.55); lichenase/endo-β-1,3-1,4-glucanase (EC 3.2.1.73) are major enzymes of the group with retaining mode of action, and glutamic acid residues at catalytic conserved site acting as nucleophile/base and proton donor. These enzymes belong to Clan GH-A of TIM-barrel glycoside hydrolase superfamily and have (β/α)8 motif in their three dimensional structure (Fig. 4e2).
Glycoside Hydrolase Family 52
Beta-xylosidase (EC 3.2.1.37) is the activity of enzymes of this group, having retaining mode of catalytic mechanism, with glutamic acid and aspartic acid residues on conserved domains acting as nucelophile/base and proton donor, respectively. They belong to clan GH-O with (α/α)6 barrels in their three dimensional structure. These enzymes are reported to be from Bacillus stearothermophilus, functioning in xylan degradation through hydrolysis of 1,4- beta-D-xylans, through successive removal of D-xylose residues from non-reducing termini. Alongside it also carries out hydrolysis of xylobiose (Baba et al. 1994) (Fig. 4f2).
Glycoside Hydrolase Family 53
Endo-β-1,4-galactanase (EC 3.2.1.89) are major activities of family 53 enzymes. They belong to clan GH-A of TIM barrel glycoside hydrolase superfamily with (β/α)8 barrels in their three dimensional structure. They undergo retaining type catalytic mechanism with glutamic acid as catalytic nucleophile/base and proton donor (Ryttersgaard et al. 2002) (Fig. 4g2).
Glycoside Hydrolase Family 54
Alpha-L-arabinofuranosidase (EC 3.2.1.55); β-xylosidase (EC 3.2.1.37) are activities of enzymes of the group. They show retaining mode of catalytic mechanism. This group of enzymes is very less explored and needs detailed study (Fig. 4h2).
Glycoside Hydrolase Family 55
Exo-β-1,3-glucanase (EC 3.2.1.58); endo-β-1,3-glucanase (EC 3.2.1.39) are activities of enzymes belonging to this family and have inverting mode of catalytic mechanism. These enzymes have prominent beta-helix in their three dimensional structure and conserved glutamic acid residue is known to be catalytic proton donor. This group of enzyme is less explored and classified is not available (Fig. 4i2).
Glycoside Hydrolase Family 56
Hyaluronidase (EC 3.2.1.35); chondroitin hydrolase (EC 3.2.1.-) are activities of enzymes of this family (eg., venom of Apis mellifera; Gmachl and Kreil 1993). They belong to TIM-barrel glycoside hydrolase superfamily with (β/α)7 motif in their three dimensional structure and are reported to be located on plasma membrane. They have retaining type of catalytic mechanism, where carbonyl oxygen of C-2 acetamido group of substrate work as nucleophile/base and conserved glutamic acid residue works as proton donor. Amino acid sequences of hyaluronidases have multiple glycosylation sites and numerous cysteines (Lathrop et al. 1990) (Fig. 4j2).
Glycoside Hydrolase Family 57
Alpha-amylase (EC 3.2.1.1); α-galactosidase (EC 3.2.1.22); amylopullulanase (EC 3.2.1.41); cyclomaltodextrinase (EC 3.2.1.54); branching enzyme (EC 2.4.1.18); 4-α-glucanotransferase (EC 2.4.1.25) are activities of enzymes of the family, they carry out retaining type catalytic mechanism and glutamic acid residues at conserved site working as nucleophile/base. Three dimensional structure has (β/α)7 motifs. This family includes highly specific enzymes (Laderman et al. 1993) (Fig. 4k2).
Glycoside Hydrolase Family 58
Endo-N-acetylneuraminidase or endo-sialidase (EC 3.2.1.129) are activities of enzymes found in this family having inverting type catalytic mechanism, water activated by carboxylate of substrate working as catalytic nucleophile/base and conserved glutamic acid working as catalytic proton donor. Three dimensional structures have sixfold β-propellers present. This group of enzyme is highly explored and needs extensive study and classification (Fig. 4l2).
Glycoside Hydrolase Family 59
Beta-galactosidase (EC 3.2.1.23) galactocerebrosidase (EC 3.2.1.46) are activities of enzymes of this family, with (β/α)8 motif in their three-dimensional structure, they are classified under clan GH-A of TIM barrel glycoside hydrolase superfamily (CL0058) and have conserved glutamic acid at catalytic region that acts as nucleophile/base and proton donor. These enzymes are responsible for lysosomal catabolism of galactolipids (Rafi et al. 1996; Luzi et al. 1995; Fukushima et al. 1998) (Fig. 4m2).
Glycoside Hydrolase Family 60
Precise data of any particular enzyme of this family is not reported.
Glycoside Hydrolase Family 61
Copper-dependent lytic polysaccharide monooxygenases were activities of enzymes belonging to this family, now reclassified in family AA9 (Auxilary activity Family 9). Classification under GH Family 61 was considered incorrect.
Glycoside Hydrolase Family 62
Alpha-L-arabinofuranosidase (EC 3.2.1.55) are activities of enzymes of this group, belonging to Clan GH-F of Beta-fructosidase/Furanosidase Superfamily. They catalyze hydrolysis of aryl alpha L arabinofuranosides by cleaving arabinosyl side chains from arabinoxylan and arabinan, but the detailed catalytic mechanism of the enzymes of this group is not known (Fig. 4n2).
Glycoside Hydrolase Family 63
Processing α-glucosidase (EC 3.2.1.106); α-1,3-glucosidase (EC 3.2.1.84); α-glucosidase (EC 3.2.1.20); mannosylglycerate α-mannosidase/mannosylglycerate hydrolase (EC 3.2.1.170); glucosylglycerate hydrolase (EC 3.2.1.208) are major enzymes of this family belonging to clan GH-G of Six-hairpin glycosidase superfamily with (α/α)6 motif in their three dimensional structure. They show inverting mode of catalytic mechanism with conserved glutamic acid and aspartic acid working as catalytic nucleophile/base and proton donor respectively. These enzymes are majorly located in endoplasmic reticulum membrane and they catalyze cleavage of non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC 3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway (Fig. 4o2).
Glycoside Hydrolase Family 64
Beta-1,3-glucanase (EC 3.2.1.39) are activities of the group, less explored and are known to show inverting mode of catalytic mechanism, with conserved aspartic acid and glutamic acid working as catalytic nucleophile/base and proton donor respectively (Fig. 4p2).
Glycoside Hydrolase Family 65
Alpha,α-trehalase (EC 3.2.1.28); maltose phosphorylase (EC 2.4.1.8); trehalose phosphorylase (EC 2.4.1.64); kojibiose phosphorylase (EC 2.4.1.230); trehalose-6-phosphate phosphorylase (EC 2.4.1.216); nigerose phosphorylase (EC 2.4.1.279); 3-O-α-glucopyranosyl-L-rhamnose phosphorylase (EC 2.4.1.282); 2-O-α-glucopyranosylglycerol: phosphate β-glucosyltransferase (EC 2.4.1.-); α-glucosyl-1,2-β-galactosyl-L-hydroxylysine α-glucosidase (EC 3.2.1.107); are activities of enzymes of this family. They consist of three structural domains; central catalytic domain, N-terminal domain and C-terminal domain. Central catalytic domain of these families belongs to Clan GH-L of Six-hairpin glycosidase superfamily and the N-terminal domain belongs to Galactose mutarotase like superfamily. They catalyze hydrolysis through inverting mode of catalytic mechanism, through phosphate for phosphorylases; water for hydrolases acting as nucleophile/base and conserved glutamic acid residue acting as proton donor. Maltose phosphorylase (MP) (dimeric enzyme) catalyzes the conversion of maltose and inorganic phosphate into beta-D-glucose-1-phosphate and glucose (Van Tilbeurgh et al. 2001) (Fig. 4q2, r2, s2).
Glycoside Hydrolase Family 66
Cycloisomaltooligosaccharide glucanotransferase (EC 2.4.1.248) and dextranase (EC 3.2.1.11) are activities of this family. They show retaining mode of catalytic mechanism with conserved aspartic acid residues acting as catalytic nucleophile/base and proton donor. They belong to TIM barrel glycoside hydrolase superfamily (CL0058) (Igarashi et al. 1995; Funane et al. 2011) (Fig. 4t2).
Glycoside Hydrolase Family 67
Alpha-glucuronidase (EC 3.2.1.139) and xylan α-1,2-glucuronidase (EC 3.2.1.131) are activities of enzymes of glycoside hydrolase family 67, catalyzing through inverting mode of catalytic mechanism and conserved glutamic acid residues as proton donor. They have (β/α)8 barrels in their three dimensional structure. Crystal structures of all the three domains of family are known, C-terminal, N-terminal and middle domain where the central domain contains invariant glutamic acid and aspartic residues making them catalytic centre. They catalyze removal of alpha-1,2 linked 4-O-methyl glucuronic acid from xylans. (Shoham et al. 2001 and Nurizzo et al. 2002b) (Fig. 4u2, v2, w2).
Glycoside Hydrolase Family 68
Levansucrase (EC 2.4.1.10) (beta-D-fructofuranosyl transferase); β-fructofuranosidase (EC 3.2.1.26); inulosucrase (EC 2.4.1.9) are major activities of the family, with retaining mode of catalytic mechanism, these enzymes use conserved aspartic acid and glutamic acid as catalytic nucleophile/base and proton donor. They are classified under clan GH-J of Beta-fructosidase/Furanosidase Superfamily. L catalyze the conversion of sucrose and (2,6-beta-D-fructosyl)(N) to glucose and (2,6-beta-D-fructosyl)(N+1), where other sugars can also act as fructosyl acceptors. Invertase, or extracellular sucrase (EC 3.2.1.26), catalyses the hydrolysis of terminal non-reducing beta-D-fructofuranoside residues in beta-D-fructofuranosides (Fig. 4x2).
Glycoside Hydrolase Family 69
No enzyme is classified under this family at present. Earlier classified enzymes have been grouped as family PL16.
Glycoside Hydrolase Family 70
Dextransucrase (EC 2.4.1.5); alternansucrase (EC 2.4.1.140); reuteransucrase (EC 2.4.1.-); α-4,6-glucanotransferase (EC 2.4.1.-); α-1,2-branched dextransucrase (EC 2.4.1.-); α-4,3-glucanotransferase (EC 2.4.1.-) are the activities reported to be classified under glycoside hydrolase family 70. They belong to clan GH-H of TIM Barrel glycoside hydrolase superfamily with (β/α)8 motif in their three dimensional structure. They follow retaining type catalytic mechanism with aspartic acid and glutamic acid residues working as catalytic nucleophile/base and proton donor. They catalyze transfer of D-glucopyramnosyl units from sucrose onto acceptor molecules (Croux et al. 1996) (Fig. 4y2).
Glycoside Hydrolase Family 71
Alpha-1,3-glucanase (EC 3.2.1.59) is the activity of enzyme belonging to this family and they are known to catalyze activity through inverting mechanism of reaction. This group of enzymes is not extensively studied. No known three dimensional structures is available.
Glycoside Hydrolase Family 72
Beta-1,3-glucanosyltransglycosylase (EC 2.4.1.-) is the activities of enzyme belonging to glycoside hydrolase family 72, clan GH-A of TIM Barrel glycoside hydrolase superfamily. They exhibit retaining type of catalytic mechanism with glutamic acid residues in the conserved domain acting as catalytic nucleophile/base and proton donor. These enzymes are glycolipid proteins anchored to membrane (Fig. 4z2).
Glycoside Hydrolase Family 73
Lysozyme (EC 3.2.1.17); mannosyl-glycoprotein endo-β-N-acetylglucosaminidase (EC 3.2.1.96); peptidoglycan hydrolase with endo-β-N-acetylglucosaminidase specificity (EC 3.2.1.-) are activities of enzymes of the group and the conserved glutamic acid acts as catalytic proton donor. They also catalyze hydrolysis of peptidoglycan (Nambu et al. 1999) (Fig. 4a3).
Glycoside Hydrolase Family 74
Endoglucanase (EC 3.2.1.4); oligoxyloglucan reducing end-specific cellobiohydrolase (EC 3.2.1.150); xyloglucanase (EC 3.2.1.151) are activities of enzymes of the group. They follow inverting type catalytic mechanism with conserved aspartic acid residue as catalytic nucleophile/base and proton donor. Three dimensional structures have sevenfold β-propeller. This group of enzymes requires detailed studies (Fig. 4b3).
Glycoside Hydrolase Family 75
Chitosanase (EC 3.2.1.132) is reported activity of enzymes of this group, with conserved aspartic acid and glutamic acid residues working as probable catalytic nucleophile/base and proton donor respectively. They catalyze through inverting mode of catalytic mechanism (Cheng et al. 2017). No known three dimensional structures are available.
Glycoside Hydrolase Family 76
Alpha-1,6-mannanase (EC 3.2.1.101) and α-glucosidase (EC 3.2.1.20) are activities of enzymes of this family. They belong to Six-hairpin glycosidase superfamily, and catalyses through retaining mode of action through conserved aspartic acid residues working as catalytic nucleophile/base and proton donor (Fig. 4c3).
Glycoside Hydrolase Family 77
Amylomaltase or 4-α-glucanotransferase (EC 2.4.1.25) are activities of enzymes of this family, they belong to clan GH-H of TIM Barrel glycoside hydrolase superfamily and have (β/α)8 motifs in their three dimensional structure. They undergo catalysis through retaining type mechanism with conserved aspartic acid and glutamic residues working as catalytic nucleophile/base and proton donor respectively. They catalyze transfer of a segment of a (1,4)-alpha-D-glucan to a new four-position in an acceptor, which may be glucose or (1,4)-alpha-D-glucan. They belong to the disproportionating family of enzymes (Okada et al. 1993) (Fig. 4d3).
Glycoside Hydrolase Family 78
Alpha-L-rhamnosidase (EC 3.2.1.40); rhamnogalacturonan α-L-rhamnohydrolase (EC 3.2.1.174); L-Rhap-α-1,3-D-Apif -specific α-1,3-L-rhamnosidase (EC 3.2.1.-) are activities of enzymes of this family belonging to Six-hairpin glycosidase superfamily having (α/α) 6 motif in their three dimensional structures (Zverlov et al. 2000). They catalyze hydrolysis through inverting mode of catalytic mechanism (Fig. 4e3).
Glycoside Hydrolase Family 79
Beta-glucuronidase (EC 3.2.1.31); hyaluronoglucuronidase (EC 3.2.1.36); heparanase (EC 3.2.1.166); baicalin β-glucuronidase (EC 3.2.1.167); β-4-O-methyl-glucuronidase (EC 3.2.1.-) are activities of enzymes reported to be classified under this family. They belong to clan GH-A of TIM Barrel glycoside hydrolase superfamily with (β/α)8 domains in its three dimensional structure. They undergo catalysis through retaining mechanism with conserved glutamic acid residues inferred to be catalytic nucleophile/base and proton donor. These enzymes play vital role in the self- assembly, insolubility and barrier properties of basement membranes and extracellular matrices and are also involved in cell migration associated with inflammation and autoimmunity (Vlodavsky et al. 2001) (Fig. 4f3).
Glycoside Hydrolase Family 80
Chitosanase (EC 3.2.1.132) are activities of enzymes belonging to this family. They belong to clan GH-I of Pectate lyase-like beta helix superfamily and process through inverting mode of catalytic mechanism (Shimono et al. 2002). Detailed catalytic conserved domain and three dimensional structures are yet to be explored (Fig. 4g3).
Glycoside Hydrolase Family 81
Endo-β-1,3-glucanase (EC 3.2.1.39) are the activities of enzyme exhibiting inverting mode of catalytic mechanism. Detailed catalytic mechanism and complete analysis of conserved motifs in the three dimensional structures is yet to be explored (Fig. 4h3).
Glycoside Hydrolase Family 82
Ι-carrageenase (EC 3.2.1.157) are the activities of enzymes belonging to the family with a prominent (β)-helix in their three dimensional structure and showing inverting mode of catalytic mechanism through conserved aspartic acid and glutamic acid working as catalytic nucleophile/base and proton donor, respectively (Fig. 4i3).
Glycoside Hydrolase Family 83
Neuraminidases (EC 3.2.1.18) are the enzyme activities reported for this family, belonging to clan GH-E of sialidase superfamily. With sixfold β-propeller motif in their three dimensional structure, these enzymes show retaining mode of catalytic mechanism (conserved tyrosine + glutamic acid working as catalytic nucleophile/base). Neuraminidase domain is the glycoside hydrolase domain of Hemagglutinin-neuraminidase (unit viral protein) containing both hemagglutinin and (endo) neuraminidase (Zaitsev et al. 2004 and Yuan et al. 2005) (Fig. 4j3).
Glycoside Hydrolase Family 84
N-acetyl β-glucosaminidase (EC 3.2.1.52); hyaluronidase (EC 3.2.1.35) and [protein]-3-O-(GlcNAc)-L-Ser/Thr β-N-acetylglucosaminidase (EC 3.2.1.169) are prominent activities of enzymes belonging to this family with (β/α)8 in their three dimensional structures. They exhibit retaining mode of catalytic mechanism where carbonyl oxygen of C-2 acetamido group of substrate acts as catalytic nucleophile/base and aspartic acid residues are proton donors (Macauley et al. 2005) (Fig. 4k3).
Glycoside Hydrolase Family 85
Endo-β-N-acetylglucosaminidases (EC 3.2.1.96) are activities of enzymes of GH family 85, belonging to clan, GH-A of TIM barrel glycoside hydrolase superfamily. They exhibit retaining type of catalytic mechanism with carbonyl oxygen of C-2 acetamido group of substrate acting as catalytic nucleophile/base and glutamic acid residues as proton donor. These enzymes are secretary enzymes and work on broad spectrum of substrates (Fig. 4l3).
Glycoside Hydrolase Family 86
Beta-agarase (EC 3.2.1.81); β-porphyranase (EC 3.2.1.178) are major activities of enzymes of this group. They belong to clan GH-A of TIM Barrel glycoside hydrolase superfamily, with (β/α)8 barrels in their three dimensional structures. Studies have reported probable retaining mode of catalytic mechanism with conserved glutamic acid residues as catalytic nucleophile/base and proton donor (Fig. 4m3).
Glycoside Hydrolase Family 87
Mycodextranase (EC 3.2.1.61) and α-1,3-glucanase (EC 3.2.1.59) are major enzymes of this family. These enzymes are not well explored need extensive study (Fig. 4n3).
Glycoside Hydrolase Family 88
Major activities of enzymes of this group involves d-4, 5-unsaturated β-glucuronyl hydrolase (EC 3.2.1.-) (Hashimoto et al. 1999). They belong to six-hairpin glycosidase superfamily (CL0059) containing (α/α)6 motif determined in their three dimensional structure. Mechanism of catalysis by the group of enzymes has not been explored and needs detailed study (Fig. 4o3).
Glycoside Hydrolase Family 89
Alpha-N-acetylglucosaminidase (EC 3.2.1.50) are activities of enzyme reported in GH Family 89, with specific (β/α)8, they belong to TIM-Barrel glycoside hydrolase superfamily (CL0058). They exhibit retaining mode of catalytic mechanism with conserved glutamic acid residues working as catalytic nucleophile/base and proton donor. Three structural domains of these enzymes are N-terminal domain (alpha-beta fold); central domain (TIM barrel fold) and C-Terminal domain (alpha-helical fold). Improper functioning of these enzymes leads to various disorders (Ficko-Blean et al. 2008; Li et al. 1999; Villani et al. 2002) (Fig. 4p3).
Glycoside Hydrolase Family 90
Endorhamnosidases (EC 3.2.1.-) are activities of enzymes of this group, with prominent (β)-helix in their three-dimensional structure. They carry out catalytic mechanism through inverting mode, where conserved domain with glutamic and aspartic acid acts as nucleophile/base and aspartic acid separately acts as proton donor (Fig. 4q3).
Glycoside Hydrolase Family 91
Inulin lyase [DFA-I-forming] (EC 4.2.2.17); inulin lyase [DFA-III-forming] (EC 4.2.2.18); difructofuranose 1,2′:2,3′ dianhydride hydrolase [DFA-IIIase] (EC 3.2.1.-) are major activities of enzymes of this family having β-helix domain in its three dimensional structure. They follow inverting mode of catalytic mechanism and was temporarily changed into PL19 until it was recently realized that these “PLs” are analogous to GH23 SLTs/lysozymes (there is at least one hydrolase in the GH91 family) and therefore are better classified in the GH category (Fig. 4r3).
Glycoside Hydrolase Family 92
Mannosyl-oligosaccharide α-1,2-mannosidase (EC 3.2.1.113); mannosyl-oligosaccharide α-1,3-mannosidase (EC 3.2.1.-); mannosyl-oligosaccharide α-1,6-mannosidase (EC 3.2.1.-); α-mannosidase (EC 3.2.1.24); α-1,2-mannosidase (EC 3.2.1.-); α-1,3-mannosidase (EC 3.2.1.-); α-1,4-mannosidase (EC 3.2.1.-) and mannosyl-1-phosphodiester α-1,P-mannosidase (EC 3.2.1.-) are activities of enzymes belonging to this family and pectate-lyase like beta-helix superfamily (CL0268). The conserved catalytic site consists of aspartic acid and glutamic acid residues working as nucleophile/base and proton donor for carrying out catalysis through inverting catalytic mechanism. These enzymes are reported to be critical for maturation of N- linked oligosaccharides and endoplasmic reticulum associated degradation (Liu et al. 1999) (Fig. 4s3).
Glycoside Hydrolase Family 93
Exo-α-L-1,5-arabinanase (EC 3.2.1.-) are activities classified under this family and with six-bladed β-propeller motif they belong to Clan GH-E of Sialidase superfamily (CL434). They carry out catalysis through retaining mode of action with conserved Glutamic acid residues working as catalytic nucleophile/base and proton donor (Fig. 4t3).
Glycoside Hydrolase Family 94
Cellobiose phosphorylase (EC 2.4.1.20); laminaribiose phosphorylase (EC 2.4.1.31); cellodextrin phosphorylase (EC 2.4.1.49); chitobiose phosphorylase (EC 2.4.1.-); cyclic β-1,2-glucan synthase (EC 2.4.1.-); cellobionic acid phosphorylase (EC 2.4.1.321); β-1,2-oligoglucan phosphorylase (EC 2.4.1.-) are activities of enzymes belonging to family and with (α/α)6 barrels they are classified under clan GH-Q. Catalysis involves inverting mode of mechanism and conserved phosphate and aspartic acid residues work as catalytic nucleophile/base and proton donor. These enzymes were formerly known as glycosyltransferase family GT36; and have been assigned to GH family by Hidaka et al. 2004 that reported evolutionary, structural and mechanistic relationship of these phosphorylases with glycoside hydrolases of clan GH-L (Fig. 4u3).
Glycoside Hydrolase Family 95
α-L-fucosidase (EC 3.2.1.51); α-1,2-L-fucosidase (EC 3.2.1.63); α-L-galactosidase (EC 3.2.1.-) are major enzymes with inverting catalytic mechanism containing conserved (α/α)6 motif in three dimensional stcrutures. Asparagine activated by aspartic acid works as catalytic nucleophile/base and Glutamic acid as proton donor (Katayama et al. 2004) (Fig. 4v3).
Glycoside Hydrolase Family 96
Alpha-agarase (EC 3.2.1.158) is activity classified under this family. No proper literature of three dimensional structure and catalytic mechanism for the same is reported.
Glycoside Hydrolase Family 97
Glucoamylase (EC 3.2.1.3); α-glucosidase (EC 3.2.1.20); α-galactosidase (EC 3.2.1.22) are the activities of the family, with (β/α)8 motif they are classified under TIM Barrel glycoside hydrolase Superfamily (CL0058). They exhibit both inverting and retaining mode of catalytic mechanism where Glutamic acid and Aspartic acid residues acts as catalytic nucleophile/base for inverting and retaining mechanisms respectively. Catalytic proton donor in both the cases is conserved glutamic acid residues (Hughes et al. 2003). N-terminal and C-terminal of the enzyme has pre-dominant beta-strands and the non catalytic domains are involved in oligomerization and carbohydrate binding (Naumoff 2005) (Fig. 4w3).
Glycoside Hydrolase Family 98
Blood-group endo-β-1,4-galactosidase (EC 3.2.1.102); blood group A- and B-cleaving endo-β-1,4-galactosidase (EC 3.2.1.-); endo-β-1,4-xylanase (EC 3.2.1.8); endo-β-1,4-xylanase (EC 3.2.1.8) are activities of enzymes belonging to this group. Two domains of these enzymes are known, of which putative catalytic domain is believed to be present at N-terminus of the non catalytic region (Anderson et al. 2005). They follow inverting catalytic mechanism with Aspartic acid and Glutamic acid diad as catalytic nucleophile/base and conserved glutamic acid residues as proton donor (Fig. 4x3).
Glycoside Hydrolase Family 99
Glycoprotein endo-α-1,2-mannosidase (EC 3.2.1.130) and mannan endo-1,2-α-mannanase (3.2.1.-) are activities of the group with (β/α)8 motifs in their three dimensional structure. They undergo retaining type catalytic mechanism with unclear, possible neighboring group participation from O2 (Thompson et al. 2012) working as catalytic nucleophile/base and conserved glutamic acid residues as proton donor (Spiro et al. 1997) (Fig. 4y3).
Glycoside Hydrolase Family 100
Alkaline and neutral invertase (EC 3.2.1.26) are activities of the enzymes classified under this family, with a (α/α)6 motif in their three dimensional structure, they belong to clan GH-G of Six-hairpin glycosidase superfamily. Catalytic mechanism takes place through inverting mode with glutamic acid and aspartic acid acting as nucleophile/base and proton donor respectively (Gallagher and Pollock 1998; Lee and Sturm 1996; Sturm 1999) (Fig. 4z3).
Glycoside Hydrolase Family 101
Endo-α-N-acetylgalactosaminidases (EC 3.2.1.97) are activities reported, and contains (β/α)8 motifs, belonging to TIM barrel Glycoside hydrolase Superfamily (CL0058). They carry retaining type of catalytic mechanism with conserved Aspartic acid and glutamic acid residues working as catalytic nucleophile/base and proton donor (Fujita et al. 2005). These enzymes can be split into several subfamilies (Naumoff 2010) (Fig. 4a4).
Glycoside Hydrolase Family 102
Peptidoglycan lytic transglycosylases (EC 3.2.1.-) are activities of enzymes belonging to GH family 102, exhibiting retaining type catalytic mechanism. These enzymes corresponds to family 2 of the peptidoglycan lytic transglycosylases (Blackburn and Clarke 2001) (Fig. 4b4).
Glycoside Hydrolase Family 103
Peptidoglycan lytic transglycosylases (EC 3.2.1.-) are activities of enzymes belonging to GH family 102, exhibiting retaining type catalytic mechanism with carbonyl oxygen of C-2 acetamido group of substrate inferred to be catalytic nucleophile/base and conserved glutamic acid residues working as proton donor. These enzymes corresponds to family 3 of the peptidoglycan lytic transglycosylases (Blackburn and Clarke 2001) (Fig. 4C4).
Glycoside Hydrolase Family 104
Peptidoglycan lytic transglycosylases (EC 3.2.1.-) are activities of enzymes belonging to GH family 102, exhibiting retaining type catalytic mechanism. These enzymes corresponds to family 4 of the peptidoglycan lytic transglycosylases (Blackburn and Clarke 2001) (Fig. 4d4).
Glycoside Hydrolase Family 105
Unsaturated rhamnogalacturonyl hydrolase (EC 3.2.1.172); d-4,5-unsaturated β-glucuronyl hydrolase (EC 3.2.1.-); d-4,5-unsaturated α-galacturonidase (EC 3.2.1.-) are activities reported belonging to this family with predominant (α/α)6 in their three dimensional structures. Catalytic mechanism for this group of enzymes is not known (Itoh et al. 2006) (Fig. 4e4).
Glycoside Hydrolase Family 106
Alpha-L-rhamnosidase (EC 3.2.1.40); rhamnogalacturonan α-L-rhamnohydrolase (EC 3.2.1.174) are activities of this family of enzymes containing (β/α)8 barrels in their three-dimensional structure. They exhibit inverting mode of catalytic mechanism (Miyata et al. 2005) (Fig. 4f4).
Glycoside Hydrolase Family 107
Sulfated fucan endo-1,4-fucanase (EC 3.2.1.-) are activities of enzymes belonging to GH Family 107, with (β/α)8 motif in their three dimensional structure, they belong to Clan GH-R of TIM Barrel Glycoside Hydrolase Superfamily. They exhibit retaining mode of catalytic mechanism with conserved Aspartic acid residues and Histidine acting as catalytic nucleophile/base and proton donor (Colin et al. 2006 and Vickers et al. 2018) (Fig. 4g4).
Glycoside Hydrolase Family 108
N-acetylmuramidase (EC 3.2.1.17) are the activities of enzymes belonging to this family and belong to Lysozyme like superfamily (CL0037). Conserved glutamic acid acts as catlytic proton donor. One pf their probable activities is activation of secretion of large proteins via the breaking and rearrangement of bacterial peptidoglycan layer during secretion (Stojković and Rothman-Denes 2007; Pei and Grishin 2005; Kondo et al. 1994; Emina and Lucia 2007) (Fig. 4h4).
Glycoside Hydrolase Family 109
Alpha-N-acetylgalactosaminidase (EC 3.2.1.49) are the activities of enzymes related to this family, with their catalytic mechanism unusually involving NAD+, catalyzing through retaining mode (Liu et al. 2007) (Fig. 4i4).
Glycoside Hydrolase Family 110
Alpha-galactosidase (EC 3.2.1.22); α-1,3-galactosidase (EC 3.2.1.-) are activities of enzymes reported for this family and they carry out catalysis through inverting mechanism (Liu et al. 2007). No structure is reported till date for enzymes of this family.
Glycoside Hydrolase Family 111
keratan sulfate hydrolase (endo-β-N-acetylglucosaminidase) (EC 3.2.1.-) are the activities reported with no further details available.
Glycoside Hydrolase Family 112
Lacto-N-biose phosphorylase or galacto-N-biose phosphorylase (EC 2.4.1.211); D-galactosyl-β-1,4-L-rhamnose phosphorylase (EC 2.4.1.247) and galacto-N-biose/lacto-N-biose phosphorylase (EC 2.4.1.-) are activities reported for this family and studies show that they undergo inverting mode of catalytic mechanism with Aspartic acid residues (through site directed mutagenesis) acting as proton donor (Fig. 4j4).
Glycoside Hydrolase Family 113
Beta-mannanase (EC 3.2.1.78) are activities of enzymes belonging to this family and with specific (β/α)8 barrels in their three dimensional structure they are classified under clan GH-A of TIM Barrel Glycoside hydrolase Superfamily. They exhibit retaining mode of catalytic mechanism with conserved glutamic acid residues working as catalytic nucleophile/base and proton donor (Zhang et al. 2008) (Fig. 4k4).
Glycoside Hydrolase Family 114
Endo-α-1,4-polygalactosaminidase (EC 3.2.1.109) are activities of enzymes belonging to this group, exhibiting retaining mode of catalytic mechanism (Tamura et al. 1995) (Fig. 4l4).
Glycoside Hydrolase Family 115
Xylan α-1,2-glucuronidase (3.2.1.131) and α-(4-O-methyl)-glucuronidase (3.2.1.-) are activities of enzymes belonging to this family and they catalyze reaction through inverting mechanism (Ryabova et al. 2009) (Fig. 4m4).
Glycoside Hydrolase Family 116
Beta-glucosidase (EC 3.2.1.21); β-xylosidase (EC 3.2.1.37); acid β-glucosidase/β-glucosylceramidase (EC 3.2.1.45) and β-N-acetylglucosaminidase (EC 3.2.1.52) are activities of enzymes classified under this family, with (α/α)6 motif in their three dimensional structure, they belong to clan GH-O of glycoside hydrolases superfamily. They exhibit retaining mode of catalytic mechanism through glutamic acid and aspartic acid residues working as catalytic nucleophile/base and proton donor (Ferrara et al. 2013) (Fig. 4n4).
Glycoside Hydrolase Family 117
Alpha-1,3-L-neoagarooligosaccharide hydrolase (EC 3.2.1.-) and α-1,3-L-neoagarobiase/neoagarobiose hydrolase (EC 3.2.1.-) are major activities of the enzymes classified under this family. These enzymes are distantly related to GH Family 43 (Fig. 4o4).
Glycoside Hydrolase Family 118
This family contains enzymes with activities of β-agarase (EC 3.2.1.81), found to exhibit inverting mode of catalytic mechanism.
Glycoside Hydrolase Family 119
Alpha-amylases (EC 3.2.1.1) are enzymes classified under GH Family 119. They are distantly related to GH family 57 and process through inverting catalytic mechanism (Watanabe et al. 2006)
Glycoside Hydrolase Family 120
Beta-xylosidase (EC 3.2.1.37) are activities of enzymes exhibiting retaining type catalytic mechanism with conserved aspartic acid and glutamic acid residues working as catalytic nucleophile/base and proton donor (Shao et al. 2010) (Fig. 4p4).
Glycoside Hydrolase Family 121
Beta-L-arabinobiosidases (EC 3.2.1.-) are activities belonging to this group and enzymes catalyze through retaining mechanism (Fujita et al. 2011).
Glycoside Hydrolase Family 122
Alpha-glucosidase (EC 3.2.1.20) is activity of enzymes belonging to this family (Comfort et al. 2008).
Glycoside Hydrolase Family 123
β-N-acetylgalactosaminidase (EC 3.2.1.53) and glycosphingolipid β-N-acetylgalactosaminidase (EC 3.2.1.-) are the activities for the family, exhibiting retaining mode of catalytic mechanism through carbonyl oxygen of C-2 acetamido group of substrate acting as catalytic nucleophile/base and conserved glutamic acid residues as proton donor (Sumida et al. 2011) (Fig. 4q4).
Glycoside Hydrolase Family 124
Endoglucanase (EC 3.2.1.4) is activity for the enzymes belonging to this family and they carry catalysis through inverting mechanism (Bras et al. 2011) (Fig. 4r4).
Glycoside Hydrolase Family 125
Exo-α-1,6-mannosidase (EC 3.2.1.-) is the activity reported for this group of enzymes belonging to clan GH-G with specific (α/α)6 of Six-hairpin glycosidase superfamily. They carry out catalysis through inverting mechanism with glutamic acid and aspartic acid residues working as catalytic nucleophile/base and proton donor (Gregg et al. 2011) (Fig. 4s4).
Glycoside Hydrolase Family 126
Alpha-amylase (EC 3.2.1.-), with (α/α)6 are classified under this family (Ficko-blean et al. 2011) (Fig. 4t4).
Glycoside Hydrolase Family 127
Beta-L-arabinofuranosidase (EC 3.2.1.185) and 3-C-carboxy-5-deoxy-L-xylose (aceric acid) hydrolase (EC 3.2.1.-) are activities of the enzymes reported under this family and with specific (α/α)6 motif, they belong to clan GH-P of Glycoside hydrolase superfamilies exhibiting retaining mode of catalytic mechanism (Fujita et al. 2011) (Fig. 4u4).
Glycoside Hydrolase Family 128
Beta-1,3-glucanase (EC 3.2.1.39) are activities of enzymes belonging to GH Family 128, with (β/α)8 motif in their three dimensional structures, they are classified under Clan GH-A of TIM barrel glycoside hydrolase superfamily (Sakamoto et al. 2011). They exhibit retaining mode of catalysis with conserved glutamic acid residues working as catalytic nucleophile/base and proton donor.
Glycoside Hydrolase Family 129
Enzymes of this family exhibit α-N-acetylgalactosaminidase (EC 3.2.1.49) with retaining mode of action and Aspartic acid residues as catalytic nucleophile/base (Kiyohara et al. 2012) (Fig. 4v4).
Glycoside Hydrolase Family 130
Activities of enzymes of this family includes β-1,4-mannosylglucose phosphorylase (EC 2.4.1.281); β-1,4-mannooligosaccharide phosphorylase (EC 2.4.1.319); β-1,4-mannosyl-N-acetyl-glucosamine phosphorylase (EC 2.4.1.320); β-1,2-mannobiose phosphorylase (EC 2.4.1-); β-1,2-oligomannan phosphorylase (EC 2.4.1.-) and β-1,2-mannosidase (EC 3.2.1.-). These enzymes have fivefold β-propeller in their three dimensional structure and exhibit inverting mode of catalytic mechanism (Senoura et al. 2011) (Fig. 4w4).
Glycoside hydrolase Family 131
Enzymes with broad specificity exo-β-1,3/1,6-glucanase with endo-β-1,4-glucanase activity (EC 3.2.1.-) belong to this family (Lafond et al. 2012) (Fig. 4x4).
Glycoside Hydrolase Family 132
Activity on β-1,3-glucan (curdlan) shown for the Aspergillus fumigatus Sun4 protein; activity on laminarioligosaccharides shown for Aspergillus fumigatus Sun4 protein and Candida albicans Sun41 protein; transglycosylation activity are activities of enzymes belonging to this family and they show retaining type of catalytic mechanism (Gastebois et al. 2013).
Glycoside Hydrolase Family 133
Enzymes of this exhibit amylo-α-1,6-glucosidase (EC 3.2.1.33) activity through inverting mechanism with conserved aspartic acid residues working as catalytic nucleophile/base and proton donor (Nakayama et al. 2001) (Fig. 4y4).
Glycoside Hydrolase Family 134
These are enzymes with endo-β-1,4-mannanase (EC 3.2.1.78) with inverting mode of action (a unique feature compared to all other beta-mannanses) (Shimizu et al. 2015; Jin et al. 2016) (Fig. 4z4).
Glycoside Hydrolase Family 135
Alpha-1,4-galactosaminogalactan hydrolase (EC 3.2.1.-) are activities of enzymes of this family, showing retaining type catalytic mechanism (Bamford et al. 2015) (Fig. 4a5).
Glycoside Hydrolase Family 136
Lacto-N-biosidase (EC 3.2.1.140) is the activity reported for the group, with prominent (β)-helix in their 3-D structure. They catalyze through retaining mode of catalytic mechanism with conserved aspartic acid residues as catalytic nucleophile base and proton donor (Yamada et al. (2017) (Fig. 4b5).
Glycoside hydrolase Family 137
Beta-L-arabinofuranosidase (EC 3.2.1.185) are major activities of the enzymes belonging to GH Family 137, having fivefold β-propeller in their three dimensional structure and conserved glutamic acid residues acting as catalytic nucleophile/base and proton donor (Ndeh et al. 2017) (Fig. 4c5).
Glycoside Hydrolase Family 138
This family of glycoside hydrolase have activities of rhamnogalacturonan α-1,2-galacturonohydrolase (EC 3.2.1.173); with enzymes having (β/α)8 barrels in their three dimensional structure, exhibiting retaining mode of catalytic mechanism with conserved glutamic acid residues acting as catalytic nucleophile/base and proton donor (Ndeh et al. 2017; Labourel et al. 2019) (Fig. 4d5).
Glycoside Hydrolase Family 139
Alpha-2-O-Me-L-fucosidase (EC 3.2.1.-) is the activity of enzymes reported for this group (Ndeh et al. 2017).
Glycoside Hydrolase Family 140
Beta-1,2-apiosidase (EC 3.2.1.-) are the activities of enzymes reported in this group, having (β/α)8 barrels in three dimensional structures and exhibiting retaining mode of catalytic mechanism (Ndeh et al. 2017) (Fig. 4e5).
Glycoside Hydrolase Family 141
Alpha-L-fucosidase (EC 3.2.1.51) and xylanases (EC 3.2.1.8) are activities of enzymes of this group, specifically containing (β)-helix in their three dimensional structures; and conserved aspartic acid acts as catalytic nucleophile/base and proton donor (Ndeh et al. 2017 and Heinz et al.) (Fig. 4f5).
Glycoside Hydrolase Family 142
Beta-L-arabinofuranosidase (EC 3.2.1.185) is the reported activity for the enzymes of this family with prominent (α/α)6 barrel in their three-dimensional structure. Conserved aspartic acid residues acts as catalytic nucleophile/base (Ndeh et al. 2017) (Fig. 4g5).
Glycoside Hydrolase Family 143
Enzymes with activity of 2-keto-3-deoxy-D-lyxo-heptulosaric acid hydrolase (EC 3.2.1.-) belong to this family and have prominent fivefold β-propeller in their three dimensional structures. They catalyze through retaining mode of reaction with conserved tyrosine and glutamic acid residues acting as nucleophile/base and individual glutamic acid acting as catalytic proton donor (Fig. 4h5).
Glycoside Hydrolase Family 144
Endo-β-1,2-glucanase (EC 3.2.1.71) and β-1,2-glucooligosaccharide sophorohydrolase (EC 3.2.1.-) are activities of enzymes belonging to this group with specific (α/α)6 motif in their three dimensional structures catalyzing enzymatic reaction through inverting mode (Abe et al. 2017) (Fig. 4i5).
Glycoside Hydrolase Family 145
L-Rhα-α-1,4-GlcA α-L-rhamnohydrolase (EC 3.2.1.-) are activities reported for this family, having seven-bladed β-propellerin their three dimensional structures. These enzymes are reported to exhibit retaining mode of catalytic mechanism with conserved Histidine residues working as catalytic nucelophile/base. They carry out hydrolysis of alpha-L-rhamnose residues linked to position 4 of a glucuronic acid. One of the sites of these enzymes which is still under study shows string resemblance to family PL25 suggesting polysaccharide lyase activity (Fig. 4j5).
Glycoside Hydrolase Family 146
Beta-L-arabinofuranosidases (EC 3.2.1.185) are activities of the enzymes having (α/α)6 barrels in their three dimensional structure, they are classified under clan GH-P with retaining type of catalytic mechanism (Luis et al. 2018) (Fig. 4k5).
Glycoside Hydrolase Family 147
Beta-galactosidase (EC 3.2.1.23) is activity reported for this group, with specific (β/α)8 (inferred) present in their three dimensional structures, they have been classified under Clan GH-A of TIM Barrel glycoside hydrolase superfamily. Catalysis is carried out through retaining mode with conserved glutamic acid residues acting as catalytic nucleophile/base and proton donor (Luis et al. 2018).
Glycoside Hydrolase Family 148
Enzymes of this family are reported to have β-1,3-glucanase (EC 3.2.1.-) activities, and with specific (β/α)8 (inferred) present in their three dimensional structures, they have been classified under Clan GH-A of TIM Barrel glycoside hydrolase superfamily. Conserved glutamic acid residues act as catalytcic nucleophile/base and proton donor (Angelov et al. 2017).
Glycoside Hydrolase Family 149
β-1,3-glucan phosphorylases (EC 2.4.1.97) are activities reported for enzymes belonging to family GH F-149, having (α/α)6 barrels in their three dimensional structures, they are classified under clan GH-Q of glycoside hydrolase superfamily sharing this family GH F-94. They carry out inverting mode of catalytic mechanism, with phosphate ion and conserved aspartic acid residues acting as catalytic nucleophile/base and proton donor (Kuhaudomlarp et al. 2018) (Fig. 4l5).
Glycoside Hydrolase Family 150
l-carrageenase (EC 3.2.1.-) are activities of enzymes belonging to this family, and have been reported to carry out catalysis through inverting mechanism (PMID:16926183).
Glycoside Hydrolase Family 151
Alpha-L-fucosidase (EC 3.2.1.51) are activities of enzymes belonging to this family (Sela et al. 2012), and have been reported to carry out catalysis through retaining mechanism (inferred through transglycosylation activity) (Benesová et al. 2013).
Glycoside Hydrolase Family 152
Beta-1,3-glucanase (EC 3.2.1.39) are activities reported of this group of enzymes with only few structural details available for the same (Sakamoto et al. 2006) (Fig. 4m5).
Glycoside Hydrolase Family 153
Poly-β-1,6-D-glucosamine hydrolase (EC 3.2.1.-) are activities of enzyme belong to this family. They have specific (β/α)8 barrels in their three dimensional structures (Little et al. 2018) (Fig. 4n5).
Glycoside Hydrolase Family GH F-154
Beta-glucuronidase (3.2.1.31) is the activity of enzymes reported for this group (Cartmell et al. 2018).
Glycoside Hydrolase Family GH F-155
This family was reported by Cartmell et al. (2018) with retaining type catalytic mechanism, but presently this family has been deleted.
Glycoside Hydrolase Family 156
Exo-α-sialidase (EC 3.2.1.18) is the reported activity for the enzymes belonging to this family, catalyzing reaction through inverting mode and have been found through functional metagenomics study of DNA from thermal springs (Chuzel et al. 2018) (Fig. 4o5).
Glycoside Hydrolase Family 157
Endo-β-1,3-glucanase (EC 3.2.1.39) and endo-β-1,3-glucanase/laminarinase (EC 3.2.1.39) are activities reported for this family, they carry out enzymatic reaction through retaining mode and conserved glutamic acid residues in their active site acts as catalytic nucleophile/base and proton donor. They belong to clan GH-A of TIM Barrel glycoside hydrolase superfamily (Helbert et al. 2019).
Glycoside Hydrolase Family 158
Like family 158, endo-β-1,3-glucanase (EC 3.2.1.39) are also activities of enzymes of family 158, belong to clan GH-A of Barrel glycoside hydrolase superfamily, carrying out catalytic mechanism through retaining mode with conserved glutamic acid residues working as catalytic nucleophile/base and proton donor (Helbert et al. 2019).
Glycoside Hydrolae Family 159
β-D-galactofuranosidases (EC 3.2.1.146) are the activities reported for this group of enzymes (Helbert et al. 2019).
Glycoside Hydrolase Family 160
Endo-β-1,4-galactosidase (EC 3.2.1.-) are the activities reported for enzymes belonging to this family and are known to have probable folding similarities with several PL families (Helbert et al. 2019).
Glycoside Hydrolase Family 161
Beta-1,3-glucan phosphorylase (EC 2.4.1.-) are the activities of enzymes reported for this family of enzymes, with (α/α)6 (inferred) motif in their three dimensional structure they are classified under clan GH-Q. They carry out catalysis through inverting mechanism with phosphate ion and conserved aspartic acid acting as catalytic nucleophile/base and proton donor respectively (Kuhaudomlarp et al. 2019).
Glycoside Hydrolase Family 162
Endo-β-1,2-glucanase (EC 3.2.1.71) is the activity of enzyme of this family, having (α/α)6 motif in their three dimensional structure, exhibiting inverting mode of catalytic mechanism, with conserved aspartic acid and glutamic acid residues working as catalytic nucleophile/base and proton donor (Tanaka et al. 2019) (Fig. 4p5).
Glycoside Hydrolase Family 163
Endo-β-N-acetylglucosaminidase cleaving GlcNAc-β-1,2-Man (EC 3.2.1.-) are the activities of enzymes reported to belong to this family (Briliute et al. 2019).
Glycoside Hydrolase Family 164
Beta-mannosidase (EC 3.2.1.25) are activities of enzymes of this family (Helbert et al. 2019) (Note: it appears alpha-mannosidase in reference due to uncorrected typo in Table 2)
Glycoside Hydrolase Family 165
Beta-galactosidase (EC 3.2.1.23) is activity of enzymes determined through metagenomics study (Cheng et al. 2017)
Glycoside Hydrolase Family 166
Alpha-1,4-galactosaminogalactan hydrolase (EC 3.2.1.-) is the activities of enzymes belonging to this family, exhibiting retaining type catalytic mechanism (Le Mauff et al. 2019) (Fig. 4q5).
Glycoside Hydrolase Family Non Classified
Group of glycoside hydrolase enzymes to which family has yet not been designated are reported here (Fig. 4r5).
Conclusions
Glycoside hydrolases, as stated earlier in this chapter, finds varied applications and this feature is much supported by the presence of diverse catalytic sites on these enzymes. They have been classified under 166 families which are grouped in 16 different superfamilies and three added clans (not specified yet under any superfamily). Being broadly studied for its action on numerous substrates, glycoside hydrolases like amylases, cellulases, chitinases, xylanases, glucanases, glucuronidases are few in humungous list finding usage in numerous technologies involved in production of various value added components. Specific region of catalytic domain, mode of action (retaining/inverting) and three dimensional structures for majority are available with added data on crystal structures of few specifically studied (like family 2, 3, 10, 11 etc.) and all figures have been reported in this chapter. In the present book few extensively studied reports on glycoside hydrolases have been brought forward, which broadly includes, structure, mechanism and applications of glycoside hydrolases, followed by a note on directed evolution of glycoside hdyrolases, its applications in degradation of lignocellulosic biomass, bioethanol production, food industries, paper and pulp industries, marine glycoside hydrolases as industrial tool and role of metagenomics in discovering industrially significant cellulases. With this minute initiative we bring to you an insight on industrial applications of glycoside hydrolases, an initial approach.
References
Abe K, Nakajima M, Yamashita T, Matsunaga H, Kamisuki S, Nihira T et al (2017) Biochemical and structural analyses of a bacterial endo-β-1,2-glucanase reveal a new glycoside hydrolase family. J Biol Chem 292:7487–7506
Adachi W, Sakihama Y, Shimizu S, Sunami T, Fukazawa T, Suzuki M, Yatsunami R, Nakamura S, Takénaka A (2004) Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17. J Mol Biol 343:785–795. https://doi.org/10.1016/j.jmb.2004.08.028
Alberto F, Bignon C, Sulzenbacher G, Henrissat B, Czjzek M (2004) The three-dimensional structure of invertase (beta-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases (PDF). J Biol Chem 279(18):18903–18910. https://doi.org/10.1074/jbc.M313911200
Aleshin AE, Firsov LM, Honzatko RB (1994) Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution. J Biol Chem 269:15631–15639
Allouch J, Jam M, Helbert W, Barbeyron T, Kloareg B, Henrissat B, Czjzek M (2003) The three-dimensional structures of two beta-agarases. J Biol Chem 278:47171–47180. https://doi.org/10.1074/jbc.M308313200
Al-Riyami B, Uestok FI, Stott K, Chirgadze DY, Christie G (2016) The crystal structure of Clostridium perfringens SleM, a muramidase involved in cortical hydrolysis during spore germination. Proteins 84:1681–1689. https://doi.org/10.1002/prot.25112
Alzari PM, Souchon H, Dominguez R (1996) The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum. Structure 4:265–275. https://doi.org/10.1016/s0969-2126(96)00031-7
Anderson KM, Ashida H, Maskos K, Dell A, Li SC, Li YT (2005) A clostridial endo-beta-galactosidase that cleaves both blood group A and B glycotopes: the first member of a new glycoside hydrolase family, GH98. J Biol Chem 280(9):7720–7728. https://doi.org/10.1074/jbc.M414099200
Angelov A, Pham VTT, Übelacker M, Brady S, Leis B, Pill N et al (2017) A metagenome-derived thermostable β-glucanase with an unusual module architecture which defines the new glycoside hydrolase family GH148. Sci Rep 7(1):17306
Ariza A, Eklöf JM, Spadiut O, Offen WA, Roberts SM, Besenmatter W, Friis EP, Skjøt M, Wilson KS, Brumer H, Davies G (2011) Structure and activity of Paenibacillus polymyxa xyloglucanase from glycoside hydrolase family 44. J Biol Chem 286(39):33890–33900. https://doi.org/10.1074/jbc.M111.262345
Aslanidis C, Schmid K, Schmitt R (1989) Nucleotide sequences and operon structure of plasmid-borne genes mediating uptake and utilization of raffinose in Escherichia coli. J Bacteriol 171(12):6753–6763. https://doi.org/10.1128/jb.171.12.6753-6763.1989
Baba T, Shinke R, Nanmori T (1994) Identification and characterization of clustered genes for thermostable xylan-degrading enzymes, beta-xylosidase and xylanase, of Bacillus stearothermophilus 21. Appl Environ Microbiol 60(7):2252–2258
Badieyan S, Bevan DR, Zhang C (2012) Study and design of stability in GH5 cellulases. Biotechnol Bioeng 109(1):31–44. https://doi.org/10.1002/bit.23280
Bairoch A (1999) Classification of glycosyl hydrolase families and index of glycosyl hydrolase entries in SWISS-PROT
Bamford NC, Snarr BD, Sheppard DC (2015) Sph3 is a glycoside hydrolase required for the biosynthesis of galactosaminogalactan in Aspergillus fumigates. J Biol Chem 290:27438–27450
Bashton M, Chothia C (2002) The geometry of domain combination in proteins. J Mol Biol 315:927–939
Bause E, Legler G (1980) Isolation and structure of a tryptic glycopeptide from the active site of beta-glucosidase A3 from Aspergillus wentii. Biochim Biophys Acta 626:459–465
Beguin P (1990) Molecular biology of cellulose degradation. Annu Rev Microbiol 44:219–48. https://doi.org/10.1146/annurev.mi.44.100190.001251
Béguin P, Cornet P, Aubert JP (1985) Sequence of a cellulase gene of the thermophilic bacterium Clostridium thermocellum. J Bacteriol 162:102–105
Benesová E et al (2013) α-L-fucosidase from Paenibacillus Thiaminolyticus: its hydrolytic and transglycosylation abilities. Glycobiology 23(9):1052–1065
Blackburn NT, Clarke AJ (2001) Identification of four families of peptidoglycan lytic transglycosylases. J Mol Evol 52:78–84
Bourne Y, Henrissat B (2001) Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr Opin Struct Biol 11(5):593–600. https://doi.org/10.1016/s0959-440x(00)00253-0
Braithwaite KL, Black GW, Hazlewood GP, Ali BR, Gilbert HJ (1995) A non-modular endo-beta-1,4-mannanase from pseudomonas fluorescens subspecies cellulosa. Biochem J 305(Pt 3):1005–1010. https://doi.org/10.1042/bj3051005
Bras JLA et al (2011) Structural insights into a unique cellulase fold and mechanism of cellulose hydrolysis. Proc Natl Acad Sci USA 108(13):5237–5242
Briliute J et al (2019) Complex N-glycan breakdown by gut bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci. Nat Microbiol 4(9):1571–1581
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. https://doi.org/10.1093/nar/gkn663
Cartmell A, Muñoz-Muñoz J, Briggs JA, Ndeh DA, Lowe EC, Baslé A, Terrapon N, Stott K, Heunis T, Gray J (2018) A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation. Nat Microbiol 3:1314–1326
Castillo RM, Mizuguchi K, Dhanaraj V, Albert A, Blundell TL, Murzin AG (1999) A six-stranded double-psi beta barrel is shared by several protein superfamilies. Structure Fold Des 7:227–236
Cheng J, Romantsov T, Engel K, Doxey AC, Rose DR, Neufeld JD, Charles TC (2017) Functional metagenomics reveals novel β-galactosidases not predictable from gene sequences. PLoS One 12(3):e0172545
Chuzel L, Ganatra MB, Rapp E, Henrissat B, Taron CH (2018) Functional metagenomics identifies an exosialidase with an inverting catalytic mechanism that defines a new glycoside hydrolase family (GH156). J Biol Chem 293(47):18138–18150. https://doi.org/10.1074/jbc.RA118.003302
Colin S, Deniaud E, Jam M, Descamps V, Chevolot Y, Kervarec N, Yvin J-C, Barbeyron T, Michel G, Kloareg B (2006) Cloning and biochemical characterization of the fucanase FcnA: definition of a novel glycoside hydrolase family specific for sulfated fucans. Glycobiology 16:1021–1032
Comfort DA, Bobrov KS, Ivanen DR, Shabalin KA, Harris JM, Kulminskaya AA et al (2007) Biochemical analysis of Thermotoga maritima GH36 α-galactosidase (TmGalA) confirms the mechanistic commonality of clan GH-D glycoside hydrolases. Biochemistry 46:3319–3330
Comfort DA, Chou CJ, Conners SB, VanFossen AL, Kelly RM (2008) Functional-genomics-based identification and characterization of open reading frames encoding alpha-glucoside-processing enzymes in the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol 74(4):1281–1283
Croux C, García JL (1991) Sequence of the lyc gene encoding the autolytic lysozyme of Clostridium acetobutylicum ATCC824: comparison with other lytic enzymes. Gene 104(1):25–31. https://doi.org/10.1016/0378-1119(91)90460-S
Croux C, Monchois V, Willemot RM, Remaud-simeon M, Monsan P (1996) Cloning and sequencing of a gene coding for a novel dextransucrase from Leuconostoc mesenteroides NRRL B-1299 synthesizing only alpha (1-6) and alpha (1-3) linkages. Gene 182(1–2):23–32. https://doi.org/10.1016/S0378-1119(96)00443-X
Davies G, Henrissat B (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3(9):853–859. https://doi.org/10.1016/S0969-2126(01)00220-9
Davies GJ, Dodson GG, Hubbard RE, Tolley SP, Dauter Z, Wilson KS, Hjort C, Mikkelsen JM, Rasmussen G, Schuelein M (1993) Structure and function of endoglucanase V. Nature 365:362–364. https://doi.org/10.1038/365362a0
Dey PM, Pridham JB (1972) Biochemistry of alpha-galactosidases. In: Advances in enzymology - and related areas of molecular biology, vol 36. Wiley, Hoboken, pp 91–120. https://doi.org/10.1002/9780470122815.ch3
Dinur T, Osiecki KM, Legler G, Gatt S, Desnick RJ, Grabowski GA (1986) Human acid beta-glucosidase: isolation and amino acid sequence of a peptide containing the catalytic site. Proc Natl Acad Sci U S A 83(6):1660–1664. https://doi.org/10.1073/pnas.83.6.1660
Divne C, Ståhlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles JK, Teeri TT, Jones TA (1994) The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science 265:524–528. https://doi.org/10.1126/science.8036495
Divne C, Ståhlberg J, Teeri TT, Jones TA (1998) High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei. J Mol Biol 275:309–325. https://doi.org/10.1006/jmbi.1997.1437
Ducros VM, Tarling CA, Zechel DL, Brzozowski AM, Frandsen TP, von Ossowski I, Schülein M, Withers SG, Davies GJ (2003) Anatomy of glycosynthesis: structure and kinetics of the Humicola insolens Cel7B E197A and E197S glycosynthase mutants. Chem Biol 10:619–628. https://doi.org/10.1016/s1074-5521(03)00143-1
Eijsink V, Hoell I, Vaaje-Kolstada G (2010) Structure and function of enzymes acting on chitin and chitosan. Biotechnol Genet Eng Rev 27(1):331–366. https://doi.org/10.1080/02648725.2010.10648156
El Hassouni M, Henrissat B, Chippaux M, Barras FJ (1992) Nucleotide sequences of the arb genes, which control b-glucoside utilization in Erwinia chrysanthemi: comparison with the Escherichia coli bgl operon and evidence for a new b-glycohydrolase family including enzymes from eubacteria, archeabacteria, and humans. J Bacteriol 174:765–777
Emina AS, Lucia BRD (2007) Coliphage N4 N-acetylmuramidase defines a new family of murein hydrolases. J Mol Biol 366:406–419
Ferrara MC et al (2013) The identification and molecular characterization of the first archaeal bifunctional exo-β-glucosidase/N-acetyl-β-glucosaminidase demonstrate that family GH116 is made of three functionally distinct subfamilies. Biochim Biophys Acta 1840:367–377
Ficko-Blean E, Stubbs KA, Nemirovsky O, Vocadlo DJ, Boraston AB (2008) Structural and mechanistic insight into the basis of mucopolysaccharidosis IIIB. Proc Natl Acad Sci U S A 105(18):6560–6565. https://doi.org/10.1073/pnas.0711491105
Ficko-Blean E, Stuart CP, Boraston AB (2011) Structural analysis of CPF_2247, a novel α-amylase from clostridium perfringens. Proteins 79:2771–2777
Fierobe HP, Bagnara-Tardif C, Gaudin C, Guerlesquin F, Sauve P, Belaich A, Belaich JP (1993) Purification and characterization of endoglucanase C from Clostridium cellulolyticum. Catalytic comparison with endoglucanase A. Eur J Biochem 217:557–565. https://doi.org/10.1111/j.1432-1033.1993.tb18277.x
Fisher KJ, Aronson NN (1989) Isolation and sequence analysis of a cDNA encoding rat liver alpha-L-fucosidase. Biochem J 264(3):695–701. https://doi.org/10.1042/bj2640695
Flach J, Pilet P-E, Jolles P (1992) What’s new in chitinase research? Experientia 48:701–716
Fleming D, Rumbaugh KP (2017) Approaches to dispersing medical biofilms. Microorganisms 5(2):15. https://doi.org/10.3390/microorganisms5020015
Fleming D, Chahin L, Rumbaugh K (2017) Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob Agents Chemother 61(2):e01998–16. https://doi.org/10.1128/AAC.01998-16
Friedberg F, Rhodes C (1988) Segments of amino acid sequence similarity in beta-amylases. Protein Seq Data Anal 1(6):499–501
Fujita et al (2005) Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-alpha-N-acetylgalactosaminidase from bifidobacterium longum. J Biol Chem 280:37415–37422
Fujita K, Sakamoto S, Ono Y, Wakao M, Suda Y, Kitahara K, Suganuma T (2011) Molecular cloning and characterization of a beta-L-Arabinobiosidase in Bifidobacterium longum that belongs to a novel glycoside hydrolase family. J Biol Chem 286(7):5143–5150
Fukushima H, Inui K, Fu L, Nishigaki T, Yanagihara I, Tatsumi N, Ozono K, Okada S, Sakai N (1998) Human galactocerebrosidase gene: promoter analysis of the 5'-flanking region and structural organization. Biochim Biophys Acta 1395(1):62–67. https://doi.org/10.1016/S0167-4781(97)00140-1
Funane K, Kawabata Y, Suzuki R, Kim YM, Kang HK, Suzuki N et al (2011) Deletion analysis of regions at the C-terminal part of cycloisomaltooligosaccharide glucanotransferase from Bacillus circulans T-3040. Biochim Biophys Acta 1814(3):428–434. https://doi.org/10.1016/j.bbapap.2010.12.009
Gallagher JA, Pollock CJ (1998) Isolation and characterization of a cDNA clone from Lolium temulentum L. encoding for a sucrose hydrolytic enzyme which shows alkaline/neutral invertase activity. J Exp Bot 49(322):789–795
Gastebois A, Aimanianda V, Bachellier-Bassi S, Nesseir A, Firon A, Beauvais A, Schmitt C, England P, Beau R, Prévost MC et al (2013) SUN proteins belong to a novel family of β-(1,3)-glucan-modifying enzymes involved in fungal morphogenesis. J Biol Chem 288(19):13387–13396
Gebler JC, Aebersold R, Withers SG (1992) Glu-537, not Glu-461, is the nucleophile in the active site of (lac Z) beta-galactosidase from Escherichia coli. J Biol Chem 267(16):11126–11130
Gilkes NR, Henrissat B, Kilburn DG, Miller RC Jr, Warren RA (1991) Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev 55:303–315
Gmachl M, Kreil G (1993) Bee venom hyaluronidase is homologous to a membrane protein of mammalian sperm. Proc Natl Acad Sci U S A 90(8):3569–3573. https://doi.org/10.1073/pnas.90.8.3569
Gonzalez-Candelas L, Ramon D, Polaina J (1990) Gene 95:31–38
Gregg Z, Hehemann J-H, Whitworth GE, Deng L, Vocadlo DJ, Boraston AB (2011) Analysis of a new family of widely distributed metal-independent alpha-mannosidases provides unique insight into the processing of n-linked glycans. J Biol Chem 286:15586–15596
Guérin DM, Lascombe MB, Costabel M, Souchon H, Lamzin V, Béguin P, Alzari PM (2002) Atomic (0.94 A) resolution structure of an inverting glycosidase in complex with substrate. J Mol Biol 316:1061–1069. https://doi.org/10.1006/jmbi.2001.5404
Hashimoto W, Kobayashi E, Nankai H, Sato N, Miya T, Kawai S et al (1999) Unsaturated glucuronyl hydrolase of Bacillus sp. GL1: novel enzyme prerequisite for metabolism of unsaturated oligosaccharides produced by polysaccharide lyases. Arch Biochem Biophys 368(2):367–374. https://doi.org/10.1006/abbi.1999.1305
He SY, Collmer A (1990) Molecular cloning, nucleotide sequence, and marker exchange mutagenesis of the exo-poly-alpha-D-galacturonosidase-encoding pehX gene of Erwinia chrysanthemi EC16. J Bacteriol 172(9):4988–4995. https://doi.org/10.1128/jb.172.9.4988-4995.1990
Heikinheimo P, Helland R, Leiros HK, Leiros I, Karlsen S, Evjen G, Ravelli R, Schoehn G, Ruigrok R, Tollersrud OK, McSweeney S, Hough E (2003) The structure of bovine lysosomal alpha-mannosidase suggests a novel mechanism for low-pH activation. J Mol Biol 327(3):631–644. https://doi.org/10.1016/S0022-2836(03)00172-4
Helbert W et al (2019) Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc Natl Acad Sci U S A 116(13):6063–6068
Henrissat B (1991a) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280(2):309–316. https://doi.org/10.1042/bj2800309
Henrissat B (1991b) Sequence homology between a beta-galactosidase and some beta-glucosidases. Protein Seq Data Anal 4:61–62
Henrissat B, Bairoch A (1996) Updating the sequence-based classification of glycosyl hydrolases. Biochem J 316(2):695–696. https://doi.org/10.1042/bj3160695
Henrissat B, Coutinho PM (1999) Carbohydrate-active enzymes server.
Henrissat B, Davies G (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3(9):853–859. https://doi.org/10.1016/S0969-2126(01)00220-9
Henrissat B, Davies G (1997) Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol 7(5):637–644. https://doi.org/10.1016/s0959-440x(97)80072-3
Henrissat B, Claeyssens M, Tomme P, Lemesle L, Mornon JP (1989) Cellulase families revealed by hydrophobic cluster analysis. Gene 81:83–95. https://doi.org/10.1016/0378-1119(89)90339-9
Henrissat B, Callebaut I, Mornon JP, Fabrega S, Lehn P, Davies G (1995) Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci U S A 92(15):7090–7094. https://doi.org/10.1073/pnas.92.15.7090
Hermans MM, Kroos MA, Van Beeumen J, Oostra BA, Reuser AJ (1991) Human lysosomal alpha-glucosidase. Characterization of the catalytic site. J Biol Chem 266(21):13507–13512
Hermoso JA, Monterroso B, Albert A, Galan B, Ahrazem O, Garcia P, Martinez-Ripoll M, Garcia JL, Menendez M (2003) Structural basis for selective recognition of pneumococcal cell wall by modular endolysin from phage Cp-1. Structure 11:1239–1249
Hettwer U, Jaeckel FR, Boch J, Meyer M, Rudolph K, Ullrich MS (1998) Cloning, nucleotide sequence, and expression in Escherichia coli of levansucrase genes from the plant pathogens Pseudomonas syringae pv. glycinea and P. syringae pv. phaseolicola. Appl Environ Microbiol 64:3180–3187
Hidaka M, Honda Y, Kitaoka M, Nirasawa S, Hayashi K, Wakagi T, Shoun H, Fushinobu S (2004) Chitobiose phosphorylase from vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold. Structure 12(6):937–947
Honda Y, Kitaoka M (2006) The first glycosynthase derived from an inverting glycoside hydrolase. J Biol Chem 281:1426–1431. https://doi.org/10.1074/jbc.M511202200
Hughes CV et al (2003) Cloning and expression of alpha-D-glucosidase and N-acetyl-beta-glucosaminidase from the periodontal pathogen, Tannerella forsythensis (Bacteroides forsythus). Oral Microbiol Immunol 18:309–312
Igarashi T, Yamamoto A, Goto N (1995) Sequence analysis of the Streptococcus mutans Ingbritt dexA gene encoding extracellular dextranase. Microbiol Immunol 39(11):853–860. https://doi.org/10.1111/j.1348-0421.1995.tb03282.x
Itoh T, Ochiai A, Mikami B, Hashimoto W, Murata K (2006) A novel glycoside hydrolase family 105: the structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide. J Mol Biol 360:573–585
Jacobson RH, Zhang XJ, DuBose RF, Matthews BW (1994) Three-dimensional structure of beta-galactosidase from E. coli. Nature 369(6483):761–766. https://doi.org/10.1038/369761a0
Jenkins J, Mayans O, Pickersgill R (1998) Structure and evolution of parallel beta-helix proteins. J Struct Biol 122:236–246
Jin Y et al (2016) A β-Mannanase with a lysozyme-like fold and a novel molecular catalytic mechanism. ACS Cent Sci 2(12):896–903. https://doi.org/10.1021/acscentsci.6b00232
Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K, Yamanoi T, Kumagai H, Yamamoto K (2004) Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J Bacteriol 186(15):4885–4893
Kim J-H et al (2013) Mechanism-based covalent neuraminidase inhibitors with broad spectrum influenza antiviral activity. Science 340(6128):71–75. https://doi.org/10.1126/science.1232552
Kinsella BT, Hogan S, Larkin A, Cantwell BA (1991) Primary structure and processing of the Candida tsukubaensis alpha-glucosidase. Homology with the rabbit intestinal sucrase-isomaltase complex and human lysosomal alpha-glucosidase. Eur J Biochem 202(2):657–664. https://doi.org/10.1111/j.1432-1033.1991.tb16420.x
Kitago Y, Karita S, Watanabe N, Kamiya M, Aizawa T, Sakka K, Tanaka I (2007) Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from clostridium thermocellum. J Biol Chem 282(49):35703–35711. https://doi.org/10.1074/jbc.M706835200
Kiyohara M et al (2012) α-N-acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J Biol Chem 287(1):693–700
Klarskov K, Piens K, Ståhlberg J, Høj PB, Beeumen JV, Claeyssens M (1997) Cellobiohydrolase I from Trichoderma reesei: identification of an active-site nucleophile and additional information on sequence including the glycosylation pattern of the core protein. Carbohydr Res 304:143–154. https://doi.org/10.1016/s0008-6215(97)00215-2
Kondo Y, Toyoda A, Fukushi H, Yanase H, Tonomura K, Kawasaki H et al (1994) Cloning and characterization of a pair of genes that stimulate the production and secretion of Zymomonas mobilis extracellular levansucrase and invertase. Biosci Biotechnol Biochem 58(3):526–530. https://doi.org/10.1271/bbb.58.526
Kopp M, Müller H, Holzer H (1993) Molecular analysis of the neutral trehalase gene from Saccharomyces cerevisiae. J Biol Chem 268(7):4766–4774
Korczynska JE, Danielsen S, Schagerloef U, Turkenburg JP, Davies GJ, Wilson KS, Taylor EJ (2010) The structure of a family GH25 lysozyme from Aspergillus fumigatus. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:973–977. https://doi.org/10.1107/S1744309110025601
Kuhaudomlarp S et al (2018) Identification of Euglena gracilis β-1,3-glucan phosphorylase and establishment of a new glycoside hydrolase (GH) family GH149. J Biol Chem 293(8):2865–2876
Kuhaudomlarp S et al (2019) Unraveling the Subtleties of β-(1→3)-glucan Phosphorylase Specificity in the GH94, GH149, and GH161 Glycoside Hydrolase Families. J Biol Chem 294(16):6483–6493
Labourel A et al (2019) Structural and functional analyses of glycoside hydrolase 138 enzymes targeting chain A galacturonic acid in the complex pectin rhamnogalacturonan II. J Biol Chem 294(19):7711–7721
Laderman KA, Davis BR, Krutzsch HC, Lewis MS, Griko YV, Privalov PL et al (1993) The purification and characterization of an extremely thermostable alpha-amylase from the hyperthermophilic archaebacterium pyrococcus furiosus. J Biol Chem 268(32):24394–24401
Lafond M et al (2012) Characterization of a broad-specificity β-Glucanase acting on β-(1,3)-, β-(1,4)-, and β-(1,6)-glucans that defines a new glycoside hydrolase family. Appl Environ Microbiol 78:8540–8546
Lal A, Schutzbach JS, Forsee WT, Neame PJ, Moremen KW (1994) Isolation and expression of murine and rabbit cDNAs encoding an alpha 1,2-mannosidase involved in the processing of asparagine-linked oligosaccharides. J Biol Chem 269(13):9872–9881
Lathrop WF, Carmichael EP, Myles DG, Primakoff P (1990) cDNA cloning reveals the molecular structure of a sperm surface protein, PH-20, involved in sperm-egg adhesion and the wide distribution of its gene among mammals. J Cell Biol 111(6):2939–2949. https://doi.org/10.1083/jcb.111.6.2939
Le Mauff F et al (2019) Molecular mechanism of Aspergillus fumigatus biofilm disruption by fungal and bacterial glycoside hydrolases. J Biol Chem 294(28):10760–10772
Lee HS, Sturm A (1996) Purification and characterization of neutral and alkaline invertase from carrot. Plant Physiol 112(4):1513–1522. https://doi.org/10.1104/pp.112.4.1513
Li HH, Yu WH, Rozengurt N, Zhao HZ, Lyons KM, Anagnostaras S, Fanselow MS, Suzuki K, Vanier MT, Neufeld EF (1999) Mouse model of Sanfilippo syndrome type B produced by targeted disruption of the gene encoding alpha-N-acetylglucosaminidase. Proc Natl Acad Sci U S A 96(25):14505–14510. https://doi.org/10.1073/pnas.96.25.14505
Linares-Pastén JA, Andersson M, Nordberg Karlsson E (2014) Thermostable glycoside hydrolases in biorefinery technologies. Curr Biotechnol 3(1):26–44. https://doi.org/10.2174/22115501113026660041
Little DJ et al (2018) PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-β(1,6)-N-acetylglucosamine and can disrupt bacterial biofilms. PLoS Pathog 14(4):e1006998
Liu Y, Choudhury P, Cabral CM, Sifers RN (1999) Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J Biol Chem 274(9):5861–5867. https://doi.org/10.1074/jbc.274.9.5861
Liu QP et al (2007) Bacterial glycosidases for the production of universal red blood cells. Nat Biotechnol 25:454–464
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. https://doi.org/10.1093/nar/gkt1178
Luis AS et al (2018) Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic bacteroides. Nat Microbiol 3(2):210–219
Luo Y, Li SC, Chou MY, Li YT, Luo M (1998) The crystal structure of an intramolecular trans-sialidase with a NeuAc alpha2-->3Gal specificity. Structure 6(4):521–530. https://doi.org/10.1016/S0969-2126(98)00053-7
Luzi P, Rafi MA, Wenger DA (1995) Structure and organization of the human galactocerebrosidase (GALC) gene. Genomics 26(2):407–409. https://doi.org/10.1016/0888-7543(95)80230-J
Macauley MS et al (2005) O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem 280:25313–25322
MacKenzie LF, Sulzenbacher G, Divne C, Jones TA, Wöldike HF, Schülein M, Withers SG, Davies GJ (1998) Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate. Biochem J 335(Pt 2):409–416. https://doi.org/10.1042/bj3350409
Michel G, Chantalat L, Duee E, Barbeyron T, Henrissat B, Kloareg B, Dideberg O (2001) The kappa-carrageenase of P. carrageenovora features a tunnel-shaped active site: a novel insight in the evolution of Clan-B glycoside hydrolases. Structure 9:513–525
Mikami B, Morita Y, Fukazawa C (1988) Primary structure and function of beta-amylase. J Jpn Biochem Soc 60(3):211–216
Miyata T et al (2005) Cloning, sequence analysis, and expression of the gene encoding Sphingomonas Paucimobilis FP2001 alpha-L –Rhamnosidase. Curr Microbiol 51:105–109
Mizuguchi K, Dhanaraj V, Blundell TL, Murzin AG (1999) N-ethylmaleimide-sensitive fusion protein (NSF) and CDC48 confirmed as members of the double-psi beta-barrel aspartate decarboxylase/formate dehydrogenase family. Structure Fold Des 7:215–216
Monzingo AF, Marcotte EM, Hart PJ, Robertas JD (1996) Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat Struct Biol 3:133–140
Murzin AG, Bateman A (1998) Distant homology recognition using structural classification of proteins. In: Latteman EE (ed) Proteins, vol 29(S1), pp 105–112
Naim HY, Niermann T, Kleinhans U, Hollenberg CP, Strasser AW (1991) Striking structural and functional similarities suggest that intestinal sucrase-isomaltase, human lysosomal alpha-glucosidase and Schwanniomyces occidentalis glucoamylase are derived from a common ancestral gene. FEBS Lett 294(1–2):109–112. https://doi.org/10.1016/0014-5793(91)81353-A
Nakayama A et al (2001) Identification of the catalytic residues of bifunctional glycogen debranching enzyme. J Biol Chem 276:28824–28828
Nambu T, Minamino T, Macnab RM, Kutsukake K (1999) Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol 181(5):1555–1561
Naumoff DG (2005) GH97 is a new family of glycoside hydrolases, which is related to the alpha-galactosidase superfamily. BMC Genomics 6:112. https://doi.org/10.1186/1471-2164-6-112
Naumoff DG (2006) Development of a hierarchical classification of the TIM-barrel type glycoside hydrolases (PDF). Proceedings of the fifth international conference on bioinformatics of genome regulation and structure, vol 1: 294–298.
Naumoff DG (2010) GH101 family of glycoside hydrolases: subfamily structure and evolutionary connections with other families. J Bioinform Comput Biol 8(3):437–451. https://doi.org/10.1142/s0219720010004628
Naumoff DG (2011) Hierarchical classification of glycoside hydrolases. Biochemistry 76(6):622–635. https://doi.org/10.1134/S0006297911060022
Naumov DG, Karreras M (2009) New program PSI protein classifier automatizes the PSI-BLAST results analysis. Mol Biol 43:709–721
Ndeh D et al (2017) Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544(7648):65–70
Nitta K, Sugai S (1989) The evolution of lysozyme and alpha-lactalbumin. Eur J Biochem 182(1):111–118. https://doi.org/10.1111/j.1432-1033.1989.tb14806.x
Nitta K, Tsuge H, Sugai S, Shimazaki K (1987) The calcium-binding property of equine lysozyme. FEBS Lett 223(2):405–408. https://doi.org/10.1016/0014-5793(87)80328-9
Nitta Y, Isoda Y, Toda H, Sakiyama F (1989) Identification of glutamic acid 186 affinity-labeled by 2,3-epoxypropyl alpha-D-glucopyranoside in soybean beta-amylase. J Biochem 105(4):573–576. https://doi.org/10.1093/oxfordjournals.jbchem.a122706
Nurizzo D, Nagy T, Gilbert HJ, Davies GJ (2002a) The structural basis for catalysis and specificity of the pseudomonas cellulosa alpha-glucuronidase, GlcA67A. Structure 10(4):547–556. https://doi.org/10.1016/s0969-2126(02)00742-6
Nurizzo D, Turkenburg JP, Charnock SJ, Roberts SM, Dodson EJ, McKie VA, Taylor EJ, Gilbert HJ, Davies GJ (2002b) Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold. Nat Struct Biol 9:665–668
Ohnishi H, Kitamura H, Minowa T, Sakai H, Ohta T (1992) Molecular cloning of a glucoamylase gene from a thermophilic Clostridium and kinetics of the cloned enzyme. Eur J Biochem 207(2):413–418. https://doi.org/10.1111/j.1432-1033.1992.tb17064.x
Okada S, Takaha T, Yanase M, Smith SM (1993) Disproportionating enzyme (4-alpha-glucanotransferase, EC 2.4.1.25) of potato. Purification, molecular cloning, and potential role in starch metabolism. J Biol Chem 268(2):1391–1396
Ori N, Sessa G, Lotan T, Himmelhoch S, Fluhr R (1990) A major stylar matrix polypeptide (sp41) is a member of the pathogenesis-related proteins superclass. EMBO J 9:3429–3436
Parkkinen T, Koivula A, Vehmaanperä J, Rouvinen J (2008) Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding. Protein Sci 17:1383–1394. https://doi.org/10.1110/ps.034488.108
Pei J, Grishin NV (2005) COG3926 and COG5526: a tale of two new lysozyme-like protein families. Protein Sci 14(10):2574–2581. https://doi.org/10.1110/ps.051656805
Peterbauer T, Mach L, Mucha J, Richter A (2002) Functional expression of a cDNA encoding pea (Pisum sativum L.) raffinose synthase, partial purification of the enzyme from maturing seeds, and steady-state kinetic analysis of raffinose synthesis. Planta 215(5):839–846. https://doi.org/10.1007/s00425-002-0804-7
Petersen L, Ardèvol A, Rovira C, Reilly PJ (2009) Mechanism of cellulose hydrolysis by inverting GH8 endoglucanases: a QM/MM metadynamics study. J Phys Chem B 113:7331–7339. https://doi.org/10.1021/jp811470d
Py B, Bortoli-German I, Haiech J, Chippaux M, Barras F (1991) Cellulase EGZ of Erwinia chrysanthemi: structural organization and importance of His98 and Glu133 residues for catalysis. Protein Eng 4(3):325–333. https://doi.org/10.1093/protein/4.3.325
Rafi MA, Wenger DA, Victoria T (1996) Cloning of the canine GALC cDNA and identification of the mutation causing globoid cell leukodystrophy in West Highland White and Cairn terriers. Genomics 33(3):457–462. https://doi.org/10.1006/geno.1996.0220
Rau A, Hogg T, Marquardt R, Hilgenfeld R (2001) A new lysozyme fold. Crystal structure of the muramidase from Streptomyces coelicolor at 1.65 A resolution. J Biol Chem 276:31994–31999. https://doi.org/10.1074/jbc.M102591200
Rothe B, Rothe B, Roggentin P, Schauer R (1991) The sialidase gene from Clostridium septicum: cloning, sequencing, expression in Escherichia coli and identification of conserved sequences in sialidases and other proteins. Mol Gen Genet 226(1–2):190–197. https://doi.org/10.1007/BF00273603
Rouvinen J, Bergfors T, Teeri T, Knowles JK, Jones TA (1990) Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science 249(4967):380–386. https://doi.org/10.1126/science.2377893
Ruttkowski E, Labitzke R, Khanh NQ, Löffler F, Gottschalk M, Jany KD (1990) Cloning and DNA sequence analysis of a polygalacturonase cDNA from Aspergillus niger RH5344. Biochim Biophys Acta 1087(1):104–106. https://doi.org/10.1016/0167-4781(90)90130-t
Ryabova O et al (2009) A novel family of hemicellulolytic alpha-glucuronidase. FEBS Lett 583:1457–1462
Ryttersgaard C, Lo Leggio L, Coutinho PM, Henrissat B, Larsen S (2002) Aspergillus aculeatus beta-1,4-galactanase: substrate recognition and relations to other glycoside hydrolases in clan GH-A. Biochemistry 41(51):15135–15143. https://doi.org/10.1021/bi026238c
Sakamoto Y et al (2006) Lentinula Edodes tlg1 encodes a thaumatin-like protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol 141(2):793–801
Sakamoto Y et al (2011) Endo-β-1,3-glucanase GLU1, from the fruiting body of lentinula edodes, belongs to a new glycoside hydrolase family. Appl Environ Microbiol 77(23):8350–8354
Sela DA et al (2012) Bifidobacterium Longum Subsp. Infantis ATCC 15697 α-fucosidases are active on fucosylated human milk Oligosaccharides. Appl Environ Microbiol 78(3):795–803
Senoura T et al (2011) New microbial mannan catabolic pathway that involves a novel mannosylglucose phosphorylase. Biochem Biophys Res Commun 408:701–706
Shao W, Xue Y, Wu A, Kataeva I, Pei J, Wu H, Wiegel J (2010) Characterization of a novel beta-xylosidase, XylC, from thermoanaerobacterium saccharolyticum JW/SL-YS485. Appl Environ Microbiol 77:719–726
Shimizu T, Kobayashi T, Ba-Thein W, Ohtani K, Hayashi H (1995) Sequence analysis of flanking regions of the pfoA gene of Clostridium perfringens: beta-galactosidase gene (pbg) is located in the 3'-flanking region. Microbiol Immunol 39(9):677–686. https://doi.org/10.1111/j.1348-0421.1995.tb03256.x
Shimizu M et al (2015) Novel β-1,4-mannanase belonging to a new glycoside hydrolase family in Aspergillus Nidulans. J Bio Chem 290(46):27914–27927
Shimono K, Shigeru K, Tsuchiya A, Itou N, Ohta Y, Tanaka K et al (2002) Two glutamic acids in chitosanase A from Matsuebacter chitosanotabidus 3001 are the catalytically important residues. J Biochem 131(1):87–96. https://doi.org/10.1093/oxfordjournals.jbchem.a003081
Shoham Y, Zaide G, Shallom D, Shulami S, Zolotnitsky G, Golan G, Baasov T, Shoham G (2001) Biochemical characterization and identification of catalytic residues in alpha-glucuronidase from Bacillus stearothermophilus T-6. Eur J Biochem 268(10):3006–3016. https://doi.org/10.1046/j.1432-1327.2001.02193.x
Sierks MR, Ford C, Reilly PJ, Svensson B (1990) Catalytic mechanism of fungal glucoamylase as defined by mutagenesis of Asp176, Glu179 and Glu180 in the enzyme from Aspergillus awamori. Protein Eng 3(3):193–198. https://doi.org/10.1093/protein/3.3.193
Sinnott ML (1990) Catalytic mechanisms of enzymatic glycosyl transfer. Chem Rev 90:1171–1202
Spiro MJ, Bhoyroo VD, Spiro RG (1997) Molecular cloning and expression of rat liver endo-α-mannosidase, an N-linked oligosaccharide processing enzyme. J Biol Chem 272:29356–29363
Stojković EA, Rothman-Denes LB (2007) Coliphage N4 N-acetylmuramidase defines a new family of murein hydrolases. J Mol Biol 366(2):406–419. https://doi.org/10.1016/j.jmb.2006.11.028
Stuart DI, Acharya KR, Walker NP, Smith SG, Lewis M, Phillips DC (1986) Alpha-lactalbumin possesses a novel calcium binding loop. Nature 324(6092):84–87. https://doi.org/10.1038/324084a0
Sturm A (1999) Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol 121(1):1–8. https://doi.org/10.1104/pp.121.1.1
Sulzenbacher G, Schülein M, Davies GJ (1997) Structure of the endoglucanase I from Fusarium oxysporum: native, cellobiose, and 3,4-epoxybutyl beta-D-cellobioside-inhibited forms, at 2.3 A resolution. Biochemistry 36:5902–5911. https://doi.org/10.1021/bi962963+
Sulzenbacher G, Bignon C, Nishimura T, Tarling CA, Withers SG, Henrissat B, Bourne Y (2004) Crystal structure of Thermotoga maritima alpha-L-fucosidase. Insights into the catalytic mechanism and the molecular basis for fucosidosis. J Biol Chem 279(13):13119–13128. https://doi.org/10.1074/jbc.M313783200
Sumida T, Fujimoto K, Ito M (2011) Molecular cloning and catalytic mechanism of a novel glycosphingolipid-degrading beta-N-acetylgalactosaminidase from Paenibacillus Sp. TS12. J Biol Chem 286:14065–14072
Tamura J-I et al (1995) Molecular cloning and sequence analysis of the gene encoding an endo α-1,4 polygalactosaminidase of Pseudomonas sp. 881. J Ferment Bioeng 80:305–310. https://doi.org/10.1016/0922-338X(95)94196-X
Tanaka N et al (2019) Identification, characterization, and structural analyses of a fungal endo-β-1,2-glucanase reveal a new glycoside hydrolase family. J Biol Chem 294(19):7942–7965
Te’o VS, Saul DJ, Bergquist PL (1995) celA, another gene coding for a multidomain cellulase from the extreme thermophile Caldocellum saccharolyticum. Appl Microbiol Biotechnol 43(2):291–296. https://doi.org/10.1007/BF00172827
Thoden JB, Holden HM (2002) High resolution X-ray structure of galactose mutarotase from Lactococcus lactis. J Biol Chem 277:20854–20861
Thompson J, Pikis A, Ruvinov SB, Henrissat B, Yamamoto H, Sekiguchi J (1998) The gene glvA of Bacillus subtilis 168 encodes a metal-requiring, NAD(H)-dependent 6-phospho-alpha-glucosidase. Assignment to family 4 of the glycosylhydrolase superfamily. J Biol Chem 273(42):27347–27356. https://doi.org/10.1074/jbc.273.42.27347
Thompson AJ et al (2012) Structural and mechanistic insight into N-glycan processing by endo-α-mannosidase. Proc Natl Acad Sci U S A 109:781–786
Totsuka A, Nong VH, Kadokawa H, Kim CS, Itoh Y, Fukazawa C (1994) Residues essential for catalytic activity of soybean beta-amylase. Eur J Biochem 221(2):649–654. https://doi.org/10.1111/j.1432-1033.1994.tb18777.x
Ubhayasekera W, Muñoz IG, Vasella A, Ståhlberg J, Mowbray SL (2005) Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors. FEBS J 272:1952–1964. https://doi.org/10.1111/j.1742-4658.2005.04625.x
Van Tilbeurgh H, Egloff MP, Uppenberg J, Haalck L (2001) Crystal structure of maltose phosphorylase from Lactobacillus brevis: unexpected evolutionary relationship with glucoamylases. Structure 9(8):689–697. https://doi.org/10.1016/S0969-2126(01)00626-8
Varghese JN, Garrett TPJ, Colman PM, Chen L, Hoj PB, Fincher GB (1994) Three-dimensional structures of two plant beta-glucan endohydrolases with distinct substrate specificities. Proc Natl Acad Sci U S A 91:2785–2789
Varghese JN, Hrmova M, Fincher GB (1999) Three-dimensional structure of a barley beta-D-glucan exohydrolase, a family 3 glycosyl hydrolase. Structure 7(2):179–190. https://doi.org/10.1016/S0969-2126(99)80024-0
Vickers C et al (2018) Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to α-L-fucosidases from GH29. J Biol Chem 293:18296–18308
Viladot JL, De Ramon E, Durany O, Planas A (1998) Probing the mechanism of Bacillus 1,3-1,4-beta-D-glucan 4-glucanohydrolases by chemical rescue of inactive mutants at catalytically essential residues. Biochemistry 37:11332–11342. https://doi.org/10.1021/bi980586q
Villani GR, Follenzi A, Vanacore B, Di Domenico C, Naldini L, Di Natale P (2002) Correction of mucopolysaccharidosis type IIIb fibroblasts by lentiviral vector-mediated gene transfer. Biochem J 364(Pt 3):747–753. https://doi.org/10.1042/BJ20011872
Vlodavsky I, Goldshmidt O, Zcharia E, Metzger S, Chajek-shaul T, Atzmon R, Guatta-rangini Z, Friedmann Y (2001) Molecular properties and involvement of heparanase in cancer progression and normal development. Biochimie 83(8):831–839. https://doi.org/10.1016/S0300-9084(01)01318-9
Vocadlo DJ, Davies GJ, Laine R, Withers SG (2001) Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate (PDF). Nature 412(6849):835–838. https://doi.org/10.1038/35090602
Wang AM, Bishop DF, Desnick RJ (1990) Human alpha-N-acetylgalactosaminidase-molecular cloning, nucleotide sequence, and expression of a full-length cDNA. Homology with human alpha-galactosidase A suggests evolution from a common ancestral gene. J Biol Chem 265(35):21859–21866
Watanabe H, Nishimoto T, Kubota M, Chaen H, Fukuda S (2006) Cloning, sequencing, and expression of the genes encoding an isocyclomaltooligosaccharide glucanotransferase and an alpha-amylase from a bacillus circulans strain. Biosci Biotechnol Biochem 70(11):2690–2702
Weaver LH, Matthews BW (1987) Structure of bacteriophage T4 lysozyme refined at 1.7 A resolution. J Mol Biol 193(1):189–199. https://doi.org/10.1016/0022-2836(87)90636-X
Withers SG, Warren RAJ, Street IP, Rupitz K, Kempton JB, Aebersold RJ (1990) Unequivocal demonstration of the involvement of a glutamate residue as a nucleophile in the mechanism of a retaining glycosidase. Am Chem Soc 112:5887–5889
Yamada C et al (2017) Molecular insight into evolution of symbiosis between breast-fed infants and a member of the human gut microbiome bifidobacterium longum. Cell Chem Biol 24:515–524
Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS (2005) Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13(5):803–815. https://doi.org/10.1016/j.str.2005.02.019
Zaitsev V, Von Itzstein M, Groves D et al (2004) Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J Virol 78(7):3733–3741. https://doi.org/10.1128/JVI.78.7.3733-3741.2004
Zhang Y et al (2008) Biochemical and structural characterization of the intracellular Mannanase AaManA of Alicyclobacillus acidocaldarius reveals a novel glycoside hydrolase family belonging to clan GH-A. J Biol Chem 283:31551–31558
Zverlov VV, Hertel C, Bronnenmeier K, Hroch A, Kellermann J, Schwarz WH (2000) The thermostable alpha-L-rhamnosidase RamA of clostridium stercorarium: biochemical characterization and primary structure of a bacterial alpha-L-rhamnoside hydrolase, a new type of inverting glycoside hydrolase. Mol Microbiol 35(1):173–179. https://doi.org/10.1046/j.1365-2958.2000.01691.x
Acknowledgement
Authors are thankful to Science and Engineering Research Board, New Delhi, India, for providing financial assistance to carry project File number: YSS/2015/002072.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Shrivastava, S. (2020). Introduction to Glycoside Hydrolases: Classification, Identification and Occurrence. In: Shrivastava, S. (eds) Industrial Applications of Glycoside Hydrolases . Springer, Singapore. https://doi.org/10.1007/978-981-15-4767-6_1
Download citation
DOI: https://doi.org/10.1007/978-981-15-4767-6_1
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-4766-9
Online ISBN: 978-981-15-4767-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)